Abstract
Antisense oligodeoxynucleotides can selectively block disease-causing genes, and cancer genes have been chosen as potential targets for antisense drugs to treat cancer. However, nonspecific side effects have clouded the true antisense mechanism of action and hampered clinical development of antisense therapeutics. Using DNA microarrays, we have conducted a systematic characterization of gene expression in cells exposed to antisense, either exogenously or endogenously. Here, we show that in a sequence-specific manner, antisense targeted to protein kinase A RIα alters expression of the clusters of coordinately expressed genes at a specific stage of cell growth, differentiation, and activation. The genes that define the proliferation-transformation signature are down-regulated, whereas those that define the differentiation-reverse transformation signature are up-regulated in antisense-treated cancer cells and tumors, but not in host livers. In this differentiation signature, the genes showing the highest induction include genes for the G proteins Rap1 and Cdc42. The expression signature induced by the exogenously supplied antisense oligodeoxynucleotide overlaps strikingly with that induced by endogenous antisense gene overexpression. Defining antisense DNAs on the basis of their effects on global gene expression can lead to identification of clinically relevant antisense therapeutics and can identify which molecular and cellular events might be important in complex biological processes, such as cell growth and differentiation.
The two isoforms of cAMP-dependent protein kinase (PKA), PKA-I and PKA-II, share a common catalytic subunit but contain distinct regulatory (R) subunits, RI and RII, respectively (1). Four different R subunits—RIα, RIβ, RIIα, and RIIβ—have been identified. Expression of the RIα subunit of PKA is increased in various human tumors and cell lines, including cancers of the breast (2–5), ovary (6, 7), lung (8), and colon (9–11). Furthermore, overexpression of the RIα subunit of PKA correlates with malignancy and poor prognosis in cancer patients (3–7). Therefore, the RIα subunit of PKA is a potential target for human cancer therapy. In the last decade, there have been increasing efforts to develop PKA-specific inhibitors as cancer therapeutic agents (12–19).
In the present study, we have investigated the sequence-specific effects of RIα antisense on global gene expression by using DNA microarray. We have used distinct antisense phosphorothioate oligonucleotides (PS-ODNs) targeted to the human RIα gene and the second-generation antisense ODN, which is a 2′-O-methyl RNA/DNA hybrid (19). We also examined the expression profile in cells endogenously overexpressing the RIα antisense gene. This system avoids the problems inherent in ODN treatment, namely, the delivery and stability of the ODN.
Methods
ODN Treatment.
RIα antisense ODNs used were PS-ODN (15), RNA/DNA ODN (19), mouse PS-ODN (15), and appropriate control ODNs (15, 19). Cells were treated with antisense/control ODNs (0.2 μM, 3 days) by using the cationic liposomal transfection reagent DOTAP (N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate) (Roche Diagnostics).
RIα Antisense Gene Overexpression.
Cells overexpressing the RIα antisense gene were generated by stable transfection with the RIα antisense gene [N-terminal 200 nt ligated into the OT1529 vector (20) (L. Tan and Y.S.C.-C., unpublished work)] and treatment with ZnSO4 (60 μM, 3 days).
RNA Preparation.
Total cellular RNA was prepared from control and antisense-exposed cells by using a RNeasy Midi Kit (Qiagen, Chatsworth, CA), electrophoresed, blotted onto nylon membrane, and subjected to Northern blot analysis (20). The specific fragments of cDNA corresponding to respective genes were generated by PCR.
cDNA Microarray Analysis.
Total RNA prepared from the antisense-exposed and control cells were used to synthesize 33P-labeled cDNAs by reverse transcription. The cDNAs were hybridized to a human cDNA microarray (2,304 elements) that was primarily derived from image consortium cDNA libraries (Research Genetics, Huntsville, AL), as described (21). Cluster analysis was performed on Z-transformed microarray data by using two separate programs available as shareware from Michael Eisen's laboratory (http://rana.lbl.gov).
Tumor Growth and Antisense Treatment.
PC3 M cells (2 × 106 cells) were inoculated s.c. into nude mice. When tumors became palpable (30–50 mg), antisense or control ODN (0.1 mg/0.1 ml saline per mouse, daily) or saline (0.1 ml per mouse) was injected i.p. into the mice (16). After 4 days of treatment, animals were killed, and tumors, livers, and spleens were removed, weighed, immediately frozen in liquid N2, and kept frozen at −80°C until used.
Results and Discussion
Parallel Expression Profiles Between Antisense ODN-Treated Cells and Cells Overexpressing the Antisense Gene.
We hypothesized that the sequence-specific effects as well as various unexpected effects of antisense might reflect previously unrecognized gene expression patterns in treated cells. We thus used DNA microarray technology (22) to identify the sequence-specific mechanism for antisense RIα action. We used a cDNA microarray containing 2,304 nonredundant clones to analyze gene expression patterns in cells treated with RIα antisense RNA/DNA mixed backbone (19) ODN. Total RNA prepared from antisense-treated and untreated control cells was used to synthesize 33P-labeled cDNAs, which were hybridized to cDNA microarray. Treatment of PC3 M prostate carcinoma cells with 0.2 μM RIα antisense for 3 days inhibited growth by 50%. We compared the expression profile induced by exogenous antisense ODN treatment with that induced by the endogenous antisense gene overexpression. To address the question of cell specificity, we also analyzed the expression profiles of LS-174T colon carcinoma cells transfected with the RIα antisense gene.
The expression levels of ≈240 cDNAs, representing 10% of the total DNA elements on the array, were altered (≥ 2-fold up-regulated or down-regulated) in antisense ODN or antisense gene-treated cells. The changes ranged from −20-fold to +27-fold (Table 1). In contrast, cells treated with the cationic lipid DOTAP (N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate) alone or DOTAP plus control ODN exhibited a minimal (< ±30% change) alteration in the expression profile, and the pattern of altered expression did not mimic that caused by antisense ODN treatment (data not shown).
Table 1.
Expression profile of genes altered in PC3M cells treated with antisense ODN or overexpressing antisense gene
| Genes | Fold change
|
|
|---|---|---|
| AS ODN | AS gene | |
| CDC42 | 20.0 | 18.9 |
| Myosin light chain, alkali | 14.4 | 14.0 |
| Kinesin heavy chain | 12.7 | 11.1 |
| Endothelin-converting enzyme 1 | 12.5 | 10.0 |
| CDC27 | 9.7 | 9.2 |
| Lysosome-associated membrane protein 1 | 7.9 | 6.4 |
| Basic transcription factor 3 | 6.9 | 8.8 |
| RAP1A | 5.4 | 6.3 |
| Ras-related protein RAB-8 | 5.2 | 5.8 |
| IL-3 receptor, α | 4.7 | 3.6 |
| MHC class II | 4.4 | 4.2 |
| Signal recognition particle receptor | 3.8 | 3.4 |
| Probable G protein-coupled receptor HM74 | 3.7 | 4.0 |
| Integrin, α-6 | 3.6 | 3.7 |
| PDE4B, cAMP specific | 2.3 | 3.2 |
| Heat shock protein HSPA2 | −2.3 | −2.3 |
| Transformation-sens. prot. IEF SSP 3521 | −2.3 | −2.9 |
| Superoxide dismutase 1 (Cu/Zn) | −2.6 | −2.6 |
| Plasminogen-like protein | −2.7 | −2.6 |
| Mitogen-act. prot. kinase kinase kinase 5 | −2.9 | −2.8 |
| Phosphatidylinositol 3-kinase associated p85 | −3.1 | −3.6 |
| YY1 transcription factor | −3.2 | −3.2 |
| Cytochrome p450 IIE1 | −3.7 | −4.3 |
| Natural killer cell protein 4 | −4.0 | −4.0 |
| Collagen, type IV, alpha-4 | −4.0 | −4.5 |
| Diacylglycerol kinase, γ (90 kDa) | −4.5 | −4.5 |
| Catalase | −6.7 | −6.7 |
| Ligase 1, DNA, ATP-dependent | −6.7 | −6.7 |
| M-phase inducer phosphatase 2 | −7.7 | −7.7 |
| Crystallin Mu | −20.0 | −20.0 |
AS ODN, antisense ODN treatment; AS gene, antisense gene overexpression. Fold change represents altered levels of expression in antisense ODN/antisense gene cells as compared to control cells.
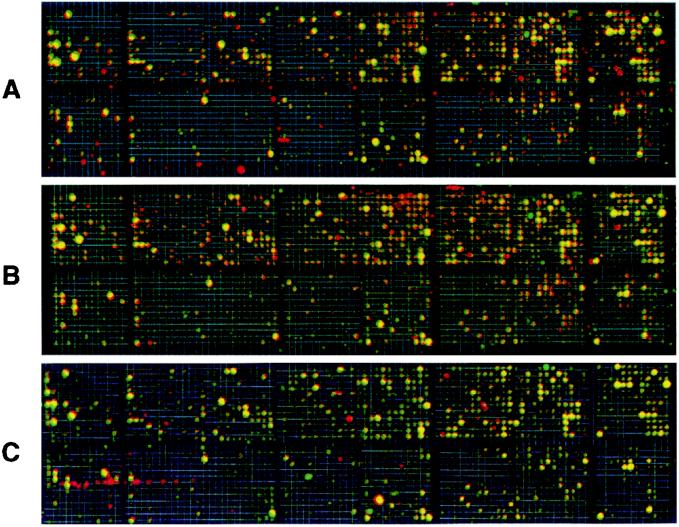
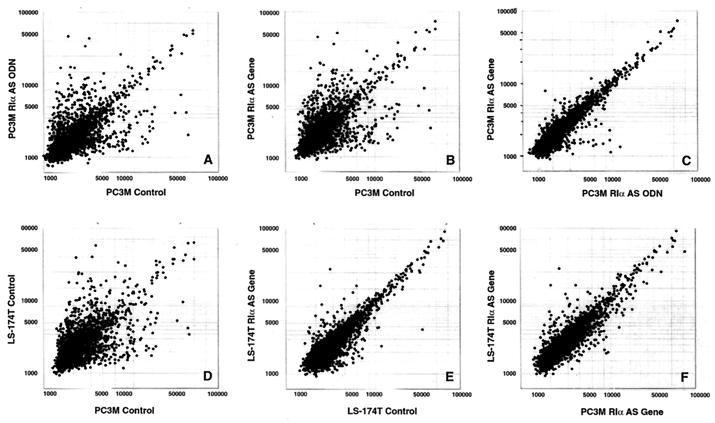
The expression profile of cells treated with antisense ODN (Figs. 1A and 2A) exhibited striking overlaps with that of cells that overexpressed the antisense gene (Figs. 1B and 2B). The same cDNA elements that were up-regulated or down-regulated in antisense ODN-treated cells were similarly up-regulated or down-regulated in the cells overexpressing antisense gene (Table 1). Less than 2% of the altered expression profiles were specific to either antisense ODN-treated cells or cells overexpressing the antisense gene (Fig. 2C). These results indicate that antisense ODN treatment and endogenous antisense gene overexpression affected nearly identical genomic pathways. Furthermore, the antisense-induced alteration in the expression profile of PC3 M cells was closely comparable to that of LS-174T cells overexpressing the antisense gene (Figs. 1 B and C and 2F), although the two cell lines exhibited large differences in their basic patterns of expression (Fig. 2D) and in the magnitude of altered expression (compare Fig. 2B with Fig. 2E). Thus, different cell types exhibit comparable expression profiles when treated with RIα antisense.
Figure 1.
Scanned phosphorimages of cDNA microarrays from RIα antisense ODN-treated cells and cells overexpressing the antisense gene, superimposed with images of respective control cells. (A) Antisense ODN-treated PC3 M cells. (B) Antisense gene overexpressing PC3 M cells. (C) LS-174T cells overexpressing the antisense gene. Genes up-regulated, compared with the control, are shown in red; down-regulated genes are shown in green, and yellow represents genes without changes in expression. The data represent one of four separate hybridizations that gave similar results.
Figure 2.
Scatter plots for expression profile comparison between antisense ODN treatment and antisense gene overexpression and between PC3 M cells and LS-174T cells. Expression profiles of untreated control versus antisense (AS) ODN/gene-targeted cells (A, B, and E), AS ODN versus AS gene treatment (C), PC3 M control cells versus LS-174T control cells (D), and PC3 M cells overexpressing the antisense gene versus LS-174T overexpressing the antisense gene (F) are shown as bivariate scatterplots of 2,304 genes from the microarray. The values are corrected intensities representing levels of expression for the DNA elements of the microarrays (21).
Northern Blotting Verifies Altered Gene Expression of Microarray.
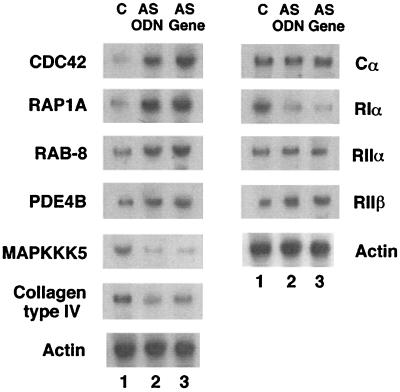
Northern blotting confirmed the microarray data, with changes in the expression profile exerted by the antisense ODN and antisense gene (that is, the direction of changes as well as the magnitude of altered expression) (Fig. 3). The Northern results further confirmed that the antisense RIα specifically down-regulated PKA RIα mRNA without affecting the expression of other PKA subunits (Fig. 3).
Figure 3.
Northern analysis of genes altered in PC3 M cells treated with antisense ODN or cells overexpressing antisense gene. C, untreated control cells; AS ODN, antisense ODN treatment; AS gene, antisense gene overexpression. The data represent one of two independent experiments that gave similar results.
Molecular Portrait of Reverted Phenotype of Prostate Carcinoma.
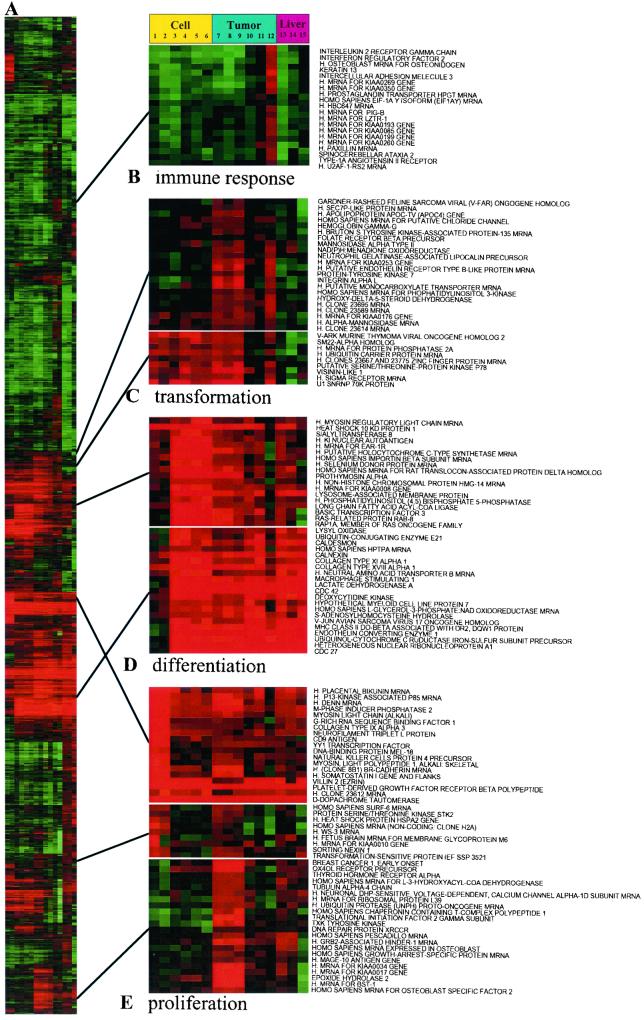
In addition to growth inhibition, RIα antisense treatment also induced changes in cell morphology, including a flat phenotype similar to the reverted phenotype of transformed cells (23). To obtain a molecular portrait of the “reverted” phenotype, we used a hierarchical clustering algorithm to group genes on the basis of similarity in their expression patterns (24). The data are presented in a matrix format (Fig. 4). Each row represents all of the hybridization results for a single DNA element of the array, and each column represents the expression levels for all genes in a single hybridization sample. The expression level of each gene is visualized, in color, relative to its median expression level across all samples. Red represents expression greater than the mean, and green represents expression less than the mean, and the intensity of the color denotes the degree of deviation from the mean (24). Distinct samples representing similar gene patterns from control cells were aligned in adjacent rows. Likewise, different samples from antisense ODN-treated or antisense gene-transfected cells were clustered in adjacent rows in our map. Also included in this map were samples from antisense-treated tumors, host livers, and various controls. The observed patterns of gene expression would thus reflect intrinsic differences between antisense-exposed cells and control cells rather than variations arising from experimental artifacts. We defined clusters of coordinately expressed genes as “signatures,” which we named on the basis of the cellular process in which the component genes participate (25).
Figure 4.
Molecular portrait of the reverted phenotype of PC3 M cells and tumors treated with antisense RIα. Data from control ODN-treated cells, antisense ODN/gene-treated cells, untreated control cells, antisense ODN- or control ODN-treated tumors, control tumors, and host livers were combined and clustered. Cluster analysis was performed on Z-transformed microarray data by using two separate programs available as shareware from Michael Eisen's lab (http://rana.lbl.gov). Each gene is represented by a single row of colored boxes; each experimental sample is represented by a single column. Columns 1 and 2, untreated control cells. Columns 3 and 4, antisense RNA/DNA hybrid ODN-treated cells. Columns 5 and 6, antisense gene overexpressing cells. Columns 7 and 8, untreated control tumors. Column 9, control ODN-treated tumor. Column 10, antisense RNA/DNA ODN-treated tumor. Column 11, mouse antisense PS-ODN-treated tumor. Column 12, antisense PS-ODN-treated tumor. Column 13, antisense PS-ODN-treated liver. Column 14, antisense RNA/DNA ODN-treated liver. Column 15, untreated control liver. The entire clustered image is shown in A. Full gene names are shown for representative clusters containing functionally related genes involved in (B) immune response, (C) transformation, (D) differentiation, and (E) proliferation. These clusters also contain uncharacterized genes and named genes not involved in these processes. The up-regulation of Cdc42 in tumors is not apparent in D because of its high basal levels. The deviations of log ratios of expression levels from controls in antisense ODN-treated cells and tumors were 4.77 and 1.84, respectively. The expression levels of genes for the wingless-type mouse mammary tumor virus integration site family and sex-determining region Y were not depicted in D. The deviations of log ratios of these genes from controls were 0.26 and 1.08, respectively, in antisense-treated cells and 1.90 and 2.04, respectively, in antisense-treated tumors. The size of the tumors compared with untreated (saline injected) control tumors were: 42%, 63%, 56%, and 98%, after 4 days of treatment with antisense RNA/DNA ODN, PS-ODN, mouse PS-ODN, and control ODN, respectively. There was no sign of systemic toxicity in treated animals, and the size of the liver and spleen remained unchanged.
Our map revealed that RIα antisense treatment affected a cluster of genes involved in proliferation and one involved in differentiation (Fig. 4). Genes that define the proliferation signature were highly expressed in untreated control cells (Fig. 4E, columns 1 and 2) and markedly suppressed in antisense-treated or -transfected cells (Fig. 4E, Top and Middle, columns 3–6). In this proliferation signature, genes encoding proteins involved in cell cycle control, DNA synthesis and regulation, transcription, and translation were predominant. Conversely, RIα antisense treatment induced genes involved in differentiation and reverse transformation (Fig. 4D). This cluster was dominated by genes encoding small G proteins, such as Cdc42 and Rap1, genes encoding transcription factors, and genes encoding regulatory proteins of the cytoskeleton, specifically the microtubules. Remarkably, the effects of RIα antisense treatment on these signatures were mirrored in cells overexpressing the antisense gene (Fig. 4, compare columns 3 and 4 with 5 and 6). The altered expression signatures generated by the RIα antisense ODN thus reflected true antisense effects rather than nonspecific antisense ODN effects described elsewhere (26).
Similar proliferation and differentiation signatures were observed in antisense-treated tumors (Fig. 4 D and E, Top and Middle, columns 10–12). However, we also observed an expression profile distinct from that observed in antisense-treated cells. For example, genes in the tumor-specific proliferation signature, such as those for TXK tyrosine kinase and Grb-2-associated protein, were markedly down-regulated in tumors, but unchanged in cells (Fig. 4E Bottom). Conversely, genes in the tumor-specific differentiation signature, such as those for developmental proteins, including wingless-type mouse mammary tumor virus integration site family and sex-determining region Y were markedly up-regulated in tumors, but not in cells (Fig. 4, legend). Likewise, genes in the transformation signature, such as oncogenes and genes for tyrosine and serine/threonine kinases that are usually overexpressed in tumors, were specifically down-regulated in tumors only, but not in cells, after antisense treatment (Fig. 4C).
To verify the specificity of the antisense effects on gene expression signatures, we used three different antisense ODNs that differed in sequence or chemical modification: a PS-ODN antisense (15), directed against codons 8–13 of human RIα; the immunosuppressive (27), less cytotoxic (18), second-generation RNA/DNA hybrid PS-ODN antisense (19); and a nonimmunostimulatory 5′-CCG-containing (28) PS-ODN antisense (15), targeted to codons 8–13 of mouse RIα, that can cross hybridize with human RIα. The immune-response signature elicited in the PS-ODN antisense-treated tumors was undetectable in the RNA/DNA hybrid antisense-treated tumors (Fig. 4B, column 10). The expression signatures of mouse RIα antisense were in close parallel with that of the RNA/DNA hybrid antisense (Fig. 4, columns 10 and 11), without immunostimulatory effect (Fig. 4B, column 11). Overall, the alterations in expression signatures described above were similarly induced by all three antisense ODNs in tumors (Fig. 4, columns 10–12), but not in host livers (Fig. 4, columns 13 and 14). By contrast, the expression signatures of antisense were not elicited by control ODN (Fig. 4, column 9), indicating that antisense modulation of the expression signatures described above was sequence-specific. These expression signatures, together with other prominent features of the antisense-induced expression profile, appear to reflect the profile of the nonmalignant or reverted phenotype, which was shared by that observed in the host livers examined.
Conclusions
Antisense technology has been applied to specifically block disease-causing genes (29, 30); therefore, its use as a gene-specific therapeutic agent is highly promising. Targeting of cancer genes by antisense ODNs could inhibit tumor growth. However, nonspecific side effects caused by antisense ODNs (26) have clouded the understanding of the single-gene targeting mechanism of action and hampered or delayed clinical development of antisense drugs. Our results have revealed a specific subset of genes in cancer cells that are coordinately regulated by antisense RIα in a sequence-specific manner. This study shows that a view of global gene expression in cancer cells exposed to antisense ODN can refine the antisense mechanism with respect to its sequence and target specificity.
Our results show that antisense acts as a multisite genomic modulator and thus goes beyond functioning as a single gene-targeting agent. RIα antisense, once in the cell, alters the expression of hundreds of different genes, including its own target gene, and these genes can be classified into subgroups, which pinpoint specific stages of cell growth, differentiation, and transformation. Importantly, the differentiation and proliferation expression signatures were altered specifically in tumor cells; these signatures were quiescent and unaltered in the host livers of antisense-treated animals.
In this differentiation signature, the genes showing the highest induction included genes for Rap1A and Cdc42, small G proteins with GTPase activity. Both Rap1A and Cdc42 are multifunctional proteins related to both cell proliferation and differentiation (31). However, the RIα antisense induction of these G proteins in cancer cells occurred concomitantly with the induction of other proteins involved in differentiation and with the down-regulation of a cluster of genes involved in cell proliferation and transformation. Thus, in this case, the induction of these G proteins appears to promote differentiation and reverse transformation rather than proliferation and transformation in cancer cells.
Although we know that RIα antisense behaves as a multisite genomic modulator, its precise molecular mechanism of action is still unknown. We speculate that cAMP response element-directed transcription (32), which is triggered by the activation of PKA via antisense depletion of RIα (19), may play a central role in this process.
Gene expression profiling presents a way of refining cancer chemotherapeutics in the future. Revisiting antisense-targeted gene expression on a genomic scale will facilitate the discovery of clinically appropriate antisense drugs as well as providing a unique perspective on the development of new chemotherapeutic combinations based on the molecular actions of these drugs.
Acknowledgments
We thank Dr. Sudhir Agrawal for providing ODNs. We also thank Dr. Frances McFarland of Palladian Partners, Inc., who provided editorial support under Contract NO2-BC-76212/C2700212 with the National Cancer Institute. The member who communicated this paper is on the Board of Directors of Hybridon, Inc., for which a patent related to RIα has been licensed.
Abbreviations
- PKA
cAMP-dependent protein kinase
- ODN
oligonucleotide
- PS-ODN
phosphorothioate ODN
References
- 1.McKnight G S, Clegg C H, Uhler M D, Chivia J C, Cadd G G, Correll L A, Otten A D. Recent Prog Horm Res. 1988;44:307–355. doi: 10.1016/b978-0-12-571144-9.50014-4. [DOI] [PubMed] [Google Scholar]
- 2.Handschin J S, Eppenberger U. FEBS Lett. 1979;106:301–304. doi: 10.1016/0014-5793(79)80519-0. [DOI] [PubMed] [Google Scholar]
- 3.Miller W R, Hulme M J, Cho-Chung Y S, Elton R A. Eur J Cancer. 1993;29:989–991. doi: 10.1016/s0959-8049(05)80207-2. [DOI] [PubMed] [Google Scholar]
- 4.Miller W R, Watson D M A, Jack W, Chetty U, Elton R A. Breast Cancer Res Treat. 1993;26:89–94. doi: 10.1007/BF00682703. [DOI] [PubMed] [Google Scholar]
- 5.Miller W R, Hulme M J, Bartlett J M, MacCallium J, Dixon J M. Clin Cancer Res. 1997;3:2399–2404. [PubMed] [Google Scholar]
- 6.Simpson B J B, Ramage A D, Hulme M J, Burns D J, Katsaros D, Langdon S P, Miller W R. Clin Cancer Res. 1996;2:201–206. [PubMed] [Google Scholar]
- 7.McDaid H M, Cairns M T, Atkinson R I, McAleer S, Harkin D P, Gilmore P, Johnston P G. Br J Cancer. 1999;79:933–939. doi: 10.1038/sj.bjc.6690149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young M R I, Montpetit M, Lozano Y, Djordjevic A, Devata S, Matthews J P, Yedavalli S. Int J Cancer. 1995;61:104–109. doi: 10.1002/ijc.2910610118. [DOI] [PubMed] [Google Scholar]
- 9.Bradbury A W, Carter D C, Miller W R, Cho-Chung Y S, Clair T. Br J Cancer. 1994;69:738–742. doi: 10.1038/bjc.1994.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bold R J, Alpard S, Ishizuka J, Townsend C M, Jr, Thompson C J. Regul Pept. 1994;53:61–70. doi: 10.1016/0167-0115(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 11.Gordge P C, Hulme M J, Clegg R A, Miller W R. Eur J Cancer. 1996;32:2120–2126. doi: 10.1016/s0959-8049(96)00255-9. [DOI] [PubMed] [Google Scholar]
- 12.Cho-Chung Y S. Cancer Res. 1990;50:7093–7100. [PubMed] [Google Scholar]
- 13.Tortora G, Yokozaki H, Pepe S, Clair T, Cho-Chung Y S. Proc Natl Acad Sci USA. 1991;88:2011–2015. doi: 10.1073/pnas.88.5.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohlff C, Glazer R I. Cell Signalling. 1995;7:431–443. doi: 10.1016/0898-6568(95)00018-k. [DOI] [PubMed] [Google Scholar]
- 15.Nesterova M V, Cho-Chung Y S. Nat Med. 1995;1:528–633. doi: 10.1038/nm0695-528. [DOI] [PubMed] [Google Scholar]
- 16.Cho-Chung Y S, Nesterova M, Kondrashin A, Noguchi K, Srivastava R, Pepe S. Antisense Nucleic Acid Drug Dev. 1997;7:217–223. doi: 10.1089/oli.1.1997.7.217. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Cai Q, Zeng X, Yu D, Agrawal S, Zhang M Q. Proc Natl Acad Sci USA. 1999;96:13989–13994. doi: 10.1073/pnas.96.24.13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H X, Marshall J L, Ness E, Martin R R, Dvorchik B, Rizvi N, Marquis J, McKinlay M, Dahut W, Hawkins M J. Clin Cancer Res. 2000;6:1259–1266. [PubMed] [Google Scholar]
- 19.Nesterova M V, Cho-Chung Y S. Antisense Nucleic Acid Drug Dev. 2000;10:423–433. doi: 10.1089/oli.1.2000.10.423. [DOI] [PubMed] [Google Scholar]
- 20.Nesterova M, Yokozaki H, McDuffie L, Cho-Chung Y S. Eur J Cancer. 1996;253:486–494. doi: 10.1111/j.1432-1033.1996.00486.x. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka T S, Jaradat S A, Lim M, Kargul G J, Wang X, Grahovac M J, Pantano S, Sano Y, Piao Y, Nagaraja R, et al. Proc Natl Acad Sci USA. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 23.Noda M, Ko M, Ogura A, Liu D G, Amano T, Takano T, Ikawa Y. Nature (London) 1985;318:73–75. doi: 10.1038/318073a0. [DOI] [PubMed] [Google Scholar]
- 24.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 26.Stein C A. Nat Med. 1995;1:1119–1121. doi: 10.1038/nm1195-1119. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Q, Yu D, Agrawal S. Bioorg Med Chem Lett. 1999;9:3453–3458. doi: 10.1016/s0960-894x(99)00635-6. [DOI] [PubMed] [Google Scholar]
- 28.Krieg A M, Yi A K, Matson S, Waldschmidt T J, Bishop G A, Teasdale R, Koretzky G A, Klinman D M. Nature (London) 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 29.Zamecnik P, Stephenson M. Proc Natl Acad Sci USA. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho-Chung Y S. Exp Opin Ther Patents. 2000;10:1711–1724. [Google Scholar]
- 31.Hall A. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 32.Roesler W J, Vandenbark G R, Hanson R W. J Biol Chem. 1988;263:9063–9066. [PubMed] [Google Scholar]