Abstract
Owl monkeys (genus Aotus) are the only taxon in simian primates that consists of nocturnal or otherwise cathemeral species. Their night vision is superior to that of other monkeys, apes, and humans but not as good as that of typical nocturnal mammals. This incomplete night vision has been used to conclude that these monkeys only secondarily adapted to a nocturnal lifestyle, or to their cathemeral lifestyle that involves high night-time activity. It is known that the rod cells of many nocturnal mammals possess a unique nuclear architecture in which heterochromatin is centrally located. This “inverted nuclear architecture”, in contrast with “conventional nuclear architecture”, provides elevated night vision by passing light efficiently to the outer segments of photoreceptors. Owl monkey rod cells exhibit an intermediate chromatin distribution, which may provide them with less efficient night vision than other nocturnal mammals. Recently, we identified three megasatellite DNAs in the genome of Azara's owl monkey (Aotus azarae). In the present study, we show that one of the three megasatellite DNAs, OwlRep, serves as the primary component of the heterochromatin block located in the central space of the rod nucleus in A. azarae. This satellite DNA is likely to have emerged in the Aotus lineage after its divergence from those of other platyrrhini taxa and underwent a rapid expansion in the genome. Our results indicate that the heterochromatin core in the A. azarae rod nucleus was newly formed in A. azarae or its recent ancestor, and supports the hypothesis that A. azarae, and with all probability other Aotus species, secondarily acquired night vision.
Keywords: nocturnality, heterochromatin, repetitive DNA, rod cell, retina, primates
Introduction
Rods and cones are photoreceptor cells in the eyes of vertebrates. These cells have an elongated shape, and light passes through the cell nucleus on its way to the outer segments of photoreceptors. Rods are more sensitive to weak light than cones. It was recently found that rod cells of many nocturnal mammals, including mice, rats, deer, cats, ferrets, and lemurs, possess a unique “inverted nuclear architecture”, in which heterochromatin localizes to the central portion of the nucleus (Solovei et al. 2009; Eberhart et al. 2013; Joffe et al. 2014). Heterochromatin is usually located adjacent to the nuclear lamina, and this chromatin distribution is called “conventional nuclear architecture”. Rod cells of diurnal mammals exhibit the conventional nuclear architecture (Solovei et al. 2009; Joffe et al. 2014). Research has also revealed that the location of heterochromatin is controlled by the expression of genes encoding the nuclear envelope proteins lamin B receptor and lamin A (Solovei et al. 2013). Refraction index measurements followed by computer simulations have shown that the inverted nuclear architecture permits light to more efficiently pass to the outer segments, thus, allowing better night vision (Solovei et al. 2013). The inverted nuclear architecture clearly represents an exquisite adaptation to nocturnal life.
It is commonly argued that the last common ancestor of all primates was nocturnal. The shift from nocturnal to diurnal life occurred in the lineage leading to simian primates after their divergence from the tarsier lineage (Kay et al. 1997; Ross and Hylander 2000; Heesy and Ross 2001). Simian primates constitute the infraorder Simiiformes, which includes New World monkeys (platyrrhini), Old World monkeys, and hominoids. All simian primates are diurnal, with one exception. The exception is owl monkeys (genus Aotus) that has a nocturnal or otherwise cathemeral lifestyle (Dyer et al. 2009; Fleagle 2012). These monkeys have better night vision than other simian primates, including humans (Jacobs 1977; Jacobs et al. 1979; Wright 1989, 1994; Young 2007). There are two possible explanations for this exceptional situation of owl monkeys. One is that the lineage leading to owl monkeys did not experience the shift from a nocturnal to a diurnal lifestyle. The alternative explanation is that owl monkeys secondarily, more recently reverted to a nocturnal/cathemeral lifestyle after their divergence from other New World monkeys. Currently, this second explanation is favored for a number of reasons. First of all, our understanding of the phylogenetic position of owl monkeys supports it. Biomolecular studies clearly show that owl monkeys are not at the root of New World monkey divergence. In widely accepted molecular phylogenetic trees (Schneider and Sampaio 2013; Perelman et al. 2011; Hodgson et al. 2009; Osterholz et al. 2009; Wildman et al. 2009), the lineage leading to Aotus does not have a basal position. Indeed, owl monkeys are nested within Cebidae (Callithrix, Callimico, Cebuella, Leontopithecus, Saguinus, Cebus, and Saimiri) that is one of three New World monkey families. The other two families are Pitheciidae (Callicebus, Pithecia, Chiropotes, Cacajao) and Atelidae (Alouatta, Ateles, Brachyteles, Lagothrix). Among these three, the divergence of Pitheciidae is estimated to have occurred earlier than the split of Cebidae and Atelidae. The secondary evolution of owl monkey adaptation to nocturnal/cathemeral life is also supported by the fact that their night vision is not as good as that of other nocturnal mammals (Wright 1989, 1994; Young 2007).
Analysis of an Aotus species showed that the rod cells have characteristics of conventional nuclear organization due to LBR expression. Thus they still manifest features of diurnal rods similar to other simian primates (Joffe et al. 2014). At the same time, heterochromatin forms a spherical block in the central space of the rod nucleus (Joffe et al. 2014). These observations suggest that the rod cells of this owl monkey were at an early stage of reacquisition of the inverted nuclear architecture, although whether full reacquisition will be attained in the future is unknown.
Up to now, there has been no true test of the two hypotheses. We do not exactly know the mechanisms or pathway that owl monkeys have followed to obtain their limited night vision. In particular, knowledge on the DNA of the nuclear architecture of owl monkey rod cells would be of considerable value. If a specific DNA sequence unique to owl monkeys is responsible for the rod cell change in the direction of the inverted nuclear architecture, the hypothesis of secondary adaptation will be strongly supported.
In previous studies, we identified three distinct megasatellite DNAs that reside in the genome of Aotus azarae (Prakhongcheep et al. 2013a, 2013b). The megasatellite DNAs are OwlAlp1, OwlAlp2, and OwlRep. We now set out to determine exactly which of these megasatellite DNAs was recruited to form the nuclear architecture of A. azarae rod cells. OwlAlp1 and OwlAlp2 occur in the centromere region of chromosomes. The nucleotide sequence of OwlAlp2 (repeat units of 344 bp) shows a good match to that of alpha satellite DNA of other New World monkeys (Suntronpong et al. 2016; Kugou et al. 2016), where alpha satellite DNA is a major centromeric repetitive DNA of simian primates. OwlAlp1 (repeat units of 185 bp) corresponds to part of OwlAlp2, suggesting that OwlAlp1 was derived from OwlAlp2 by a deletion mutation. Interestingly, OwlAlp1 occupies the primary constriction regions of all chromosomes, whereas OwlAlp2 is present in the pericentric regions of many, but not all, chromosomes. On the basis of these observations, we proposed that OwlAlp1 replaced OwlAlp2 as the main centromeric repetitive DNA (Prakhongcheep et al. 2013a). Owl monkeys carry a relatively large number of acrocentric chromosomes (Torres et al. 1998; Pieczarka et al. 1998), and OwlRep (repeat units of 187 bp) constitutes the entire short arms of these chromosomes in A. azarae (Prakhongcheep et al. 2013b). The origin of OwlRep is still unknown because no nucleotide sequence exhibiting similarity to OwlRep has yet been detected in any organism other than A. azarae.
Our future goal is to test the two hypotheses concerning the origin of owl monkey night vision: Long-time maintenance or reacquisition. In the present study, we examined a retinal tissue sample of A. azarae in which the three megasatellite DNAs had been found. The main goal of our research was to determine which of the three satellite DNAs constitutes the heterochromatin block located in the central region of A. azarae rod cells. To accomplish this goal, we examined the spatial distribution of the three satellite DNAs in the rod nucleus by conducting a three-dimensional fluorescence in situ hybridization (3D-FISH) analysis of a retina sample. We found that OwlRep is the primary component of the heterochromatin block.
Materials and Methods
Ethics
Experiments using primate samples were performed in accordance with the Guidelines for Care and Use of Nonhuman Primates (Version 3; June 2010) published by Kyoto University Primate Research Institute (KUPRI). This study included an animal experiment, and the Animal Experimentation Committee of Kyoto University approved in advance our application (approval number 2014-135-03). This study also included a recombinant DNA experiment, and the Recombinant DNA Experiment Safety Committee of Kyoto University approved in advance our application (approval number 140112).
Animals
An adult male of A. azarae was sacrificed after deep anesthesia, and most parts of the body were used for multiple purposes, including that of the present study, or frozen for future use. The retina was taken from an eye, fixed in 4% paraformaldehyde, and used for tissue section FISH analysis. Information about the owl monkey animal is as follows: Individual identification number, A54; sex, male; age at the time of sample collection, 12 years and 1 month; date of birth, October 11, 2002; place of birth, KUPRI; parents, adults collected in Bolivia and legally imported to Japan in 1977. The retina sample of the common marmoset (Callithrix jacchus) was taken, as part of multiple usage, from an adult female that was sacrificed for another project. Information about the marmoset individual is as follows: Individual identification number, Cj477; sex, female; age at the time of sample collection, 4 years and 1 month; date of birth, March 04, 2011; place of birth, KUPRI; parents, individuals of a laboratory line maintained in KUPRI for >20 years.
For FISH analysis of chromosomes, we used the same cell line of a female A. azarae (individual identification number A34) as described in our previous studies (Prakhongcheep et al. 2013a, 2013b; Sujiwattanarat et al. 2015). We used the already established cell line of A34 because cell culturing from the skin tissue of A54 failed due to bacterial contamination. Detailed information about A34 is described previously (Prakhongcheep et al. 2013a).
Materials from another species (Aotus lemurinus) were also used in the present study for confirmation of the results obtained from A. azarae. The cell line of A. lemurinus was obtained from the Foundation Biomedical Primate Research Center, the Netherlands as previously reported (Capozzi et al. 2016).
Three-Dimensional FISH Analysis of the Retina Sample
The retina sample fixed with 4% paraformaldehyde was embedded in paraffin and sliced to a 20 µm thickness, using an HM325 Rotary Microtome (Thermo Fisher Scientific Inc.). A slice placed on a slide glass was deparaffinized, washed with water, and then air dried. Hybridization mixture containing probe DNAs in 50% formamide/2× SSC was loaded on the slice. The probe DNAs had been fluorescently labeled by the nick translation method (OwlAlp1 with DEAC-dUTP, OwlAlp2 with Cy3, OwlRep with SpectrumGreen, and MarAlp with DEAC-dUTP). The slide glass was incubated at 80 °C for 10 min for DNA denaturation, and then incubated at 37 °C for 16 h for DNA hybridization. After the hybridization, the tissue slice was washed at 37 °C with 50% formamide/2× SSC, and further with 1× SSC. Subsequently to the hybridization with the DNA probes, immunostaining of the sample was conducted. The sample was exposed to a solution of 5% skim milk/PBS containing an antibody for bovine rhodopsin (1/5,000 dilution of ab5417; Abcam Inc.) at 37 °C for 1 h, washed with 0.1% NP-40/PBS, exposed to Cy5-labeled anti-IgG antibody, washed again, stained with DAPI, and then sealed under a cover glass. The cell nuclei were scanned with an axial distance of 200 nm using a confocal laser scanning microscope (LSM 510 meta or LSM880; Carl Zeiss Co. Ltd.) equipped with a 63×/1.4 Plan-Apochromat objective. For each optical section, images were recorded sequentially for all fluorochromes with 12-bit image stacks. The image stacks were processed with the ZEN2 and Imaris 7.4 softwares and 3D reconstructions were obtained by the Amira 3.2 TGS software.
FISH Analysis of Chromosome Spreads
Chromosome spreads were prepared as described previously (Hirai et al. 2005), and hybridized with the OwlRep probe (labeled with SpectrumGreen) by the same method as that for the 3D-FISH analysis, with one exception. The DNA denaturation step was conducted at 68 °C for 5 min instead of incubation at 80 °C for 10 min. This milder condition was intended to avoid detachment of the chromosome sample from the slide glass. This difference is considered to have little influence on the results because this mild condition was always sufficient for complete DNA denaturation in our routine experiments.
Results
BLAST Searches of Genome Sequence Databases for OwlRep
OwlRep was identified as megasatellite DNA present in the genome of A. azarae by our comparative genomic hybridization experiments (Prakhongcheep et al. 2013b). In this previous study, we did not find OwlRep or similar sequences in a wide range of vertebrates in searches of genome sequence databases (NCBI collection of April, 2013). In the present study, we performed the same searches (against databases in NCBI of May, 2017) and found several sequences of A. lemurinus that exhibited >99% nucleotide identities (for example, GenBank file JQ933005). However, no significant hits were obtained from organisms other than this owl monkey species. Thus, OwlRep is likely to be unique to some or all species of the genus Aotus.
Chromosomal Locations of OwlRep
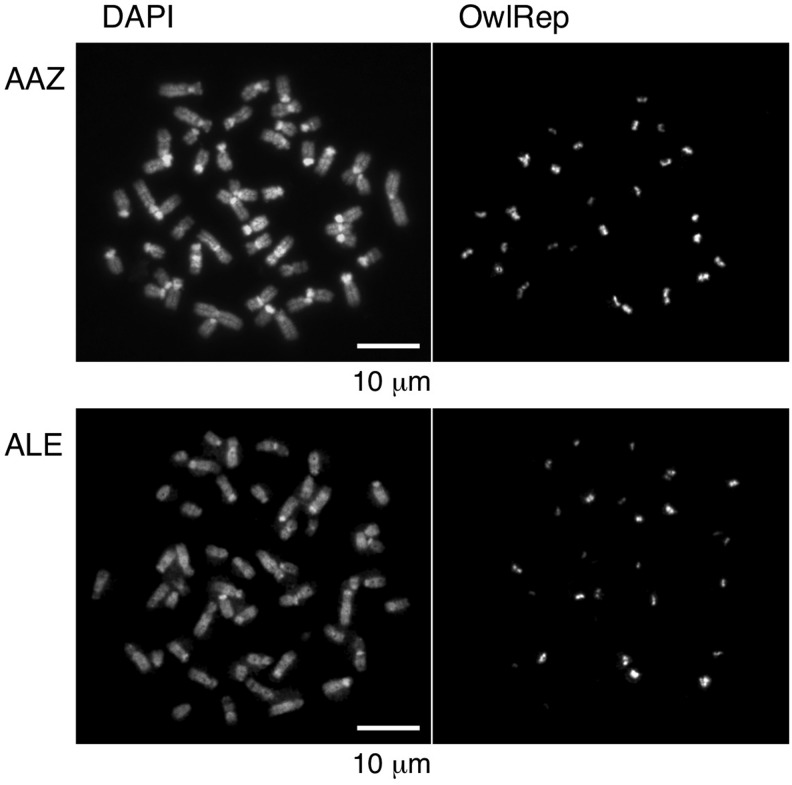
Many species of the genus Aotus are known to contain a relatively large number of acrocentric chromosomes (Torres et al. 1998; Pieczarka et al. 1998). A FISH analysis of chromosome spreads of A. azarae showed that the entire short arms of acrocentric chromosomes are composed mainly of OwlRep (Prakhongcheep et al. 2013b). OwlRep was also found in pericentric regions of a few of the biarmed chromosomes (Prakhongcheep et al. 2013b). We conducted the same FISH analysis of the chromosomes of A. lemurinus to examine whether OwlRep is distributed in the same fashion in this species. We also included an A. azarae sample in this analysis for accurate, direct comparison. The results obtained are shown in figure 1. The distribution pattern of OwlRep signals observed with the A. azarae sample was consistent with our previous results (Prakhongcheep et al. 2013b), and the A. lemurinus sample exhibited an identical distribution pattern.
Fig. 1.—
FISH analysis of chromosomes. Hybridization was conducted on chromosome spreads of Aotus azarae (AAZ) and Aotus lemurinus (ALE), using OwlRep as a probe.
FISH Analysis of Retinal Sections
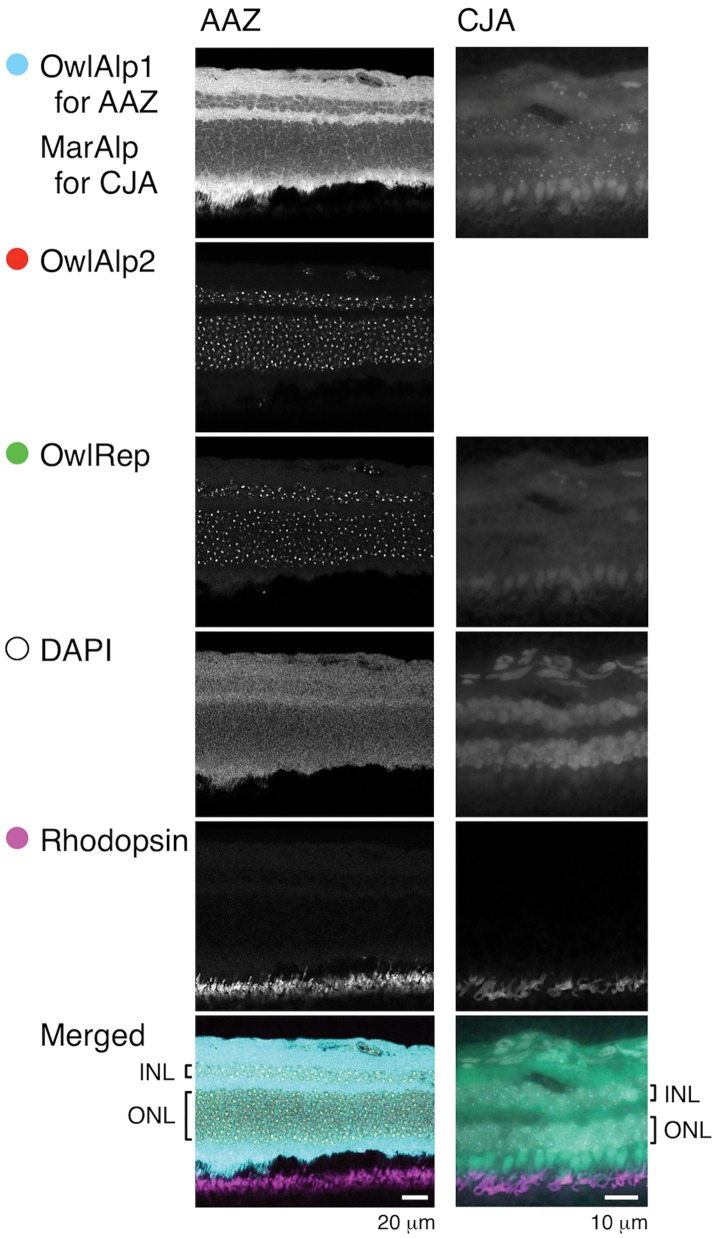
We prepared cross-sections of the retina obtained from an adult of A. azarae. The sample was laid on a slide glass and a FISH assay was performed using OwlAlp1, OwlAlp2, and OwlRep as DNA probes that were labeled using different fluorochromes. We also performed an assay against a retina section from an adult of the common marmoset (a diurnal New World monkey), using MarAlp (alpha satellite DNA of the marmoset) and OwlRep as probes. In both assays, subsequent to the FISH reaction, the retina sample was immunostained with the rhodopsin antibody, and stained with DAPI for nuclear visualization. Figure 2 shows images obtained from a peripheral region of the retina where the great majority of the photoreceptor cells was considered to be rods. Rhodopsin signals indicated that the side where they appear was the outer side of the retina. We regarded the layer containing the nuclei and positioned closest to the rhodopsin signal region as the outer nuclear layer, which consisted of the cell bodies of photoreceptor cells. In the outer nuclear layer of the owl monkey retina sample, positive signals were observed for OwlAlp1, OwlAlp2, and OwlRep. In comparison to the signals of OwlAlp1, those of OwlAlp2 and OwlRep were packed and distinct. In the outer nuclear layer of the marmoset retina sample, no clear signals were found for OwlRep, and spot signals were obtained for MarAlp. The MarAlp signals (in the marmoset sample) differed from the OwlAlp1 and OwlAlp2 signals (in the owl monkey sample) in that the spots were clearer and the size relative to the nucleus was smaller, respectively.
Fig. 2.—
Two-dimensional analysis of a retina section. Hybridization was conducted with three DNA probes (OwlAlp1, OwlAlp2, and OwlRep) on an Aotus azarae sample (AAZ), and two DNA probes (MarAlp and OwlRep) on a common marmoset sample (CJA). The samples were further treated for immunostaining with rhodopsin antibody and DAPI staining. Merged photographs are shown at the bottom, in which signals were colored as indicated with the circles. INL and ONL indicate the inner nuclear layer and outer nuclear layer, respectively, of the retina.
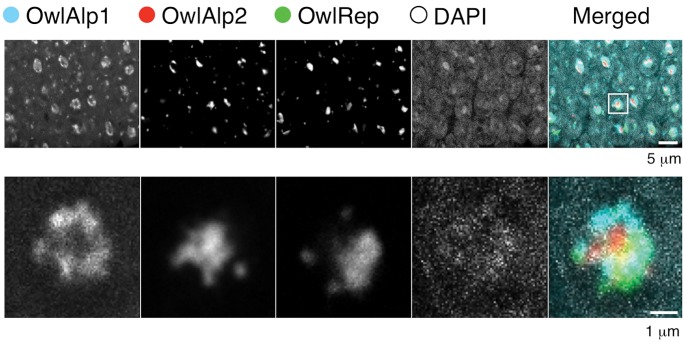
To analyze rod nuclei at higher magnifications, we conducted a multi-layer scan of the outer nuclear layer region of the A. azarae retina, with a z axis interval of 200 nm. A collection of the images along the z axis is shown in supplementary figure S1, Supplementary Material online, and an example layer is shown in figure 3. A typical nucleus is magnified in supplementary figure S2, Supplementary Material online and its example layer is shown in figure 3. A heterochromatin block was observed in the central region of the majority of the nuclei. The block was composed of all three satellite DNAs. OwlRep occupied a single central space with a nonuniform shape due to its rugged surface. In each cell, OwlAlp2 formed one or multiple smaller clumps. As revealed in the merged photographs, OwlAp2 clumps were located mostly in the hollows on the surface of the OwlRep space. Signals of OwlAlp2 rarely overlapped those of OwlRep. Spaces consisting of OwlRep and OwlAlp2 exhibited forms closer to a sphere than those occupied only by OwlRep. Signals of OwlAlp1 were mostly located in the outer regions of the space occupied by OwlRep and OwlAlp2. In addition, small signals were observed in association with the nearly spherical heterochromatin blocks located in the center of the nucleus. The numbers of these small signals were mostly 0–3 per cell, and they were often found on the inner surface of the nuclear membrane. OwlAlp1 and OwlRep, and to a lesser extent OwlAlp2, were responsible for these small signals.
Fig. 3.—
Distribution of the three megasatellite DNAs. The upper panels show the distribution in the retina tissue. Photographs of multiple cross-sections along the z axis are shown in supplementary figure S1, Supplementary Material online, and layer 11 is shown here as an example. A typical single nucleus, indicated by the square in the merged photograph, was picked up and its magnified photographs are shown in the lower panels. Photographs of multiple cross-sections are shown in supplementary figure S2, Supplementary Material online.
Discussion
Rod cells of an unidentified Aotus species were previously shown to exhibit a chromatin distribution that is intermediate between the conventional and inverted nuclear architecture (Joffe et al. 2014). In agreement with these previous results, heterochromatin was found adjacent to the nuclear lamina, and additionally, a relatively large heterochromatin block was observed in the central space of the nucleus. The heterochromatin adjacent to the nuclear lamina indicates that the owl monkey rod cells maintain the molecular mechanisms to build the conventional nuclear architecture. The large heterochromatin block in the central space of the nucleus is similar to the inverted nuclear architecture. In the present study, we found that OwlRep forms a single block in the central region of the nucleus, and that OwlAlp1 and OwlAlp2 do not overlap the space occupied by OwlRep. The A. azarae genome does not contain other satellite DNAs of comparable scale or larger than OwlAlp1, OwlAlp2, or OwlRep. We reached this conclusion by isolating these DNAs by the genomic hybridization method, in which satellite DNAs of larger scales are identified more efficiently, irrespective of their nucleotide sequences (Koga et al. 2012; Hara et al. 2012). Altogether, these data indicate that OwlRep is the primary component of the heterochromatin block that occupies the central space of the A. azarae rod nucleus. OwlRep is present as megasatellite DNA in all Aotus species of which genome sequence databases or experimental materials were available (A. azarae, Aotus nancymaae, and A. lemurinus), but similar sequences were never found in any other organism we examined. It is thus likely that OwlRep was generated and amplified in the Aotus lineage after its split from other New World monkeys. The final goal of our research will test the two hypotheses about the origin of the owl monkey night vision: Long-time maintenance or reacquisition. Here as an initial step, we examined the retinal tissue of A. azarae. Our results, which indicate that the centrally located heterochromatin block is composed primarily of a satellite DNA that is unique to Aotus, support the reacquisition hypothesis. Obviously, our direct experimental data mainly concerns A. azarae. We intend to examine other species when we have an opportunity to obtain a fresh eye sample, to test whether the conclusion about A. azarae can be extended to other Aotus species which seems likely.
Aotus azarae is currently considered to be a cathemeral species. However, a recent analysis of activity patterns of primates suggested that the last common ancestor of all extant Aotus species, including A. azarae, was nocturnal. It is hypothesized that some Aotus species now considered cathemeral evolved by expanding their activity to include extra daytime activity (Santini et al. 2015). This is certainly the case of A. azarae. This species has the most southern distribution of all owl monkeys (farthest from the original habitat). It can be argued that this species became more cathermeral to cope with the conditions of its extreme southern range and more recently habitat disturbance due to human encroachment. We can conclude with some confidence that the cathemeral lifestyle of A. azarae resulted from an expansion of their nocturnal life to include extra daytime activity (Fernández-Duque et al. 2010; Santini et al. 2015). Further, an analysis of long-time, 24-h activity records showed that A. azarae is still more active in the night than in the day (Fernández-Duque et al. 2010). In addition, while the inverted nuclear architecture positively affects night-time activity, its possession or maintenance is not considered to have deleterious effects on the daytime activity in which cones play a significant role. The reason we used A. azare in the present study was that an old individual long past the reproductive stage was available at our institute. Currently, we have no immediate intentions to examine other Aotus species because we hesitate to sacrifice additional rare animals. We are, however, prepared to replicate the same experiments when we have an opportunity to obtain a fresh eye sample due to natural deaths.
The rod nuclei of many nocturnal mammals have an inverted nuclear architecture (Solovei et al. 2009; Joffe et al. 2014). However, it has not been reported that satellite DNAs found in nocturnal mammals share specific features. Apparently, satellite DNAs of any nucleotide sequence can serve as a component of the heterochromatin core in the inverted nuclear architecture. In A. azarae, three satellite DNAs participate in the formation of the heterochromatin core. Among these, OwlRep is the primary component because it occupies a single, relatively large space at the very center of the nucleus. In contrast, OwlAlp2 is distributed at the periphery of the OwlRep space as multiple, relatively small blocks and OwlAlp1 is located even more externally. OwlAlp2 was probably present as a large-scale satellite DNA ever since the owl monkey lineage diverged from other New World monkeys. In addition, although the scale of OwlAlp1 may have been small when it was derived from OwlAlp2, it grew into a large-scale satellite DNA over time. This raises the question of why OwlRep, rather than OwlAlp2 or OwlAlp1, was recruited as the primary component of the heterochromatin core in A. zarae or its ancestor. One possible answer is that, even though any satellite DNA has the potential to serve as a component, their efficiencies may vary especially in the initial stage of the shift in the direction of the inverted nuclear architecture. A key point may not be the nucleotide sequence but another attribute, such as higher-order structure, chromosomal location, original function, speed of amplification, or interaction with coexisting satellite DNAs. When the common ancestor of A. azarae and other owl monkeys began the transition to a nocturnal lifestyle, OwlRep may have been preferred as the primary component because of one or more of its attributes. It is not presently clear exactly how OwlRep emerged because we have not found similar sequences in organisms other than some Aotus species. The answer to this question may be found when further genome sequence data are obtained for other taxa.
FISH analysis of A. azarae chromosomes in our previous study (Prakhongcheep et al. 2013a) showed that OwlAlp1 occupies the primary constriction regions of all chromosomes, whereas OwlAlp2 is present in the pericentric regions of many, but not all, chromosomes. These results suggest that the centromere function has been completely transferred to OwlAlp1; in other words, OwlAlp2 was freed from the centromere function. OwlAlp2 is, however, still present on a large scale comparable to that of OwlAlp1 (Prakhongcheep et al. 2013a). In relation to these results, it is noteworthy that OwlRep occupies an amorphous single space at the center of the nucleus and OwlAlp2 forms one or multiple smaller clumps that fill hollows on the surface of the OwlRep space. Thus, in the past, OwlAlp2 experienced two events: Loss of the centromere function, and participation in the formation of the heterochromatin block as a component. It is of evolutionary interest whether these two events occurred independently and, if no, which one caused the other.
In conclusion, the centrally located heterochromatin block of the A. azarae rod nucleus is composed primarily of co-opted OwlRep which is unique to Aotus, and these results support the hypothesis that the night vision of A. azarae evolved secondarily. Our results represent a unique example of how a newly acquired megasatellite DNA contributed to the adaptation of its host organism to exploit an ecological niche.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author Contributions
A.K. and H.H. conceived and designed the experiments. A.K., H.T., M. I., and Y.H. performed the molecular biology experiments. H.I., T.O., and R.S. arranged for the animals and cell lines. A.K. and R.S. wrote the report. All authors approved the final version of the manuscript.
Supplementary Material
Acknowledgments
This work was supported by Grants-in-Aid from the MEXT of Japan (grant numbers 15H04427, 15K14435, 26251040, and 23114005 to A.K.). The work of R.S. was supported by a grant from the Italian Ministry of Universities and Research (PRIN 2015). We are grateful to Masanaru Takai (Kyoto University) and Takehiko Kobayashi (University of Tokyo) for helpful discussions and Yuki Enomoto (Kyoto University) for technical assistance.
Literature Cited
- Capozzi O, Archidiacono N, Lorusso N, Stanyon R, Rocchi M.. 2016. The 14/15 association as a paradigmatic example of tracing karyotype evolution in New World monkeys. Chromosoma 125(4):747–756. [DOI] [PubMed] [Google Scholar]
- Dyer MA, et al. 2009. Developmental sources of conservation and variation in the evolution of the primate eye. Proc Natl Acad Sci U S A. 106(22):8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart A, et al. 2013. Epigenetics of eu- and heterochromatin in inverted and conventional nuclei from mouse retina. Chromosome Res. 21(5):535–554. [DOI] [PubMed] [Google Scholar]
- Fernández-Duque E, de la Iglesia H, Erkert HG.. 2010. Moonstruck primates: owl monkeys (Aotus) need moonlight for nocturnal activity in their natural environment. PLoS ONE. 5(9):e12572.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleagle JG. 2012. Perspectives in primate evolution. Evol Anthropol. 21(6):207.. [DOI] [PubMed] [Google Scholar]
- Hara T, Hirai Y, Jahan I, Hirai H, Koga A.. 2012. Tandem repeat sequences evolutionarily related to SVA-type retrotransposons are expanded in the centromere region of the western hoolock gibbon, a small ape. J Hum Genet. 57:760–765. [DOI] [PubMed] [Google Scholar]
- Heesy CP, Ross CF.. 2001. Evolution of activity patterns and chromatic vision in primates: morphometrics, genetics and cladistics. J Hum Evol. 40(2):111–149. [DOI] [PubMed] [Google Scholar]
- Hirai H, et al. 2005. A whole-arm translocation (WAT8/9) separating Sumatran and Bornean agile gibbons, and its evolutionary features. Chromosome Res. 13(2):123–133. [DOI] [PubMed] [Google Scholar]
- Hodgson JA, et al. 2009. Successive radiations, not stasis, in the South American primate fauna. Proc Natl Acad Sci U S A. 106(14):5534–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs GH. 1977. Visual capacities of the owl monkey (Aotus trivirgatus) – II. Spatial contrast sensitivity. Vis Res. 17(7):821–825. [DOI] [PubMed] [Google Scholar]
- Jacobs GH, Tootell RB, Blakeslee B.. 1979. Visual capacities of the owl monkey (Aotus trivirgatus): temporal contrast sensitivity. Folia Primatol (Basel). 32:193–199. [DOI] [PubMed] [Google Scholar]
- Joffe B, Peichl L, Hendrickson A, Leonhardt H, Solovei I.. 2014. Diurnality and nocturnality in primates: an analysis from the rod Photoreceptor nuclei perspective. Evol Biol. 41:1–11. [Google Scholar]
- Kay RF, Ross C, Williams BA.. 1997. Anthropoid origins. Science 275(5301):797–804. [DOI] [PubMed] [Google Scholar]
- Koga A, Hirai Y, Hara T, Hirai H.. 2012. Repetitive sequences originating from the centromere constitute large-scale heterochromatin in the telomere region in the siamang, a small ape. Heredity 109(3):180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugou K, Hirai H, Masumoto H, Koga A.. 2016. Formation of functional CENP-B boxes at diverse locations in repeat units of centromeric DNA in New World monkeys. Sci Rep. 6:27833.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterholz M, Walter L, Roos C.. 2009. Retropositional events consolidate the branching order among New World monkey genera. Mol Phylogenet Evol. 50(3):507–513. [DOI] [PubMed] [Google Scholar]
- Perelman P, et al. 2011. A molecular phylogeny of living primates. PLoS Genet. 7(3):e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieczarka JC, Nagamachi CY, Muniz JA, Barros RM, Mattevi MS.. 1998. Analysis of constitutive heterochromatin of Aotus (Cebidae, Primates) by restriction enzyme and fluorochrome bands. Chromosome Res. 6(2):77–83. [DOI] [PubMed] [Google Scholar]
- Prakhongcheep O, et al. 2013a. Two types of alpha satellite DNA in distinct chromosomal locations in Azara‘s owl monkey. DNA Res. 20(3):235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakhongcheep O, et al. 2013b. Heterochromatin blocks constituting the entire short arms of acrocentric chromosomes of Azara's owl monkey: formation processes inferred from chromosomal locations. DNA Res. 20:461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CF, Hylander W.. 2000. Electromyography of the anterior temporalis and masseter muscles of owl monkeys (Aotus trivirgatus) and the function of the postorbital septum. Am J Phys Anthropol. 112(4):455–468. Review. [DOI] [PubMed] [Google Scholar]
- Santini L, Rojas D, Donati G.. 2015. Evolving through day and night: origin and diversification of activity pattern in modern primates. Behav Ecol. 26:789–796. [Google Scholar]
- Schneider H, Sampaio I.. 2013. The systematics and evolution of New World primates – a review. Mol Phylogenet Evol. 82:348–357. [DOI] [PubMed] [Google Scholar]
- Solovei I, et al. 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137(2):356–368. [DOI] [PubMed] [Google Scholar]
- Solovei I, et al. 2013. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152(3):584–598. [DOI] [PubMed] [Google Scholar]
- Sujiwattanarat P, et al. 2015. Higher-order repeat structure in alpha satellite DNA occurs in New World monkeys and is not confined to hominoids. Sci Rep. 5(1):10315.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntronpong A, et al. 2016. CENP-B box, a nucleotide motif involved in centromere formation, occurs in a New World monkey. Biol Lett. 12(3):20150817.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres OM, Enciso S, Ruiz F, Silva E, Yunis I.. 1998. Chromosome diversity of the genus Aotus from Colombia. Am J Primatol. 44(4):255–275. [DOI] [PubMed] [Google Scholar]
- Wildman DE, Jameson NM, Opazo JC, Yi SV.. 2009. A fully resolved genus level phylogeny of neotropical primates (Platyrrhini). Mol Phylogenet Evol. 53(3):694–702. [DOI] [PubMed] [Google Scholar]
- Wright PC. 1989. The nocturnal primate niche in the New World. J Hum Evol. 18(7):635–658. [Google Scholar]
- Wright PC. 1994. The behavior and ecology of the owl monkey In: Baer JH, Weller RE, Kakoma I, editors. Aotus: the owl monkey. Elsevier (Amsterdam) p. 97–112. ISBN 0-12-072405-7. [Google Scholar]
- Young A. 2007. Night creatures. Learning Media (Wellington). ISBN 978-0-7903-1915-5.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.