ABSTRACT
In the large majority of cases, HIV infection is established by a single variant, and understanding the characteristics of successfully transmitted variants is relevant to prevention strategies. Few studies have investigated the viral determinants of mother-to-child transmission. To determine the impact of Gag-protease-driven viral replication capacity on mother-to-child transmission, the replication capacities of 148 recombinant viruses encoding plasma-derived Gag-protease from 53 nontransmitter mothers, 48 transmitter mothers, and 47 infected infants were assayed in an HIV-1-inducible green fluorescent protein reporter cell line. All study participants were infected with HIV-1 subtype C. There was no significant difference in replication capacities between the nontransmitter (n = 53) and transmitter (n = 44) mothers (P = 0.48). Infant-derived Gag-protease NL4-3 recombinant viruses (n = 41) were found to have a significantly lower Gag-protease-driven replication capacity than that of viruses derived from the mothers (P < 0.0001 by a paired t test). High percent similarities to consensus subtype C Gag, p17, p24, and protease sequences were also found in the infants (n = 28) in comparison to their mothers (P = 0.07, P = 0.002, P = 0.03, and P = 0.02, respectively, as determined by a paired t test). These data suggest that of the viral quasispecies found in mothers, the HIV mother-to-child transmission bottleneck favors the transmission of consensus-like viruses with lower viral replication capacities.
IMPORTANCE Understanding the characteristics of successfully transmitted HIV variants has important implications for preventative interventions. Little is known about the viral determinants of HIV mother-to-child transmission (MTCT). We addressed the role of viral replication capacity driven by Gag, a major structural protein that is a significant determinant of overall viral replicative ability and an important target of the host immune response, in the MTCT bottleneck. This study advances our understanding of the genetic bottleneck in MTCT by revealing that viruses transmitted to infants have a lower replicative ability as well as a higher similarity to the population consensus (in this case HIV subtype C) than those of their mothers. Furthermore, the observation that “consensus-like” virus sequences correspond to lower in vitro replication abilities yet appear to be preferentially transmitted suggests that viral characteristics favoring transmission are decoupled from those that enhance replicative capacity.
KEYWORDS: human immunodeficiency virus, mother-to-child transmission, transmission bottleneck, viral replication capacity
INTRODUCTION
It is well known that 80 to 90% of heterosexual HIV-1 transmissions are established by a single viral variant due to a genetic bottleneck (1, 2). Understanding the characteristics of successfully transmitted viral variants may aid in the development of HIV prevention strategies. Overall, transmitted HIV-1 strains tend to display CCR5 coreceptor usage (3). Moreover, HIV-1 strains transmitted heterosexually tend to have shorter envelope variable loops with fewer glycosylation sites (thereby making them more neutralization sensitive [3, 4]) and tend to exhibit enhanced interferon alpha resistance (5, 6) (although this may not be the case for all HIV-1 subtypes [7]). Heterosexual HIV-1 transmission also tends to select for transmitted viruses with greater genetic similarity to the HIV-1 subtype consensus sequence (7, 8) but not an enhanced viral replicative capacity (7). Importantly, the strength of the transmission bottleneck, and, thus, the selection bias for certain viral characteristics, differs significantly by mode of transmission: the selection bias for consensus-like founder variants is more relaxed in the case of male-to-female transmission than vice versa (8), and there are distinct genetic footprints of the transmitted/founder virus that distinguish between heterosexual versus men-who-have-sex-with-men (MSM) transmissions (9).
While a relatively large number of studies have investigated the role of host factors, including HLA allele sharing, placental CCR5 expression, polymorphisms in chemokine and other immunomodulatory genes, and maternal HIV-specific antibodies, in mother-to-child transmission (MTCT) risk (10–16), relatively little is known regarding the role of viral factors in MTCT. Kong and colleagues found that viral replication capacity determined by the V1-to-V5 region of Env was a determinant of MTCT from chronically infected mothers to their infants; specifically, the faster-replicating variants were preferentially transmitted (17). No studies have yet investigated the effect of Gag-driven viral replication capacity on MTCT, although it has been found to be a significant determinant of adult disease progression (18–20). Gag is an important structural protein crucial for viral replication (21) and has been shown to contribute significantly to whole-isolate replication capacity (22, 23), explaining as much as 72% of the variability in HIV-1 replication capacity (24), although another recent study did not observe this relationship (25). Furthermore, Gag has been shown to be an important target of CD8+ T cell (cytotoxic T lymphocyte [CTL]) immune responses (26, 27), and CTL escape mutations in critical Gag epitopes have been shown to significantly reduce viral replication capacity (28–32).
The purpose of this study was to investigate viral determinants of MTCT in HIV-1 subtype C, the most prevalent subtype globally. The specific aims were 2-fold: first to determine whether the Gag-protease-driven replication capacity of viruses in mothers is a determinant of transmission to their infants and second to determine whether viruses that establish infection in infants have a different Gag-protease-driven replication capacity than that of viruses present in their mothers. We studied totals of 53 nontransmitter mothers, 48 transmitter mothers, and 41 mother-infant pairs (MIPs). Gag-protease-driven replication capacity did not distinguish between transmitter and nontransmitter mothers; however, the viruses that were successfully transmitted to the infants displayed significantly lower replication capacities than those of viruses from the mothers. Sequence analysis determined that the transmitted viruses were biased toward the consensus sequence.
RESULTS
Transmitter mothers transmit viruses with lower replication capacity to their infants.
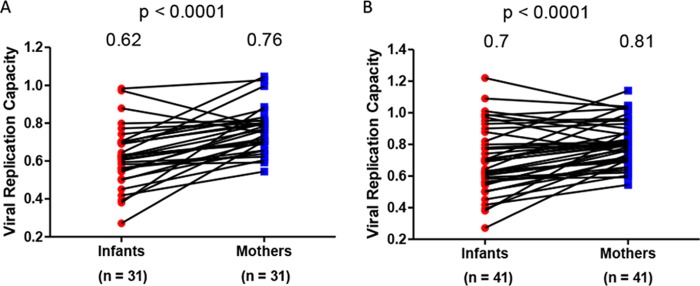
The MTCT bottleneck was characterized by comparing the viral replication capacities of mothers and infants. The chronically HIV-1-infected mothers harbor a diverse pool of viruses that consists of a heterogeneous mixture of variants. MTCT acts as a bottleneck and allows very few variants to be transmitted to the infant (33). Indeed, sequencing of Gag-protease from the transmitter mothers and their infants revealed that the mothers have more mixtures (median = 10; interquartile range [IQR], 5 to 15.5) than their infants (median = 2; IQR, 0 to 4) (P = 0.0003 as determined by a paired t test) (data not shown). It was hypothesized that viral variants with a higher replication capacity would be transmitted. The viral replication capacity driven by Gag-protease was determined for 31 MIPs by constructing recombinant NL4-3 viruses encoding patient-derived Gag-protease (using bulk PCR products) for the mothers and their infants and then comparing the replication capacity of these viruses in a green fluorescent protein (GFP) reporter T cell line. Contrary to our hypothesis, the viral replication capacity of viruses infecting the infants (mean = 0.62) was significantly lower than that of viruses infecting the mothers (mean = 0.76) (P < 0.0001 as determined by a paired t test) (Fig. 1A).
FIG 1.
Gag-protease-mediated viral replication capacities in HIV-infected mother-infant pairs. (A) Data from 31 mother-infant pairs (MIPs). (B) Data from the 31 MIPs in panel A and an additional 10 MIPs for which analyses were undertaken in a separate laboratory. Gag-protease NL4-3 recombinant viruses derived from the infants had significantly lower viral replication capacities than those of viruses their mothers (P < 0.0001 as determined by a paired t test). Replication capacity was assayed in GXR cells by using flow cytometry and normalized to the growth of the wild-type NL4-3 virus (replication capacity value of 1).
As this finding was unexpected, the experiments were repeated in an independent laboratory on a subset of 19 MIPs and an additional 10 MIPs from the same cohort. There was an excellent correlation between the replication capacity data on the same 19 MIPs that were generated in both laboratories (Pearson's correlation [r] = 0.83 and P < 0.0001) (data not shown), although the scales of these values differed between laboratories, likely reflecting differences in the laboratory-adapted NL4-3 reference stock used for normalization purposes. The repeated subset of 19 MIPs yielded the same observation that viruses transmitted to the infants had lower replication capacities (mean = 0.76) than those of viruses from their mothers (mean = 0.86) (P = 0.03 as determined by a paired t test) (data not shown). When the additional 10 MIPs were included in the original analysis of the 31 MIPs (n = 41), the replication capacities of the infants (mean = 0.7) still remained significantly lower than those of their mothers (mean = 0.81) (P < 0.0001 as determined by a paired t test) (Fig. 1B).
Next, we wished to investigate whether the mode of transmission may influence the difference in viral replication capacities between the mothers and their infants. This was investigated in the 41 MIPs. We hypothesized that in utero (IU) transmission may present a greater bottleneck than intrapartum (IP) transmission. Therefore, we performed the same comparison of viral replication capacities between mothers and infants in MIPs classified as either IU or IP transmission. A significant difference in viral replication capacities between mothers and their infants was observed in both transmission groups (P = 0.0012 [IU] and P = 0.043 [IP] as determined by a paired t test) (data not shown). The mean differences in replication capacities between viruses derived from mothers and those derived their infants were similar in both the IU and IP transmission groups (0.11 versus 0.1, respectively), indicating no evidence for a greater bottleneck in IU versus IP transmission in terms of viral replication capacity.
No difference in viral replication capacities were observed between transmitter and nontransmitter mothers.
Although we observed that viruses with lower replication capacities were selected for transmission, we wanted to investigate whether viral replication capacity in the mother was a factor determining transmission, with the initial hypothesis that a higher viral replication capacity in the mother would likely favor transmission. Therefore, we compared viral replication capacities in 44 transmitter mothers and 53 nontransmitter mothers who were matched for CD4 counts and viral loads. No significant difference in viral replication capacities between transmitter (mean = 0.76) and nontransmitter (mean = 0.74) mothers was found (P = 0.48 as determined by a t test) (Fig. 2). This suggests that Gag-protease-driven replication capacity does not determine whether mothers transmit the virus to their infants.
FIG 2.
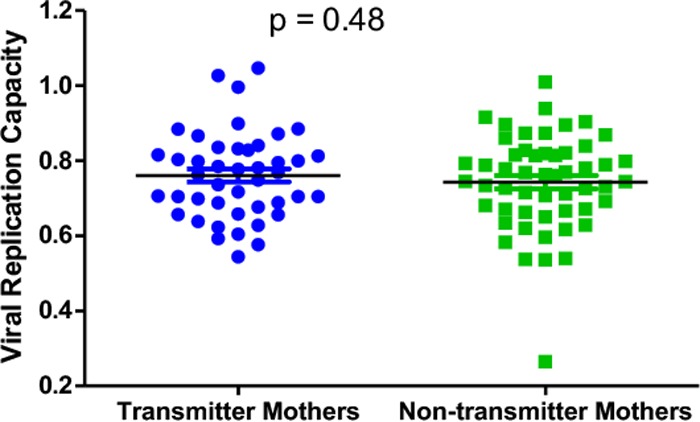
Gag-protease-mediated viral replication capacities in transmitter and nontransmitter mothers. There is no significant difference in Gag-protease-mediated replication capacities between viruses derived from mothers that transmit the virus to their infants and those derived from mothers that do not transmit the virus to their infants (t test).
Selection of consensus viruses for MTCT.
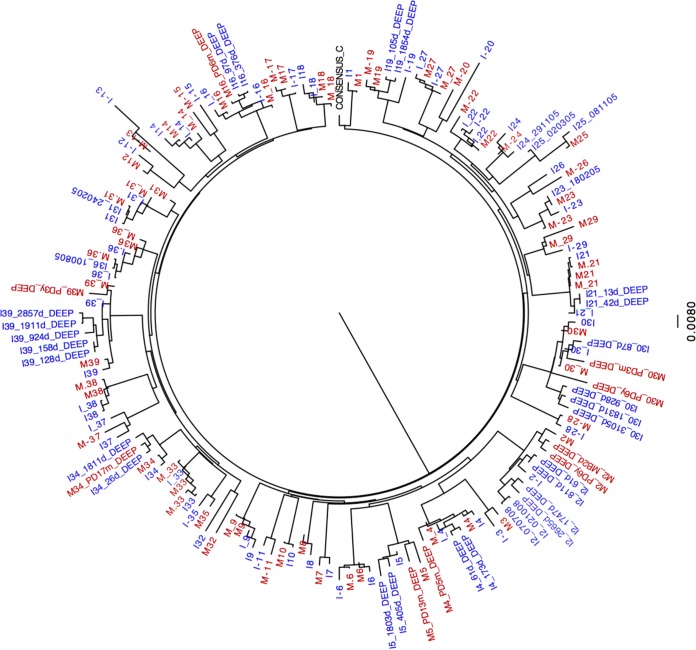
The sequencing of Gag-protease from bulk PCR products was successful for 28 out of the 31 MIPs for which replication capacities were determined. Sequences were analyzed to investigate genetic determinants of the differences in viral replication capacities between mothers and their infants. The phylogenetic relatedness within each MIP was confirmed (Fig. 3). Gag-protease from a subset of recombinant viruses was also sequenced; in all cases, these sequences clustered with their respective bulk PCR sequences, as expected (data not shown).
FIG 3.
Viral phylogenetic relatedness between mother and child. Replication capacities were successfully determined for 41 MIPs, including 31 MIPs in the Durban laboratory and an additional 10 MIPs from an independent laboratory. Out of the 41 pairs, sequencing data were successful for 39 pairs. A maximum likelihood phylogenetic tree was constructed by using gag sequences, generated by Sanger and ultradeep sequencing, from 39 mother-infant pairs and the subtype C reference sequence. Sequences from the mothers are highlighted in red, and sequences from the infants are highlighted in blue. Viral sequences from the mothers and infants in each of the pairs clustered together, confirming the phylogenetic relatedness between viruses isolated from the mother and infant in each pair.
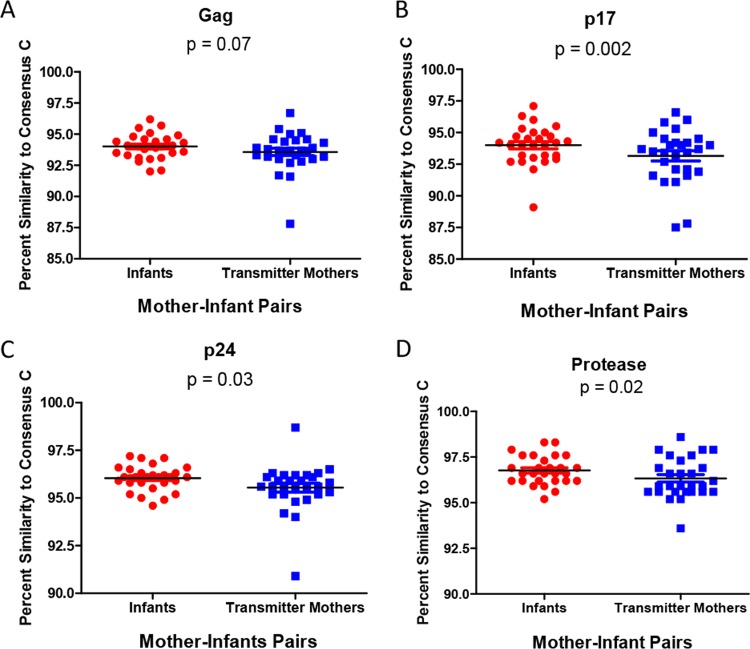
Previous studies of Gag-protease recombinant viruses and infectious molecular clones found an inverse relationship between sequence similarity to the consensus and in vitro replication capacity (7, 18). Furthermore, it was demonstrated that there is a transmission bias for sequences that are consensus-like, using Gag, Pol, and Nef sequencing of 137 adult subtype C-infected transmission pairs (8), and this finding was corroborated in a whole-genome analysis of 6 transmission pairs from the same cohort (7). Therefore, we analyzed the relationship between similarity to the consensus sequence and replication capacity in our cohort and investigated whether similarities to the consensus were significantly different between mothers and their infants. We found an inverse relationship between the percent amino acid similarity of Gag, and each of the individual Gag proteins, to the consensus sequence and viral replication capacity, although this was statistically significant for only Gag p17 (r = −0.41 and P = 0.0003) (data not shown). This was found to be in line with data from other studies (18). The differences in Gag-protease percent amino acid similarities to the consensus between sequences derived from mothers and those derived from infants were then compared. There was a trend that infants had higher percent amino acid similarities to the consensus sequence than their mothers for Gag (P = 0.07 as determined by a paired t test) (Fig. 4A). The infants had significantly higher percent amino acid similarity to the consensus than their mothers for p17 (P = 0.002) (Fig. 4B), p24 (P = 0.03) (Fig. 4C), and protease (P = 0.02) (Fig. 4D).
FIG 4.
Differences in percent similarities to the consensus subtype C sequence between Gag-protease sequences derived from mothers and those derived from infants. Sequences derived from infants had higher percent amino acid similarities to the consensus than did those derived from their mothers, for the Gag (A), Gag p17 (B), Gag p24 (C), and protease (D) regions (paired t test).
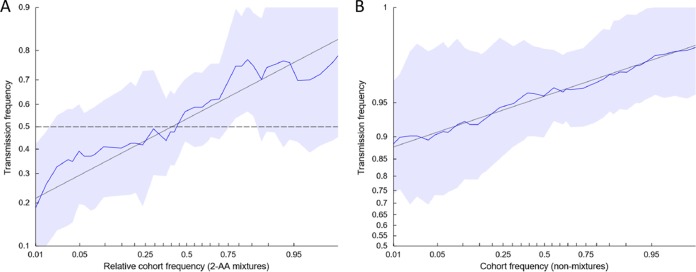
However, this analysis does not account for the higher quantity of mixtures found in the mothers than in the infants. To further evaluate whether there was a bias for the transmission of consensus-like viruses while controlling for mixtures, per-site empirical transmission frequencies were estimated as a function of site-specific amino acid frequencies of circulating viruses, as previously described (8). A significant relationship (P < 1e−10) between amino acid frequency and transmission probability was observed, both at sites where a mixture of amino acids was observed in the mother (Fig. 5A) and at sites where a single residue was observed (Fig. 5B), indicating that consensus amino acids were more frequently transmitted. These results are consistent with the circulating amino acid frequency as a correlate of transmission fitness.
FIG 5.
Relationship between amino acid frequency of circulating viruses and mother-to-child transmission. The odds that the mother's amino acid will be transmitted to the infant is a function of the empirical frequency of amino acids among circulating viruses. Each plot shows the empirical transmission probability (odds on a log10 scale) of a variant as a function of the frequency of the variant among 929 consensus Gag sequences isolated from chronically infected, antiretroviral-naive patients from Durban, South Africa. Empirical transmission probabilities (solid colored lines) are estimated by counting the proportion transmitted within a continuous sliding window with a 1-log-odds width with respect to the value on the abscissa. All log-odds values are smoothed by adding a pseudocount equal to 1% of the number of Durban sequences used to estimate the cohort frequency. Gray lines represent a linear fit to the sliding-window averages; shaded areas represent 95% confidence intervals estimated by using the percentile-t method on 1,000 multilevel bootstraps. Sites in which a mixture was observed in the infant founder virus were excluded. (A) Among 13,441 nonmixture sites from 29 mothers, the odds of transmission is associated with the frequency of the amino acid (AA) in the Durban cohort. (B) Among 281 sites containing two-amino-acid mixtures from 26 mothers, the probability of transmission is associated with the relative cohort frequency of the amino acid. Transmission probability is calculated with respect to a randomly chosen member of the mixture; the abscissa represents the relative frequency of that amino acid in the Durban cohort compared to the other amino acid in the mixture. For both plots, the slope is significantly greater than 0 (P <2e−10, as estimated by a multilevel logistic regression model [see Materials and Methods]).
Therefore, using two independent approaches, we observed that Gag-protease from infant-derived viruses was more consensus-like than viruses derived from the mothers. Although these results appear to contrast with lower in vitro replicative capacities among infants, they are consistent with data from previous reports of an inverse correlation between similarity to the consensus and in vitro replicative capacity (18), which were corroborated here.
Early escape in infants does not explain lower viral replication capacity.
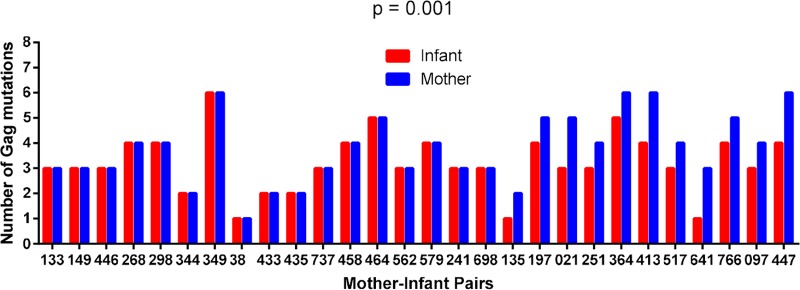
CTL escape mutations in critical Gag epitopes, particularly those at conserved residues, have been shown to significantly reduce viral replication capacity (18, 28–32). Furthermore, a significant negative correlation between Gag-protease-driven replication capacity and the number of Gag epitope mutations was observed, when considering 20 CTL escape mutations within defined Gag CTL epitopes (Table 1) (34). We first considered whether the numbers of these known Gag epitope mutations differed significantly between paired mother and infant sequences, irrespective of the HLA profile of the participants. In 17 MIPs, the numbers of these mutations were identical in the mother and infant sequences, but in the remaining 11 pairs, the sequences from the infants had one or two fewer mutations than the sequences from their mothers (P = 0.001 as determined by a Wilcoxon matched-pairs signed-rank test) (Fig. 6). Furthermore, for the 17 MIPs with the same number of Gag epitope mutations, infants were still infected with the virus with a lower replication capacity than that of viruses from their mothers (P = 0.0003 as determined by a paired t test). Therefore, the number of Gag mutations in defined epitopes does not account for the lower replication capacity of viruses transmitted to the infants. We extended our analysis of escape to include all HLA-associated footprints in Gag identified previously (8) to further investigate whether there is evidence of early escape in the infants that may be contributing to the lower replication capacity of their viruses. However, only two infant-derived viruses contained footprints (two in total), which corresponded to an HLA type expressed in the infant but not in the mother.
TABLE 1.
List of Gag epitope mutations in defined Gag CTL epitopesa
| HLA restriction | Epitope | Gag variant |
|---|---|---|
| HLA-B*42:01 | RPGGKKHYM | H28X |
| HLA-B*42:01 | RPGGKKHYM | M30X |
| HLA-B*57 | AISPRTLNAW | A146X |
| HLA-B*57 | AISPRTLNAW | I147X |
| HLA-B*57:03 | KAFSPEVIPMF | A163X |
| HLA-B*57:03 | KAFSPEVIPMF | S165X |
| HLA-B*81:01/42:01 | TPQDLNTML | Q182X |
| HLA-B*81:01 | TPQDLNTML | T186X |
| HLA-B*81:01 | TPQDLNTMLNT | T190X |
| HLA-B*57/58:01 | TSTLQEQIAW | T242X |
| HLA-B*57:03 | TSTLQEQIAW | I247X |
| HLA-B*35:01 | NPPIPVGDIY | D260E |
| HLA-B*27:05 | KRWIILGLNK | R264X |
| HLA-B*27:05 | KRWIILGLNK | L268X |
| HLA-B*14:01 | DRFFKTLRA | K302R |
| HLA-B*58:01 | QATQDVKNW | T310S |
| HLA-B*44:03 | AEQATQDVKNW | D312E |
| HLA-B*08:01 | NANPDCKTIL | K331X |
| HLA-B*39:10 | NPDCKTILRAL | T332X |
| HLA-B*07:02 | GPSHKARVL | S357X |
The underlined amino acids represent the consensus sequences in HIV-1 subtype C, with corresponding escape variants shown in column 3.
FIG 6.
Differences in the numbers of Gag mutations between Gag sequences derived from mothers and those derived from infants. CD8+ T cell escape mutations in defined Gag epitopes (30) in mother-infant pairs were enumerated. In 17 MIPs, the numbers of mutations were identical in the mother and infant sequences, but in the remaining 11 pairs, the infants had one or two fewer mutations than the sequences from the mothers (P = 0.001 as determined by a Wilcoxon matched-pairs signed-rank test).
Single-amino-acid variants do not explain replication capacity differences between mothers and their infants.
We performed an exploratory analysis to identify amino acids in Gag and protease that have an impact on replication capacity in order to then investigate whether these amino acids might differ between mothers and their infants and explain differences in replication capacities. To do this, we used a data set derived from 37 infants and 39 mothers (which included 28 MIPs, 9 unpaired infants, and 11 unpaired mothers) for whom Gag-protease sequencing and viral replication capacity measurements had been successful. Using this data set, 30 amino acids in Gag associated with decreased or increased replication capacity were identified, 5 of which, 7V, 7I, 28H, 62T, and 90K, were significant after correction for multiple comparisons (P < 0.05 as determined by a Mann-Whitney U test; q < 0.2) (Table 2). We then sought to determine whether the frequencies of these 5 amino acids differed significantly between mothers and infants, using all available sequences (39 infants and 42 mothers), and found that they did not. However, there was a trend that a higher proportion of mothers were infected with viruses that had glutamine (Q) at position 28 of p17 (P = 0.05). This residue was one of 30 residues originally found to be associated with increased replication capacity (P < 0.04; q = 0.62), although it was not among the 5 that remained significant after correction for multiple comparisons. We speculate that H28Q potentially partially contributes to the higher replication capacity of viruses from the transmitter mothers; however, overall, we did not convincingly find evidence of single-amino-acid variants that consistently contribute to differences in replication capacities between mothers and their infants.
TABLE 2.
Proportions of infants and mothers harboring amino acid residues in Gag significantly associated with altered replication capacity (P < 0.05; q < 0.2)
| Protein | Type of association with replication capacity | Codon | Amino acid | Consensus sequence amino acid | P value determined by a Mann-Whitney U test | q value | No. of infants harboring amino acid/total no. of infants | No. of transmitter mothers harboring amino acid/total no. of transmitter mothers | P value determined by Fisher's exact test |
|---|---|---|---|---|---|---|---|---|---|
| Gag p17 | Positive | 7 | V | I | 0.00008 | 0.01572 | 11/46 | 12/45 | 0.81 |
| Gag p17 | Negative | 7 | I | I | 0.00008 | 0.01572 | 34/46 | 32/45 | 0.82 |
| Gag p17 | Negative | 28 | H | H | 0.00209 | 0.164274 | 21/46 | 20/45 | 1 |
| Gag p17 | Positive | 62 | T | K | 0.00101 | 0.13231 | 5/46 | 6/45 | 0.76 |
| Gag p17 | Positive | 90 | K | E | 0.00175 | 0.164274 | 11/46 | 13/45 | 0.64 |
| Gag p17 | Positive | 28 | Q | H | 0.04306 | 0.623167 | 3/46 | 9/45 | 0.07 |
DISCUSSION
While much progress has recently been made in characterizing the viruses that establish infection following adult HIV-1 transmission (7–9), little is known about the viral factors involved in MTCT. It has been reported that viruses transmitted to infants have a higher Env-driven replication capacity than do viruses infecting their mothers (17); however, the effect of other genes that contribute significantly to the overall whole-isolate viral replication capacity, such as Gag (24), on MTCT remains unknown. Here we investigated the role of Gag-protease-driven replication capacity in MTCT in HIV-1 subtype C infection.
Although samples from the infants were obtained 6 to 8 weeks after birth, previous literature described viral populations in HIV-infected infants to be genetically homogeneous (35) and with low sequence-based diversity 6 to 8 weeks after birth (36). Gounder and colleagues also found low intrapatient diversity during acute infection in the case of HIV-infected adults, which had increased at 1 year postinfection (37). Goonetilleke and colleagues analyzed the evolution of transmitted viruses in 3 adult patients from the time of infection (38). The earliest escape that they detected in Gag was at 45 days postinfection, which is after 6 weeks of infection. Therefore, we can conclude that the virus in the infants at the time of sampling would be similar to the transmitted virus.
We observed that while the Gag-protease-driven replication capacity in the mothers was not a determinant of MTCT, there was selection for the transmission of viruses with a lower Gag-protease-driven replication capacity to the infants. This finding is in contrast to data from a previous study showing higher Env-driven replication capacities in viruses transmitted to infants than that of viruses in the mothers for 7 MIPs (17), suggesting that the selective forces associated with the transmission bottleneck may differ between Gag and Env. While this finding contradicted our initial hypothesis that transmitter mothers would transmit viruses with higher Gag-protease-driven replication capacities to their infants, it is in line with data from previous reports. Declines in the replicative fitness of RNA viruses following passage across a transmission bottleneck (vertical transmission in particular) were described previously in the literature (39–41); moreover, Adland et al. recently reported a trend toward lower HIV-1 Gag-protease-driven replication capacities in infants than in their mothers for 43 MIPs (although the infant-derived viruses studied were sampled months to years after infection in that study) (34). Furthermore, a study of HIV-1 infectious molecular clones from six adult heterosexual transmission pairs reported no selection for the transmission of viruses with higher replication capacities in peripheral blood mononuclear cells (PBMCs) (7).
We compared the virus sequences from the mothers and those from their infants in order to investigate the genetic determinants of the lower Gag-protease-driven replication capacity in the infants. Since CTL escape mutations in key Gag epitopes have been linked to lower viral replication capacity (7, 8, 18, 28–32, 40), the numbers of these mutations in paired mother-infant sequences were compared. It was observed that Gag epitope escape mutations were frequently transmitted, as has been reported in studies of viruses from early HIV-1 infection in adults (8, 19, 37), but overall, infant-derived viruses had fewer Gag epitope escape mutations than did viruses from the mothers, and therefore, the frequency of these mutations did not explain the difference in replication capacities between mothers and their infants. We also investigated the hypothesis that early escape in the infant-derived viruses was driving the lower replication capacity by analyzing sequences for HLA-associated footprints in Gag, but we found no evidence that this was the case.
Since previous studies of heterosexual transmission have identified a selection bias for the transmission of consensus-like viruses (7, 8), we compared the percent amino acid similarities to the consensus of the viruses derived from infants and their mothers and also modeled the odds of MTCT based on the amino acid frequency of viruses circulating in southern Africa. Our study is the first to show that viruses transmitted to infants are significantly more similar to the subtype consensus than are those from their mothers and that there is a bias for the transmission of consensus-like viruses from mothers to their infants.
Consistent with our finding of lower Gag-protease-driven replication capacities in the infant-derived viruses, our group and others previously showed that Gag-protease NL4-3 recombinant viruses and Gag-MJ4 recombinant viruses that have a higher percent similarity to the consensus Gag sequence have a lower replication capacity, and this was most pronounced for the Gag p17 region in analyses of Gag-protease NL4-3 recombinant viruses (18, 20). Using a much smaller sample size, we also find a significant inverse relationship between the percent similarity to the consensus Gag p17 sequence and replication capacity, which is consistent with data from those previous studies. Furthermore, a recent study of infectious molecular clones found an inverse relationship between similarity to the consensus sequence across the whole genome and replication capacity in PBMCs (7). Taken together with data from those previous studies, these findings suggest that viruses that are more consensus-like have a lower intrinsic replication capacity (although they are likely more fit in the host in the face of immune pressure, as they reflect the version of the virus best adapted to the population [42]) and that these viruses are favored in both heterosexual transmission and MTCT; i.e., they have a higher transmission fitness.
Two alternative interpretations merit mention. First, it is possible that parts of the virus other than Gag, for example, Env, play a greater role in transmission. Indeed, the in vitro Env-driven replication capacity was greater in infant-derived viruses then in those from the mothers in 7 MIPs (17), although it should be noted that the majority of infants in that study were likely infected via breastfeeding. The mode of transmission may alter the selection bias and could possibly explain the different findings of Kong et al. (17) showing that viruses with a higher Env-driven replication capacity were favored in transmission. Since Claiborne et al. (24) showed a strong correlation (r2 = 0.72 and P = 0.03) between the replication capacities of infectious molecular clones and Gag recombinant viruses from the same individuals, and it has been demonstrated that viruses with a lower replication capacity are more consensus-like and are also favored in heterosexual transmission (7), it seems more probable that the viruses transmitted to infants in the present study had a lower overall in vitro replication capacity, although genes other than Gag also make a significant contribution to the overall replication capacity (43, 44). A second possibility is that viral replication capacity is not intrinsic but rather cell type dependent; it is conceivable, for example, that a given viral variant could exhibit a low replication ability in peripheral cells but a high replication ability in mucosal cells infected during transmission, as a consequence of the differential expression of specific host factors in these cell types (33, 34, 39–46, 50). However, the viruses derived from infants who were infected in utero also exhibited significantly lower replication capacities than those of viruses from their mothers, arguing against this as an explanation for the study findings.
Finally, the observation that consensus-like virus sequences exhibit a lower in vitro replication ability yet appear to be preferentially transmitted suggests that the viral characteristics favoring transmission are largely decoupled from those that enhance replicative capacity. Nevertheless, it is still possible that replicative (“intrinsic”) fitness and transmission fitness are largely synonymous. In vitro replicative capacity and sequence-based comparisons to circulating viruses may simply be noisy estimates that measure different aspects of intrinsic fitness. This hypothesis is supported in principle by the correlations between these measures and viral loads (18, 20), which are weak enough to simultaneously permit positive correlations with viral loads yet negative correlations between each other. Thus, intrinsic fitness may be only approximately estimated by these two independent, and at times conflicting, measures, while there would be a few sites in which transmission and intrinsic fitness are fundamentally different. Our cautious interpretation of the identification of 28H within Gag p17 as a consensus residue that has been linked with lower in vitro replicative capacity (19) and yet may confer increased transmission fitness provides a possible example. Notably, this site is in the region of p17 that has been implicated in binding the cytoplasmic tail of gp41 and facilitating virion incorporation (51) and thus may suggest that selection in Env is driving the selection of this site through an epistatic interaction. Conversely, an epistatic interaction with Env may suggest an artifact in this in vitro system, which does not use autologous Env proteins, and is thus an assay limitation.
The assay used in the present study is designed to capture the Gag-protease-driven replication capacity of the quasispecies infecting an individual, and previously, this measure was found to correlate significantly with the replication capacity of whole isolates derived from the same individuals (22). However, a potential limitation is that, in the mothers, the variants in the quasispecies with the highest replication capacity may grow out in culture (although it should be noted that about 60% of the mixtures in the bulk PCR products are represented in the virus stocks obtained in this assay [18]). If the variant in the infant is a random selection of the mother's variants, it would be expected that, across the cohort, the replication capacities of the infants' viruses would on average be lower than those of the viruses from the mothers. However, in this scenario, it is unlikely that we would observe with such high consistency, in 29 out of 31 MIPs, that the viruses in the infants had lower replication capacities, as, by chance, a greater proportion of the infant variants should have had a replication capacity at least equal to that of the virus in the mother. Furthermore, in this scenario, if transmission favored the variants with the highest replication capacities (our initial hypothesis, disputed by the data here), in the majority of the MIPs, equal replication capacities of the mother and infant variants would be observed. Finally, if the infant virus represented the consensus of the maternal variants and there was selection in culture for the most fit maternal variant, this may have driven the results observed here; however, in 20% of cases where there were nucleotide mixtures in the maternal virus, the dominant nucleotide in the mother was not transmitted to the infant, indicating that the virus transmitted to the infant is not simply the consensus of the maternal variants. Rather, while accounting for the higher number of mixtures in the maternal sequences, we demonstrated here that there is a bias for the transmitted virus to be closer to the consensus of the circulating population variants. Therefore, we conclude that it is improbable that this assay limitation was responsible for our finding of transmission of variants with low Gag-protease-driven replication capacities.
It is furthermore important to note the assay limitation of subtype mismatch between the Gag-protease insert and the NL4-3 backbone. Nevertheless, in previous studies, the assay yielded data that are clinically relevant (18, 52–54), and in a recent comparison of Gag-protease-driven replication between different subtypes, the subtype-specific hierarchy remained consistent regardless of whether a subtype C (pZM246-F10) or a subtype B (NL4-3) backbone was used (22).
In conclusion, MTCT is characterized by a transmission bottleneck that favors the transmission of viruses that have lower Gag-protease-driven replication capacities and are closer to the circulating consensus from mother to infant out of their quasispecies of the virus. While the consensus virus sequence corresponds to a lower in vitro Gag-protease-driven replication ability, it favors virus transmission and thus reflects increased transmission fitness. Further work is required to fully dissect intrinsic, transmission, and posttransmission fitness.
MATERIALS AND METHODS
Study population.
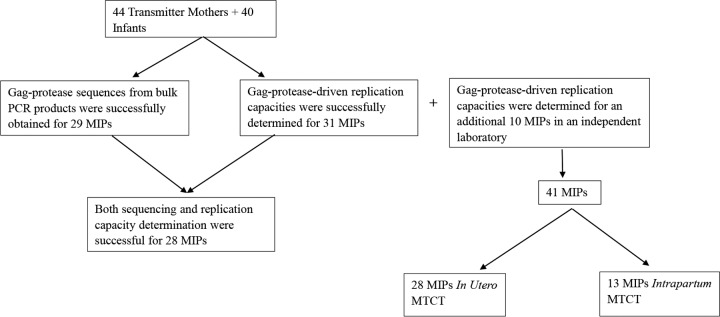
The study population was a pediatric cohort in Durban, South Africa, which was described previously (55–57). Gag-protease-driven viral replication capacities were investigated in 53 HIV-1-positive nontransmitting mothers, 44 HIV-1-positive transmitting mothers, and 40 infants infected with HIV-1 subtype C who were antiretroviral therapy naive except for a single dose of nevirapine, a reverse transcriptase inhibitor (58). Studies have shown that tenofovir (also a reverse transcriptase inhibitor) gel has no effect on Gag-protease-driven replication capacity (59), and therefore, it is unlikely that single-dose nevirapine would impact Gag-protease-driven replication capacity. The nontransmitter and transmitter mothers were matched for CD4 counts and viral loads. Plasma samples from the mothers and infants were obtained at medians of 5 weeks (IQR, 3.8 to 6 weeks) before birth and 6 weeks (IQR, 4 to 8 weeks) after birth, respectively. Overall, there were 31 MIPs (Fig. 7). Replication capacities for 19 of these 31 MIPs were also determined in an independent laboratory, along with an additional 10 MIPs from the same cohort that were available to that laboratory but not available to the first laboratory. Of the 41 MIPs, 28 were IU transmission cases, defined as infants that seroconverted within 48 h after birth, and 13 were IP transmission cases (Fig. 7), defined as infants that seroconverted more than 48 h after birth. Demographic and clinical data for all mothers and infants are shown in Tables 3 and 4, respectively.
FIG 7.
Study population. Plasma samples were obtained for 44 transmitter mothers and 40 infants. Gag-protease was successfully sequenced for 29 MIPs, and Gag-protease-driven replication capacities were successfully determined for 31 MIPs. However, altogether, replication capacity assays and sequencing were successful for 28 MIPs. Altogether, Gag-protease-driven replication capacities were determined for 41 MIPs (31 from the Durban laboratory and 10 from an independent laboratory). The mode of transmission within 28 out of the 41 MIPs was in utero mother-to-child transmission (MTCT). The mode of transmission for the remaining 13 MIPs was intrapartum transmission.
TABLE 3.
Clinical and demographic characteristics of HIV-1-infected transmitter and nontransmitter mothers
| Parameter | Value for group |
P value | |
|---|---|---|---|
| Transmitter mothers | Nontransmitter mothers | ||
| Median CD4+ T cell count (cells/mm3) (IQR) | 325 (188–443) | 289 (147–422) | 0.32 |
| Median viral load (log10 copies/ml) (IQR) | 5 (4.4–5.4) | 4.7 (3.9–5.3) | 0.23 |
TABLE 4.
Clinical and demographic characteristics of HIV-1-infected infants
Generation of Gag-protease NL4-3 recombinant viruses.
Gag-protease NL4-3 recombinant viruses were generated according to methods described previously (18). Briefly, gag-protease was amplified from viral RNA extracted from plasma by performing nested PCR using the primers and conditions described previously (18). The pNL4-3Δgag-protease plasmid was generated (52) and prepared (18) as described previously. Ten micrograms of BstEII-digested pNL4-3Δgag-protease and ∼80 μl of the gag-protease PCR product were cotransfected into 2.5 million CEM-GXR25 cells (GXR cells) (60) in 800 μl R10 medium via electroporation at 300 V and 500 μF (18, 37). The electroporated cells were allowed to recover for 1 h before the contents of each cuvette were transferred to 25-cm2 flasks containing 4 ml medium. Five milliliters of medium was added to each of the flasks after 5 days. Flow cytometry of the transfected cells was performed from day 12 onwards to measure the percentage of cells infected by recombinant viruses. Culture supernatants were harvested once the cells were 25 to 30% infected and were stored in 0.5- to 1-ml aliquots at −80°C for use in subsequent titration and replication assays.
Titration and replication capacity assays of Gag-protease recombinant viruses.
Titration and replication capacity assays were performed as described previously (45, 46, 52). GXR cells were infected at a multiplicity of infection (MOI) of 0.003. The replication capacity of the recombinant viruses determined by Gag-protease function is indicated by the mean slope of exponential growth from days 3 to 6, which was determined by using the semilog method in Excel. These values were normalized to the values for the positive control, wild-type NL4-3, which was included in each assay. The replication capacity assays were done in duplicate. If the difference in the replication capacities of a sample between the two assays exceeded 0.1, a third replication assay was done for those samples. The final replication capacity for each sample was calculated as an average of the replication capacities determined by all assays done for that sample.
Gag-protease sequencing and phylogenetic analysis.
The gag-protease gene was sequenced as described previously (18). The gag-protease PCR product was diluted 1:15 in diethyl pyrocarbonate-treated water and sequenced by using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) (47), using primers described previously (18) in addition to the following primers: 5′-TGACTAGCGGAGGCTAGAA-3′ (corresponding to HIV-1 subtype B genomic reference strain HXB2 nucleotides 763 to 781), 5′-AGAGAACCAAGGGGAAGTGA-3′ (nucleotides 1474 to 1493), 5′-ACAGGCTAATTTTTTAGGGA-3′ (nucleotides 2076 to 2095), 5′-CTAATACTGTATCATCTGCTCCTGT-3′ (nucleotides 2353 to 2328), 5′-TCCAATTCCTCCTATCATTTTTGG-3′ (nucleotides 2405 to 2382), and 5′-TCTTCTGTCAATGGCCATTG-3′ (nucleotides 2635 to 2616). Sequences were run on the ABI 3130xl genetic analyzer (Applied Biosystems) and edited in Sequencher (Gene Codes) (47). The REGA HIV subtyping tool was used to confirm that all sequences were subtype C gag-protease sequences (http://bioafrica.mrc.ac.za/). Nucleotides for each gene were aligned manually in Se-Al v.2.0a11. Maximum likelihood phylogenetic trees were generated by using PhyML (http://www.hiv.lanl.gov/) and visualized by using Figtree v.1.2.2 (http://tree.bio.ed.ac.uk/software/figtee/). For the calculation of similarity to the consensus, sequences were aligned to the HXB2 sequence by using Gene cutter (http://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html), and insertions relative to HXB2 were removed by using BioEdit 7.0. The percent amino acid similarities of Gag and protease to the most recent (2004) consensus subtype C sequence were calculated by using the sequence identity matrix function in BioEdit 7.0.
Modeling the odds of transmission based on amino acid frequency in southern African chronic sequences.
Per-site empirical transmission frequencies were estimated as a function of site-specific amino acid frequencies of circulating viruses as described previously (8). Briefly, for 28 matched mother-child pairs (for whom gag-protease sequencing was successful), mother and child bulk gag amino acid sequences were aligned to a reference panel of circulating Durban gag sequences (8). We then identified each codon in the mother's bulk gag sequence where the dominant amino acid was also present in the child (sites in which an amino acid mixture was observed in the child, or a mixture of more than two amino acids was observed in the mother, were excluded). The frequency of each amino acid at the corresponding site in the Durban alignment was then used as a reference; frequencies were smoothed with a 1% “pseudocount.” To visualize the probability of transmission as a function of the frequency of a variant in the Durban cohort, we used a sliding-window approach in which we measured the observed proportion of sites that were transmitted within a given window. We used a window size of 1 on the log-odds scale and included only windows with at least 20 observations. To estimate 95% confidence intervals for the empirical transmission probability curves, we used a block bootstrap approach using the percentile-t method (48). For each of 1,000 bootstrap replicates, we sampled, with replacement, the mother-child pairs and then the sites within each sampled pair. We then estimated the empirical transmission probability for each cohort frequency value observed in the complete data set. The percentile-t 95% confidence interval was then estimated independently for each cohort frequency value. To estimate P values, we used a multilevel logistic regression model, in which the dependent variable is whether the site was transmitted and the fixed effect is the (smoothed) log odds of the frequency of the variant in the Durban cohort (or of the relative frequency in the case of the 2-amino-acid mixtures). Random offsets were estimated for each protein within Gag (p17, p24, and p15) and for each mother-child pair. We report the P value for the null hypothesis that the fixed effect was equal to 0.
Statistical analysis.
The Gag-protease-driven replication capacities of viruses infecting transmitting mothers and their infants were compared by performing the paired t test for MIPs, and the Student t test was used when transmitter and nontransmitter mothers were compared. The percent amino acid similarities of Gag, Gag p17, Gag p24, and protease to the consensus C sequence were compared between transmitter mothers and their infants within MIPs by using the paired t test. The relationship between the cohort frequency of amino acids and transmission was assessed by using a multilevel logistic regression model to determine significance, as described in detail above. Gag-protease sequences from mothers and infants were analyzed by using the Mann-Whitney U test to identify specific single codons in Gag-protease where amino acid variants occurring at a frequency of ≥5 were associated with increased or decreased replication capacity. q values were determined to account for multiple comparisons (49). Associations were considered to be significant if the P value was <0.05 and the q value was <0.2. Two-tailed Fisher's exact test was done to determine if the proportions of replication capacity-associated residues in the mothers and infants were significantly different.
Ethical considerations.
This study is a subset of a larger study, which is entitled “Augmentation of HIV-specific T cell immunity by highly active antiretroviral therapy (HAART) instituted in acute paediatric HIV infection followed by supervised treatment interruptions (STI)” (http://hpp.ukzn.ac.za/Cohorts.aspx). Full ethical approval, from the Biomedical Research Ethics Committee of the Nelson R. Mandela School of Medicine, University of KwaZulu-Natal (reference no. E065/01), and informed patient consent were obtained for this study.
ACKNOWLEDGMENTS
This research was funded by the Howard Hughes Medical Institute, the South African Department of Science and Technology/National Research Foundation Research Chairs Initiative, and the Victor Daitz Foundation. Partial funding was received from the National Institutes of Health through the IMPAACT Network. This work was also supported in part through the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative (grant number DEL-15-006). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS) Alliance for Accelerating Excellence in Science in Africa (AESA) and is supported by the New Partnership for Africa's Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (grant number 107752/Z/15/Z) and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of the AAS, the NEPAD Agency, the Wellcome Trust, or the UK government. Z.L.B. is supported by a scholar award from the Michael Smith Foundation for Health Research. M.A.B. holds a Canada Research Chair in Viral Pathogenesis and Immunity.
We thank Taryn Green, Mary van der Stok, and Zenele Mncube for technical assistance; Johannes Viljoen and the Africa Centre laboratory for providing access to tissue culture and sequencing facilities; and the Durban clinic staff and the management of St Mary's and Prince Mshiyeni Hospitals for their support of the PEHSS cohort. Finally, we thank and acknowledge the study participants.
REFERENCES
- 1.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrahams M-R, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping L-H, Athreya GS, Treurnicht FK, Keele BF, Wood N, Salazar-Gonzalez JF, Bhattacharya T, Chu H, Hoffman I, Galvin S, Mapanje C, Kazembe P, Thebus R, Fiscus S, Hide W, Cohen MS, Karim SA, Haynes BF, Shaw GM, Hahn BH, Korber BT, Swanstrom R, Williamson C, CAPRISA Acute Infection Study Team, Center for HIV-AIDS Vaccine Immunology Consortium. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol 83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sagar M. 2010. HIV-1 transmission biology: selection and characteristics of infecting viruses. J Infect Dis 202(Suppl 2):S289–S296. doi: 10.1086/655656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 26:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 5.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP Jr, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A 110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, Aasa-Chapman MM, Pellegrino P, Williams I, Cohen MS, Gao F, Shaw GM, Hahn BH, Ochsenbauer C, Kappes JC, Borrow P. 2013. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology 10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deymier MJ, Ende Z, Fenton-May AE, Dilernia D, Kilembe W, Allen SA, Borrow P, Hunter E. 2015. Heterosexual transmission of subtype C HIV-1 selects consensus-like variants without increased replicative capacity or interferon-alpha resistance. PLoS Pathog 11:e1005154. doi: 10.1371/journal.ppat.1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, Deymier MJ, Ende ZS, Klatt NR, DeZiel CE, Lin TH, Peng J, Seese AM, Shapiro R, Frater J, Ndung'u T, Tang J, Goepfert P, Gilmour J, Price MA, Kilember W, Heckerman D, Goulder PJ, Allen TM, Allen S, Hunter E. 2014. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science 345:1254031. doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tully DC, Ogilvie CB, Batorsky RE, Bean DJ, Power KA, Ghebremichael M, Bedard HE, Gladden AD, Seese AM, Amero MA, Lane K, McGrath G, Bazner SB, Tinsley J, Lennon NJ, Henn MR, Brumme ZL, Norris PJ, Rosenberg ES, Mayer KH, Jessen H, Kosakovsky Pond SL, Walker BD, Altfeld M, Carlson JM, Allen TM. 2016. Differences in the selection bottleneck between modes of sexual transmission influence the genetic composition of the HIV-1 founder virus. PLoS Pathog 12:e1005619. doi: 10.1371/journal.ppat.1005619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matt C, Roger M. 2001. Genetic determinants of pediatric HIV-1 infection: vertical transmission and disease progression among children. Mol Med 7:583–589. [PMC free article] [PubMed] [Google Scholar]
- 11.Lehman DA, Farquahar C. 2007. Biological mechanisms of vertical human immunodeficiency virus (HIV-1) transmission. Rev Med Virol 17:381–403. doi: 10.1002/rmv.543. [DOI] [PubMed] [Google Scholar]
- 12.Behbahani H, Popek E, Garcia P, Andersson J, Spetz AL, Landay A, Flener Z, Patterson BK. 2000. Up-regulation of CCR5 expression in the placenta is associated with human immunodeficiency virus-1 vertical transmission. Am J Pathol 157:1811–1818. doi: 10.1016/S0002-9440(10)64819-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh KK, Spector S. 2009. Host genetic determinants of human immunodeficiency virus infection and disease progression in children. Pediatr Res 65:55R–63R. doi: 10.1203/PDR.0b013e31819dca03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackelprang RD, John-Stewart G, Carrington M, Richardson B, Rowland-Jones S, Gao X, Mbori-Ngacha D, Mabuka J, Lohman-Payne B, Farquhar C. 2008. Maternal HLA homozygosity and mother-child HLA concordance increase the risk of vertical transmission of HIV-1. J Infect Dis 197:1156–1161. doi: 10.1086/529528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winchester R, Pitt J, Charurat M, Magder LS, Göring HH, Landay A, Read JS, Shearer W, Handelsman E, Luzuriaga K, Hillyer GV, Blattner W. 2004. Mother-to-child transmission of HIV-1: strong association with certain maternal HLA-B alleles independent of viral load implicates innate immune mechanisms. J Acquir Immune Defic Syndr 36:659–670. doi: 10.1097/00126334-200406010-00002. [DOI] [PubMed] [Google Scholar]
- 16.Overbaugh J. 2014. Mother-infant HIV transmission: do maternal HIV-specific antibodies protect the infant? PLoS Pathog 10:e1004283. doi: 10.1371/journal.ppat.1004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong X, West JT, Zhang H, Shea DM, M'soka TJ, Wood C. 2008. The human immunodeficiency virus type 1 envelope confers higher rates of replicative fitness to perinatally transmitted viruses than to nontransmitted viruses. J Virol 82:11609–11618. doi: 10.1128/JVI.00952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright JK, Brumme ZL, Carlson JM, Heckerman D, Kadie CM, Brumme CJ, Wang B, Losina E, Miura T, Chonco F, van der Stok M, Mncube Z, Bishop K, Goulder PJ, Walker BD, Brockman MA, Ndung'u T. 2010. Gag protease-mediated replication capacity in HIV-1 subtype C chronic infection: associations with HLA type and clinical parameters. J Virol 84:10820–10831. doi: 10.1128/JVI.01084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright JK, Novitsky V, Brockman MA, Brumme ZL, Brumme CJ, Carlson JM, Heckerman D, Wang B, Losina E, Leshwedi M, van der Stok M, Maphumulo L, Mkhwanazi N, Chonco F, Goulder PJ, Essex M, Walker BD, Ndung'u T. 2011. Influence of Gag-protease-mediated replication capacity on disease progression in individuals recently infected with HIV-1 subtype C. J Virol 85:3996–4006. doi: 10.1128/JVI.02520-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prince JL, Claiborne DT, Carlson JM, Schaefer M, Yu T, Lahki S, Prentice HA, Yue L, Vishwanathan SA, Kilembe W, Goepfert P, Price MA, Gilmour J, Mulenga J, Farmer P, Derdeyn CA, Tang J, Heckerman D, Kaslow RA, Allen SA, Hunter E. 2012. Role of transmitted Gag CTL polymorphisms in defining replicative capacity and early HIV-1 pathogenesis. PLoS Pathog 8:e1003041. doi: 10.1371/journal.ppat.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawamura M, Shimano R, Inubushi R, Amano K, Ogasawara T, Akari H, Adachi A. 1997. Functional domain mapping of HIV-1 Gag proteins. Biochem Biophys Res Commun 241:317–320. doi: 10.1006/bbrc.1997.7814. [DOI] [PubMed] [Google Scholar]
- 22.Kiguoya MW, Mann JK, Chopera D, Gounder K, Lee GQ, Hunt PW, Martin JN, Ball BB, Kimani J, Brumme ZL, Brockman MA, Ndung'u T. 19 April 2017. Subtype-specific differences in Gag-protease-driven replication capacity are consistent with intersubtype differences in HIV-1 disease progression. J Virol doi: 10.1128/JVI.00253-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prado JG, Prendergast A, Thobakgale C, Molina C, Tudor-Williams G, Ndung'u T, Walker BD, Goulder P. 2010. Replicative capacity of human immunodeficiency virus type 1 transmitted from mother to child is associated with pediatric disease progression rate. J Virol 84:492–502. doi: 10.1128/JVI.01743-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claiborne DT, Prince JL, Scully E, Macharia G, Micci L, Lawson B, Kopycinski J, Deymier MJ, Vanderford TH, Nganou-Makamdop K, Ende Z, Brooks K, Tang J, Yu T, Lakhi S, Kilembe W, Silvestri G, Douek D, Goepfert PA, Price MA, Allen SA, Paiardini M, Altfeld M, Gilmour J, Hunter E. 2015. Replicative fitness of transmitted HIV-1 drives acute immune activation, proviral load in memory CD4+ T cells, and disease progression. Proc Natl Acad Sci U S A 112:E1480–E1489. doi: 10.1073/pnas.1421607112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selhorst P, Combrinck C, Ndabambi N, Ismail SD, Abrahams MR, Lacerda M, Samsunder N, Garrett N, Abdool Karim Q, Abdool Karim SS, Williamson C. 2017. Replication capacity of viruses from acute infection drives HIV-1 disease progression. J Virol 91:e01806-16. doi: 10.1128/JVI.01806-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuñiga R, Lucchetti A, Galvan P, Sanchez S, Sanchez C, Hernandez A, Sanchez H, Frahm N, Linde CH, Hewitt HS, Hildebrand W, Altfeld M, Allen TM, Walker BD, Korber BT, Leitner T, Sanchez J, Brander C. 2006. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J Virol 80:3122–3125. doi: 10.1128/JVI.80.6.3122-3125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rolland M, Heckerman D, Deng W, Rousseau CM, Coovadia H, Bishop K, Goulder PJ, Walker BD, Brander C, Mullins JI. 2008. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS One 3:e1424. doi: 10.1371/journal.pone.0001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Merino V, Nie S, Luzuriaga K. 2005. HIV-1-specific CD8+ T cell responses and viral evolution in women and infants. J Immunol 175:6976–6986. doi: 10.4049/jimmunol.175.10.6976. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, McNevin J, Cao J, Zhao H, Genowati I, Wong K, Mclaughlin S, McSweyn MD, Diem K, Stevens CE, Maenza J, He H, Nickle DC, Shriner D, Holte SE, Collier AC, Corey L, McElrath MJ, Mullins JI. 2006. Selection on the human immunodeficiency virus type 1 proteome following primary infection. J Virol 80:9519–9529. doi: 10.1128/JVI.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Picado J, Prado J, Fry EE, Pfafferott K, Leslie A, Chetty S, Thobakgale C, Honeyborne I, Crawford H, Matthews P, Pillay T, Rousseau C, Mullins JI, Brander C, Walker BD, Stuart DI, Kiepiela P, Goulder P. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol 80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Troyer RM, McNevin J, Liu Y, Zhang SC, Krizan RW, Abraha A, Tebit DM, Zhao H, Avila S, Lobritz MA, McElrath MJ, Le Gall S, Mullins JI, Arts EJ. 2009. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog 5:e1000365. doi: 10.1371/journal.ppat.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright JK, Naidoo VL, Brumme ZL, Prince JL, Claiborne DT, Goulder PJ, Brockman MA, Hunter E, Ndung'u T. 2012. Impact of HLA-B*81-associated mutations in HIV-1 Gag on viral replication capacity. J Virol 86:3193–3199. doi: 10.1128/JVI.06682-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolinsky SM, Wike CM, Korber BT, Hutto C, Parks WP, Rosenblum LL. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 34.Adland E, Paioni P, Thobakgale C, Laker L, Mori L, Muenchhoff M, Csala A, Clapson M, Flynn J, Novelli V, Hurst J, Naidoo V, Shapiro R, Huang KH, Frater J, Prendergast A, Prado JG, Ndung'u T, Walker BD, Carrington M, Jooste P, Goulder PJ. 2015. Discordant impact of HLA on viral replicative capacity and disease progression in pediatric and adult HIV infection. PLoS Pathog 11:e1004954. doi: 10.1371/journal.ppat.1004954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verhofstede C, Demecheleer E, De Cabooter N, Gaillard P, Mwayumba F, Claeys P, Chohan V, Mandaliya K, Temmerman M, Plum J. 2003. Diversity of the human immunodeficiency virus type 1 (HIV-1) env sequence after vertical transmission in mother-child pairs infected with HIV-1 subtype A. J Virol 77:3050–3057. doi: 10.1128/JVI.77.5.3050-3057.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Towler WI, James MM, Ray SC, Wang L, Donnell D, Mwatha A, Guay L, Nakabiito C, Musoke P, Jackson JB, Eshleman S. 2010. Analysis of HIV diversity using a high resolution melting assay. AIDS Res Hum Retroviruses 26:913–918. doi: 10.1089/aid.2009.0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gounder K, Padayachi N, Mann JK, Radebe M, Mokgoro M, van der Stok M, Mkhize L, Mncube Z, Jaggernath M, Reddy T, Walker BD, Ndung'u T. 2015. High frequency of transmitted HIV-1 Gag HLA class I-driven immune escape variants but minimal immune selection over the first year of clade C infection. PLoS One 10:e0119886. doi: 10.1371/journal.pone.0119886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goonetilleke N, Liu MKP, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, Weinhold KJ, Moore S, CHAVI Clinical Core B, Letvin N, Haynes BF, Cohen MS, Hraber P, Bhattacharya T, Borrow P, Perelson AS, Hahn BH, Shaw GM, Korber BT, McMicheal AJ. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med 206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergstrom CT, McElhany P, Real LA. 1999. Transmission bottlenecks as determinants of virulence in rapidly evolving pathogens. Proc Natl Acad Sci U S A 96:5095–5100. doi: 10.1073/pnas.96.9.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lázaro E, Escarmis C, Pérez-Mercader J, Manrubia SC, Domingo E. 2003. Resistance of virus to extinction on bottleneck passages: study of a decaying and fluctuating pattern of fitness loss. Proc Natl Acad Sci U S A 100:10830–10835. doi: 10.1073/pnas.1332668100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elena SF, González-Candelas F, Novella IS, Duarte EA, Clarke DK, Domingo E, Holland JJ, Moya A. 1996. Evolution of fitness in experimental populations of vesicular stomatitis virus. Genetics 142:673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenzer S, Crawford H, Pymm P, Gifford R, Sreenu VB, Weimershaus M, de Oliveira T, Burgevin A, Gerstoft J, Akkad N, Lunn D, Fugger L, Bell J, Schild H, van Endert P, Iversen AK. 2014. HIV-1 adaptation to antigen processing results in population-level immune evasion and affects subtype diversification. Cell Rep 7:448–463. doi: 10.1016/j.celrep.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rangel HR, Weber J, Chakraborty B, Gutierrez A, Marotta ML, Mirza M, Kiser P, Martinez MA, Este JA, Quinones-Mateu ME. 2003. Role of the human immunodeficiency virus type 1 envelope gene in viral fitness. J Virol 77:9069–9073. doi: 10.1128/JVI.77.16.9069-9073.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell TB, Schneider K, Wrin T, Petropoulos CJ, Connick E. 2003. Relationship between in vitro human immunodeficiency virus type 1 replication rate and virus load in plasma. J Virol 77:12105–12112. doi: 10.1128/JVI.77.22.12105-12112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brockman MA, Schneidewind A, Lahaie M, Schmidt A, Miura T, DeSouza I, Ryvkin F, Derdeyn CA, Allen S, Hunter E, Mulenga J, Goepfert PA, Walker BD, Allen TM. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J Virol 81:12608–12618. doi: 10.1128/JVI.01369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneidewind A, Brockman M, Yang R, Adam I, Li B, Le Gall S, Rinaldo CR, Craggs SL, Allgaier RL, Power KA, Kuntzen T, Tung CS, LaBute MX, Mueller SM, Harrer T, McMichael AJ, Goulder PJ, Aiken C, Brander C, Kelleher AD, Allen TM. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol 81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gene Codes Corporation. 2009. Sequencher version 4.9 DNA sequence analysis software. Gene Codes Corporation, Ann Arbor, MI. [Google Scholar]
- 48.Efron B. 1981. Nonparametric standard errors and confidence intervals. Can J Stat 9:139–158. doi: 10.2307/3314608. [DOI] [Google Scholar]
- 49.Storey JD, Tibshirani R. 2001. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Back NK, Nijhuis M, Keulen W, Boucher CA, Oude Essink BO, van Kuilenburg AB, van Gennip AH, Berkhout B. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to processivity defect of reverse transcriptase enzyme. EMBO J 15:4040–4029. [PMC free article] [PubMed] [Google Scholar]
- 51.Cosson P. 1996. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J 15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 52.Miura T, Brockman M, Schneidewind A, Lobritz M, Pereyra F, Rathod A, Block BL, Brumme ZL, Baker B, Rothchild AC, Li B, Trocha A, Cutrell E, Frahm N, Brander C, Toth I, Arts EJ, Allen TM, Walker BD. 2009. HLA B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte recognition. J Virol 83:2743–2755. doi: 10.1128/JVI.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brockman MA, Brumme ZL, Brumme CJ, Miura T, Sela J, Rosato PC, Kadie CM, Carlson JM, Markle TJ, Streeck H, Kelleher AD, Harrigan PR, Heckerman D, Walker BD, Allen TM. 2010. Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated for in chronic infection. J Virol 84:11937–11949. doi: 10.1128/JVI.01086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang KH, Goedhals D, Carlson JM, Brockman MA, Mishra S, Brumme ZL, Hickling S, Tang CS, Miura T, Seebregts C, Heckerman D, Ndung'u T, Walker B, Klenerman P, Steyn D, Goulder P, Phillips R, Bloemfontein-Oxford Collaborative Group, van Vuuren C, Frater J. 2011. Progression to AIDS in South Africa is associated with both reverting and compensatory viral mutations. PLoS One 6:e19018. doi: 10.1371/journal.pone.0019018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mphatswe W, Blanckenberg N, Tudor-Williams G, Prendergast A, Thobakgale C, Mkhwanazi N, McCarthy N, Walker BD, Kiepiela P, Goulder P. 2007. High frequency of rapid immunological progression in African infants infected in the era of perinatal HIV prophylaxis. AIDS 21:1253–1261. doi: 10.1097/QAD.0b013e3281a3bec2. [DOI] [PubMed] [Google Scholar]
- 56.Thobakgale CF, Ramduth D, Reddy S, Mkhwanazi N, de Pierres C, Moodley E, Mphatswe W, Blanckenberg N, Cengimbo A, Prendergast A, Tudor-Williams G, Dong K, Jeena P, Kindra G, Bobat R, Coovadia H, Kiepiela P, Walker BD, Goulder PJ. 2007. Human immunodeficiency virus-specific CD8+ T-cell activity is detectable from birth in the majority of in utero-infected infants. J Virol 81:12775–12784. doi: 10.1128/JVI.00624-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thobakgale CF, Prendergast A, Crawford H, Mkhwanazi N, Ramduth D, Reddy S, Molina C, Mncube Z, Leslie A, Prado J, Chonco F, Mphatswe W, Tudor-Williams G, Jeena P, Blanckenberg N, Dong K, Kiepiela P, Coovadia H, Ndung'u T, Walker PD, Goulder PJ. 2009. Impact of HLA in mother and child on disease progression of pediatric human immunodeficiency virus type 1 infection. J Virol 83:10234–10244. doi: 10.1128/JVI.00921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guay LA, Musoke P, Fleming T, Bagenda D, Allen M, Nakabiito C, Sherman J, Bakaki P, Ducar C, Deseyve M, Emel L, Mirochnick M, Fowler MG, Mofenson L, Miotti P, Dransfeld K, Bray D, Mmiro F, Jackson JB. 1999. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet 354:795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 59.Chopera DR, Mann JK, Mwimanzi P, Omarjee S, Kuang XT, Ndabambi N. 2013. No evidence for selection of HIV-1 with enhanced Gag-protease or Nef function among breakthrough infections in the CAPRISA 004 Tenofovir Microbicide Trial. PLoS One 8:e71758. doi: 10.1371/journal.pone.0071758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brockman MA, Tanzi GO, Walker BD, Allen TM. 2006. Use of a novel GFP reporter cell line to examine replication capacity of CXCR4- and CCR5-tropic HIV-1 by flow cytometry. J Virol Methods 131:134–142. doi: 10.1016/j.jviromet.2005.08.003. [DOI] [PubMed] [Google Scholar]