ABSTRACT
NAD (NAD+) is a cofactor related to many cellular processes. This cofactor is known to be unstable, especially at high temperatures, where it chemically decomposes to nicotinamide and ADP-ribose. Bacteria, yeast, and higher organisms possess the salvage pathway for reconstructing NAD+ from these decomposition products; however, the importance of the salvage pathway for survival is not well elucidated, except for in pathogens lacking the NAD+ de novo synthesis pathway. Herein, we report the importance of the NAD+ salvage pathway in the thermophilic bacterium Thermus thermophilus HB8 at high temperatures. We identified the gene encoding nicotinamidase (TTHA0328), which catalyzes the first reaction of the NAD+ salvage pathway. This recombinant enzyme has a high catalytic activity against nicotinamide (Km of 17 μM, kcat of 50 s−1, kcat/Km of 3.0 × 103 s−1 · mM−1). Deletion of this gene abolished nicotinamide deamination activity in crude extracts of T. thermophilus and disrupted the NAD+ salvage pathway in T. thermophilus. Disruption of the salvage pathway led to the severe growth retardation at a higher temperature (80°C), owing to the drastic decrease in the intracellular concentrations of NAD+ and NADH.
IMPORTANCE NAD+ and other nicotinamide cofactors are essential for cell metabolism. These molecules are unstable and decompose, even under the physiological conditions in most organisms. Thermophiles can survive at high temperatures where NAD+ decomposition is, in general, more rapid. This study emphasizes that NAD+ instability and its homeostasis can be one of the important factors for thermophile survival in extreme temperatures.
KEYWORDS: NAD+, Thermus thermophilus, nicotinamidase, salvage synthesis
INTRODUCTION
NAD (NAD+) is an essential molecule for cellular metabolism. It serves as an electron donor/acceptor for many redox reactions in cellular metabolism. It also serves as a precursor for NADP+ and a substrate for both bacterial DNA ligases and ADP ribosyl transferases (1, 2). Due to its importance as a cofactor for many oxidoreductases, NAD+ is also an essential compound in biotechnology applications (3); however, NAD+ is known to be chemically unstable, especially at high temperatures, where it nonenzymatically decomposes to ADP-ribose and nicotinamide (4, 5).
Cells need to maintain a certain concentration of NAD+ for metabolism, and different organisms possess different pathways for synthesizing NAD+, such as the de novo biosynthesis pathway and the salvage pathway (6). In the salvage pathway, decomposition products from NAD+ are used for reconstructing the molecule. Many bacteria possess the salvage pathway initiated from the deamination of nicotinamide to nicotinate by nicotinamidase (2). Nicotinate is further converted to nicotinate mononucleotide (NaMN), nicotinate adenine dinucleotide (NaAD), and finally to NAD+ via the Preiss-Handler pathway (7, 8) (Fig. 1).
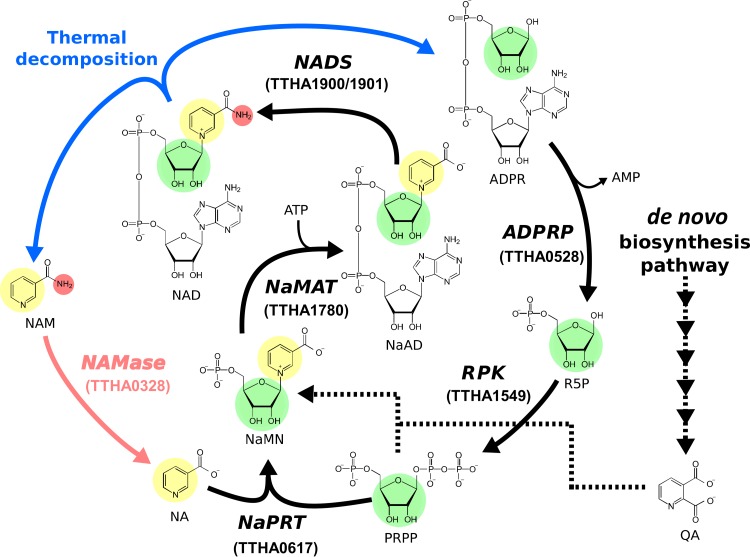
FIG 1.
Salvage pathway for NAD+. The moieties used for the salvage synthesis of NAD+ are represented with different colors (pyridine, yellow; ribose, green; amino group, red). The de novo biosynthesis pathway for NAD+ is also shown with dashed lines. NAD, NAD+; NAM, nicotinamide; NA, nicotinate; ADPR, ADP-ribose; R5P, ribose 5-phosphate; PRPP, phosphoribosyl diphosphate; NaMN, nicotinate mononucleotide; NaAD, nicotinate adenine dinucleotide; QA, quinolinate; NAMase, nicotinamidase; NaMAT, nicotinate mononucleotide adenylyltransferase; NaPRT, nicotinate phosphoribosyltransferase; ADPRP, ADPR pyrophosphatase; RPK, ribose-phosphate pyrophosphokinase; NADS, NAD+ synthase.
In our previous studies, we recognized that the decomposition of NAD+ and the consequent decrease in NAD+ concentrations at high temperatures are major obstacles for the biocatalytic manufacturing of value-added chemicals, in particular, with thermophilic enzymes (9, 10). As a solution, our group constructed an “in vitro synthetic pathway” for the salvage synthesis of NAD+ from its decomposition products (5). For the pathway construction, five of six enzymes required for the salvage pathway were predicted from the Thermus thermophilus HB8 genome information, and the catalytic activities of these enzymes were confirmed by in vitro experiments (Fig. 1); however, nicotinamidase, which catalyzes the first reaction of this pathway, was not found in the genome annotation data for this organism.
T. thermophilus HB8 is a thermophilic Gram-negative bacterium that has an optimum growth temperature between 65°C and 72°C and can grow at temperatures up to 85°C (11). Considering the thermal instability of NAD+, the NAD+ salvage synthesis is expected to be one of the important strategies for coping with the thermal degradation of the cofactor, especially for thermophiles. To our knowledge, however, the importance of the salvage pathway in vivo at high temperatures has not been described. Therefore, we predicted that T. thermophilus would possess a complete set of enzymes required for this pathway and hypothesized that the salvage pathway in T. thermophilus would play an important role for the homeostasis of NAD+ availability at high temperatures.
In this study, we identified and characterized the nicotinamidase of T. thermophilus HB8 that catalyzes the first reaction of the salvage synthesis of NAD+ and confirmed that T. thermophilus HB8 possesses the functional salvage pathway for NAD+. Additionally, we showed that this salvage pathway is essential for maintaining the intracellular concentration of NAD+/NADH and therefore is important for cell growth of T. thermophilus HB8, especially at high temperatures.
RESULTS
Prediction of the gene encoding nicotinamidase in T. thermophilus HB8.
A homology search of nicotinamidase was performed against the protein databases of T. thermophilus HB8 using BLASTP. As a query sequence, we used the nicotinamidase from Saccharomyces cerevisiae (UniProt accession number P53184), whose physiological function as a nicotinamidase has been experimentally confirmed (12). As a result, the protein encoded by TTHA0328, which was annotated as a probable isochorismatase, showed significant similarities in the amino acid sequences (93% query coverage, 39% identity, and E value of 3E−28).
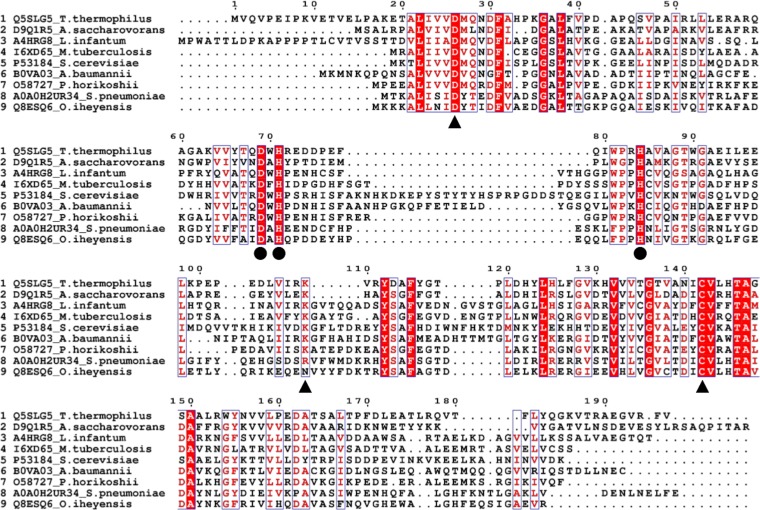
Multiple sequence alignments with previously described nicotinamidases revealed that the product of TTHA0328 has several amino acid residues that are well conserved among the known nicotinamidase enzymes, six of which are reported to be important for the catalytic activity of nicotinamidase (D26, K109, and C142 for the catalytic triad motif, and D69, H71, and H84 for the metal ion-binding motif) (13–17) (Fig. 2).
FIG 2.
Multiple-sequence alignment of TTHA0328 and other nicotinamidases. The multiple-sequence alignment was performed with ClustalOmega and visualized by ESPript. UniProt accession numbers and organisms from which proteins were sourced are shown on the left side. The position of amino acid residues based on T. thermophilus TTHA0328 product (Q5SLG5) is shown. The complete or partially conserved amino acids are colored in red. Triangles and circles indicate the amino acid residues important for the catalytic triad motif and the metal ion-binding motif, respectively.
Characterization of the product of TTHA0328.
Based on the prediction, TTHA0328 was overexpressed in Escherichia coli Rosetta 2 (DE3) pLysS as a recombinant protein. After heat treatment, the crude lysate was separated by size exclusion chromatography. The activity was confirmed in several fractions by high-pressure liquid chromatography (HPLC) analysis, as described in Materials and Methods. The protein homogeneity of high-activity fractions was confirmed by SDS-PAGE (see Fig. S1 in the supplemental material). These fractions were pooled and used for the characterization of this protein.
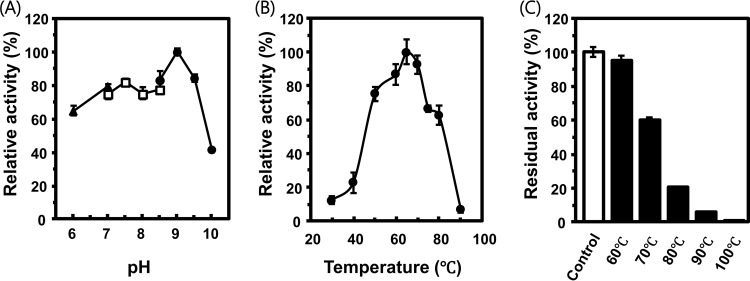
The specific activity toward nicotinamide was determined at different pHs and temperatures. The enzyme showed maximum activity at pH 9.0, and above 50% of the maximum activity was confirmed at a broad range of pHs from 6.0 to 9.5 (Fig. 3A). Activity dropped quickly at pH 10.0, which is higher than the previously reported lethal pH for T. thermophilus HB8 (11). The optimal temperature for enzyme activity was determined to be 65°C, which is comparable to the optimal temperature for T. thermophilus HB8 cell growth (65°C to 72°C) (11) (Fig. 3B). The enzyme was active at relatively high temperatures. At 80°C, the enzyme displayed ∼60% of the activity observed at the optimal temperature of 65°C. The thermostability of the enzyme was quantified by incubating the enzyme at different temperatures for 1 h. The enzyme maintained 95% activity after incubation at 60°C, while the activity decreased to 20% after incubation at 80°C (Fig. 3C).
FIG 3.
Optimal pH and temperature for the activity and thermotolerance of the enzyme. (A) Nicotinamidase activities at different pHs are shown. Triangles, squares, and circles represent activities in different buffers (MES, HEPES, and CHES, respectively). Activities are normalized to the maximum activity at pH 9 and are shown as percentages. (B) Activities at different temperatures are shown. Activities were normalized to the maximum activity at 65°C and are shown as percentage. (C) Residual activities after heat treatment at different temperatures are shown. These activities were normalized by the enzymatic activity without heat treatment (control) and are shown as percentages. Averages and the standard errors from triplicates are shown.
Based on the result above, the enzyme kinetic parameters were determined at pH 9 at 65°C. This enzyme has a relatively low Km value of 17 μM and high kcat value of 50 s−1 against nicotinamide (see Fig. S2). The kinetic parameters of nicotinamidase have been studied in different bacteria (Mycobacterium tuberculosis [17], Streptococcus pneumoniae [18], and Oceanobacillus iheyensis [16]), archaea (Acidilobus saccharovorans [19]), and eukaryotes (Saccharomyces cerevisiae [12]). The kinetic parameters determined in this study are comparable to those of nicotinamidases previously reported from other organisms (Table 1).
TABLE 1.
Kinetic parameters of nicotinamidases from different organisms
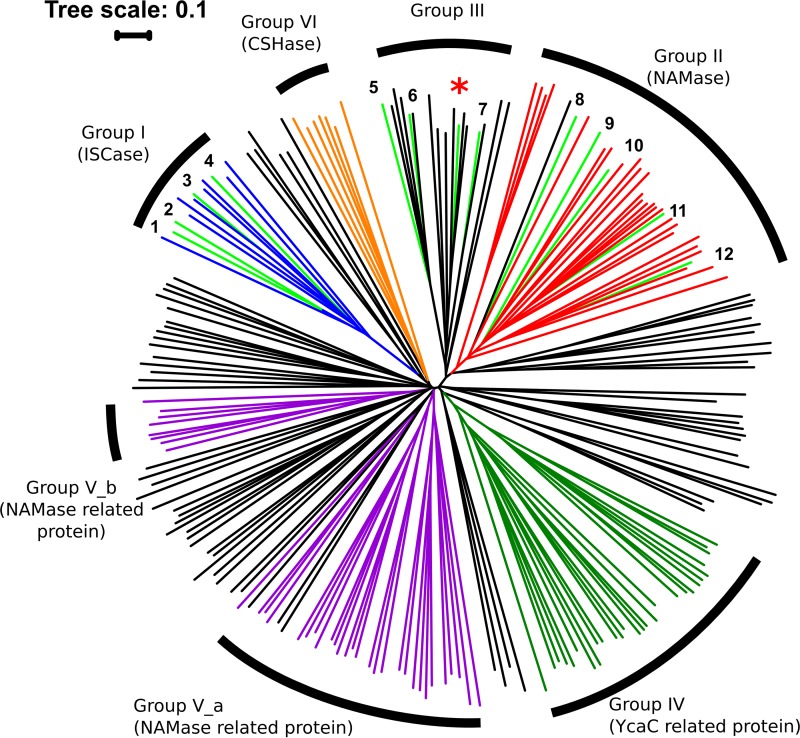
The product of TTHA0328 was annotated as isochorismatase from its amino acid sequence. Nicotinamidases belongs to the cysteine hydrolase superfamily, along with other subfamilies, including isochorismatase (EC 3.3.2.1), N-carbamoylsarcosine amidohydrolase (EC 3.5.1.59), nicotinamidase-related protein, and a functionally unknown YcaC-related protein (20). The classification of these proteins is challenging because of the high similarity in their amino acid sequences (2). In the phylogenetic tree, a high similarity of proteins from the cysteine hydrolases superfamily was observed (Fig. 4). Proteins experimentally characterized as isochorismatase belong to group I. On the other hand, proteins experimentally characterized as nicotinamidase belong to two distinct groups, one described as nicotinamidase (group II) and the other described as cysteine hydrolase not belonging to any specific subfamilies (group III) (20). The nicotinamidase characterized in this study belongs to group III in the phylogenetic tree, along with other proteins functioning as nicotinamidases.
FIG 4.
Phylogenetic tree of nicotinamidases and other cysteine hydrolases. The light green line with an asterisk represents the product of TTHA0328. Lines in light green indicate the proteins that were experimentally characterized as isochorismatase (no. 1 to 4) or nicotinamidase (no. 5 to 10). Blue, orange, red, dark green, and purple lines indicate the protein species belonging to the subfamilies isochorismatase (ISCase), nicotinamidase (NAMase), YcaC-related proteins, NAMase-related proteins, and N-carbamoylsarcosine amidohydrolase (CSHase), respectively, according to the NCBI conserved protein domain family. Black lines indicate the proteins belonging to the cysteine hydrolase superfamily but not to specific subfamilies. 1, P0C6D3 from V. cholerae (ISCase); 2, Q6W4P9 from V. anguillarum (ISCase); 3, Q7DC80 from Pseudomonas aeruginosa (ISCase); 4, P0ADI4 from E. coli (ISCase); 5, A0A0H2UR34 from S. pneumoniae (NAMase); 6, Q8ESQ6 from O. iheyensis (NAMase); 7, D9Q1R5 from A. saccharovorans (NAMase); 8, P53184 from S. cerevisiae (NAMase); 9, A4HRG8 from L. infantum (NAMase); 10, O58727 from P. horikoshii (NAMase); 11, B0VA03 from A. baumannii (NAMase); 12, I6XD65 from M. tuberculosis (NAMase).
Considering the catalytic activity at a wide range of pHs as well as at high temperatures, the kinetic parameters, and the similarity of its amino acid sequence with other nicotinamidases, we showed that the product of TTHA0328 is a physiologically functional nicotinamidase in T. thermophilus HB8.
A single nicotinamidase in T. thermophilus.
To investigate other candidates of nicotinamidase in T. thermophilus, a deletion experiment and the following biochemical assay were performed. The deletion strain of TTHA0328 (ΔTTHA0328) was constructed, as described in Materials and Methods (see Fig. S3). The wild-type strain of T. thermophilus HB8 (WT) and the ΔTTHA0328 deletion strain were both cultured in a complex medium, and crude lysates were prepared for determining the residual nicotinamidase activity. The lysate of the ΔTTHA0328 deletion strain showed no detectable nicotinamidase activity, while that of the WT showed the activity of 0.111 ± 0.002 U/mg of total protein (see Fig. S4). This result showed that the product of TTHA0328 was the only enzyme functioning as a nicotinamidase in T. thermophilus HB8.
Effects of nicotinamidase gene deletion on cell growth at different temperatures.
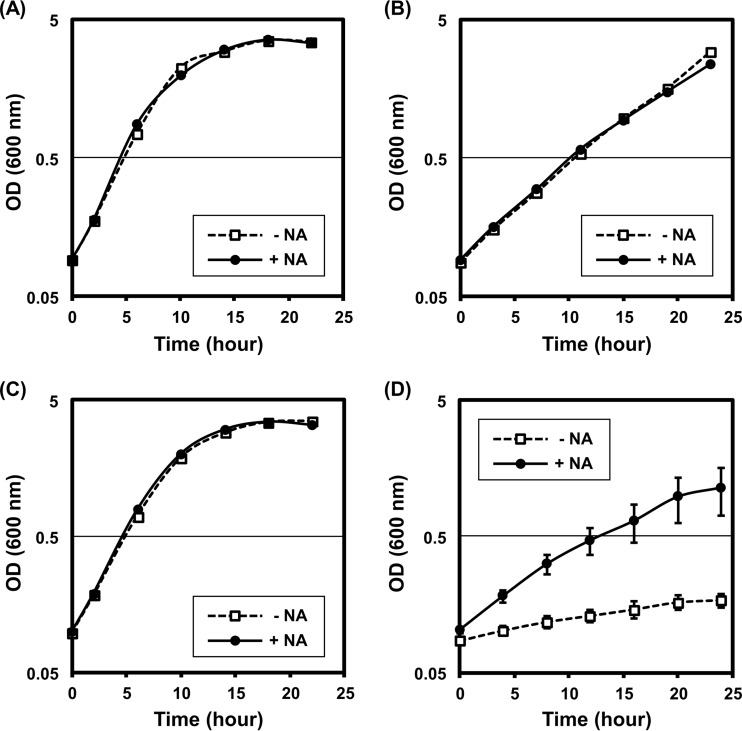
The salvage pathway for NAD+ initiates from the deamination of nicotinamide to nicotinate in most bacteria; therefore, the deletion of nicotinamidase disrupts the salvage pathway by halting the nicotinate supply (Fig. 1). We evaluated the influence of TTHA0328 deletion on cell growth in minimal medium with or without nicotinate supplement. Two different temperatures were used for the evaluation: 70°C, which is within the optimal growth temperature of T. thermophilus HB8, and 80°C, which is higher than the optimal temperature. First, we confirmed that the addition of nicotinate to the medium did not influence the cell growth of the WT at either 70°C or 80°C (Fig. 5A and B). At 70°C, the ΔTTHA0328 deletion strain showed little or no difference in cell growth with or without nicotinate supplementation, and the cell growth was comparable to that of the WT strain (Fig. 5A and C). On the contrary, the growth of the ΔTTHA0328 deletion strain was strongly suppressed when the temperature was elevated from 70°C to 80°C. This growth suppression of the ΔTTHA0328 deletion strain at 80°C was restored by nicotinate supplementation (Fig. 5D). These results indicated that the nicotinate supply to the NAD+ salvage pathway is limiting for cell growth of the ΔTTHA0328 deletion strain at 80°C.
FIG 5.
Cell growth of T. thermophilus WT and ΔTTHA0328 deletion strains at different temperatures. The cell growth of the WT strain (A and B) and the ΔTTHA0328 deletion strain (C and D) without nicotinate supplementation (−NA) and with nicotinate supplementation (+NA) are shown. The cultivation temperatures are 70°C (A and C) and 80°C (B and D). The y axes are in the logarithmic scale. Averages and standard errors from biological triplicates are shown.
Intracellular concentration of NAD+/NADH of the ΔTTHA0328 deletion strain at different temperatures.
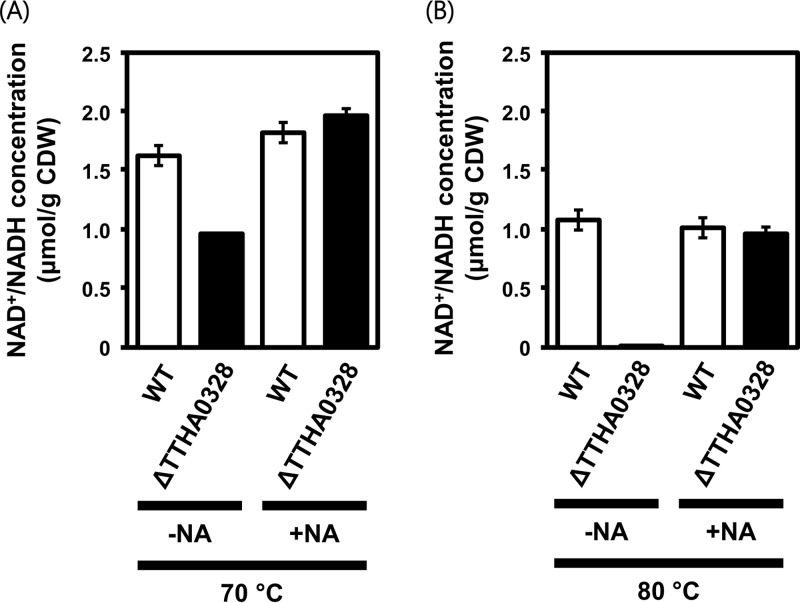
To evaluate the effect of TTHA0328 deletion on the salvage synthesis of NAD+, we measured the total concentration of NAD+/NADH inside cells of the WT and the ΔTTHA0328 deletion strain at two different temperatures of 70°C and 80°C. Although TTHA0328 deletion did not markedly affect cell growth at 70°C, its effect on the intracellular concentration of NAD+/NADH was already significant at this temperature (Fig. 6A). At 80°C, the NAD+/NADH concentration drastically decreased in the ΔTTHA0328 deletion strain to 0.09 ± 0.05 μmol/g (dry weight) cells (Fig. 6B). Both at 70°C and at 80°C, the addition of nicotinate could cancel the effect of TTHA0328 deletion. These results demonstrated that the product of TTHA0328 and the NAD+ salvage pathway in T. thermophilus HB8 play an essential role for maintaining the NAD+/NADH availability in cells, especially at high temperatures.
FIG 6.
Intracellular concentration of NAD+/NADH. The intracellular concentration of NAD+/NADH is shown for the WT strain and the ΔTTHA0328 deletion strain grown at 70°C (A) and 80°C (B). The concentration was determined without nicotinate addition (−NA) and with nicotinate addition (+NA). Averages and standard errors from triplicates are shown. CDW, cell dry weight.
DISCUSSION
The availability of NAD+ is essential for cell growth. For example, the growth of Escherichia coli was severely suppressed under limited availability of intracellular NAD+ (21). Many organisms possess distinct types of pathways for the salvage synthesis of NAD+, in which chemical and/or enzymatic degradants of NAD+ (e.g., nicotinamide and nicotinate) can be directly utilized as building blocks of the cofactor resynthesis (2, 6). Some nicotinate-auxotrophic yeasts (22) and pathogenic microorganisms (23–25) are known to grow even without a de novo synthetic pathway for NAD+, indicating the importance of the salvage pathway in these organisms. In addition, the growth of thermophiles also appears to be highly dependent on the salvage pathway because of the thermally unstable nature of NAD+. Nevertheless, the contribution of the salvage pathway to NAD+ availability in thermophiles, including T. thermophilus, has not been fully examined. In this study, we demonstrated that TTHA0328 encodes a functional nicotinamidase, which catalyzes the initial reaction of the NAD+ salvage pathway, in T. thermophilus HB8 and that the salvage pathway is crucial for the bacterial growth at high temperatures.
To cope with a hot environment, thermophiles are equipped with a variety of molecular mechanisms, such as the stabilization of DNA, RNA, proteins, and lipid membranes (26–28). Reverse gyrase is known to be a protein unique to thermophilic organisms (29), and its essentiality at high temperature was reported in Thermococcus kodakarensis (30) and Pyrococcus furiosus (31). Although nicotinamidases are conserved not only in thermophiles but also in mesophilic organisms, our study highlighted that this enzyme and the NAD+ salvage pathway is essential for a thermophilic model bacterium, T. thermophilus HB8, as a strategy to live with the thermal instability of NAD+ at high temperatures.
Homology and literature searches revealed that nicotinamidases are widely distributed in thermophilic organisms, regardless of whether they are bacteria/archaea or aerobic/anaerobic. For example, a nicotinamidase was identified in the anaerobic archaeon Pyrococcus horikoshii (13), and the catalytic activity of a nicotinamidase from the aerobic archaeon Thermoplasma acidophilum was experimentally confirmed (5). Furthermore, the presence of a nicotinamidase gene was predicted in many thermophilic organisms, such as the anaerobic archaeon T. kodakarensis, the aerobic archaeon Aeropyrum pernix, the anaerobic bacterium Thermotoga maritima, and the aerobic bacterium Aquifex aeolicus (2). On the other hand, several thermophiles, such as Nanoarchaeum equitans, Thermosynechococcus elongates, Methanobacterium thermoautotrophicum, and Methanocaldococcus jannaschii, are reported to have no nicotinamidase homologue (2). N. equitans is an obligate symbiont of the Ignicoccus genus and expected to obtain NAD+ or its precursors from the symbiont (32, 33). T. elongates possesses a gene encoding a putative nicotinate phosphoribosyltransferase (NaPRT), which is involved in the salvage synthesis of NAD+ and therefore may have an alternative route to supply nicotinate to the salvage pathway. By contrast, genes involved in NAD+ salvage synthesis are not found in thermophilic methanogenic archaea M. thermoautotrophicum and M. jannaschii (2, 6). These facts indicate that the NAD+ salvage pathway is not necessarily conserved among all thermophiles. However, considering the importance of NAD+ for cell metabolisms and its thermal instability, it would be reasonable to expect that these thermophiles have an alternative molecular apparatus to manage the thermal degradation of NAD+. Further studies would be needed to elucidate the mechanisms underlying the intracellular cofactor homeostasis of these organisms.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides used in this study.
Bacterial strains, plasmids, and oligonucleotides and their sequences are listed in Table 2. The E. coli Rosetta 2 (DE3) pLysS strain and the DH5α strain were purchased from Merck Millipore, Germany, and TaKaRa BIO, Japan, respectively. T. thermophilus HB8 was obtained from BioResource Center, RIKEN, Japan. The expression plasmid, pET11a-TTHA0328, as well as the plasmid encoding highly thermostable kanamycin nucleotidyltransferase (HTK) for the kanamycin resistance marker were obtained from the T. thermophilus HB8 gene expression library (BioResource Center, RIKEN, Japan) (34, 35). All oligonucleotides were purchased from Integrated DNA Technologies, USA.
TABLE 2.
Bacterial strains, plasmids, and oligonucleotides
| Designation | Description | Source or purpose |
|---|---|---|
| Strains and mutations | ||
| Rosetta 2 (DE3) pLysS | E. coli used for protein expression | Merck Millipore, Germany |
| DH5α | E. coli used for cloning | TaKaRa, Japan |
| T. thermophilus HB8 | Wild type | BioResource Center, RIKEN, Japan. |
| ΔTTHA0328 | TTHA0328 deletion strain of T. thermophilus | This work |
| Plasmids | ||
| pET11a-TTHA0328 | Expression plasmid for nicotinamidase | BioResource Center, RIKEN, Japan |
| pUC19-HTK | Source of thermostable kanamycin resistance cassette | BioResource Center, RIKEN, Japan |
| pUC19-ΔTTHA0328 | Plasmid for TTHA0328 deletion, containing HTK gene | This work |
| Oligonucleotides | ||
| U-F | 5′-ATTCGAGCTCGGTACCCGGGGGAGACCCGGTCCCCGTA-3′ | PCR for upstream region of TTHA0328 |
| U-R | 5′-GCCGTCAACGCACCTGGACCATGCCCTC-3′ | PCR for upstream region of TTHA0328 |
| D-F | 5′-GGTCCAGGTGCGTTGACGGCGGATATGG-3′ | PCR for downstream region of TTHA0328 |
| D-R | 5′-AAACAAACCGCGTAACCAACATGATTAAGAATTATTAGAGG-3′ | PCR for downstream region of TTHA0328 |
| HTK-F | 5′-GTTGGTTACGCGGTTTGTTTGAGCACCCCG-3′ | PCR for HTK |
| HTK-R | 5′-CCTGCAGGTCGACTCTAGAGCCTCGCCGAAGGCGGGAA-3′ | PCR for HTK |
| Seq_F | 5′-GCGTTGTTGAAGAAGAGCTGGG-3′ | PCR for sequencing |
| Seq_R | 5′-CTTTAGGGGGAGGACAGGCTTC-3′ | PCR for sequencing |
Multiple-sequence alignment.
Multiple-sequence alignment was performed by Clustal Omega and visualized by ESPript (36). Nicotinamidase sequences from different organisms were used for comparison (UniProt accession number P53184 from S. cerevisiae, A4HRG8 from Leishmania infantum, O58727 from P. horikoshii, D9Q1R5 from A. saccharovorans, I6XD65 from M. tuberculosis, A0A0H2UR34 from S. pneumoniae, B0VA03 from Acinetobacter baumannii, and Q8ESQ6 from O. iheyensis).
Protein expression and purification by size exclusion chromatography.
For protein expression, Rosetta 2 (DE3) pLysS E. coli cells were transformed with pET11a-TTHA0328 and grown in lysogeny broth (LB) medium (37) at 37°C at 200 rpm to an optical density at 600 nm (OD600) of 0.5. Protein expression was induced by the addition of 0.4 mM isopropyl-β-d-1-thiogalactopyranoside, and cells were further incubated at 37°C at 200 rpm for 4 h. Cells were harvested by centrifugation, resuspended in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, pH 7.5), and disrupted by sonication. Cell debris was removed by centrifugation at 15,000 × g for 10 min at 4°C. The supernatants were incubated at 60°C for 30 min to deactivate proteins derived from E. coli. They were then recentrifuged at 15,000 × g for 10 min at 4°C, and the supernatants were used as the crude lysates.
The lysates were purified by gel filtration chromatography using ÄKTA protein purification systems (GE Healthcare Life Sciences, USA). HiLoad Superdex 200 pg (120 ml, 1.6 cm by 60 cm; GE Healthcare Life Sciences, USA) was equilibrated with 3 column volumes of the mobile phase (50 mM Tris HCl, 150 mM NaCl, pH 7.5). The crude lysates were injected and eluted with the mobile phase at the flow rate of 0.8 ml/min. Fractions of 1 ml were collected.
Purified protein concentrations were determined using the Bio-Rad protein assay kit (Bio-Rad, USA). The homogeneity of the purified proteins was confirmed by 12% SDS-PAGE, which were then used in subsequent experiments.
Detection of nicotinamidase activity by HPLC analysis.
Nicotinamidase activity was determined using HPLC analysis by measuring the conversion of nicotinamide to nicotinate (38). The Prominence HPLC system (Shimadzu, Japan) was used. The samples (10 μl) were separated using COSMOSIL 5 C18 AR-II columns (Nacalai Tesque, Japan) with the mobile phase (9% of methanol, 1% of acetic acid, and 0.05 M KH2PO4) at the flow rate of 0.6 ml/min. The absorbance profile at 264 nm was monitored, and the nicotinate concentration was determined by integrating the area of the corresponding peak.
Enzyme assay.
The enzymatic activity was determined using 2 nM enzyme and 1 mM nicotinamide. The samples were collected every 5 min and the reactions were quenched by the addition of an equal volume of 0.6 M HCl. The conversion of 1 μmol of substrate per minute was defined as one unit of activity, and the specific activity was described as units per milligram of purified protein.
To determine the optimal pH for enzymatic activity, the enzyme reactions were performed at 60°C in buffer solutions with different pHs {50 mM MES [2-(N-morpholino)ethanesulfonic acid] and 150 mM NaCl for pH 6.0 and 7.0; 50 mM HEPES and 150 mM NaCl for pH 7.0, 7.5, 8.0, and 8.5; and 50 mM CHES (N-cyclohexyl-2-aminoethanesulfonic acid) and 150 mM NaCl for pH 8.5, 9.0, 9.5, and 10.0}. To determine the optimal temperature, enzymatic activity was measured in 50 mM CHES and 150 mM NaCl (pH 9.0) at different temperatures (30, 40, 50, 60, 65, 70, 75, 80, and 90°C). For thermostability, the enzyme was preincubated for 1 h at different temperatures (60, 70, 80, 90, and 100°C), and then the activity was measured in 50 mM CHES and 150 mM NaCl at 65°C. The kinetic parameters of the enzyme were determined using 0.2 nM enzyme in 50 mM CHES and 150 mM NaCl at 65°C with different concentrations of nicotinamide (10, 20, 30, 40, 50, 80, 100, and 200 μM).
Phylogenetic tree.
The phylogenetic tree was constructed using either of the three types of sequences: (i) model proteins in the cd00431 sequence cluster, cysteine hydrolases (1), (ii) protein sequences for which the activity was experimentally characterized as nicotinamidase (also used for multiple alignment), or (iii) protein sequences for which the activity was experimentally characterized as isochorismatase (UniProt accession number P0ADI4 from E. coli, Q7DC89 from Pseudomonas aeruginosa, P0C6D3 from Vibrio cholerae, and Q6W4P9 from Vibrio anguillarum). A neighbor-joining phylogenetic tree was visualized using iTOL (39) after clustering by Clustal Omega.
Deletion of TTHA0328 in T. thermophilus HB8.
Deletion of TTHA0328 in T. thermophilus was performed as previously described (40). For construction of the deletion plasmid, the gene encoding HTK and both the upstream and downstream genomic regions flanking TTHA0328 were applied by PCR with the following oligonucleotide pairs: HTK-F/HTK-R, U-F/U-R, and D-F/D-R. These DNA fragments were assembled and inserted into a BamHI-digested pUC19 plasmid (TaKaRa Bio, Japan) by the In-Fusion HD cloning kit (Clontech, USA). The DNA mixture was used for the transformation of E. coli DH5α, and the plasmid was extracted from the ampicillin-resistant cells.
For the gene deletion, T. thermophilus cells were cultured at 70°C at 160 rpm in TT (T. thermophilus) liquid medium (0.8% polypeptone, 0.4% yeast extract, 0.2% NaCl, 0.4 mM MgCl2, 0.4 mM CaCl2, pH 7.2). For the transformation, 500 μl of this culture was supplemented with 20 μl of the deletion plasmid (∼5 μg) at an OD600 of 0.6 and further cultured for 2 h. The transformants were selected on TT gellan gum plates (1.5% gellan gum) with 50 μg/ml of kanamycin. For confirmation of the gene deletion in the chromosome, genomic DNA was extracted with the Wizard genomic DNA purification kit (Promega, USA) from kanamycin resistant transformants and then amplified with the oligonucleotide pair Seq-F/Seq-R. PCR products were purified with the Wizard Plus SV Minipreps DNA purification system (Promega, USA) and then sequenced with the oligonucleotides Seq-F and Seq-R (FASMAC, Japan) for confirmation of the gene deletion.
Enzyme activity of the crude extracts from T. thermophilus.
T. thermophilus cells were grown in TT liquid medium at 70°C at 160 rpm. Since this organism is reported as a polyploid bacterium (41), the cultivation of the ΔTTHA0328 deletion strain was performed with the supplementation of 50 μg/ml of kanamycin to avoid the occurrence of cells without the TTHA0328 deletion. Cells were harvested by centrifugation at 4,000 × g, were washed once, and were then resuspended in the reaction buffer (50 mM Tris-HCl, 150 mM NaCl, pH 7.5) at a concentration of 200 mg (wet weight) of cells/ml. Cells were disrupted by sonication and centrifuged again, and the crude lysate was collected. The reaction mixtures (containing 50 μl of the lysate and 1 mM nicotinamide in the reaction buffer) were incubated at 65°C, and samples were collected every 5 min. Enzymatic activity was determined by HPLC analysis, and the specific activity was described as units per milligram of total protein.
Growth experiment for T. thermophilus.
T. thermophilus WT and ΔTTHA0328 deletion strains were precultured at 70°C in the chemically defined medium (42) (see Table S1 in the supplemental material) and inoculated at an OD600 of 0.1 into fresh medium with or without supplementation of 100 μM nicotinate. For cultivation of the ΔTTHA0328 deletion strain, 50 μg/ml of kanamycin was used. The OD600 was measured with appropriate dilution.
Measurement of intracellular concentration of NAD+/NADH at different temperatures.
The intracellular concentration of NAD+/NADH was measured using the cycling assay as previously described with modifications (21). Briefly, cells were grown in the chemically defined medium with or without supplementation of 100 μM nicotinate and harvested at the late-exponential phase (OD600 of 1) by centrifugation at 15,000 × g for 1 min at 4°C. Cells were washed twice with ice-cold phosphate-buffered saline (pH 7.4) and stored at −80°C until further use. For the NAD+/NADH quantification assay, cells were resuspended with deionized water to an OD600 of 10 and incubated on ice for 5 min. Cell debris was removed by centrifugation at 15,000 × g for 5 min at 4°C, and 10 μl of the supernatants was used for quantification. The assay was performed on a 96-well microplate using the microplate reader GENios (TECAN, Switzerland). The reaction mixtures (200 μl) were composed of 100 mM BICIN buffer (pH 8.0), 10% (vol/vol) ethanol, 4 mM EDTA (pH 8.0), 0.42 mM 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), 3.32 mM phenazine methosulfate, 5 U yeast alcohol dehydrogenases II (Oriental Yeast Co., Ltd., Japan), and the sample. The increase of absorption at 595 nm was continually monitored for 5 min, and the concentration of NAD+/NADH was calculated based on the NAD+ standard curve.
Supplementary Material
ACKNOWLEDGMENTS
This work was partly supported by the Japan Science and Technology Agency (JST), A-STEP programs, and the Japan Society for the Promotion of Science (JSPS) KAKENHI grant (26450088).
S.S. and P.C. appreciate the support from Japan Student Services Organization (JASSO).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00359-17.
REFERENCES
- 1.Berger F, Ramírez-Hernández MH, Ziegler M. 2004. The new life of a centenarian: signalling functions of NAD(P). Trends Biochem Sci 29:111–118. doi: 10.1016/j.tibs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Gazzaniga F, Stebbins R, Chang SZ, McPeek MA, Brenner C. 2009. Microbial NAD metabolism: lessons from comparative genomics. Microbiol Mol Biol Rev 73:529–541. doi: 10.1128/MMBR.00042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K. 2012. Engineering the third wave of biocatalysis. Nature 485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 4.Chenault HK, Whitesides GM. 1987. Regeneration of nicotinamide cofactors for use in organic synthesis. Appl Biochem Biotechnol 14:147–197. doi: 10.1007/BF02798431. [DOI] [PubMed] [Google Scholar]
- 5.Honda K, Hara N, Cheng M, Nakamura A, Mandai K, Okano K, Ohtake H. 2016. In vitro metabolic engineering for the salvage synthesis of NAD+. Metab Eng 35:114–120. doi: 10.1016/j.ymben.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Gossmann TI, Ziegler M, Puntervoll P, de Figueiredo LF, Schuster S, Heiland I. 2012. NAD+ biosynthesis and salvage—a phylogenetic perspective. FEBS J 279:3355–3363. doi: 10.1111/j.1742-4658.2012.08559.x. [DOI] [PubMed] [Google Scholar]
- 7.Preiss J, Handler P. 1958. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J Biol Chem 233:488–492. [PubMed] [Google Scholar]
- 8.Preiss J, Handler P. 1958. Biosynthesis of diphosphopyridine nucleotide. II. Enzymatic aspects. J Biol Chem 233:493–500. [PubMed] [Google Scholar]
- 9.Krutsakorn B, Honda K, Ye X, Imagawa T, Bei X, Okano K, Ohtake H. 2013. In vitro production of n-butanol from glucose. Metab Eng 20:84–91. doi: 10.1016/j.ymben.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Ninh PH, Honda K, Sakai T, Okano K, Ohtake H. 2015. Assembly and multiple gene expression of thermophilic enzymes in Escherichia coli for in vitro metabolic engineering. Biotechnol Bioeng 112:189–196. doi: 10.1002/bit.25338. [DOI] [PubMed] [Google Scholar]
- 11.Oshima T, Imahori K. 1974. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int J Syst Evol Microbiol 24:102–112. doi: 10.1099/00207713-24-1-102. [DOI] [Google Scholar]
- 12.Ghislain M, Talla E, François JM. 2002. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast 19:215–224. doi: 10.1002/yea.810. [DOI] [PubMed] [Google Scholar]
- 13.Du X, Wang W, Kim R, Yakota H, Nguyen H, Kim SH. 2001. Crystal structure and mechanism of catalysis of a pyrazinamidase from Pyrococcus horikoshii. Biochemistry 40:14166–14172. doi: 10.1021/bi0115479. [DOI] [PubMed] [Google Scholar]
- 14.French JB, Cen Y, Vrablik TL, Xu P, Allen E, Hanna-Rose W, Sauve AA. 2010. Characterization of nicotinamidases: steady state kinetic parameters, classwide inhibition by nicotinaldehydes, and catalytic mechanism. Biochemistry 49:10421–10439. doi: 10.1021/bi1012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu G, Taylor AB, McAlister-Henn L, Hart PJ. 2007. Crystal structure of the yeast nicotinamidase Pnc1p. Arch Biochem Biophys 461:66–75. doi: 10.1016/j.abb.2007.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sánchez-Carrón G, García-García MI, Zapata-Pérez R, Takami H, García-Carmona F, Sánchez-Ferrer A. 2013. Biochemical and mutational analysis of a novel nicotinamidase from Oceanobacillus iheyensis HTE831. PLoS One 8:e56727. doi: 10.1371/journal.pone.0056727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Deng J-Y, Bi L-J, Zhou Y-F, Zhang Z-P, Zhang C-G, Zhang Y, Zhang X-E. 2008. Characterization of Mycobacterium tuberculosis nicotinamidase/pyrazinamidase. FEBS J 275:753–762. doi: 10.1111/j.1742-4658.2007.06241.x. [DOI] [PubMed] [Google Scholar]
- 18.French JB, Cen Y, Sauve AA, Ealick SE. 2010. High-resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into the catalytic mechanism and inhibition by aldehydes. Biochemistry 49:8803–8812. doi: 10.1021/bi1012436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stekhanova TN, Bezsudnova EY, Mardanov AV, Osipov EM, Ravin NV, Skryabin KG, Popov VO. 2014. Nicotinamidase from the thermophilic archaeon Acidilobus saccharovorans: structural and functional characteristics. Biochemistry 79:54–61. doi: 10.1134/S0006297914010088. [DOI] [PubMed] [Google Scholar]
- 20.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Wang L, Yang F, Lin X, Zhang S, Zhao ZK. 2011. Determining the extremes of the cellular NAD(H) level by using an Escherichia coli NAD+-auxotrophic mutant. Appl Environ Microbiol 77:6133–6140. doi: 10.1128/AEM.00630-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y-F, Bao W-G. 2007. Why do some yeast species require niacin for growth? Different modes of NAD synthesis. FEMS Yeast Res 7:657–664. doi: 10.1111/j.1567-1364.2007.00231.x. [DOI] [PubMed] [Google Scholar]
- 23.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 24.Sorci L, Blaby IK, Rodionova IA, De Ingeniis J, Tkachenko S, de Crécy-Lagard V, Osterman AL. 2013. Quinolinate salvage and insights for targeting NAD biosynthesis in group A streptococci. J Bacteriol 195:726–732. doi: 10.1128/JB.02002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gazanion E, Garcia D, Silvestre R, Gérard C, Guichou JF, Labesse G, Seveno M, Cordeiro-Da-Silva A, Ouaissi A, Sereno D, Vergnes B. 2011. The Leishmania nicotinamidase is essential for NAD+ production and parasite proliferation. Mol Microbiol 82:21–38. doi: 10.1111/j.1365-2958.2011.07799.x. [DOI] [PubMed] [Google Scholar]
- 26.Fukuchi S, Nishikawa K. 2001. Protein surface amino acid compositions distinctively differ between thermophilic and mesophilic bacteria. J Mol Biol 309:835–843. doi: 10.1006/jmbi.2001.4718. [DOI] [PubMed] [Google Scholar]
- 27.Koga Y. 2012. Thermal adaptation of the archaeal and bacterial lipid membranes. Archaea 2012:e789652. doi: 10.1155/2012/789652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakashima H, Fukuchi S, Nishikawa K. 2003. Compositional changes in RNA, DNA and proteins for bacterial adaptation to higher and lower temperatures. J Biochem 133:507–513. doi: 10.1093/jb/mvg067. [DOI] [PubMed] [Google Scholar]
- 29.Lulchev P, Klostermeier D. 2014. Reverse gyrase—recent advances and current mechanistic understanding of positive DNA supercoiling. Nucleic Acids Res 42:8200–8213. doi: 10.1093/nar/gku589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atomi H, Matsumi R, Imanaka T. 2004. Reverse gyrase is not a prerequisite for hyperthermophilic life. J Bacteriol 186:4829–4833. doi: 10.1128/JB.186.14.4829-4833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipscomb GL, Hahn EM, Crowley AT, Adams MWW. 2017. Reverse gyrase is essential for microbial growth at 95°C. Extremophiles 21:603–608. doi: 10.1007/s00792-017-0929-z. [DOI] [PubMed] [Google Scholar]
- 32.Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. 2002. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 33.Waters E, Hohn MJ, Ahel I, Graham DE, Adams MD, Barnstead M, Beeson KY, Bibbs L, Bolanos R, Keller M, Kretz K, Lin X, Mathur E, Ni J, Podar M, Richardson T, Sutton GG, Simon M, Söll D, Stetter KO, Short JM, Noordewier M. 2003. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci U S A 100:12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoseki J, Yano T, Koyama Y, Kuramitsu S, Kagamiyama H. 1999. Directed evolution of thermostable kanamycin-resistance gene: a convenient selection marker for Thermus thermophilus. J Biochem 126:951–956. doi: 10.1093/oxfordjournals.jbchem.a022539. [DOI] [PubMed] [Google Scholar]
- 35.Yokoyama S, Hirota H, Kigawa T, Yabuki T, Shirouzu M, Terada T, Ito Y, Matsuo Y, Kuroda Y, Nishimura Y, Kyogoku Y, Miki K, Masui R, Kuramitsu S. 2000. Structural genomics projects in Japan. Nat Struct Mol Biol 7:943–945. doi: 10.1038/80712. [DOI] [PubMed] [Google Scholar]
- 36.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 38.Ghosh MK. 2012. HPLC methods on drug analysis. Springer Science & Business Media, Berlin, Germany. [Google Scholar]
- 39.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hashimoto Y, Yano T, Kuramitsu S, Kagamiyama H. 2001. Disruption of Thermus thermophilus genes by homologous recombination using a thermostable kanamycin-resistant marker. FEBS Lett 506:231–234. doi: 10.1016/S0014-5793(01)02926-X. [DOI] [PubMed] [Google Scholar]
- 41.Ohtani N, Tomita M, Itaya M. 2010. An extreme thermophile, Thermus thermophilus, is a polyploid bacterium. J Bacteriol 192:5499–5505. doi: 10.1128/JB.00662-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida T, Lorence RM, Choc MG, Tarr GE, Findling KL, Fee JA. 1984. Respiratory proteins from the extremely thermophilic aerobic bacterium, Thermus thermophilus. Purification procedures for cytochromes c552, c555,549, and c1aa3 and chemical evidence for a single subunit cytochrome aa3. J Biol Chem 259:112–123. [PubMed] [Google Scholar]
- 43.Smith BC, Anderson MA, Hoadley KA, Keck JL, Cleland WW, Denu JM. 2012. Structural and kinetic isotope effect studies of nicotinamidase (Pnc1) from Saccharomyces cerevisiae. Biochemistry 51:243–256. doi: 10.1021/bi2015508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seiner DR, Hegde SS, Blanchard JS. 2010. Kinetics and inhibition of nicotinamidase from Mycobacterium tuberculosis. Biochemistry 49:9613–9619. doi: 10.1021/bi1011157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.