Abstract
Type IV secretion systems (T4SSs) are commonly used secretion machineries in Gram-negative bacteria. They are used in the infection of human, animal, or plant cells and the propagation of antibiotic resistance. The T4SS apparatus spans both membranes of the bacterium and generally is composed of 12 proteins, named VirB1–11 and VirD4 after proteins of the canonical Agrobacterium tumefaciens T4SS. The periplasmic core complex of VirB8/VirB10 structurally and functionally links the cytoplasmic NTPases of the system with its outer membrane and pilus components. Here we present crystal structures of VirB8 of Brucella suis, the causative agent of brucellosis, and ComB10, a VirB10 homolog of Helicobacter pylori, the causative agent of gastric ulcers. The structures of VirB8 and ComB10 resemble known folds, albeit with novel secondary-structure modifications unique to and conserved within their respective families. Both proteins crystallized as dimers, providing detailed predictions about their self associations. These structures make a substantial contribution to the repertoire of T4SS component structures and will serve as springboards for future functional and protein–protein interaction studies by using knowledge-based site-directed and deletion mutagenesis.
Keywords: protein/DNA transport, Gram-negative bacteria, structural biology, crystallography
Type IV secretion systems (T4SSs) are used by Gram-negative bacteria to transport protein toxins and other virulence factors into host cells (1, 2). They play an essential part in a variety of human infectious diseases such as stomach ulcers, whooping cough, and Legionnaire's disease, caused by Helicobacter pylori, Bordetella pertussis, and Legionella pneumophila, respectively (3–5). Bacterial conjugation, which is responsible for the rapid spread of antibiotic resistance genes through bacterial populations, is also a T4SS-mediated process (6). Additionally, a T4SS is used by Agrobacterium tumefaciens to transform plant cells with DNA and is the primary means by which genetically modified plants are created (7).
The most common T4SSs comprise 12 proteins that can be identified as homologs of the VirB1–11 and VirD4 proteins of the A. tumefaciens Ti plasmid transfer system (8). T4SSs span both membranes of Gram-negative bacteria, using a specific transglycosylase, VirB1, to digest the intervening murein (9, 10). Three NTPases (VirB4, -B11, and -D4) comprise the cytoplasmic component of the system and are closely associated with the inner membrane (2). VirB6 has multiple transmembrane helices and forms an inner membrane complex with VirB8 and VirB10 (11). VirB8 and VirB10, however, are largely periplasmic, each possessing a short cytoplasmic N-terminal tail, a single transmembrane helix, and a large conserved periplasmic C-terminal domain separated from the helix by a nonconserved linker sequence. VirB9 and the lipoprotein VirB7 form an outer membrane complex (12). VirB2 and VirB5 are the major and minor pilus components, respectively (13, 14). T4SSs may be composed of subsets of these 12 proteins. For example, in systems that function in DNA uptake or release, such as the H. pylori ComB system, only VirB7–10 homologs and a single NTPase have been identified (15).
VirB8 and VirB10 are crucial structural and functional components of the T4SS. In A. tumefaciens, VirB8 contacts the T-complex substrate (VirD2 and the single-stranded T-DNA) directly and is required for passage of the substrate from the cytoplasm to the periplasm (16). VirB10 is essential for the transfer of the substrate from the inner to the outer membrane but does not directly contact the substrate (16). Instead, it acts as an energy-sensing bridge between the inner and outer membranes (17). Interactions between VirB8 and many other T4SS proteins have been reported, including VirB10 (18, 19), VirB9 (19), VirB1, VirB4, and VirB11 (20), as well as with itself (18–20). In addition to its VirB8 association, VirB10 interacts with VirB9 (19, 21), VirB4 (20), VirD4 (22, 23), VirB1, and VirB11 (20), as well as itself (18, 19). VirB8 and VirB10, therefore, are keystone components at the heart of the T4SS machinery, providing the structural and functional link between the cytoplasmic/inner membrane assembling and powering components (VirB6, VirB4, VirB11, and VirD4) and the outer membrane/pilus subassembly (VirB7, VirB9, VirB2, and VirB5). We present here the crystal structures of the periplasmic domains of VirB8 (VirB8 from Brucella suis) and VirB10 (ComB10 from H. pylori) homologs. These two structures constitute structural prototypes for the VirB8 and VirB10 families of proteins and provide fundamental insight into T4SS architecture.
Methods
Cloning and Protein Preparation. Native and selenomethionine-substituted strepII-tagged B. suis VirB8 (residues 77–239) was purified by using affinity chromatography, followed by tag cleavage by Factor Xa and purified to homogeneity (99%) by using gel filtration. H. pylori His-6-tagged ComB10 (residues 144–376) was purified by using affinity chromatography, followed by tag cleavage by TEV protease and further purified by using gel filtration.
Crystallization and Data Collection. Crystals were grown by using vapor diffusion in hanging drops by using 0.7 M K2HPO4/40 mM NaH2PO4 and 8–14% (wt/vol) polyethylene glycol 8000/100 mM sodium cacodylate, pH 6.0/200 mM magnesium acetate as reservoir conditions for VirB8 (28 mg/ml) and ComB10 (30 mg/ml), respectively. Crystals of VirB8 belonged to the space group I4122, with cell dimensions of a = 203.8 Å, b = 203.8 Å, and c = 103.1 Å, and diffracted to a resolution of 2.4 Å. Crystals of native ComB10 belonged to the space group P212121, with cell dimensions of a = 69.8 Å, b = 139.5 Å, and c = 168.6 Å and diffracted to a resolution of 3.0 Å.
Structure Determination. For VirB8, 14 selenium sites were located by shelxd (24) by using the peak dataset SeMet-1 (Tables 1, 2, 3). mlphare (25) was used to generate initial single-wavelength anomalous dispersion phases, which were improved by density modification and the noncrystallographic symmetry (NCS) averaging in resolve (26). Initial building of a few helices led to the definition of the NCS relating each chain. The five NCS operators derived from the coordinates of these helices were used in resolve to produce a map (Fig. 4A, which is published as supporting information on the PNAS web site) that was used to build most residues of all five molecules in the asymmetric unit. The refinement [cns (27)] converged to yield a model with R and Rfree factors of 24.5% and 27.5% with good geometry (Tables 1, 2, 3). For ComB10, two xenon derivative datasets were used (Tables 1, 2, 3). Six Xe atoms sites were found (shelxd), and a modification of the siras method was used [sharp (28)] to generate an initial set of experimental phases at 3.2 Å. NCS averaging together with density modification [profess, dm (25)] resulted in an improved density map. Refinement [refmac (25)] using each chain as a translation/libration/screw (TLS) group with tight NCS constraints applied to all six molecules in the asymmetric units, except for the helical regions where loose NCS constraints were applied, converged to a model with R and Rfree factors of 25.8% and 29.6% with good geometry. Both structures have been deposited in the Protein Data Bank (ID codes 2BHM and 2BHV). Details of the methods can be found in Supporting Text, which is published as supporting information on the PNAS web site.
Table 1. VirB8 data collection.
| SeMet-1 | SeMet-2 | Native | |
|---|---|---|---|
| Wavelength, Å | 0.9795 | 0.9794 | 0.9763 |
| Resolution, Å | 3.0 | 2.5 | 2.4 |
| Reflections, total | 158,701 | 257,231 | 1,055,412 |
| Unique | 21,902 | 35,315 | 44,402 |
| Completeness, % | 99.9 (100.0) | 98.5 (96.3) | 98.8 (98.8) |
| Multiplicity | 7.2 (7.4) | 7.1 (6.9) | 7.3 (7.2) |
| Rsym,* % | 14.9 (51.1) | 5.9 (28.2) | 9.5 (43.8) |
| 〈I/σ(I)〉 | 4.8 (1.5) | 10.2 (2.7) | 6.3 (2.2) |
Rsym = Σ|I—<I>|/ΣI, where I equals observed intensity and <I> equals average intensity for symmetry-related reflections.
Table 2. ComB10 data collection.
| Xe-1 | Xe-2 | Native | |
|---|---|---|---|
| Wavelength, Å | 1.7712 | 1.7712 | 0.9793 |
| Resolution, Å | 3.2 | 3.7 | 3.0 |
| Reflections, total | 302,294 | 114,799 | 122,735 |
| unique | 28,039 | 18,337 | 32,829 |
| Completeness, % | 99.9 (100.0) | 99.8 (100.0) | 97.4 (98.0) |
| Multiplicity | 10.8 (10.7) | 6.3 (6.3) | 3.7 (3.8) |
| Anomalous completeness, % | 99.9 (99.9) | 99.9 (100.0) | N/A |
| Anomalous multiplicity | 5.6 (5.5) | 3.3 (3.3) | N/A |
| Rsym,* % | 10.5 (40.1) | 11.3 (42.9) | 9.1 (61.0) |
| 〈I/σ(I)〉 | 19.3 (5.0) | 14.2 (4.1) | 11.0 (2.3) |
N/A, not applicable.
Rsym = Σ|I—<I>|/ΣI, where I equals observed intensity and <I> equals average intensity for symmetry-related reflections.
Table 3. Refinement statistics.
| VirB8 | ComB10 | |
|---|---|---|
| Resolution, Å | 26.6-2.4 | 20.0-3.0 |
| Reflections, working/test | 39,271/2,083 | 29,714/1,674 |
| R/Rfree | 24.5/27.5 | 25.8/29.6 |
| Total atoms/solvent | 5,321/61 | 8,768/0 |
| rms deviation | ||
| Bonds, Å | 0.008 | 0.014 |
| Angles, ° | 1.3 | 1.3 |
| B values (main/side), Å2 | 1.5/3.2 | 0.4/0.7 |
Results
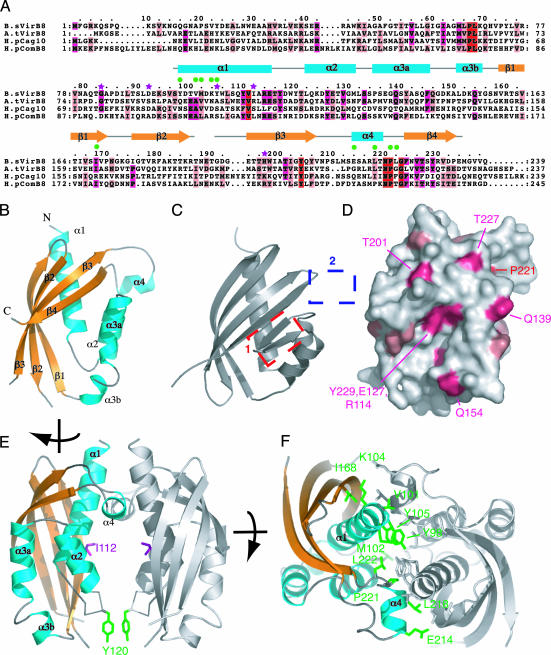
Structure of VirB8. We present here the structure of a fragment of B. suis VirB8 (residues 77–239) encompassing the periplasmic domain (termed pVirB8). pVirB8 is known to contain the site of interaction with VirB10 and also the site of VirB8 self assembly (20, 29). The structure of pVirB8 consists of a large extended β sheet (β1, β2, β3, and β4) juxtaposed against five α-helices (α1, α2, α3a, α3b, and α4) (Fig. 1 A and B). As shown in Fig. 1B, the four-stranded antiparallel β-sheet wraps around one side of helix α1. Helices α2 and α3 pack against the C terminus of α1 and form one edge of a deep surface groove along with β3 and β4, which form the other edge. Helix α4 is less closely associated with the rest of the domain and is positioned at a right angle to α1 but, as shown in Fig. 1E, protrudes further out from the domain. Helix α3 is interrupted toward its C terminus by an unusual bulge between residues 146–150. This arrangement of secondary structures is likely conserved among all VirB8 proteins. Indeed, the sequence alignment shown in Fig. 1A indicates a strong conservation of structural residues, i.e., residues buried in the fold. Thus, the structure presented here can be seen as a prototype for all periplasmic domains of VirB8 proteins.
Fig. 1.
Crystal structure of B. suis pVirB8. (A) Sequence alignment of VirB8 proteins and secondary structure assignment. Amino acids of four representative homologs were aligned, from B. suis (B.sVirB8), A. tumefaciens (A.tVirB8, 21% identical to B.sVirB8), H. pylori Cag [H.pCag10 (HP0530, 14% identity], and ComB [H.pComB8 (HP0030), 18% identity] systems (38). Strictly conserved, strongly conserved, and conserved residues are marked in red, magenta, and light pink, respectively. The modeled region (residues 97–188 and 191–234) is shown as a gray line above the sequence for nonregular structure or as cyan boxes and yellow arrows for α-helices and β-strands, respectively. Green dots mark residues involved in VirB8 self association. Purple stars indicate residues mutated in previous functional studies. (B) Overall fold of pVirB8. Secondary structure representation and labels are as in A. (C) Structure of NTF2, most similar fold to pVirB8. Boxes mark the two major points of difference between the NTF2 and pVirB8 fold, the addition of α4 (blue box), and the loss of two β strands (red box). (D) Surface representation of VirB8 coloring side chains by degree of conservation, as shown in A. Orientation is as in B. (E) pVirB8 dimer. Both monomers are shown in ribbon representation with the monomer on the left shown as in B but turned ≈90° clockwise. The other monomer is in gray. Ile-112 and Tyr-120 are shown in stick representation and colored in magenta and green, respectively. (F) Top-down view of pVirB8 dimer showing side chains involved in interface as marked in A. For clarity, one pVirB8 chain is colored gray, and the other is colored as in B. Residues at the interface are in stick representation, color-coded in green, and labeled. The figure was produced by using pymol, http://pymol.sourceforge.net.
The pVirB8 fold is overall most similar to the NTF2-like fold. The closest matches found by dali (30) were the association domain of CaMKII (31), the nuclear transport factor NTF2 (32), and enzymes of the steroid δ-isomerase family (33). For comparison, the pVirB8 and NTF2 structures are shown side by side in Fig. 1 B and C, respectively. The pVirB8 fold differs from these other structures in that it (i) lacks two short β-strands between α2 and α3 (Fig. 1C, red box) and (ii) has an additional short α-helix between strands β3 and β4 (Fig. 1C, blue box).
Potential Protein–Protein Interactions of VirB8. As shown in Fig. 1A, the pattern of conserved residues between VirB8 homologs shows several hotspots. As expected, most of the conserved residues reflect structural requirements for the VirB8 fold, such as those involved in the hydrophobic core. The region of VirB8 showing the most conservation among surface-exposed residues is shown in Fig. 1D. Most striking is the patch of highly conserved side chains (Tyr-229, Glu-127, and Arg-114) at the base of a deep groove. This groove is much less pronounced in NTF2-like molecules where it is filled in by two short β-strands to form a less-extensive pocket. In NTF2-like molecules exhibiting this pocket, it is either used for protein–protein interactions, as is the case for NTF2 (32) and CamKII (31), or forms the active site of a large class of lipid and steroid enzymes (33). This important feature of VirB8 could accommodate an α-helix or pair of β-strands and is, especially given the degree of surface conservation, a likely site of protein–protein interaction.
The most conserved motif among VirB8 homologs is the “220NPxG” sequence, which lies between α4 and β4. It adopts a sharp-turn conformation that could almost certainly not be maintained were any of the three key residues to be mutated. The sharp turn positions α4 close to and approximately perpendicular to α1. Although the position of α4 looks somewhat odd, sticking out from and only loosely associated with the rest of the pVirB8 monomer, it could be explained when considering the crystal packing. pVirB8 crystallized with five molecules in the asymmetric unit, providing five independent snapshots of the structure. Each of the five pVirB8 chains is found to dimerize as shown in Fig. 1 E and F, two dimers are formed by NCS between chains C and E and B and D, whereas chain A forms a dimer through a crystallographic twofold axis, making three dimers in total. The relative orientation of the three dimers is very similar (0.27- to 0.38-Å rms deviation in Cα atoms). Over 1,700 Å2 of surface area is buried, making this interface the largest and most conserved crystal packing interaction. The interface is largely between the N terminus of α1 and the region encompassing α4 and the sharp turn between α4 and β4 (Fig. 1 A and E). That this interaction depends on α4, one of the novel features of the VirB8, and is identical in all five independent molecules strongly supports our belief that this interaction reflects a physiological self association of VirB8.
Viewed in the orientation shown in Fig. 1E, the interaction is strikingly asymmetric: all but one contacts are in the upper half of the dimer. A channel of ≈10 Å in diameter runs through the center of the dimer. Ile-112 is exposed in this channel, equivalent to Arg-107 in the A. tumefaciens protein, a residue that has been implicated in interactions of VirB8 with VirB9 and VirB10.
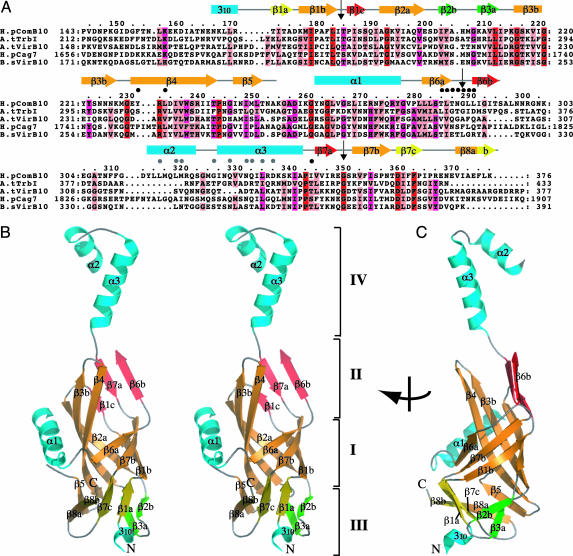
Structure of ComB10. The structure of a fragment of H. pylori ComB10 encompassing the conserved, periplasmic, C-terminal domain (termed pComB10) was determined (Fig. 2A). The structure of pComB10 comprises an extensively modified β-barrel, opened on one side and extended by a flexible α-helical antenna at the top. In fact, the structure deviates so significantly from a “canonical” β-barrel fold that no significant structural homolog could be found by using the program dali. The structure of pComB10 can be subdivided into four parts. The central part (I in Fig. 2B), composed of the β1b, β2a, β3b, β4, β6a, and β7b strands, forms a β-barrel opened between strands β4 and β6a. Indeed, the main chain of β4 and β6a does not hydrogen bond, and β6a splays open the β-barrel by directing the next strand, β6b, toward a different part of the structure (II in Fig. 2B). Another prominent feature of the structure in this region is helix α1 (Fig. 2B). Helix α1 precedes strand β6 and sits at the side of the central β-barrel, perpendicular to strands β3b and β4. Part II encompasses a β-sheet composed of the β6b, β7a, and β1c strands (red in Fig. 2B), partially stacked onto an extension of the central β-barrel consisting of strands β4, β3b, and β2a. The β6b-β7a-β1c sheet is pulled apart from a possible β-barrel arrangement with β4-β3a-β2a because of three bulges, between β6a and β6b, β7a and β7b and between β1b and β1c (indicated by arrows in Fig. 2A). As a result, this part of the structure contains a large groove circumscribed by strands β6a and β6b on one side and the N terminus of strand β4 on the other (Figs. 2B and 3A). The lower part of the structure (part III in Fig. 2B) is composed of three different and loosely connected β-sheets (β5-β8a, β8b-β7c-β1a, and β3a-β2b in orange, yellow, and green, respectively, in Fig. 2B). The β8 strand is shared between the two first β-sheets; also, the β5-β8a sheet forms an extension of the central β2a-β3b-β4 β-barrel strands, thus the β5-β8a and β8b-β7c-β1a sheets can be considered extensions of the central β-barrel. However, the β3a-β2b sheet (in green in Fig. 2B) is clearly apart and forms a flap structure sealing the β-barrel at the bottom (better seen in Fig. 2C). Finally, part IV of the structure is formed by an extended helix–loop–helix (α2, α3, and the connecting loop) structure that protrudes at the top of the barrel and extends the domain to >70 Å long. It is a very flexible part, and the connection between strand β6b and helix α2 could not be traced. It also has the least-conserved sequence (Fig. 2A).
Fig. 2.
Crystal structure of H. pylori pComB10. (A) Sequence alignment of the conserved C-terminal region of VirB10 proteins and secondary structure assignment. Amino acids of five representative homologs were aligned, from H. pylori ComB [H.pComB10, (HP0041/0042)] and Cag [H.pCag7 (HP0527), 25% identical to H.pComB10 over region shown] systems, from A. tumefaciens VirB (A.tVirB10, 21% identity) and Trb (A.tTrbI, 24% identity) systems, and from B. suis VirB (B.sVirB10, 24% identity) system. Strictly conserved, strongly conserved, and conserved residues are marked in red, magenta, and light pink, respectively. The modeled region (amino acids 166–253, 261–294, and 311–376) is shown as a gray line above the sequence for nonregular structure or as cyan boxes for α-helices and yellow, orange, red, or green arrows for β-strands. The “bulge” regions that lie between the central (orange) and platform (red) β-sheets are marked with black arrows. Black and gray dots mark residues in the crystal packing interface. (B) Stereo diagram showing overall fold of pComB10. Representation, color-coding, and labeling of β-strands and α-helices are as in A. The four regions referred to in the text are labeled I, II, III, and IV. (C) View of pComB10 rotated through 90° in the vertical axis. The figure was produced by using pymol.
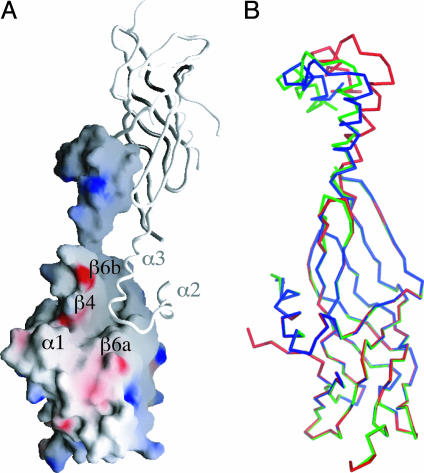
Fig. 3.
Dimer interface and flexible helical region of pComB10. (A) Crystallographic dimer of pComB10. One monomer (in same orientation as Fig. 2B) is shown as a surface representation of charge potential. The second monomer is shown as a ribbon. α1, β4, β6a, and β6b are shown. (B) Superposition of three representative chains shows conformational flexibility in the protruding helical region. B was produced by using pymol, and A was produced by using grasp (39).
The sequence alignment shown in Fig. 2A indicates that the overall fold is likely conserved among VirB10 proteins. Thus, the structure of ComB10 can be seen as a prototype for all periplasmic domains of VirB10 proteins.
Potential Protein–Protein Interactions of ComB10. We have noted above the presence of a large groove formed by a depression in the structure between the β4 strand and the β6a and β6b strands. This groove continues around the molecule between the top of helix α1 and strands β3b and β4 (compare Figs. 2B and 3A). This continuous groove is certainly a prominent feature of the surface of ComB10. It is also the site of a large crystal-packing interface. The asymmetric unit of the ComB10 crystals contained six molecules that form three dimers, each burying a 3,000-Å2 surface area (Fig. 3A). Residues in the α2 and α3 helices (gray dots, Fig. 2A) interact primarily with residues in the β6a and β6b strands and the intervening bulge region (black dots, Fig. 2A) but also with residues in the groove (in β4 and the α3-β7a linker). Although the interactions are symmetrical, with both molecules in the dimer making identical contacts with its partner, the overall appearance of each dimer is not symmetrical because of the intrinsic flexibility of the linker between the barrel (part II) and the protruding helical region (part IV). Indeed, the position of helices α2 and α3 with respect to the rest of the structure varies. One representative chain from each group is shown in Fig. 3B (in red, green, and blue, respectively). The hinge for this rotation is at residues Ala-341 and Pro-342 in the linker between α3 and β7a and results in a maximum displacement of almost 10 Å at the protrusion tip.
Discussion
Bacterial T4SSs are transport machineries dedicated to the traffic of macromolecules (proteins and DNA) through the double membrane of Gram-negative bacteria. Several studies have shown that a core protein complex consisting of VirB8, VirB9, and VirB10 spans the periplasmic space allowing substrate delivery via direct contact [VirB8 and VirB9 (16)] or via a possible energy-sensing process [VirB10 (17)]. We have determined the structures of the periplasmic parts of two of the core components of the T4SS channel. These structures are important, because they provide a clear definition of the domain structure of these proteins and also of the surfaces that may be used not only for self assembly but also for assembly with each other. We anticipate that these structures will serve as springboards for a thorough functional and protein–protein interaction study using site-directed and deletion mutagenesis. The structures of VirB8 and VirB10 resemble known folds, albeit with novel modifications unique to and conserved within their respective families. These modifications are most likely sites of functionality, because they are not required for the basic fold but are conserved features of the modified family-specific fold. Each protein has two modifications in common: (i) the insertion of an α-helix between β-strand elements (α4 in VirB8; α1 in VirB10) that protrudes from the side of the molecule and generates a novel surface for protein–protein interactions; in the case of VirB8, it appears this could be used for homodimerization (Fig. 1 E and F); (ii) the removal or reduction of β-strands that produces a marked groove on the protein surface (Figs. 1D and 3A) in the case of ComB10; this could be involved in homodimerization (Fig. 3A). Because the modifications are not essential for the basic integrity of the fold, they are amenable to mutation and deletion for functional and protein–protein interaction studies.
Several point mutations of A. tumefaciens VirB8 have been identified that reduce virulence (29): Gly-78AT (Gly-83BS) to Ser, Ser-87AT (Asp-92BS) to Leu, Ala-100AT (Tyr-105BS) to Val, Arg-107AT (Ile-112BS) to Pro, Arg-107AT to Ala, Thr-192AT (His-197BS) to Met (residues in A. tumefaciens and B. suis are labeled with the superscripts AT and BS, respectively, and are indicated by purple stars in Fig. 1A). The equivalent residues to two of these, Gly-78AT (Gly-83BS) and Ser-87AT (Asp-92BS), were not observed in the electron density map and are therefore in flexible regions. Tyr-105BS (Ala-100AT) is at the interface of the VirB8 dimer (Fig. 1F) and, although the mutant appeared to maintain VirB8 self-interaction by yeast two-hybrid assay (29), it was not tested whether the interaction was attenuated in the context of the entire T4SS in bacteria. The Arg-107AT to Pro mutant was shown to be defective in VirB9 and VirB10 interactions by two-hybrid assay. The equivalent residue Ile-112BS is exposed in the channel between the subunits of the pVirB8 crystallographic dimer (Fig. 1E). Mutation to proline would probably have a distorting effect on the structure (29), and so it is not clear whether the reduced two-hybrid interactions merely reflect this. However, mutation to alanine would have very limited structural consequences and thus this residue is likely crucially involved in type IV secretion. Thr-192AT (His-197BS) is exposed to solvent and also appears to have a vital function in type IV secretion.
The entire C-terminal perisplasmic domain was unequivocally shown to be essential for dimerization of VirB10 (18). The interface in the pVirB10 dimer is substantial and clearly points to likely protein–protein interaction hotspots. But could the dimer observed in the crystal reflect an actual functional state of VirB10? At first sight, the relative orientation of the monomers does not seem to make sense as their N termini are at opposite ends of the dimer, ≈110 Å apart. Because VirB10 is thought to bridge the periplasm, its N terminus in the cytoplasm and its contact with the outer membrane complex somewhere in its C-terminal portion, it would seem more likely that a VirB10 dimer would form with the N termini on the same side and the long axes of the monomers parallel. In a recent paper, Cascales and Christie (17) identified a conformational change in VirB10 associated with energy sensing of the molecule, suggesting a functional similarity between the periplasmic energy transducer TonB and VirB10. In the presence of ATP and the energizing components of the T4SS, VirB10 adopts a protease-sensitive conformation. However, when the cell is depleted of ATP and/or the energizing components of the T4SS are disabled, then VirB10 is protease resistant. Proteolysis removes 8 kDa from the protein, consistent with the cleavage occurring in the protruding helical antennae (region IV in Fig. 2B). Thus, the dimer observed in the crystal may reflect the protease-resistant form of VirB10. The action of ATP and of the energizing NTPases would then dissociate this dimer, releasing the protruding helices for interaction with VirB9 and making them prone to proteolysis. It is also noteworthy that a proteolytic fragment of VirB10 of 40 kDa is observed in cell extracts even without addition of protease. This fragment may correspond to the fragment of VirB10 released as a result of cycles of energization/deenergization. Note that, although TonB and VirB10 may have similar functional properties and are both extended molecules with similar domain arrangements (17), the 3D structures of their periplasmic domains are unrelated (see ref. 34 for TonB structure). The fragment of ComB10 crystallized lacked a poorly conserved periplasmic region that is predicted to form an α-helical coiled-coil (15) and that is generally proline-rich in other VirB10 homologs and in TonB (17). We cannot exclude that these additional regions may be involved in additional protein–protein interactions resulting in higher degrees of oligomerization.
The work presented here makes a substantial contribution to the repertoire of T4SS component structures (35–37). Future work will focus on interactions among components of the T4SS. This will require production of multiple components from the same system in quantity and purity suitable for x-ray crystallographic studies. In the case of the VirB8 and VirB10 homologs of B. suis and H. pylori, this has proven difficult (B. suis pVirB10 was unstable, whereas H. pylori ComB8 was insufficiently soluble, and coexpression did not overcome these problems). However, the structures presented here will facilitate further investigations of other T4SSs where both components may be amenable for crystallization as complexes. They will not only inform the functional work in the years to come but will also encourage structure-aided drug design efforts aiming at discovering novel antimicrobials against important bacterial pathogens using T4SSs for pathogenicity.
Supplementary Material
Acknowledgments
L.T. thanks D. Nurizzo (European Sychrotron Radiation Facility, Grenoble, France) for advice on xenon derivatives. This work was funded by Wellcome Trust Grant 065932 and National Institutes of Health Grant R01 AI49950 (to G.W.) and Canadian Institutes of Health Research Grant MOP-64300 (to C.B.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: T4SS, type IV secretion system; NCS, noncrystallographic symmetry. Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2BHM and 2BHV).
References
- 1.Burns, D. L. (2003) Curr. Opin. Microbiol. 6, 29–34. [DOI] [PubMed] [Google Scholar]
- 2.Cascales, E. & Christie, P. J. (2003) Nat. Rev. Microbiol. 1, 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Censini, S., Lange, C., Xiang, Z., Crabtree, J. E., Ghiara, P., Borodovsky, M., Rappuoli, R. & Covacci, A. (1996) Proc. Natl. Acad. Sci. USA 93, 14648–14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss, A. A., Johnson, F. D. & Burns, D. L. (1993) Proc. Natl. Acad. Sci. USA 90, 2970–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel, J. P., Andrews, H. L., Wong, S. K. & Isberg, R. R. (1998) Science 279, 873–876. [DOI] [PubMed] [Google Scholar]
- 6.Lanka, E. & Wilkins, B. M. (1995) Annu. Rev. Biochem. 64, 141–169. [DOI] [PubMed] [Google Scholar]
- 7.Zupan, J., Muth, T. R., Draper, O. & Zambryski, P. (2000) Plant J. 23, 11–28. [DOI] [PubMed] [Google Scholar]
- 8.Christie, P. J. & Vogel, J. P. (2000) Trends Microbiol. 8, 354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koraimann, G. (2003) Cell Mol. Life Sci. 60, 2371–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baron, C., Llosa, M., Zhou, S. & Zambryski, P. C. (1997) J. Bacteriol. 179, 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakubowski, S. J., Krishnamoorthy, V., Cascales, E. & Christie, P. J. (2004) J. Mol. Biol. 341, 961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez, D., Dang, T. A., Spudich, G. M., Zhou, X. R., Berger, B. R. & Christie, P. J. (1996) J. Bacteriol. 178, 3156–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt-Eisenlohr, H., Domke, N., Angerer, C., Wanner, G., Zambryski, P. C. & Baron, C. (1999) J. Bacteriol. 181, 7485–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai, E. M. & Kado, C. I. (1998) J. Bacteriol. 180, 2711–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofreuter, D., Odenbreit, S. & Haas, R. (2001) Mol. Microbiol. 41, 379–391. [DOI] [PubMed] [Google Scholar]
- 16.Cascales, E. & Christie, P. J. (2004) Science 304, 1170–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cascales, E. & Christie, P. J. (2004) Proc. Natl. Acad. Sci. USA 101, 17228–17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding, Z., Zhao, Z., Jakubowski, S. J., Krishnamohan, A., Margolin, W. & Christie, P. J. (2002) J. Bacteriol. 184, 5572–5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das, A. & Xie, Y. H. (2000) J. Bacteriol. 182, 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward, D. V., Draper, O., Zupan, J. R. & Zambryski, P. C. (2002) Proc. Natl. Acad. Sci. USA 99, 11493–11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beaupre, C. E., Bohne, J., Dale, E. M. & Binns, A. N. (1997) J. Bacteriol. 179, 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilmour, M. W., Gunton, J. E., Lawley, T. D. & Taylor, D. E. (2003) Mol. Microbiol. 49, 105–116. [DOI] [PubMed] [Google Scholar]
- 23.Llosa, M., Zunzunegui, S. & De La Cruz, F. (2003) Proc. Natl. Acad. Sci. USA 100, 10465–10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider, T. R. & Sheldrick, G. M. (2002) Acta Crystallogr. D 1772–1779. [DOI] [PubMed]
- 25.CCP4 (1994) Acta Crystallogr. D 50, 760–763. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger, T. C. (2002) Acta Crystallogr. D 58, 1937–1940. [DOI] [PubMed] [Google Scholar]
- 27.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 28.de La Fortelle, E. & Bricogne, G. (1997) Methods Enzymol. 276, 472–494. [DOI] [PubMed] [Google Scholar]
- 29.Kumar, R. B. & Das, A. (2001) J. Bacteriol. 183, 3636–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm, L. & Sander, C. (1993) J. Mol. Biol. 233, 123–138. [DOI] [PubMed] [Google Scholar]
- 31.Hoelz, A., Nairn, A. C. & Kuriyan, J. (2003) Mol. Cell 11, 1241–1251. [DOI] [PubMed] [Google Scholar]
- 32.Stewart, M., Kent, H. M. & McCoy, A. J. (1998) J. Mol. Biol. 277, 635–646. [DOI] [PubMed] [Google Scholar]
- 33.Lundqvist, T., Rice, J., Hodge, C. N., Basarab, G. S., Pierce, J. & Lindqvist, Y. (1994) Structure (London) 2, 937–944. [DOI] [PubMed] [Google Scholar]
- 34.Chang, C., Mooser, A., Pluckthun, A. & Wlodawer, A. (2001) J. Biol. Chem. 276, 27535–27540. [DOI] [PubMed] [Google Scholar]
- 35.Yeo, H. J., Savvides, S. N., Herr, A. B., Lanka, E. & Waksman, G. (2000) Mol. Cell 6, 1461–1472. [DOI] [PubMed] [Google Scholar]
- 36.Gomis-Ruth, F. X., Moncalian, G., Perez-Luque, R., Gonzalez, A., Cabezon, E., de la Cruz, F. & Coll, M. (2001) Nature 409, 637–641. [DOI] [PubMed] [Google Scholar]
- 37.Yeo, H. J., Yuan, Q., Beck, M. R., Baron, C. & Waksman, G. (2003) Proc. Natl. Acad. Sci. USA 100, 15947–15952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buhrdorf, R., Forster, C., Haas, R. & Fischer, W. (2003) Int. J. Med. Microbiol. 293, 213–217. [DOI] [PubMed] [Google Scholar]
- 39.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins Struct. Funct. Genet. 11, 281–296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.