Abstract
Target directed proteolysis allows specific processing of proteins in vivo. This method uses tobacco etch virus (TEV) NIa protease that recognizes a seven-residue consensus sequence. Because of its specificity, proteins engineered to contain a cleavage site are proteolysed, whereas other proteins remain unaffected. Therefore, this approach can be used to study the structure and function of target proteins in their natural environment within living cells. One application is the conditional inactivation of essential proteins, which is based on the concept that a target containing a recognition site can be inactivated by coexpressed TEV protease. We have previously identified one site in the secretion factor SecA that tolerated a TEV protease site insert. Coexpression of TEV protease in the cytoplasm led to incomplete cleavage and a mild secretion defect. To improve the efficiency of proteolysis, TEV protease was attached to the ribosome. We show here that cleaving SecA under these conditions is one way of increasing the efficiency of target directed proteolysis. The implications of recruiting novel biological activities to ribosomes are discussed.
Keywords: membrane protein insertion, ribosome attachment, transport
Protein translocation across the cytoplasmic membrane is a highly conserved process. Common features include the recognition of targeting signals of the secretory protein by dedicated cellular factors on the cis side of a membrane, a heterooligomeric channel that gates across and into the membrane, a peripherally associated component providing energy and a folding system on the trans side of the membrane (for review, see ref. 1). In bacteria, classical secretory proteins are synthesized with a N-terminal signal peptide. These preproteins are normally transferred across the cytoplasmic membrane in an extended conformation. The translocation channel or Sec machinery is a well studied heterooligomeric complex consisting of at least three integral membrane proteins, SecY, SecE, and SecG, and the peripherally bound cytoplasmic ATPase SecA (2, 3).
The posttranslational targeting pathway of soluble proteins is well understood. In the cytoplasm, cleavable signal sequences are recognized by the chaperone SecB and the essential SecA protein. SecA is a multifunctional component that acts as a targeting factor, provides energy for translocation through its ATPase activity, and acts as a chaperone excluding nonsecretory proteins from translocation (4–6). Targeting of cytoplasmic membrane proteins depends on transmembrane segments that function as internal and noncleavable signal and stop-transfer sequences. Therefore, transmembrane segments do not only function in targeting but are also involved in determining the transmembrane topology (7–10). Recent evidence suggests that targeting of integral membrane proteins is cotranslational, involving the components of the signal recognition particle (SRP) system. This targeting includes the SRP itself, composed of the Ffh protein and the 4.5S RNA, and the SRP receptor FtsY (reviewed in ref. 11). In some cases, the additional requirement of SecA in membrane insertion has been demonstrated, but it is believed that in this case, SecA is not involved in targeting but in the translocation of periplasmic domains of the membrane proteins (for review, see ref. 12).
Because many components involved in secretion or membrane insertion are essential for viability, their conditional inactivation presents experimental problems. The most common techniques involve depletion (by using tight promoters that can be efficiently shut off) or point mutations conferring to conditional phenotypes, such as cold or temperature sensitivity. To extend the portfolio of approaches, allowing controlled and conditional inactivation of essential proteins, we have developed target-directed proteolysis (TDP) that allows proteolytic inactivation of target proteins in vivo (13). TDP uses the highly specific tobacco etch virus (TEV) NIa protease, which recognizes a seven-residue target sequence, ExxYXQS, where X can be several amino acyl residues (14). Cleavage occurs between the conserved Gln and Ser residues (15). Because of its specificity, TEV protease can be used to process target proteins in vivo. Besides the conditional inactivation of essential proteins, other applications of TDP include, for example, the determination of cellular localization and the topology of membrane proteins (16, 17). Efficient processing of TDP substrates depends on the accessibility of the cleavage sites. When investigating TDP sites in SecA, it was found that sites inserted after residues 252 or 830 were efficiently processed (13, 17), whereas a site inserted after residue 195 (SecA195) was only incompletely cleaved (13). We reasoned that the accessibility of the TDP site in SecA195 might be improved when proteolytic processing would occur before folding of SecA. To test this model, TEV protease was attached to the ribosome. Colocalization of nascent chains and protease might thus enhance cleavage of the target protein. We compared the effects of SecA depletion with TDP by studying membrane insertion or secretion of model substrates. Ffh and FtsY depletion strains that are known to affect membrane insertion served as a control of the reporter system. Our results suggest that TEV protease can be attached to the ribosome without interfering with viability and represents one way to increase the efficiency of TDP.
Methods
Bacterial Strains. E. coli strains used were the Ffh depletion strain WAM113 (18), the FtsY depletion strain N4156::Para14-ftsY′ (19) and MC4100 (20). The bacteriophage λInCh was used to transfer plasmid-encoded secA constructs to the attB site on the E. coli chromosome as described in ref. 21. Chromosomal wild-type secA was eliminated by using λ Red recombinase (22). Each newly constructed strain was verified through PCR by using whole-cell DNA as a template. Unless stated otherwise, cells were grown at 37°C in rich media containing 10 g of NZ amine A, 10 g of yeast extract, and 7.5 g of NaCl per liter.
Expression of TEV Protease. Expression of TEV protease was tightly regulated by using low copy number plasmids conferring to spectinomycin resistance and the tet promoter that was induced with 100 ng/ml anhydrotetracycline. pTH9 expresses cytoplasmic TEV protease with a N-terminal glutatione S-transferase tag. It was constructed from pZS*2tet0–1 (23) by exchanging the neo by a spectinomycin resistance cassette with flanking AclI sites. pTH10 is as pTH9 except that it has an insert expressing Tig144-TEV protease. Here, GST was replaced by a trigger factor fragment encoding the N-terminal 144 residues, which was PCR amplified and cloned into pTH9 by using KpnI and MluI sites. DNA sequencing of the relevant inserts verified all newly constructed plasmids.
Expression of Reporter Proteins. A plasmid expressing FtsQ-PSBT reporter protein is described in ref. 24. This plasmid was used to construct two sets of plasmids. One set was based on pBAD18-cm (25) carrying the relevant inserts. In these plasmids, the expression of the Propionibacterium shermanii transcarboxylase (PSBT) reporter constructs depended on the presence of arabinose in the growth medium. The second set of plasmids was derived from pVC19, a pACYC184 derivative of pCS19 (26). Here, expression was under the control of the tac promoter. In both sets of plasmids, the relevant regulatory genes araC and lacIQ, respectively, were present on the plasmids, allowing tight control of expression levels. To construct similar plasmids encoding alkaline phosphatase (AP)-PSBT and MalF274-PSBT, respectively, wild-type phoA and the relevant fragment of wild-type malF were PCR amplified and used to replace the ftsQ-PSBT insert.
Detection of Biotinylated Proteins by Immunoblotting. Proteins containing the biotinylation domain from the 1.3S subunit of PSBT were detected by Western blotting with streptavidin-alkaline phosphatase by following standard procedures (27, 28). To detect SecA, trigger factor, and TEV protease, polyclonal anti-rabbit antibodies were used.
Cell Fractionation. Cells were grown to OD600 = 0.6. After harvest, cells were lysed in buffer 1 (15% sucrose/10 mM MgCl2/100 mM Tris·HCl, pH 7.5/100 mM NaCl). Cell debris was removed by centrifugation. Either 100 or 500 mM KCl was added to supernatants. Subsequently, sucrose cushion centrifugation (20% sucrose/20 mM Tris·HCl, pH 7.5/100 mM or 500 mM KCl/10 mM MgCl2/5 mM 2-mercaptoethanol) was performed as described in ref. 29. The cytoplasmic fraction was in the supernatant while pellets contained ribosomes that were resuspended in 10 mM Tris·HCl, pH 7.5/6 mM MgCl2/30 mM NH4Cl/4 mM 2-mercaptoethanol. These samples were used for Western blotting.
Results
To study the effects of SecA inactivation on protein translocation, we compared TDP with well established depletion approaches. We used depletion strains for SecA, Ffh, FtsY, and a system that monitors translocation of reporter substrates in vivo. This reporter system is based on fusing a biotinylation domain from PSBT to the C terminus of the protein to be translocated (27). When translocation is impaired, PSBT remains in the cytoplasm and is thus biotinylated by cytoplasmic biotin ligase, a native E. coli enzyme. Biotinylation occurs at a lysine residue that is located 35 amino acids from the C terminus of the PSBT domain. The only native biotinylated protein in E. coli is biotin carboxyl carrier protein. This small cytoplasmic protein of ≈16 kDa is visible on all Western blots incubated with streptavidin-AP. When PSBT is translocated across the cytoplasmic membrane, it is no longer accessible to biotin ligase and is thus not biotinylated. Therefore, biotinylation serves as a sensitive reporter system to monitor the efficiency of translocation in vivo (24, 27, 28).
To show that the reporter system gives the expected results, we initially examined the effects of Ffh and FtsY depletion on membrane insertion by using FtsQ-PSBT and MalF274-PSBT. PSBT was fused to the last codon of ftsQ. FtsQ has a cytoplasmic N terminus, one transmembrane segment, and a periplasmic C terminus (24). In MalF274, the PSBT domain was fused to the C terminus of the large second periplasmic loop of ≈180 amino acids. Thus, FtsQ-PSBT and MalF274-PSBT should only become biotinylated when membrane insertion is inefficient.
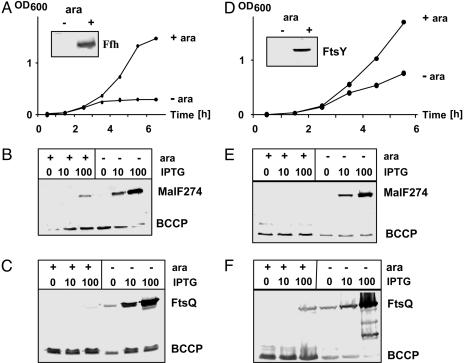
Ffh and FtsY Depletion. The effects of Ffh depletion on membrane insertion was followed by examining biotinylation of MalF274-PSBT and FtsQ-PSBT in strain WAM113 (18). This strain contains ffh::kan, a chromosomal knockout mutation plus a chromosomal copy of the ffh gene under control of the arabinose promoter ParaBAD. In arabinose-free medium, growth ceased after ≈3 h, and Ffh was no longer detectable on a Western blot (Fig. 1A). At this time point, production of the PSBT reporter proteins was induced for 1 h. Subsequently, samples were taken for Western blot analysis to detect biotinylated proteins. Both membrane proteins exhibited a similar biotinylation pattern as a result of Ffh depletion, and biotinylation increased with higher concentrations of isopropyl β-d-thiogalactoside (IPTG) (Fig. 1 B and C). A small amount of protein was biotinylated even under nondepletion conditions when cells were induced with 100 μM IPTG. This result suggests that the efficiency of membrane insertion is reduced when the reporter membrane proteins are overproduced.
Fig. 1.
Effects of Ffh and FtsY depletion on membrane insertion. (A and D) Growth of Ffh (A) and FtsY (D) depletion strains in NZA rich medium with or without 0.2% arabinose. (A and D Insets) Western blot with anti-Ffh or FtsY antibody. Samples were taken after 3 h of growth in medium with or without 0.2% arabinose. (B, C, E, and F) Biotinylation of MalF274-PSBT (B and E) or FtsQ-PSBT (C and F). Cells either were depleted (no arabinose) or contained Ffh or FtsY (0.2% arabinose). Expression of reporter proteins was either not induced or induced with 10 or 100 μM IPTG. Samples were taken after 1 h of induction for Western blot analysis by using streptavidin-AP. BCCP, biotin carboxyl carrier protein.
Strain N4156::Para14-ftsY′ contains the ftsY gene under the control of ParaBAD (19). Without arabinose, growth of the culture ceased. However, the growth defect was less pronounced compared with Ffh depletion even though FtsY was undetectable on a Western blot after 3 h of growth in arabinose-free medium (Fig. 1D). Biotinylation patterns of MalF274-PSBT and FtsQ-PSBT were very similar to those after Ffh depletion. Depletion of FtsY caused biotinylation of the PSBT domain of both membrane proteins (Fig. 1 E and F). When FtsQ-PSBT was overexpressed with 100 μM IPTG under nondepletion conditions, a small amount of biotinylated protein was visible on the FtsQ Western blots, again indicating that overproduction of membrane proteins leads to a saturation of the translocation machinery.
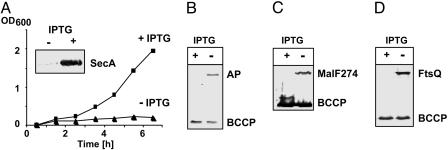
SecA Depletion. To investigate SecA depletion, we used a strain in which chromosomal wild-type secA was deleted, and a secA derivative was present at the attB site on the chromosome. This secA derivative encoded SecA830, which has a TDP site inserted after residue 830 (17). Expression of secA830 was under control of the Ptac promoter, and because the lacIq gene was present on plasmid pTHS19, SecA could be depleted by removing IPTG from the medium. It should be noted that the TDP site in SecA830 does not interfere with SecA function because it is introduced into a nonessential region. SecA was depleted for 6.5 h before assaying biotinylation of reporter proteins. Under these conditions, SecA levels were drastically reduced (Fig. 2A). Because SecA is involved in translocation of periplasmic proteins, we used AP carrying a C-terminal PSBT domain as a reporter for secretion defects. Biotinylation of AP-PSBT was detected after SecA830 depletion, even when AP production was not induced with arabinose (Fig. 2B). Translocation of MalF274-PSBT was also affected as detected by biotinylation as a consequence of SecA depletion (Fig. 2C). In the case of FtsQ, induction of FtsQ-PSBT with arabinose was required to detect SecA-dependent biotinylation (Fig. 2D). The reason for the requirement of higher FtsQ levels for detection of insertion defects is unknown. One explanation might be that membrane insertion of FtsQ depends less on SecA. Another explanation that is equally likely is that SecA has a higher affinity for FtsQ. Here, overproduction is required for insertion defects to occur.
Fig. 2.
Effects of SecA depletion on translocation. (A) Growth of SecA depletion strain in NZA medium with (▪) or without (▴) 100 μM IPTG. (Inset) Western blot with an anti-SecA antibody. Samples were taken after 7 h of growth in medium with or without IPTG. (B) Biotinylation of AP-PSBT. Cells containing pTHS19 and a plasmid producing AP-PSBT were either depleted (no IPTG) or contained SecA (IPTG). Expression of AP-PSBT was not induced. Samples were taken after 6.5 h of growth for Western blot analysis with streptavidin-AP. BCCP, biotin carboxyl carrier protein. (C) Biotinylation of MalF274-PSBT was carried out as in B. (D) Biotinylation of FtsQ-PSBT was done as in B except that cultures were induced with 0.2% arabinose.
TDP of SecA Through Cytoplasmic and Ribosome-Attached TEV Protease. Previous experiments indicated that a TDP site after residue 195 of SecA (SecA195) is not completely cleaved by TEV protease, leading to a mild secretion defect (13). This result suggested that if complete processing of SecA195 could be achieved, inactivation of SecA through TDP would be feasible. We reasoned that inefficient cleavage of SecA195 could be due to limited surface accessibility of the TDP site. If this model would be correct, cleaving SecA before folding should increase cleavage efficiency. To test this model, we attached TEV protease to the exit channel of the ribosome. This procedure should allow TEV protease to interact with SecA before completion of folding and should thus improve proteolytic processing. Ribosome attachment can be achieved by using the well defined attachment domain of trigger factor that interacts with L23, a protein of the ribosomal exit channel (30).
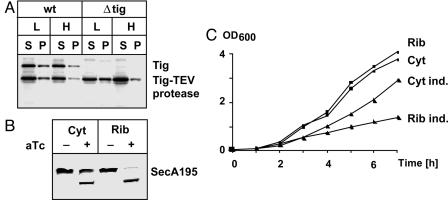
To investigate processing of SecA195 by cytoplasmic and ribosome-attached TEV protease, we used a secA tig double mutant strain in which secA195 was placed at the attB site of the chromosome. Cytoplasmic TEV protease was expressed under control of the tet promoter from low copy number plasmid pTH9. Ribosome-attached TEV protease was expressed from pTH10. This plasmid is a derivative of pTH9, expressing a fusion of the N-terminal 144 residues of trigger factor and TEV protease because this part of the trigger factor is sufficient for ribosomal attachment (29). Both plasmids did not interfere with the growth of cultures, even under inducing conditions. This result indicates that expression of TEV protease either in the cytoplasm or when attached to the ribosome has no effect on viability (data not shown). Ribosome attachment of Tig-TEV protease was similar to the wild-type trigger factor as shown when comparing levels of these proteins in cytoplasmic and ribosomal fractions of wild-type cells and trigger factor mutants (Fig. 3A). It should be noted that trigger factor itself is not exclusively localized at the ribosome but is also found in the cytoplasm (29).
Fig. 3.
TDP with cytoplasmic and ribosome-attached SecA. (A) Cellular localization of TEV protease. Wild-type (wt) and Δtig cells expressing Tig144-TEV protease were fractionated into cytoplasmic (S) and ribosomal fractions (P) by using low (L) and high (H) salt conditions for Western blotting. Trigger factor was visualized by using anti-trigger factor antibodies and Tig144-TEV protease by anti-TEV protease antibodies. (B) Proteolysis of SecA195 by cytoplasmic and ribosomal TEV protease. Western blot analysis was performed with anti-TEV antibody. Samples were taken after 5 h of growth in NZA medium. Expression of TEV protease was induced with 100 ng/ml aTc. (C) Growth of isogenic strains expressing either cytoplasmic (Cyt) or ribosome-attached (Rib) TEV protease. Cultures were grown with or without inducer (ind.) (100 ng/ml aTc).
The efficiency of SecA195 processing was compared by coexpression of cytoplasmic and ribosome-attached TEV protease (Fig. 3B). Although cytoplasmic TEV protease cleaved only a fraction of SecA195, ribosome-attached TEV protease was able to almost completely process SecA. Therefore, ribosomal attachment appears to be one way of improving TDP. Improved processing of SecA195 resulted in a decrease in growth of the relevant bacterial culture, indicating that SecA was inactivated by TDP (Fig. 3C).
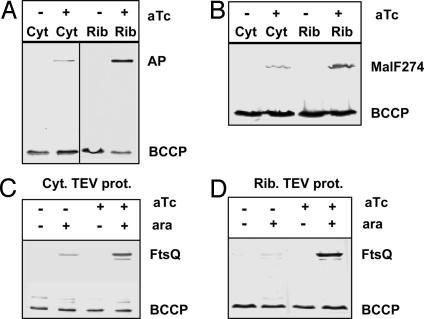
Effects of SecA Inactivation on Translocation. To monitor translocation defects, biotinylation of reporter proteins was assayed under conditions where SecA was either inactivated or not inactivated by TDP. AP-PSBT was biotinylated when SecA195 was inactivated by ribosome-attached Tig-TEV protease (Fig. 4A). Similarly, biotinylation of MalF274-PSBT was observed under these conditions (Fig. 4B). These translocation defects were significantly weaker when cytoplasmic TEV protease was coexpressed. The insertion of FtsQ was also affected by SecA195 inactivation. However, as seen above, when investigating SecA depletion, biotinylation of FtsQ-PSBT was only observed when expression was induced with arabinose (Fig. 4C). In contrast to AP-PSBT and MalF274-PSBT, biotinylation of FtsQ-PSBT was less affected by the subcellular localization of TEV protease, suggesting that under these conditions, even incomplete inactivation of SecA by cytoplasmic TEV protease affected membrane insertion of FtsQ-PSBT.
Fig. 4.
Effects of TDP of SecA on translocation. (A and B) Detection of biotinylated AP-PSBT (A) and MalF274-PSBT (B). Cultures of cells expressing cytoplasmic TEV protease (Cyt) or ribosome attached TEV protease (Rib) were grown with or without 100 ng/ml aTc (to induce TEV protease). Samples were taken after 5 h for Western blot analysis with streptavidin-AP. BCCP, biotin carboxyl carrier protein. Detection of biotinylated of FtsQ-PSBT (C) was done as above except that ftsQ was induced with or without 0.2% arabinose.
Several additional controls were performed. To exclude the possibility that the inducer anhydrotetracycline (aTc) was responsible for the observed effects, we monitored growth of cells not expressing TEV protease in the presence and absence of aTc. These tests showed that aTc itself has no effect on cell viability. Also, the addition of aTc to the growth medium did not promote biotinylation in strains not expressing TEV protease. Finally, depletion did not change the levels of the reporter proteins produced, which is in agreement with published data (28). This control indicates that changes in biotinylation are reflecting changes in translocation efficiency and not of protein levels (data not shown).
Discussion
We describe the attachment of the highly specific TEV protease to ribosomes in E. coli and show that this approach improves processing of a cleavage site in SecA that is only partially processed by cytoplasmic TEV protease (Fig. 5). In principle, there are two options to attach TEV protease to ribosomes. One is to tether it directly to ribosomal proteins that construct or line the exit channel of nascent polypeptides. Provided that no proteolytic processing of the linker between the ribosomal protein and TEV protease would occur, this method would ensure that TEV protease is exclusively localized to the ribosome. We have chosen an alternative method by using trigger factor, an accessory protein of the ribosome, as a tool to promote attachment. This method may have several advantages. TEV protease is not localized too close to the exit channel, which should reduce the possibility of interference with proper translation. Also, trigger factor is a nonessential protein in E. coli that can be deleted without loss of viability, whereas at least some ribosomal proteins that form the exit channel are essential (30). Another advantage might be that trigger factor is not exclusively localized to the ribosome, but significant amounts are also present in the cytoplasm. This nonexclusive localization might be beneficial because cytoplasmic TEV protease can move around and cleave substrates that have either escaped processing during translation or have been synthesized before induction of TEV protease.
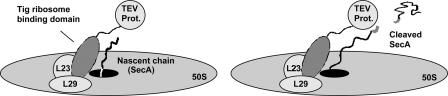
Fig. 5.
Schematic representation of ribosome-attached TEV protease. TEV protease is fused to the ribosome-binding domain of trigger factor, interacting with ribosomal proteins L23 and L29 that are lining the exit channel for nascent chains.
Biochemical and mutational evidence indicates that the N-terminal domain of trigger factor mediates attachment to ribosomal protein L23 (30). Recent structural analyses of trigger factor cocrystallized with the 50S subunit of the ribosome revealed that this N-terminal domain is composed of a four-stranded antiparallel β-sheet and three α-helices. Interaction of trigger factor with the ribosome is mediated by the short helix 2 (31). These data suggest that TEV protease is positioned closely to the exit channel of the ribosome. As tethering TEV protease to the ribosome attachment domain of trigger factor did not result in detectable toxicity, we conclude that TEV protease expression per se is well tolerated by E. coli.
When comparing two approaches of SecA inactivation, depletion and TDP, we saw a stronger and more rapid effect on cell growth under depletion conditions compared with TDP inactivation. At first sight, this result might suggest that proteolytic inactivation of SecA195 is not as efficient as SecA depletion. However, similar translocation defects were observed when comparing levels of biotinylation of AP, FtsQ, and MalF after depletion and proteolytic inactivation, suggesting that TDP produces significant secretion phenotypes. It should also be considered that healthier cells are expected to yield more direct phenotypes than cells that have stopped dividing completely.
The observed translocation defects resulting from SecA, Ffh, and FtsY depletion or inactivation suggest that each of these factors is implicated in membrane insertion, which is in agreement with published evidence (24, 28, 32–34). To further analyze the effects of depletion, we carried out DNA array analysis (data not shown). Effects seen with DNA arrays that could be confirmed by Western blotting included up-regulation of DnaK, GroEL, and HslS after Ffh and FtsY depletion, which is, in part, consistent with published evidence (35). A different pattern was observed after SecA depletion. Here, a down-regulation of the degP gene was observed. This finding might indicate an interesting link between translocation and protein quality control in the cell envelope. This model is in agreement with previously published evidence showing that overproduction of secreted proteins activates the unfolded protein response that controls degP promoter activity (36), indicating that DegP levels are adjusted to the needs of the cell.
The general implications of our results are that biological activities can be recruited to the ribosome. The experimental principle involves making use of the ribosome attachment domains of the various nonessential accessory proteins that bind to ribosomes. Biological activities of interest might, for example, include any type of posttranslational modification or molecular chaperones. One might also consider attaching less-specific proteases to the ribosome that could readily remove proteins that misfold during cotranslational folding. It might be advantageous to remove such proteins from ribosomes early to prevent entry into the aggregation pathway, a strategy that might potentially lead to healthier strains and, thus, to optimized production of recombinant proteins.
Acknowledgments
We thank Jon Beckwith (Harvard Medical School) for providing plasmids and Roland Freudl (Forschungszentrum Jülich GmbH, Jülich, Germany) for antibodies against SecA. This work was supported by grants from the Biotechnology and Biological Sciences Research Council to M.E. and from the German–Israeli Foundation for Scientific Research and Development to E.B. and M.E.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AP, alkaline phosphatase; aTc, anhydrotetracycline; IPTG, isopropyl β-D-thiogalactoside; PSBT, Propionibacterium shermanii transcarboxylase; TDP, target-directed proteolysis.
References
- 1.Rapoport, T. A., Jungnickel, B. & Kutay, U. (1996) Annu. Rev. Biochem. 65, 271–303. [DOI] [PubMed] [Google Scholar]
- 2.Van den Berg, B., Clemons, W. M., Jr., Collinson, I., Modis, Y., Hartmann, E., Harrison, S. C. & Rapoport, T. A. (2004) Nature 427, 36–44. [DOI] [PubMed] [Google Scholar]
- 3.Driessen, A. J., Fekkes, P. & van der Wolk, J. P. (1998) Curr. Opin. Microbiol. 1, 216–222. [DOI] [PubMed] [Google Scholar]
- 4.Oliver, D. B. (1993) Mol. Microbiol. 7, 159–165. [DOI] [PubMed] [Google Scholar]
- 5.Hunt, J. F., Weinkauf, S., Henry, L., Fak, J. J., McNicholas, P., Oliver, D. B. & Deisenhofer, J. (2002) Science 297, 2018–2026. [DOI] [PubMed] [Google Scholar]
- 6.Eser, M. & Ehrmann, M. (2003) Proc. Natl. Acad. Sci. USA 100, 13231–13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrmann, M., Boyd, D. & Beckwith, J. (1990) Proc. Natl. Acad. Sci. USA 87, 7574–7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrmann, M. & Beckwith, J. (1991) J. Biol. Chem. 266, 16530–16533. [PubMed] [Google Scholar]
- 9.McGovern, K., Ehrmann, M. & Beckwith, J. (1991) EMBO J. 10, 2773–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uhland, K., Ehrle, R., Zander, T. & Ehrmann, M. (1994) J. Bacteriol. 176, 4565–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herskovits, A. A., Bochkareva, E. S. & Bibi, E. (2000) Mol. Microbiol. 38, 927–939. [DOI] [PubMed] [Google Scholar]
- 12.Driessen, A. J., Manting, E. H. & van der Does, C. (2001) Nat. Struct. Biol. 8, 492–498. [DOI] [PubMed] [Google Scholar]
- 13.Mondigler, M. & Ehrmann, M. (1996) J. Bacteriol. 178, 2986–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dougherty, W. G., Cary, S. M. & Parks, T. D. (1989) Virology 171, 356–364. [DOI] [PubMed] [Google Scholar]
- 15.Carrington, J. C. & Dougherty, W. G. (1988) Proc. Natl. Acad. Sci. USA 85, 3391–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faber, K. N., Kram, A. M., Ehrmann, M. & Veenhuis, M. (2001) J. Biol. Chem. 276, 36501–36507. [DOI] [PubMed] [Google Scholar]
- 17.Ehrmann, M., Bolek, P., Mondigler, M., Boyd, D. & Lange, R. (1997) Proc. Natl. Acad. Sci. USA 94, 13111–13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips, G. J. & Silhavy, T. J. (1992) Nature 359, 744–746. [DOI] [PubMed] [Google Scholar]
- 19.Luirink, J., ten Hagen-Jongman, C. M., van der Weijden, C. C., Oudega, B., High, S., Dobberstein, B. & Kusters, R. (1994) EMBO J. 13, 2289–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casadaban, M. J. (1976) J. Mol. Biol. 104, 541–555. [DOI] [PubMed] [Google Scholar]
- 21.Boyd, D., Weiss, D., Chen, J. & Beckwith, J. (2000) J. Bacteriol. 182, 842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datsenko, K. & Wanner, B. (2000) Proc. Natl. Acad. Sci. USA 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herskovits, A. A., Seluanov, A., Rajsbaum, R., ten Hagen-Jongman, C. M., Henrichs, T., Bochkareva, E. S., Phillips, G. J., Probst, F. J., Nakae, T., Ehrmann, M., et al. (2001) EMBO Rep. 2, 1040–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian, H., Boyd, D. & Beckwith, J. (2000) Proc. Natl. Acad. Sci. USA 97, 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman, L.-M., Belin, D., Carson, M. & Beckwith, J. (1995) J. Bacteriol. 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spiess, C., Beil, A. & Ehrmann, M. (1999) Cell 97, 339–347. [DOI] [PubMed] [Google Scholar]
- 27.Jander, G., Cronan, J. J. & Beckwith, J. (1996) J. Bacteriol. 178, 3049–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian, H. & Beckwith, J. (2002) J. Bacteriol. 184, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hesterkamp, T., Deuerling, E. & Bukau, B. (1997) J. Biol. Chem. 272, 21865–21871. [DOI] [PubMed] [Google Scholar]
- 30.Kramer, G., Rauch, T., Rist, W., Vorderwulbecke, S., Patzelt, H., Schulze-Specking, A., Ban, N., Deuerling, E. & Bukau, B. (2002) Nature 419, 171–174. [DOI] [PubMed] [Google Scholar]
- 31.Ferbitz, L., Maier, T., Patzelt, H., Bukau, B., Deuerling, E. & Ban, N. (2004) Nature 431, 590–596. [DOI] [PubMed] [Google Scholar]
- 32.Froderberg, L., Houben, E., Samuelson, J., Chen, M., Park, S., Phillips, G., Dalbey, R., Luirink, J. & De Gier, J. (2003) Mol. Microbiol. 47, 1015–1027. [DOI] [PubMed] [Google Scholar]
- 33.Neumann-Haefelin, C., Schafer, U., Muller, M. & Koch, H. G. (2000) EMBO J. 19, 6419–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi, H. & Bernstein, H. (1999) J. Biol. Chem. 274, 8993–8997. [DOI] [PubMed] [Google Scholar]
- 35.Bernstein, H. D. & Hyndman, J. B. (2001) J. Bacteriol. 183, 2187–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raivio, T. L. & Silhavy, T. J. (2001) Annu. Rev. Microbiol. 55, 591–624. [DOI] [PubMed] [Google Scholar]