Abstract
We explored the epidemic history of HIV-1 subtype B in the United Kingdom by using statistical methods that infer the population history of pathogens from sampled gene sequence data. Phylogenetic analysis of HIV-1 pol gene sequences from Britain showed at least six large transmission chains, indicating a genetically variable, but epidemiologically homogeneous, epidemic among men having sex with men. Through coalescent-based analysis, we showed that these chains arose through separate introductions of subtype B strains into the United Kingdom in the early to mid-1980s. After an initial period of exponential growth, the rate of spread generally slowed in the early 1990s, which is more likely to correlate with behavior change than with reduced infectiousness resulting from highly active antiretroviral therapy. Our results provide insights into the complexity of HIV-1 epidemics that must be considered when developing HIV monitoring and prevention initiatives.
Keywords: epidemic history, phylogenetics
More than 57,700 people have been infected with HIV type 1 (HIV-1) in the United Kingdom (U.K.) since the first identification of AIDS in 1982 (www.hpa.org.uk). Despite a recent increase in heterosexually acquired infections within the U.K., predominantly originating in sub-Saharan Africa, the most prevalent clade of virus within the country remains subtype B, from the main group of HIV-1, which is mainly transmitted through sex between men (1). To date, very little is known about how subtype B successfully invaded the British population and, more importantly, how the virus has subsequently spread and evolved.
Phylogenies reconstructed from sampled viral gene sequences hold valuable and unique information about the past structure of a population and can be used to understand the course of a viral epidemic over time (2, 3). Hence, the history of a pathogen population can be inferred from the genealogy of randomly sampled strains (as represented by a phylogenetic tree) by using the coalescent theory of population genetics (4, 5). By this means, one can reconstruct the changing number of infected individuals through time and estimate the demographic parameters that shape the epidemic, such as the rate of growth in the number of infections and the date of introduction of a lineage into a host population (6). Molecular data on HIV-1 within the U.K. have become increasingly available since the introduction of routine HIV-1 gene sequencing for drug-resistance monitoring. The genetic variability of the envelope (env) gene has previously made it attractive for evolutionary studies. However, we have recently demonstrated that the polymerase (pol) gene encodes sufficient variation to reconstruct transmission events despite the potential bias conferred by emergence of drug resistance-associated mutations (7). Moreover, although the coalescent framework assumes neutral evolution, the HIV-1 pol gene is known to be under positive and negative selection (8–11). However, selection on HIV genes within infected individuals does not appear to generate nonneutral genealogies at the epidemiological (among-individual) level (12) and, therefore, should not significantly bias coalescent estimates. Importantly, previous coalescent analyses have yielded similar demographic estimates from different HIV-1 genes that are under considerably different selection pressures (13).
Using a previously uncharacterized statistical framework, we reconstructed the history of the HIV-1 subtype B epidemic in the U.K. from a large data set of contemporary pol gene sequences. We characterized separate subepidemics of HIV-1 within a defined risk group, dating the introduction of epidemiologically significant viral lineages and estimating their rates of spread. Our analysis, with U.K. data, illustrates the complexity of HIV-1 epidemics that is applicable to other transmission groups and geographic regions.
Methods
Study Population. HIV-1 subtype B pol gene sequences were generated from plasma samples collected in the U.K. by the Health Protection Agency's Antiviral Susceptibility Reference Unit. The samples were submitted for routine genotypic drug resistance testing between 1999 and 2003 and included samples from acute infections, chronic but drug-naïve infections, and from patients at the time of therapy failure. The sequences were 952 bp long, including the full protease gene and the first 218 codons of the reverse transcriptase gene. Approximately 85% of these sequences were from men who had sex with men (MSM).
Phylogenetic Reconstruction. To identify HIV-1 lineages derived from single independent introductions of the virus into the U.K. population, a neighbor-joining phylogenetic tree was constructed from 3,429 HIV-1 subtype B pol gene sequences (1,645 U.K. isolates plus 1,784 subtype B reference sequences from throughout the world) (14). The tree was estimated under the Hasegawa–Kishino–Yano model of nucleotide substitution (15). The non-U.K. sequences used for the study were extracted from GenBank (www.ncbi.nlm.nih.gov) and the Los Alamos HIV Sequence Database (www.hiv.lanl.gov). The size of the sequence alignment and the computational power required prevented the use of a more complex evolutionary model.
After identification of U.K. transmission clusters, sequences of non-U.K. origin were removed and the phylogenies of the clusters were reestimated with the program paup* by using a maximum likelihood approach (16). The trees were constructed under the General Time Reversible model of nucleotide substitution (17), with proportion of invariable sites and substitution rate heterogeneity, as selected by the program modeltest (18). Each U.K. cluster was rooted by using a subtype D pol sequence from our database. The statistical robustness of the maximum likelihood topologies was assessed by bootstrapping with 1,000 replicates (19). The sequences in the transmission clusters are deposited in GenBank under the accession numbers AY669865–AY670087.
Estimation of HIV-1 Subtype B pol Gene Rate of Nucleotide Substitution. To work within a calendar time scale (i.e., years), the genealogies were rescaled by applying a constant rate of nucleotide substitution μ (units are nucleotide substitutions per site per year) to the branches of the phylogenies. Preliminary analyses demonstrated that the time span covered by our U.K. samples (i.e., five years) was not sufficient to reliably estimate μ. The rate of nucleotide substitution was therefore estimated from an independent data set of 106 subtype B pol gene sequences. The sequences used to estimate μ were sampled between 1983 and 2000 from MSM and injecting drug users participating in cohort studies at the Academic Medical Centre of Amsterdam (20). The sequences were 804 bp long, including the entire protease gene (294 bp) and the first 510 bp of the reverse transcriptase gene. GenBank entries for these sequences are available in the original publication. A posterior distribution for substitution rate was estimated by Bayesian Markov Chain Monte Carlo (MCMC) inference (21) with a MCMC chain of 10 million states sampled every 100th generation, as implemented in the program beast (evolve.zoo.ox.ac.uk/beast). The estimated posterior distribution was subsequently used as an empirical prior distribution in the coalescent analyses that follow.
Estimation of Demographic History and Population Dynamics. The investigation of the epidemic history of the six U.K. clusters involved two steps. First, several models of demographic history, each of which illustrate effective numbers of infections through time, were compared to select the model that best describes the epidemiological history of the U.K. transmission clusters. The demographic models were evaluated by the likelihood ratio test from likelihoods calculated by the program genie (22). The five models tested in this study were constant population size, exponential growth, piecewise logistic (exponential growth followed by constant population size), piecewise expansion (constant population size followed by exponential growth), and piecewise con-exp-con (constant growth periods flanking an exponential growth phase). See ref. 16 for more details of these models. To fit a constant molecular clock framework, as required for coalescent analyses, the program tipdate (23) was used to rescale each transmission tree under the Single Rate Dated Tip (SRDT) model.
Second, the demographic and evolutionary parameters of the epidemic, together with their confidence intervals, were estimated by Bayesian MCMC inference by using a chain of 10 million states sampled at every 100th generation, as implemented in the program beast. The estimated parameters include the date of the most recent common ancestor of the cluster, the effective number of infections at the most recent time of sampling Ne (i.e., the effective number of prevalent infections), and the growth rate during the exponential phase r. The Bayesian MCMC results were used to calculate a marginal posterior distribution of the demographic model for each cluster, a graphical representation of the effective number of infections through time, generated by using the program tracer (http://evolve.zoo.ox.ac.uk/software.html?id=tracer).
Results
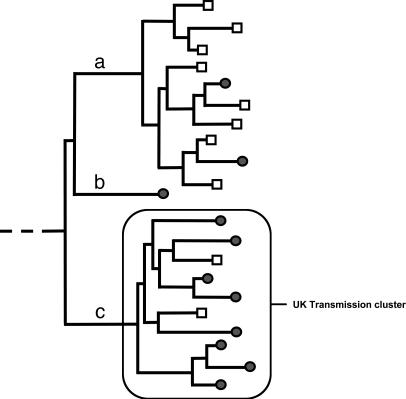
Introduction of HIV-1 Subtype B into the U.K. The initial neighbor-joining phylogenetic tree constructed from 3,429 U.K. and worldwide subtype B pol sequences is too large to display here (Data Set 1, which is published as supporting information on the PNAS web site). A schematic representation of the clustering patterns seen within the phylogeny is presented in Fig. 1. Three clustering patterns were distinguished, namely sporadic U.K. sequences, non-U.K. transmission clusters, and U.K. transmission clusters. Sporadic U.K. sequences (i.e., those that do not group with other U.K. lineages in the tree) probably represent single, independent introductions of the virus without subsequent spread. Transmission clusters were identified as clades of sequences from a particular location that descend from a common ancestor, indicating spread of the virus in that region. U.K. transmission clusters were differentiated from non-U.K. clusters on the basis of the size of the clade and the proportion of U.K. sequences within it: U.K. transmission clusters were defined as those clades with >25 sequences, 90% or more of which were of U.K. origin. A minimum clade size of 25 was used because smaller sample sizes are unlikely to give reliable coalescent estimates under complex demographic models. A minimum fraction of 90% U.K. sequences was chosen to ensure that the clusters that were identified represent chains of transmission that have overwhelmingly occurred in the U.K. However, we note that this methodology probably underestimates the number of transmission chains identified.
Fig. 1.
Schematic representation of the phylogeny generated from 3,429 U.K. and worldwide HIV-1 subtype B pol sequences. Filled circles represent sequences from the U.K., and open squares represent non-U.K. sequences. Three branching patterns were distinguished: non-U.K. transmission clusters (a), sporadic U.K. infections (b), and U.K. transmission clusters (c). Transmission clusters are clades of sequences from a particular location that descend from a common ancestor, indicating a successful spread of the virus in that location. U.K. transmission clusters are defined as those clades that include at least 25 sequences, 90% or more of which are of U.K. origin.
Most of the U.K. sequences represented sporadic lineages (86%) scattered among sequences from other geographical areas, suggesting much geographical mixing and migration of subtype B strains on a worldwide scale. Nonetheless, six U.K. transmission clusters were identified, involving 45, 62, 29, 26, 27, and 34 sequences. These transmission chains were distinct (i.e., reciprocally monophyletic), indicating that at least six independent introductions of subtype B HIV-1 have succeeded in sustaining onward transmission within the U.K. over time and until the present. Each transmission chain contained sequences from a variety of locations within the U.K., and no obvious geographic correlations were observed. The robustness of the clusters within the overall tree could not be statistically evaluated because of the huge size of the data set. Nonetheless, the branching patterns of the six U.K. lineages showed statistical robustness when compared with subsets of worldwide control sequences by using bootstrap analyses (neighbor-joining method with 1,000 replicates, as implemented in the program paup*; data not shown). To further explore the history of these six successful viral lineages, sequences of non-U.K. origin were removed from the six clusters, and the phylogenetic histories of the U.K. sequences were rigorously reestimated by using a maximum likelihood approach. The maximum likelihood trees are available from the authors on request.
Estimation of the Rate of Evolution for the HIV-1 Subtype B pol Gene. The rate of evolution for the subtype B HIV-1 pol gene was calculated by using an independent data set of 106 sequences, sampled between 1983 and 2000 in Amsterdam (20). Using a Bayesian MCMC method, this rate was estimated to be 2.55 × 10–3 substitutions per nucleotide site per year (95% confidence intervals: 1.74 × 10–3 to 3.51 × 10–3). In comparison, previous estimates of HIV-1 evolution rates have typically relied on partial env gene sequences and have ranged from 2.4 × 10–3 to 6.7 × 10–3 substitutions per site per year (24–26). Our estimate is consistent with the order of magnitude of 10–3 expected for an HIV-1 gene. The phylogenetic trees in Fig. 2 are thus shown on a time scale of years.
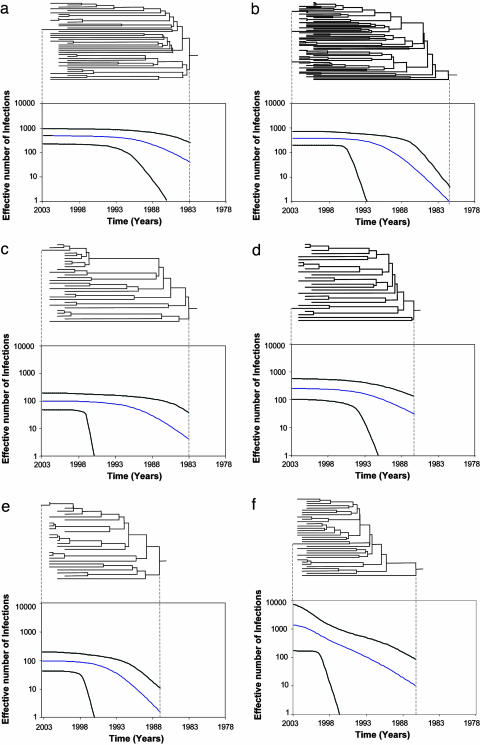
Fig. 2.
Phylogenetic trees of the six U.K. transmission clusters and their corresponding estimated epidemic histories (all shown on the same time scale). The trees represent the ancestral relationships of sequences belonging to each cluster. (a) Cluster 1. (b) Cluster 2. (c) Cluster 3. (d) Cluster 4. (e) Cluster 5. (f) Cluster 6. The demographic histories were estimated by Bayesian MCMC inference with a model of logistic growth (see text for details) and show change in the effective number of infections through time (time scale in calendar years). The blue line shows the median estimate of the effective number of infections, whereas the black lines show the 95% confidence limits of the estimate.
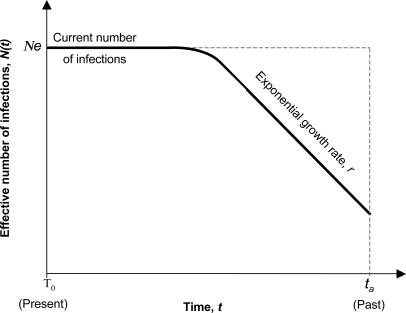
Epidemic History and Parameter Estimation. For each of the six clusters, a model of logistic population growth best fitted the demographic information contained in the tree topologies (likelihoods shown in Table 2, which is published as supporting information on the PNAS web site). Under the logistic model, the effective number of infections Ne grows exponentially at rate r from time ta (time of the most recent common ancestor of the cluster) then decreases in growth rate toward the present. A schematic representation of the logistic model is given in Fig. 3. Note that Ne reflects the number of infections contributing to new infections, rather than the total number of prevalent infections within the transmission cluster.
Fig. 3.
Schematic representation of the logistic model of population growth. According to this model, the number of infections population grows exponentially at rate r from time ta (time of the most recent common ancestor of the sampled sequences). The growth rate slows as time moves toward the present, such that Ne represents the effective number of infections at the present. Ne can be thought of as the number of infections contributing to new infections rather than the total number of prevalent infections within the cluster.
The demographic parameters that determine the shape of the logistic growth curve were estimated by Bayesian MCMC inference (Table 1), and the epidemic histories of the six clusters were reconstructed with appropriate confidence limits (Fig. 2). Our estimates suggest that three of the six genealogies originated in the early 1980s (1981 for cluster 2 and 1983 for clusters 1 and 3), whereas the remaining clusters were introduced later in the same decade (1986 for clusters 4 and 6 and 1987 for cluster 5). Although the initial exponential growth phase clearly ended in the early 1990s for clusters 1–5 (see Fig. 2 a–e), the growth rate decrease is more tentative for cluster 6 and is only apparent very recently (see Fig. 2f), such that cluster 6 appears to also fit a model of exponential growth. To explore this issue further, we estimated the epidemic doubling time of each transmission cluster at the most recent sampling time, year 2003 (by rearrangement of the model in Pybus et al., ref. 27). This “current” epidemic doubling time is considerably shorter for cluster 6 than for clusters 1–5; specifically, the current doubling time for cluster 6 is significantly more likely to be <20 years (equal to an exponential growth rate >0.035 years–1) in comparison with the other clusters (data not shown). In marked contrast, the exponential growth rates at the time of initiation of each cluster (r) are very similar, with an average of 0.80 years–1. Finally, the current effective number of infections Ne varied from cluster to cluster, ranging from 94 (cluster 5) to 1,350 (cluster 6) effective infections.
Table 1. Parametric estimates under the logistic growth demographic model for the six lineages.
| Cluster | μ | Ne, no. of infections | r, yr-1 | Origin of the tree, yr |
|---|---|---|---|---|
| 1 | 2.55 × 10-3 (0.0017-0.0035) | 493 (201-833) | 1.08 (0.66-2.56) | 1983 (1978-1988) |
| 2 | 2.55 × 10-3 (0.0017-0.0035) | 386 (190-655) | 0.47 (0.30-0.95) | 1981 (1976-1987) |
| 3 | 2.55 × 10-3 (0.0017-0.0035) | 98 (42-171) | 0.50 (0.19-4.62) | 1983 (1977-1988) |
| 4 | 2.55 × 10-3 (0.0017-0.0035) | 250 (88-483) | 1.38 (0.63-2.50) | 1986 (1982-1991) |
| 5 | 2.55 × 10-3 (0.0017-0.0035) | 94 (36-85) | 0.68 (0.35-2.10) | 1987 (1983-1991) |
| 6 | 2.55 × 10-3 (0.0017-0.0035) | 1,350 (109-5489) | 0.67 (0.37-3.85) | 1986 (1981-1991) |
| U.S.* | 6.7 × 10-3 (ND) | 4,830 (1,995-26,750) | 0.834 (0.72-0.945) | 1968 (1966-1970) |
Numbers in parentheses are the range with 95% confidence intervals. μ, rate of nucleotide substitution estimated from an independent data set of subtype B pol sequences. r, rate of exponential growth.
Data from Robbins et al. (26).
Discussion
Our estimates suggest that the HIV-1 subtype B epidemic currently circulating within the U.K. is comprised of at least six established chains of transmission, introduced in the early and mid-1980s. This finding demonstrates the existence of distinct, possibly nonoverlapping sexual networks within the predominant MSM risk group and argues against the hypothesis that one initial entry of HIV-1 was responsible for the spread of the subtype B epidemic. It also emphasizes the role of migration in the HIV-1 epidemic in Britain, as illustrated by the overwhelming prevalence of sporadic lineages (86% of the total U.K. samples) in the genealogy, representing viruses arising from outside the U.K. that have failed to establish a large outbreak.
The transmission clusters we characterized had similar epidemic curves and geographic distributions within the U.K., indicating a concurrent spread under similar demographic pressures, at least during the early stages of the epidemic. The introduction of the earlier viruses in the early 1980s (i.e., clusters 1–3) seems to coincide with the explosion of new infections reported by epidemiological data at the time (www.hpa.org.uk). The coupling of HIV strain “immigration” with epidemiological changes is likely to have favored the emergence and persistence of the transmission chains presently circulating amongst MSM.
However, the first U.K. cases of AIDS were reported in 1982 (www.who.int/emc-hiv/fact_sheets), and these individuals were probably infected within a window of 10 years before that time; hence, the currently circulating strains may not represent the first HIV-1 lineages identified within the U.K. If earlier strains existed, they may have been unsuccessful in sustaining transmission chains until the present and may no longer be of epidemiological significance. However, the absence of older strains could also reflect a sampling bias.
For all six transmission clusters, the exponential growth phase coincides with a reported augmentation of newly acquired HIV-1 infections within MSM and injecting drug users in the U.K. (www.hpa.org.uk). The average growth rate during the initial exponential phase was estimated to be 0.80 years–1 (ranging from 0.47 to 1.38), approximating a doubling time of 1 year. This value is similar to that estimated for the United States subtype B epidemic (0.83 years–1, 0.72–0.94), suggesting that the two epidemics follow similar trends at the macroepidemiological scale (26). This idea is supported by the effective number of infections estimated for the two epidemics. Despite a wide variation in Ne across the six U.K. transmission clusters, the average effective number of infections among the six U.K. clusters is 445, which is ≈2.5% of the infected population. This result is remarkably similar to the values for the United States epidemic, where the effective number of infections and prevalence in 1995 reached 5,000 and 200,000 infections, respectively. Ne represents the number of infections contributing to onward transmission, rather than the larger number of actual infections. Importantly, we observe that the population represented by cluster 6 exhibits a faster doubling time in 2003 than the other five clusters, suggesting a difference in current growth rate among clusters. Current surveillance data for the U.K. reports a very recent increase in infections in MSM (www.hpa.org.uk), and it is reasonable to suppose that the lineage we have identified as cluster 6 has contributed to this recent upturn in infection.
Since 1990, there have been important changes in Britain's demographic structure, social attitude, and awareness of HIV-1/AIDS (28). Despite a very recent increase in high-risk behavior among MSM (such as the number of sexual partners or concurrent partnerships), a significant increase in consistent condom use has been reported since 1990. Such a change in sexual health, coupled to large-scale educational campaigns over the past decade, could explain the equilibrium reached by the effective number of prevalent infections. The effect of antiretroviral therapy on past epidemic dynamics should also be considered: although such therapy is instituted primarily to reduce progression of disease, it may also impact on transmission through reduction of infectivity. If so, we would expect evidence of a growth rate decrease in the late (rather than early) 1990s, the time that highly active antiretroviral therapy became widely used. In fact, Health Protection Agency data suggests no significant changes in the incidence of HIV-1 within gay men since the late 1980s and an actual increase over the past three years (29). We therefore suggest that antiviral therapy has not had a significant impact on the growth of the epidemic; indeed, some studies suggest that the epidemic is driven by transmissions in primary infection (30–32), before therapy is usually initiated. The current increase in new infections is too recent to be reflected in the growth dynamics of any of the six populations identified by our analysis. Ongoing analyses of the type undertaken here will clarify whether the recent increase in new subtype B infections derive from longstanding viral lineages or newly introduced viruses.
In conclusion, we show that currently circulating HIV-1 subtype B strains entered the U.K. in the mid 1980s and that the rate of spread of these lineages slowed in the early 1990s. It is often assumed that the HIV-1 epidemic within the U.K. is composed of smaller, independent epidemics defined by risk group. We demonstrate here the existence of multiple subepidemics (at least six) within MSM that obey similar demographic constraints during their early stages, yet exhibit differences in their more recent rates of spread. The identification of these multiple lineages within the predominant risk group of the HIV-1 epidemic in the U.K. suggests the existence of subepidemics within groups of MSM, and it is reasonable to assume that this structure exists in comparable risk groups in other countries. Such heterogeneity must therefore be considered when developing HIV monitoring prevention and treatment initiatives.
Supplementary Material
Acknowledgments
We thank Dr. Patricia Cane from the Health Protection Agency Antiretroviral Susceptibility Reference Unit and Dr. Vladimir Lukashov from the Academic Medical Center of Amsterdam for kindly providing the sequences used in this study, Dr. Andy Rambaut for support in the phylogenetic analyses, and Prof. Robin Weiss and Dr. Paul Kellam for useful comments on the manuscript. This work was funded by the Health Protection Agency, U.K. O.G.P. was funded by the Wellcome Trust and The Royal Society.
Author contributions: S.H., D.P., and O.G.P. designed research; S.H. and O.G.P. performed research; S.H., D.P., J.P.C., and O.G.P. analyzed data; and S.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HIV-1, type 1 HIV; MCMC, Markov Chain Monte Carlo; MSM, men who had sex with men; U.K., United Kingdom.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY669865–AY670087).
References
- 1.Murphy, G., Charlett, A., Jordan, L. F., Osner, N., Gill, O. N. & Parry, J. V. (2004) AIDS 18, 265–272. [DOI] [PubMed] [Google Scholar]
- 2.Holmes, E. C., Nee, S., Rambaut, A., Garnett, G. P. & Harvey, P. H. (1995) Philos. Trans. R. Soc. London B 349, 33–40. [DOI] [PubMed] [Google Scholar]
- 3.Nee, S., Holmes, E. C., Rambaut, A. & Harvey, P. H. (1995) Philos. Trans. R. Soc. London B 349, 25–31. [DOI] [PubMed] [Google Scholar]
- 4.Kingman, J. F. (2000) Genetics 156, 1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffiths, R. C. & Tavare, S. (1994) Philos. Trans. R. Soc. London B 344, 403–410. [DOI] [PubMed] [Google Scholar]
- 6.Kuhner, M. K., Yamato, J. & Felsenstein, J. (1995) Genetics 140, 1421–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hué, S., Clewley, J. P., Cane, P.A. & Pillay, D. (2004) AIDS 18, 719–728. [DOI] [PubMed] [Google Scholar]
- 8.Leal, E. d. S., Holmes, E. C. & Zanotto, P. M. (2004) Virology 325, 181–191. [DOI] [PubMed] [Google Scholar]
- 9.Richman, D. D., Havlir, D., Corbeil, J., Looney, D., Ignacio, C., Spector, S. A., Sullivan, J., Cheeseman, S., Barringer, K., Pauletti, D., et al. (1994) J. Virol. 68, 1660–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frost, S. D., Nijhuis, M., Schuurman, R., Boucher, C. A. & Brown, A. J. (2000) J. Virol. 74, 6262–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rouzine, I. M. & Coffin, J. M. (1999) J. Virol. 73, 8167–8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenfell, B. T., Pybus, O. G., Gog, J. R., Wood, J. L., Daly, J. M., Mumford, J. A. & Holmes, E. C. (2004) Science 303, 327–332. [DOI] [PubMed] [Google Scholar]
- 13.Lemey, P., Pybus, O. G., Wang, B., Saksena, N. K., Salemi, M. & Vandamme, A.-M. (2003) Proc. Natl. Acad. Sci. USA 100, 6588–6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa, M., Kishino, H. & Yano, T. (1985) J. Mol. Evol. 22, 160–174. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein, J. (1973) Am. J. Hum. Genet. 25, 471–492. [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, Z. (1994) J. Mol. Evol. 39, 105–111. [DOI] [PubMed] [Google Scholar]
- 18.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817–818. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein, J. (1985) Evolution 39, 783–791. [DOI] [PubMed] [Google Scholar]
- 20.Lukashov, V. V. & Goudsmit, J. (2002) J. Mol. Evol. 54, 680–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drummond, A. J., Nicholls, G. K., Rodrigo, A. G. & Solomon, W. (2002) Genetics 161, 1307–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pybus, O. G. & Rambaut, A. (2002) Bioinformatics 18, 1404–1405. [DOI] [PubMed] [Google Scholar]
- 23.Rambaut, A. (2000) Bioinformatics 16, 395–399. [DOI] [PubMed] [Google Scholar]
- 24.Korber, B., Muldoon, M., Theiler, J., Gao, F., Gupta, R., Lapedes, A., Hahn, B. H., Wolinsky, S. & Bhattacharya, T. (2000) Science 288, 1789–1796. [DOI] [PubMed] [Google Scholar]
- 25.Leitner, T., Escanilla, D., Franzen, C., Uhlen, M. & Albert, J. (1996) Proc. Natl. Acad. Sci. USA 93, 10864–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbins, K. E., Lemey, P., Pybus, O. G., Jaffe, H. W., Youngpairoj, A. S., Brown, T. M., Salemi, M., Vandamme, A. M. & Kalish, M. L. (2003) J. Virol. 77, 6359–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pybus, O. G., Charleston, M. A., Gupta, S., Rambaut, A., Holmes, E. C. & Harvey, P. H. (2001) Science 292, 2323–2325. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, A. M., Mercer, C. H., Erens, B., Copas, A. J., McManus, S., Wellings, K., Fenton, K. A., Korovessis, C., Macdowall, W., Nanchahal, K., et al. (2001) Lancet 358, 1835–1842. [DOI] [PubMed] [Google Scholar]
- 29.Brown, A. E., Sadler, K. E., Tomkins, S. E., McGarrigle, C. A., LaMontagne, D. S., Goldberg, D., Tookey, P. A., Smyth, B., Thomas, D., Murphy, G., et al. (2004) Sex. Transm. Infect. 80, 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koopman, J. S., Jacquez, J. A., Welch, G. W., Simon, C. P., Foxman, B., Pollock, S. M., Barth-Jones, D., Adams, A. L. & Lange, K. (1997) J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 14, 249–258. [DOI] [PubMed] [Google Scholar]
- 31.Jacquez, J. A., Koopman, J. S., Simon, C. P. & Longini, I. M., Jr. (1994) J. Acquired Immune Defic. Syndr. 7, 1169–1184. [PubMed] [Google Scholar]
- 32.Yerly, S., Vora, S., Rizzardi, P., Chave, J. P., Vernazza, P. L., Flepp, M., Telenti, A., Battegay, M., Veuthey, A. L., Bru, J. P., et al. (2001) AIDS 15, 2287–2292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.