Abstract
To find their DNA target sites in complex solution environments containing excess heterogeneous DNA, sequence-specific DNA-binding proteins execute various translocation mechanisms known collectively as facilitated diffusion. For proteins harboring a single DNA contact surface, long-range translocation occurs by jumping between widely spaced DNA segments. We have configured biosensor-based surface plasmon resonance to directly measure the affinity and kinetics of this intersegmental jumping by the ETS-family transcription factor ETS variant 6 (ETV6). To isolate intersegmental target binding in a functionally defined manner, we pre-equilibrated ETV6 with excess salmon sperm DNA, a heterogeneous polymer, before exposing the nonspecifically bound protein to immobilized oligomeric DNA harboring a high-affinity ETV6 site. In this way, the mechanism of ETV6–target association could be toggled electrostatically through varying NaCl concentration in the bulk solution. Direct measurements of association and dissociation kinetics of the site-specific complex indicated that 1) freely diffusive binding by ETV6 proceeds through a nonspecific-like intermediate, 2) intersegmental jumping is rate-limited by dissociation from the nonspecific polymer, and 3) dissociation of the specific complex is independent of the history of complex formation. These results show that target searches by proteins with an ETS domain, such as ETV6, whose single DNA-binding domain cannot contact both source and destination sites simultaneously, are nonetheless strongly modulated by intersegmental jumping in heterogeneous site environments. Our findings establish biosensors as a general technique for directly and specifically measuring target site search by DNA-binding proteins via intersegmental translocation.
Keywords: DNA-binding protein, DNA-protein interaction, ETS transcription factor family, surface plasmon resonance (SPR), translocation
Introduction
A longstanding area of inquiry in protein–DNA interactions that continues to spark considerable interest is the mechanisms by which DNA-binding proteins with specific sequence preferences find their target sites. In experimental investigations of site-specific DNA-binding proteins typically involving short oligonucleotides harboring a cognate sequence, the protein finds its target site directly by free diffusion from bulk solvent. In the presence of excess nonspecific DNA, other modes of target search become available. Indeed, under genomic conditions, target search by DNA-binding proteins occurs by facilitated diffusion rather than direct binding from free diffusion (1, 2). Once associated with nonspecific DNA, a protein can scan the DNA sequence locally by sliding or hop discontinuously over short segments of DNA. Both mechanisms allow DNA-binding proteins to find their target sites much more rapidly in heterogeneous sequence environments than by free diffusion alone. Longer-range translocation between DNA segments mitigates unproductive local trapping of protein due to back-and-forth sliding and hopping without returning to bulk solvent. Intersegmental translocation is typically referred to as “transfer” for DNA-binding proteins that can bridge two DNA duplexes simultaneously (3–6), whereas proteins that present only a single binding surface execute a “jumping” mechanism between DNA segments (7).
Functionally, facilitated diffusion contributes to the processivity of DNA-modifying enzymes, such as restriction endonucleases (8–11), methyltransferases (12, 13), and repair enzymes (7, 9, 14). Beyond their biological significance, enzymes are attractive model systems of DNA translocation in ensemble studies because their DNA-binding behavior can be inferred from the apparent acceleration of their enzyme kinetics on substrates harboring multiple copies of target sites. Although transcription factors also perform target search by facilitated diffusion, their lack of enzymatic activities means that translocation must be measured directly (15–17). To this end, we have found biosensor–surface plasmon resonance (SPR)4 to be a useful technique for directly observing site-specific target search (18) and have been working on developing this technique to directly measure the effect of facilitated diffusion on site-specific recognition.
Inspired by knowledge that facilitated translocation by lac repressor to its operator site could be toggled from free diffusion via bulk salt concentration (19, 20), we devised an approach to directly measure the affinity and kinetics of DNA recognition by intersegmental jumping via biosensor–SPR. We have previously found that by including excess nonspecific DNA in the flow solution, a site-specific protein could be presented to immobilized DNA in a nonspecifically bound state (18), a condition that effectively mimicked intersegmental translocation, because the nonspecific site and specific site are not on the same strand. This setup mechanistically insulates intersegmental translocation from sliding and short-range hopping that cannot be excluded if the binding site is embedded within a long DNA fragment. We applied this method to study the thermodynamics and kinetics of transfer binding by the DNA-binding domain of the transcription factor ETV6. Details on how ETS-family proteins, such as ETV6, as well as other species bearing only a single DNA contact surface execute intersegmental translocation are poorly known. The quantitative salt dependence of binding in the presence and absence of nonspecific DNA confirmed that excess nonspecific DNA in the flow solution isolated the mechanism of site-specific binding to intersegmental jumping. Moreover, it was possible to toggle discretely between jumping and free diffusion to the immobilized specific site by varying the salt concentration of the flow solution. Kinetic measurements showed that dissociation from nonspecific DNA dominated the kinetics of site-specific association by intersegmental translocation, highlighting the potential for this mechanism to modulate target search by ETV6 and structurally homologous transcription factors in complex DNA environments.
Results
Site-specific binding by the ETS domain of ETV6 is strongly salt-sensitive
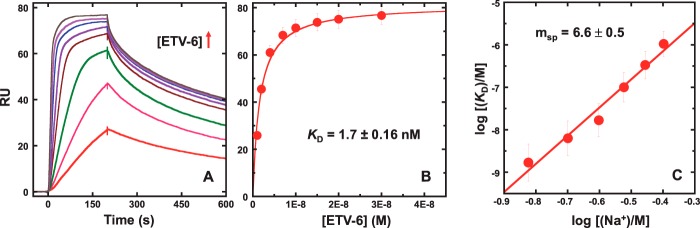
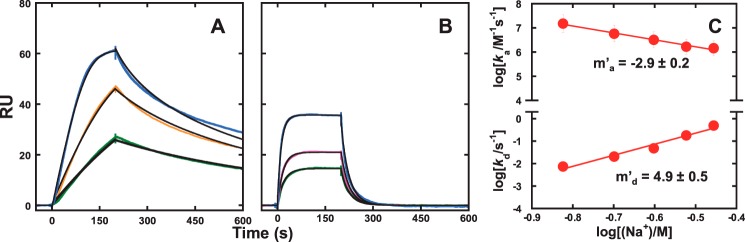
Cognate binding sites for ETS proteins consist of at least one helical turn of DNA harboring a central 5′-GGA(A/T)- 3′ core consensus sequence. In this study, we used biotinylated immobilized 23-bp hairpin oligomers harboring the sequence 5′-GCCGGAAGTG-3′ (DNAsp; Table 1), an established high-affinity site for ETV6 (21–24). Strong binding between the minimal DNA-binding (ETS) domain of ETV6 and DNAsp was observed under these conditions. The biosensor was exposed to graded concentrations of ETV6 for a sufficient period either to achieve apparent steady state or, at the lowest protein concentrations (≤4 nm), to predict the steady-state signal by kinetic analysis (Fig. 1A). In all cases, the observed SPR signal corresponds to the calculated value for a 1:1 complex (∼80 relative units (RU)) (25), and the data were analyzed with a 1:1 binding model (Fig. 1B) in accordance with the co-crystal structure (22). Our observed KD of 1.7 ± 0.2 nm at 0.15 m Na+ agreed quantitatively with published values obtained by electrophoretic mobility shift under similar solution conditions (24). Due to the strong binding at low salt concentrations, we bracketed the salt range between 0.15 and 0.4 m Na+ to remain within instrumental limits for kinetic and affinity determinations. Over this salt range, KD increased sharply to over 10−6 m, showing that the specific interaction was highly sensitive to salt concentration. The ionic coupling of KD was interpreted according to the polyelectrolyte theory (26),
| (Eq. 1) |
where Δm is the number of monovalent cations that are thermodynamically released (negative) upon complex formation and is related to the number of neutralized DNA backbone phosphates z by ψc = 0.76 (for counter-ion condensation) and ψs = 0.12 (a screening factor) for double-stranded B-form DNA. Analysis of ETV6–DNAsp binding to Equation 1 gave Δmsp = −(∂log KDsp/∂log[Na+]) = −6.6 ± 0.5 (Fig. 1C), corresponding to the neutralization of z = −Δm/ψs + ψc) = 7.5 ± 0.6 DNA backbone phosphates upon site-specific complex formation.
Table 1.
Immobilized DNA sequences used in this study
DNAsp is an established high-affinity binding affinity sequence for ETV6. SD1 and SD2 are two non-cognate sequences in which the 5′-GGAA-3′ core consensus has been isomerically mutated.
| DNA | Sequences |
|---|---|
| DNAsp | 5′-CGGCCAAGCCGGAAGTGAGTGCCC |
| 3′-GCCGGTTCGGCCTTCACTCACGGT | |
| SD1 | 5′-CGCAAAAGCTGAGATGGCGTGCCC |
| 3′-GCGTTTTCGACTCTACCGCACGGT | |
| SD2 | 5′-CCAAATAAAAGAGATGGCAACCAAGC |
| 3′-GGTTTATTTTCTCTAGGCTTGGTTCT |
Figure 1.
Quantitative analysis of site-specific DNA binding by the ETS domain of ETV6 by biosensor–SPR. A, sensorgrams of DNAsp binding by graded concentrations of ETV6 from 2 to 30 nm in the presence of 0.15 m Na+. B, nonlinear least-square fit of the steady-state data obtained in plateau regions of the sensorgrams by a 1:1 binding model. Parametric values are given in supplemental Table S1. C, salt dependence of the equilibrium dissociation constant of ETV6 with DNAsp, increasing sharply from 10−9 to 10−6 m over ∼3-fold Na+ concentrations. The slope is 6.6 ± 0.5. Error bars, S.E.
Nonspecific DNA binding by ETV6 is strongly differentiated between oligomeric and polymeric DNA
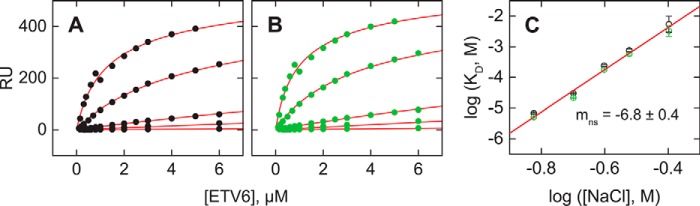
To define experimental conditions that achieve DNA target site search by intersegmental translocation, nonspecific DNA binding by ETV6 was measured by SPR at 0.15–0.4 m Na+. Initially, we examined two biotinylated immobilized nonspecific oligomers, termed SD1 and SD2, of equal length as DNAsp (23 bp) in which the cognate sequences had been scrambled (Table 1). Both oligomers bound ETV6 indistinguishably across the range of salt concentration tested, implying that they indeed exhibited nonspecific binding (Fig. 2, A and B). The data were analyzed according to the McGhee–von Hippel equation (see Equation 8 under “Experimental procedures”) to account for the effect of site exclusion in nonspecific binding to a DNA lattice. McIntosh and co-workers (22) previously analyzed the binding of the minimal ETS domain of ETV6 to a nonspecific 15-bp oligomer at 0.05 mm Na+, using Equation 8 without cooperativity (ω = 1), and determined a binding site size of 5.2 ± 0.1 bp, which we adopted here. Global analysis of the binding data for SD1 and SD2 with Equation 8 afforded an excellent fit that was not improved by allowing ω to float over fixing it at unity (Fisher's F-test on sums of squares, p = 0.05). The statistics therefore indicated that ETV6 also did not bind these nonspecific DNA oligomers with apparent cooperativity. Although the nonspecific binding affinities were 102-fold weaker than their site-specific counterparts at corresponding Na+ concentrations, the ionic coupling was quantitatively similar to specific binding with Δmns = −(∂log KDns/∂log[Na+]) = −6.8 ± 0.4 (Fig. 2C).
Figure 2.
Nonspecific DNA binding by the ETS domain of ETV6 to oligomeric DNA. Steady-state titration of immobilized 23-bp non-cognate sequences SD1 (A) and SD2 (B) with the ETS domain of ETV6 at 0.15, 0.20, 0.25, 0.30, and 0.4 m Na+ (from bottom o top). Curves represent the global fit of each data set to the McGhee–von Hippel equation for oligomeric DNA (Equation 8) with ω fixed at unity. Parametric values are given in supplemental Table S2. C, salt dependence of the equilibrium dissociation constant of ETV6 for SD1 (black) and SD2 (green). The line represents a linear fit of the aggregate data with a slope of 6.8 ± 0.4. Error bars, S.E.
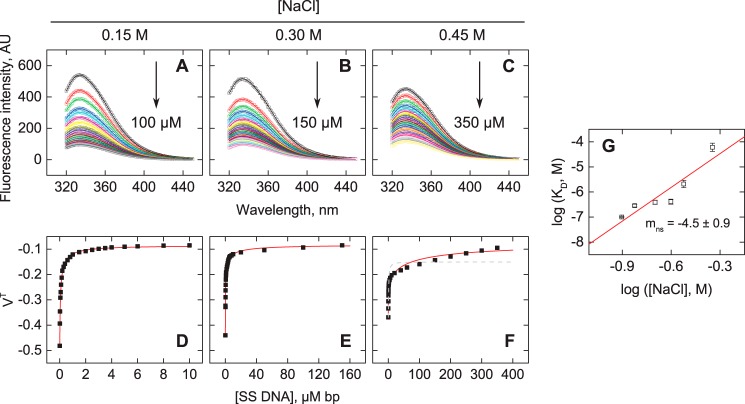
The apparent salt dependence of nonspecific binding to oligomeric DNA indicated a level of DNA backbone phosphate neutralization (∼8) near the limit for the apparent site size (∼10). The most likely explanation was that binding at or near the DNA ends or hairpin represented a different mode than binding at the interior of the oligomers. The apparent neutralization of 8 phosphates therefore represented the overall effect of different binding modes occurring to comparable extents on a relatively short nonspecific oligomer. We therefore investigated nonspecific binding by ETV6 to a polymeric DNA lattice, for which such end effects could be neglected. More precisely, we used salmon sperm DNA (SS DNA), a mixed-sequence polymer (average 2 kbp) that contained cognate ETS binding sites at statistically negligible frequencies, to titrate the intrinsic tryptophan fluorescence of the ETV6 ETS domain. We acquired fluorescence intensity spectra from 320 to 450 nm at a fixed concentration of ETV6 (200 nm) in the presence of graded concentrations of SS DNA (Fig. 3, A–C) and constructed binding profiles in which the lattice concentration N (in bp) was taken as independent variable (Fig. 3, D–F). The data were analyzed with the McGhee–von Hippel equation for an infinite lattice (see Equations 8–11 under “Experimental procedures”).
Figure 3.
Nonspecific binding of the ETS domain of ETV6 to polymeric DNA. Representative fluorescence emission spectra of ETV6 (A–C) in the absence (black) or presence of SS DNA (colored) up to the indicated concentrations in bp. Symbols and curves represent the observed intensity and reconstructed spectra using the first singular value following SVD decomposition. D–F, titration profiles corresponding to the first singular value (symbols). Curves represent a fit of the McGhee-von Hippel isotherm for infinite lattices to the data (Equation 12). Parametric values are given in supplemental Table S3. The gray curve in F is a fit with ω fixed at unity. G, salt dependence for the full set of polymeric nonspecific titrations from 0.125 to 0.45 m Na+. The slope is 4.5 ± 0.9. Error bars, S.E.
In contrast to nonspecific binding to oligomeric DNA, ETV6 binding to polymeric SS DNA under otherwise identical solution conditions was significantly stronger. The polymeric binding profiles also exhibited positive cooperativity (ω > 1) that increased with Na+ concentration. Qualitatively, von Hippel and co-workers (27) had pointed out that, with lattice as titrant, Equation 8 produces a characteristic biphasic behavior for positively cooperative systems. Specifically, rather than a tight lattice saturation, as when ligand is titrated, free ligand is depleted aggressively by lattice in a steep initial phase (with affinity ω/KD when ligand is in excess) until a point is reached where the residual ligand is bound much less tightly (with affinity 1/KD) by even a vast excess of lattice. Although ω values are known to be numerically challenging to estimate to high precision (28), particularly when titrated by lattice, this biphasic behavior was qualitatively manifest at 0.45 m Na+. We note that the final slope in the observed fluorescence (representing >30% of the total signal change) was not due to dilution (<10%) of the protein as SS DNA was added. An explicit two-dimensional analysis of the binding curve at 0.45 m, with both N and Lt (the nominal total ETV6 concentration after each addition of SS DNA) as independent variables, yielded a poor fit to the data with ω = 1 (Fig. 3F).
The polymeric nonspecific binding data gave a salt dependence, corresponding to z = 5.1 ± 1.0 (Fig. 3G), that was lower than that observed for the 23-bp oligomer and consistent with the nonspecific binding site size of ∼5 bp. Because this size was determined with oligomeric DNA (22), it was possible that it represented an average for binding modes occupying differently sized sites. As a check, refitting the data with a fixed site size of 10 bp (as per crystal structures) negligibly altered the estimates of KD and ω (<10%). Thus, nonspecific binding at or near ends was strongly differentiated electrostatically from binding at interior sites, an observation that highlights potential limitations to extrapolating features of nonspecific binding between oligomeric and polymeric DNA.
Configuring biosensor–SPR for probing intersegmental target search by ETV6
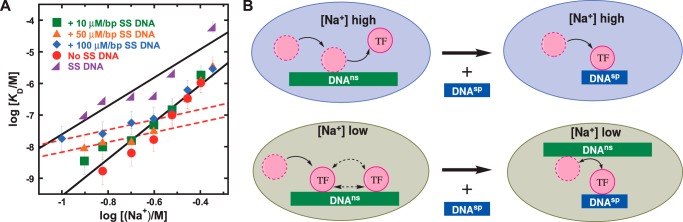
Given the significant end effects on nonspecific binding by ETV6, we chose excess SS DNA as the initial reservoir to model the intersegmental target search of ETV6 from a nonspecifically bound state. SS DNA at concentrations from 10 to 100 μm bp was added to the flow solution containing ETV6 30 min before exposure to immobilized DNAsp target. Under these conditions, ETV6 was quantitatively sequestered in nonspecifically bound complexes with the excess SS DNA prior to detection, and we observed that site-specific binding by ETV6 was modulated by bulk NaCl in an unusual manner (Fig. 4A). Specifically, at Na+ concentrations below ∼0.25 m, binding was attenuated at increasing concentrations of SS DNA; at 0.15 m Na+, binding was ∼10-fold weaker in the presence of 100 μm bp SS DNA (Table 2). Interestingly, the salt dependence of the DNAsp binding when SS DNA was present was lower than that in the absence of SS DNA. Above ∼0.25 m Na+, SS DNA at up to 100 μm had no apparent effect on site-specific binding relative to SS-free conditions. Thus, the addition of SS DNA induced two salt-dependent regimes in site-specific binding by ETV6: a low-salt regime, which exhibited reduced salt dependence, and a high-salt one that was indistinguishable from the absence of SS DNA. The transition between the two regimes depended on the (constant) concentration of SS DNA in the flow buffer but appeared to occur abruptly with respect to salt concentration.
Figure 4.
Bulk salt toggles intersegmental target search by the ETS domain of ETV6. The salt dependence of site-specific DNA binding (with and without SS DNA) and nonspecific binding by ETV6 was quantified by linear least-square analysis. Parametric values are given in supplemental Table S1. Data sets or subsets with statistically indistinguishable values were fitted globally. For site-specific binding, the dashed line describes log KD values at low salt concentrations in the presence of 50 and 100 μm SS DNA. Shown are ETV6–DNAsp without SS DNA (●), with 10 μm SS DNA (■), with 50 μm SS DNA (▴), and with 100 μm SS DNA (♦). Data from Fig. 3G are also given as SS DNA ( ). Limiting instrumental resolution prevented measurements of affinities at the very low Na+ concentrations required to define the low-salt regime for 10 μm SS DNA. B, schematic model of electrostatic toggle of target search by free diffusion at high salt and intersegmental jumping at low salt. Error bars, S.E.
). Limiting instrumental resolution prevented measurements of affinities at the very low Na+ concentrations required to define the low-salt regime for 10 μm SS DNA. B, schematic model of electrostatic toggle of target search by free diffusion at high salt and intersegmental jumping at low salt. Error bars, S.E.
Table 2.
Salt gradients of equilibrium dissociation constants for ETV6–DNA binding
The sensitivity of the apparent equilibrium dissociation constant for site-specific and nonspecific DNA binding by ETV6 was estimated by linear least-square analysis of the log–log gradients with respect to Na+ concentrations. ▴ and ◆ represent the data acquired in the presence of 50 and 100 μm SS DNA, respectively. The values at the reference state of 1 m Na+ were extrapolated from the linear fit. The nonspecific binding data for oligomeric DNA (SD1 and SD2) are omitted here for clarity (cf. Fig. 2).
| Experimental conditions | Slope = −Δm | z | KD at [Na+] = 0.15 m | Estimated KD at [Na+] = 1 m |
|---|---|---|---|---|
| m | m | |||
| ETV-6 + DNAsp | 6.6 ± 0.5 | 7–8 | (1.7 ± 0.2) × 10−9 | 3.2 × 10−4 |
| ETV-6 + SS DNA | 4.5 ± 0.9 | 5 | (0.29 ± 0.16) × 10−6 | 7.6 × 10−4 |
| ETV6 + DNAsp + SS DNA; [Na+] ≤ 0.25 m (dotted line) | 1.6 ± 0.5 (▴) | 2 | (2.0 ± 0.1) × 10−8 | 2.5 × 10−7 (▴) |
| 1.6 ± 0.3 (◆) | (2.5 ± 0.2) × 10−8 | 6.7 × 10−7 (◆) | ||
| ETV6 + DNAsp +/− SS DNA; [Na+] > 0.25 m (solid line) | 5.9 ± 0.4 | 6 | 2.0 × 10−4 |
To model the salt-dependent behavior of site-specific search in terms of intersegmental translocation, we first considered the site-specific and nonspecific DNA binding of ETV6 by free diffusion.
| (Eq. 2) |
| (Eq. 3) |
With only ETV6 in the flow solution, binding to immobilized cognate DNA represents direct site search by free diffusion as indicated by Equation 2. The inclusion of SS DNA in the flow solution establishes a pre-equilibrium of nonspecific DNA binding, as indicated by Equation 3, before biosensor exposure. The observed salt dependence of site-specific binding in the presence of SS DNA is obtained from subtracting Equation 3 from Equation 2,
| (Eq. 4) |
where Δm Δmsp − mns, and KDtr = KDsp/KDns. Based on the salt dependence of nonspecific ETV6 binding to polymeric SS DNA (not oligomeric SD1 or SD2), cognate binding in the presence of 50 and 100 μm SS DNA yielded an intermediate salt dependence between 0.15 and 0.25 m Na+, in close agreement with Equation 4: Δm = msp − mns = (6.6 ± 0.5) − (4.5 ± 0.9) = (2.1 ± 1.0) (Fig. 4 and Table 2). This agreement provided independent evidence that the mixed-sequence salmon sperm DNA indeed acted as a nonspecific polymer, and supported SPR-detected cognate binding in the presence of this polymer as a direct measurement of intersegmental target search at physiologically relevant Na+ concentrations.
Δmsp − mns, and KDtr = KDsp/KDns. Based on the salt dependence of nonspecific ETV6 binding to polymeric SS DNA (not oligomeric SD1 or SD2), cognate binding in the presence of 50 and 100 μm SS DNA yielded an intermediate salt dependence between 0.15 and 0.25 m Na+, in close agreement with Equation 4: Δm = msp − mns = (6.6 ± 0.5) − (4.5 ± 0.9) = (2.1 ± 1.0) (Fig. 4 and Table 2). This agreement provided independent evidence that the mixed-sequence salmon sperm DNA indeed acted as a nonspecific polymer, and supported SPR-detected cognate binding in the presence of this polymer as a direct measurement of intersegmental target search at physiologically relevant Na+ concentrations.
Biosensor–SPR reveals the kinetics of intersegmental jumping
In addition to affinity measurements, a strength of SPR is its real-time capabilities for kinetics measurements on the second-to-minute time scale. Because the excess SS DNA and the high flow rate used in the SPR experiments ensured that ETV6 was quantitatively trapped in nonspecific complexes when not interacting with the biosensor, as manifest through the sharpness with which the low- and high-salt affinity regimes were defined (cf. Fig. 4A), direct transfer by intersegmental translocation represented the only pathways to and from the immobilized oligomeric DNA (Fig. 4B). The observed kinetics of association and dissociation therefore represented direct indicators of ETV6 binding and unbinding by intersegmental translocation. Between 0.15 and 0.35 m Na+, the kinetics of site-specific association and dissociation of ETV6 in the absence of SS DNA fell within the temporal resolution of the Biacore instrument. In the second time scale, the observed kinetics were well described with a 1:1 model, as the representative sensorgrams for 0.15 and 0.25 m of Na+ in Fig. 5 (A and B). Both the association and dissociation rate constants yielded linear log-log salt gradients over 0.15–0.35 m Na+. Increasing Na+ concentrations reduced the apparent association rate constants and increased the dissociation rate constants (Fig. 5C). The higher sensitivity of dissociation to salt resulted in an overall drop in affinity. Empirically, the salt dependence of the rate and equilibrium constants are related as follows.
| (Eq. 5) |
Figure 5.
Kinetic characterization of site-specific ETV6–DNA binding via free diffusion by biosensor–SPR. Shown are representative sensorgrams for ETV6 (1, 2, and 4 nm ETV6, from bottom to top) with DNAsp at 0.15 m (A) and 0.25 m (B) Na+ in the absence of SS DNA. Black curves, kinetic fits by a 1:1 binding model with mass transfer correction. Parametric values are given in supplemental Table S1. C, salt dependence of the kinetic rate constants, where m′a and m′d are the log–log salt gradients of the association and dissociation rate constants, respectively. Error bars, S.E.
The equilibrium salt dependence inferred from the rate constants, −(2.9 ± 0.2) − (4.9 ± 0.5) = −7.8 ± 0.5, was close to the value obtained from steady-state measurements (−6.6 ± 0.5). Agreement was significantly improved if the 150 mm data, in which dissociation did not progress fully within the time frame of the experiment, were excluded. Nonetheless, the kinetic data clearly showed that the inhibitory effect of salt on ETV6–DNA binding by free diffusion was mediated through a prohibitive effect on association and an overall destabilizing effect on the formed complex.
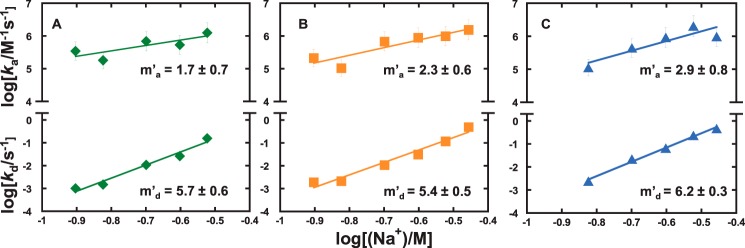
Direct measurement of the time-dependent association and dissociation of site-specific ETV6–DNA complex formation in the presence of SS DNA showed starkly contrasting kinetic profiles relative to target site binding by free diffusion. Specifically, the salt dependence of the association rate constant was positive at all concentrations of SS DNA tested, increasing in magnitude with abundance of SS DNA (Fig. 6). Thus, target site search by intersegmental translocation was kinetically promoted by salt. Previously, the dissociation rate constant was more sensitive to salt than that observed in free diffusion, but its magnitude exhibited no significant dependence on SS DNA abundance.
Figure 6.
Direct kinetic measurement of intersegmental translocation by the ETS domain of ETV6. The apparent rate constants for the apparent association to and dissociation from DNAsp at the indicated Na+ concentrations were determined in the presence of 10 μm (♦, A), 50 μm (■, B), and 100 μm SS DNA (▴, C). Lines represent linear fits of the log–log salt gradients with slopes m′a and m′d, respectively. Error bars, S.E.
Because the SPR-detected ETV6–DNA kinetics represented the overall progress of multiple microscopic steps, the rate constants and their salt dependence reflected the rate-limiting step in the association and dissociation mechanism. The dependence of the association rate constant on SS DNA concentration as well as the apparent uptake of Na+ upon binding therefore suggested that the rate-limiting step in intersegmental transfer to the target site involved the dissociation of nonspecifically bound DNA. The slow dissociation from polymeric nonspecific DNA contrasted sharply with the very fast dissociation (beyond instrumental dead time) from the oligomeric nonspecific sites SD1 and SD2 (supplemental Fig. S1). Because dissociation from the DNA oligomers, which were immobilized at low density, was limited to unbinding from the DNA, this difference reflected the sequestrative effect of additional translocation mechanisms (sliding, hopping, and transfer among duplexes) available to the protein on the polymeric SS DNA. This interpretation is also consistent with the higher apparent nonspecific affinity for polymeric versus oligomeric DNA (cf. Figs. 2 and 3). In summary, the kinetics of intersegmental site association are dominated by dissociation dynamics (Fig. 7).
Figure 7.
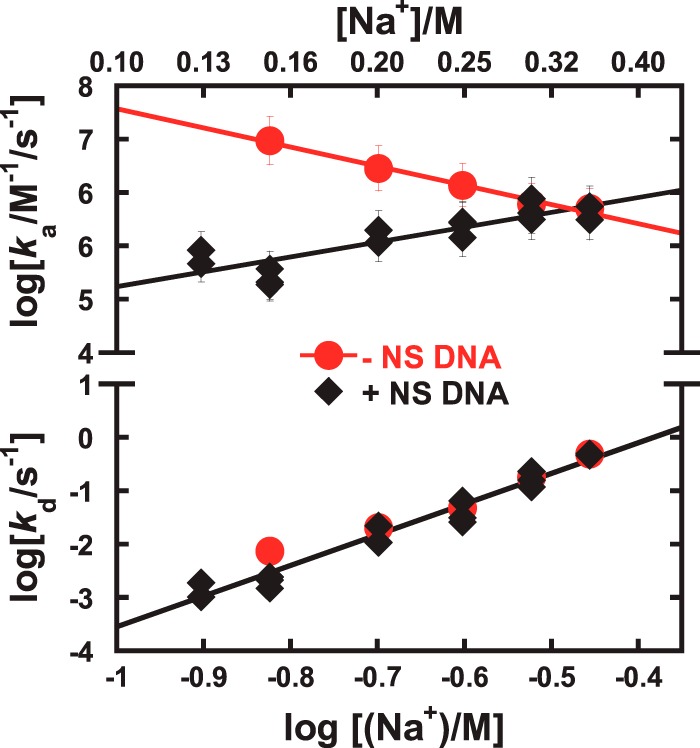
Kinetic summary of site-specific DNA binding by the ETS domain of ETV6 via free diffusion or intersegmental transfer as revealed by ionic perturbations. Comparison of the salt effect on the kinetics of ETV6–DNAsp binding in the absence (●) and presence of SS DNA (10, 50, and 100 μm, all shown as ♦). The data with or without SS DNA was globally fitted by linear least-square analysis. In the absence of SS DNA, the association rates decrease upon increasing Na+ concentration. In the presence of SS DNA, association rates increase upon increasing salt concentration. In contrast, there are no significant differences in the salt dependence of the dissociation rate constant with or without SS DNA. Error bars, S.E.
Discussion
To better understand ETV6–DNA interactions and, more generally, the structurally homologous family of ETS proteins, we have carried out a detailed analysis of the affinity and kinetics of site-specific binding by ETV6 by intersegmental translocation, as measured by biosensor–SPR, and perturbing the interaction with NaCl in the bulk solution. In the absence of nonspecific DNA, the affinity of ETV6 for site-specific DNA was strongly and monotonically sensitive to bulk NaCl concentrations typical of DNA-binding proteins that are capable of multiple charge–charge interactions (Fig. 1). Quantitatively, the log–log salt gradient of KD indicated zsp = 7.5 thermodynamically active phosphate interactions between ETV6 and the target DNA backbone, in good agreement with the ETV6–DNA co-crystal structure (22) (Fig. 8A), which shows five DNA phosphates that are directly neutralized by or hydrogen-bonded to the protein and three additional phosphates that are hydrogen-bonded to the protein via ordered bridging water molecules.
Figure 8.
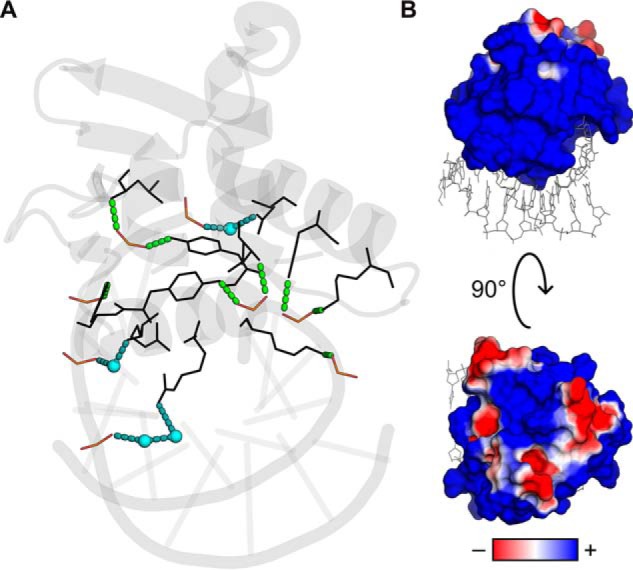
Electrostatic interactions in the site-specific ETV6–DNA complex. A, the co-crystal structure (Protein Data Bank entry 4MHG) was examined for polar and charged interactions emanating from DNA backbone phosphates. Direct contacts with protein backbone or side chains (5 phosphates) are shown as green dashes, all within 3.0 Å. Water-mediated contacts (3 phosphates) are shown as blue dashes (<3.3 Å) together with the bridging water molecules as spheres. To maximize clarity, only contacting DNA phosphates (red and orange) and protein residues (black) are shown explicitly as lines. B, surface electrostatic potential of the protein as computed from the Poisson–Boltzmann equation using APBS (39) using ionic parameters corresponding to aqueous solution containing 0.15 m NaCl.
To define the experimental conditions that isolate target search by ETV6 by intersegmental translocation, we equilibrated preset concentrations of protein and excess SS DNA before injecting the mixture over the sensor chip with immobilized cognate DNA (Fig. 4). Because the nonspecific and specific sites are not on the same strand, and protein is presented to a specific site in a nonspecific DNA-bound state, the experimental setup is designed to include only intersegmental translocation as a possible mechanism of binding to the immobilized target site. The results under these conditions are quite different from those in Fig. 1. At relatively low NaCl concentrations, binding of ETV6 to SS DNA results in a nonspecific complex with the release of ∼4 Na+ ions. In a coupled translocation step, analogous to a scheme proposed by Bresloff and Crothers (29) for ethidium binding to polymeric DNA, ETV6 dissociates from the nonspecific complex, with attendant rebinding of Na+ to the SS DNA, and binds to the cognate site with dissociation of Na+ ions characteristic of that interaction. The salt dependence of the equilibrium constant in the presence of SS DNA therefore represents the difference in Na+ ions released in forming the cognate DNA complex and recondensed on dissociation of the nonspecific complex, as indicated by Equation 4. At high salt concentrations, the gradient is quantitatively indistinguishable from that observed in the absence of SS DNA, indicating that ETV6 binds the immobilized specific site only by direct diffusion. This occurs if nonspecific destabilization by Na+ occurs to an extent such that ETV6 is quantitatively dissociated from the SS DNA in the flow solution. The sharpness of the SS DNA-induced transition as well as the constancy in the gradients of the two regimes indicate that the mechanism of site-specific binding changes in an essentially all-or-none manner when perturbed by Na+. Increasing concentrations of SS DNA moderately raised the salt threshold to revert site-specific binding to free diffusion but did not alter the salt dependence of the translocation regime (Fig. 4). This observation indicated that excess nonspecific polymer perturbed the apparent affinity of site-specific binding but not the structure of the site-specific complex itself.
Site-specific binding by ETV6 involves a kinetic intermediate at the cognate sequence
The real-time biosensor–SPR signal represents a direct probe of the kinetics of protein–DNA interactions on the second-to-minute time scale. The temporal range accessible to biosensor–SPR at fast time scales is bounded by the instrument's temporal resolution (<1 s) and the dead time required to inject a sample, flush the biosensor flow cell, and obtain a stable reading on the one hand. On the other hand, the measurement of very slow kinetics (∼10 min) is limited by the time required to obtain sufficient dissociation of the complex to fit the curve (preferably >50% but at least 20% dissociation). Within these bounds, biosensor–SPR offers advantages over competing techniques, such as solution NMR (15), in terms of signal sensitivity, compatible with heterogeneous DNA in the flow solution, sample volume requirements through its microfluidic architecture, and capability to multiplex several samples in a single experiment to maximize comparability.
In the absence of SS DNA, the salt gradient of the association rate constant was negative, and that of the dissociation rate constant was positive (Figs. 5 and 7). When free diffusion is the only available mechanism of site binding in a monovalent salt solution, Record and co-workers (30) and Lohman (31) established that, in the absence of a significant kinetic intermediate, the association kinetics would be governed by the weak screening effect of bulk monovalent ions.
| (Eq. 6) |
Deviation from Equation 6 would therefore indicate the presence of a kinetic intermediate en route to the final complex. From our binding data on ETV6–DNAsp binding in the absence of SS DNA, zsp = −(1/ψ)msp = −(1/ψ) × (dlog Kdsp/dlog[Na+]) = 7.5 (Fig. 1C) gives −ψszsp = −0.9 ± 0.1 ≠ m′a = −2.9 ± 0.2 (Fig. 5C). The observed association kinetic rate constant is therefore far more salt-sensitive than the screening-controlled mechanism requires. We therefore conclude that site-specific binding by ETV6 by direct diffusion involves a kinetic intermediate that appears to resemble the nonspecific complex. This interpretation is supported by the ionic properties of this intermediate (30),
| (Eq. 7) |
which coincides with zns = 3.8 ± 0.7 for nonspecific binding to SS DNA. The agreement between the predicted salt dependence of the dissociation rate constant of this intermediate, (dlog kd/dlog[Na+]) = ψ(zsp − zint) = 3.3 ± 0.8, and the observed value of m′a in the presence of SS DNA (2.9 ± 0.8) further bolsters this view.
The kinetics and thermodynamics of nonspecific DNA binding by ETV6
Although site-specific association by ETV6 by free diffusion is rapid, the observation of dissociation from the nonspecific SS DNA polymer as the rate-limiting step is not immediately expected, given the extremely rapid dissociation from immobilized nonspecific oligomers. The strong difference in the dissociation kinetics between polymeric and oligomeric nonspecific DNA must therefore reflect the additional translocation mechanisms available to the protein only when bound to polymeric DNA. More specifically, sliding and hopping represent kinetically favorable paths for the nonspecific ETV6–DNA complex relative to direct diffusion out of the DNA lattice. Thus, the apparent association rate constant by intersegmental translocation decreases detectably with increasing concentrations of SS DNA. The clear failure of oligomeric DNA to recapitulate nonspecific binding to polymeric DNA, as we have observed with ETV6, is an important aspect of nonspecific binding that has not received adequate attention. Several previous reports on intersegmental translocation from oligomeric DNA have found that the association rate constant is promoted by high DNA concentration (4, 15) and might have found rather different results had polymeric DNA been used as the nonspecific reservoir.
Thermodynamically, the emergence of positive cooperativity in nonspecific binding to SS DNA indicated that electrostatic interactions were operative not only at the protein–DNA interface (release of condensed counter-ions), but also in protein–protein interactions. At 0.45 m Na+, the cooperativity parameter was estimated to be on the order of ω ∼103, which is large but in line with other reported DNA-binding proteins, such as the T4 bacteriophage gene 32 protein under low-affinity conditions (27, 32). This positive cooperativity may arise from the screening of electrostatic repulsion that offsets favorable interactions as proteins become arrayed along the DNA lattice. This notion is plausible in view of several clusters of charged surfaces on ETV6 in addition to the strongly cationic DNA-binding surface (Fig. 8B). Coupled conformational changes that unpair salt bridges or otherwise expose uncompensated charges may also play a role.
Biological relevance of intersegmental translocation for proteins with a single DNA-binding interface
At concentrations of ETV6 examined (up to 10−7 m), ETV6 bound specific DNA strictly as a monomer. The significant affinity and contrasting kinetic differences in site-specific ETV6–DNA binding by free diffusion versus intersegmental jumping are therefore probably representative of DNA-binding proteins harboring a single DNA-binding interface. Specifically, the rate-limiting role of dissociation from the nonspecifically bound state agrees with the physical intuition that, unlike oligomeric proteins with multiple DNA-binding surfaces, ETV6 must dissociate first from one DNA duplex before engaging the next. However, as the present data show, the transfer mechanism asserts itself strongly in the effective affinity and kinetics of association under conditions that functionally mimic the DNA-dense environment encountered in the genome. That physiological salt conditions (equivalent to 0.15 m Na+) lie firmly in the translocation regime further underscores the biological relevance of the modulatory potential of intersegmental jumping in the case of single-interface proteins. The slower association kinetics when intersegmental jumping is the only binding mechanism suggests that net acceleration in target search in vivo must receive compensatory contributions from short-range sliding and hopping that were purposefully excluded from the short site-specific target used in the present biosensor–SPR configuration.
In the presence of excess SS DNA, the positive salt gradient in the site-specific association rate constant of ETV6 bears similarity to the operator-binding properties of lac repressor in the presence of excess non-operator phage DNA. Like ETV6, low salt concentrations also promote the association rate constant for lac repressor–operator binding (19). However, in the context of an operator site embedded in heterogeneous phage DNA on the same strand, short-distance sliding and hopping mechanisms dominate (20). As a result, whereas facilitated diffusion by lac repressor was kinetically competitive with direct binding under physiologic conditions (<4-fold difference in apparent ka at 0.15 m Na+ despite a 107-fold higher affinity for operator binding) (19), site-specific ETV6 association by intersegmental jumping was ∼100 times slower than direct diffusion at the same Na+ concentration (cf. Figs. 5 and 6), even though KDns/KDsp = 102.
In conclusion, using biosensor–SPR, we showed that two distinct mechanisms of target search, namely free diffusion and intersegmental translocation, could be directly probed and toggled with salt in the presence of excess nonspecific polymeric DNA. This represents a broadly applicable approach for interrogating DNA-binding proteins, such as transcription factors that lack enzymatic activities, as experimental probes of DNA localization. Importantly, even for monomeric proteins, such as ETV6, harboring only a single DNA contact interface for both specific and nonspecific interactions, intersegmental jumping represents a major molecular event that significantly modifies the thermodynamics and kinetics of DNA target search. The need for and utility of facile experimental avenues for probing intersegmental translocation in a defined, isolated manner is apparent.
Experimental procedures
Nucleic acids
DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) and annealed to form duplex binding sites as described previously (18). DNA purity was routinely verified by the manufacturer by electrospray ionization–mass spectrometry. Salmon sperm DNA was purchased from Sigma-Aldrich (D1626) and used without further purification. A full-length clone of human ETV6 (HsCD00338954) was purchased from the Harvard Plasmid Repository.
Molecular cloning
A construct encoding the minimal DNA-binding (ETS) domain of ETV6 (human residues 331–426) was amplified from a full-length clone in pCMV-SPORT6 by PCR and ligated into the NcoI/XhoI sites of pET28b using our reported cloning strategy described previously (33). The cloned construct consists of the open reading frame for the ETV6 ETS domain plus additional sequences encoding a C-terminal thrombin cleavage site followed by a His6 tag. The recombinant plasmid was transformed into DH5α Escherichia coli and sequence-verified by Sanger sequencing.
Protein expression and purification
Recombinant pET28b-ETV6 plasmid was propagated in BL21*(DE3) E. coli and cultured as described previously (34, 35). In brief, cells induced for 4 h with 0.5 mm isopropyl β-d-1-thiogalactopyranoside were harvested by centrifugation, lysed by sonication, and partially purified by immobilized metal affinity chromatography on cobalt-nitrilotriacetic acid resin. The C-terminal His6 tag was cleaved with thrombin overnight at room temperature and polished by cation exchange chromatography on Sepharose SP (GE Healthcare). All buffers used during purification contained 0.5 mm TCEP to maintain a lone cysteine. Following purification, the protein was stored at ∼100 μm in single-use (20-μl) aliquots at −20 °C. Protein concentrations were determined by UV absorption at 280 nm using an extinction coefficient of 25,440 m−1 cm−1.
Biosensor–SPR
SPR measurements were performed with a 4-channel Biacore T200 biosensor with a 4-channel system. 5′-Biotinylated DNA sequences of interest were immobilized on CM4 chips on flow cells 2–4 at low density (∼150 RU). Flow cell 1 was used as a reference cell. The experimental buffer was 25 mm Na2HPO4/NaH2PO4, pH 7.4, containing 1 mm EDTA, 1 mm freshly dissolved dithiothreitol, 0.05% P20 surfactant, and additional NaCl to achieve the required total Na+ concentration. Flow rate was maintained at 50 μl/min to minimize mass transfer between bulk and local environments on the sensor chip surface. Reference-subtracted sensorgrams for site-specific DNA binding were analyzed based on a 1:1 binding model to extract apparent association and dissociation kinetic rate constants (ka and kd) from time-dependent data or apparent equilibrium constants KD from steady-state data as described previously (18, 36). Nonspecific binding was analyzed using the McGhee–von Hippel equation,
| (Eq. 8) |
where
| (Eq. 9) |
| (Eq. 10) |
| (Eq. 11) |
where KDns is the intrinsic nonspecific equilibrium dissociation constant, s is the binding site size (in bp), and ω is the cooperativity parameter (ratio of the equilibrium constants for isolated and juxtaposed sites). The concentration of ETV6-bound DNA as observed is related to the binding density vns in Equation 8 by [DNA]b = svns (27). For nonspecific binding to oligomeric DNA (of length l in bp), Equation 8 is extended by Record and co-workers (37) as follows.
| (Eq. 12) |
Equilibrium fluorescence titrations
The quenching of the intrinsic tryptophan fluorescence of ETV6 upon binding to salmon sperm DNA was measured with a Cary Eclipse instrument (Agilent) in the same buffer as the SPR experiments. Both emission and excitation slits were set to 10 nm. Fluorescence were measured in a starting volume of 1 ml at an initial ETV6 concentration of 200 nm. The sample was excited at 286 nm, and the fluorescence spectrum was recorded from 320 to 450 nm. The concentration of salmon sperm DNA needed at each step was calculated and was corrected for dilution. Each spectrum was recorded in triplicate, averaged, and subtracted from the reference scan of buffer alone. To maximize the spectral content in our analysis, we performed singular value decomposition (SVD) on each set of spectra. The spectra at each Na+ concentration were encoded as column vectors a of a matrix A and subjected to SVD using Mathematica (Wolfram Research), as described previously (38). In brief, SVD of A yielded a product of three matrices, U, S, and V,
| (Eq. 13) |
where S is a diagonal matrix with ordered (from largest to smallest) singular values. The spectrum corresponding to each SS DNA concentration was reconstructed using the largest singular value λ1 by the following.
| (Eq. 14) |
Binding isotherms were generated by mapping the elements in ViT to the SS DNA concentrations used in the titration experiment. The fractional DNA-bound ETV6 (Fb) was related to the observed signal S by the following,
| (Eq. 15) |
where Smax and Smin are the signal intensities corresponding to the estimated saturated and unbound states. For nonspecific binding, the McGhee–von Hippel equation and its modifications were solved numerically by the Newton–Raphson method or via an optimized equation solver from the NAG library (c05sdc) within the Origin C programming environment (Originlab, Northampton, MA). Parametric estimates are given ± S.E. following nonlinear least-square analysis.
Author contributions
T. V. and S. W. conducted the SPR and fluorescence titration experiments. T. V., G. M. K. P., and W. D. W. analyzed the results. G. M. K. P. and W. D. W. conceived the idea for the project and wrote the paper with TV. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by National Science Foundation Grant MCB 15451600 (to G. M. K. P.) and National Institutes of Health Grant GM 111749 (to W. D. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1–S3 and Fig. S1.
- SPR
- surface plasmon resonance
- RU
- relative units
- SVD
- singular value decomposition
- SS
- salmon sperm.
References
- 1. Berg O. G., Winter R. B., and von Hippel P. H. (1981) Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry 20, 6929–6948 [DOI] [PubMed] [Google Scholar]
- 2. Halford S. E., and Marko J. F. (2004) How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 32, 3040–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doucleff M., and Clore G. M. (2008) Global jumping and domain-specific intersegment transfer between DNA cognate sites of the multidomain transcription factor Oct-1. Proc. Natl. Acad. Sci. U.S.A. 105, 13871–13876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lieberman B. A., and Nordeen S. K. (1997) DNA intersegment transfer, how steroid receptors search for a target site. J. Biol. Chem. 272, 1061–1068 [DOI] [PubMed] [Google Scholar]
- 5. Fried M. G., and Crothers D. M. (1984) Kinetics and mechanism in the reaction of gene regulatory proteins with DNA. J. Mol. Biol. 172, 263–282 [DOI] [PubMed] [Google Scholar]
- 6. Ruusala T., and Crothers D. M. (1992) Sliding and intermolecular transfer of the lac repressor: kinetic perturbation of a reaction intermediate by a distant DNA sequence. Proc. Natl. Acad. Sci. U.S.A. 89, 4903–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hedglin M., Zhang Y., and O'Brien P. J. (2013) Isolating contributions from intersegmental transfer to DNA searching by alkyladenine DNA glycosylase. J. Biol. Chem. 288, 24550–24559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jeltsch A., Alves J., Wolfes H., Maass G., and Pingoud A. (1994) Pausing of the restriction endonuclease EcoRI during linear diffusion on DNA. Biochemistry 33, 10215–10219 [DOI] [PubMed] [Google Scholar]
- 9. Terry B. J., Jack W. E., and Modrich P. (1985) Facilitated diffusion during catalysis by EcoRI endonuclease: nonspecific interactions in EcoRI catalysis. J. Biol. Chem. 260, 13130–13137 [PubMed] [Google Scholar]
- 10. Rau D. C., and Sidorova N. Y. (2010) Diffusion of the restriction nuclease EcoRI along DNA. J. Mol. Biol. 395, 408–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halford S. E. (2001) Hopping, jumping and looping by restriction enzymes. Biochem. Soc. Trans. 29, 363–374 [DOI] [PubMed] [Google Scholar]
- 12. Pollak A. J., Chin A. T., Brown F. L. H., and Reich N. O. (2014) DNA looping provides for “intersegmental hopping” by proteins: a mechanism for long-range site localization. J. Mol. Biol. 426, 3539–3552 [DOI] [PubMed] [Google Scholar]
- 13. Pollak A. J., and Reich N. O. (2015) DNA adenine methyltransferase facilitated diffusion is enhanced by protein-DNA “roadblock” complexes that induce DNA looping. Biochemistry 54, 2181–2192 [DOI] [PubMed] [Google Scholar]
- 14. Hedglin M., and O'Brien P. J. (2008) Human alkyladenine DNA glycosylase employs a processive search for DNA damage. Biochemistry 47, 11434–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iwahara J., and Clore G. M. (2006) Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J. Am. Chem. Soc. 128, 404–405 [DOI] [PubMed] [Google Scholar]
- 16. Kozlov A. G., and Lohman T. M. (2002) Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochemistry 41, 6032–6044 [DOI] [PubMed] [Google Scholar]
- 17. Kemme C. A., Esadze A., and Iwahara J. (2015) Influence of quasi-specific sites on kinetics of target DNA search by a sequence-specific DNA-binding protein. Biochemistry 54, 6684–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Munde M., Poon G. M., and Wilson W. D. (2013) Probing the electrostatics and pharmacological modulation of sequence-specific binding by the DNA-binding domain of the ETS family transcription factor PU.1: a binding affinity and kinetics investigation. J. Mol. Biol. 425, 1655–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barkley M. D. (1981) Salt dependence of the kinetics of the lac repressor-operator interaction: role of nonoperator deoxyribonucleic acid (DNA) in the association reaction. Biochemistry 20, 3833–3842 [DOI] [PubMed] [Google Scholar]
- 20. Berg O. G., and Blomberg C. (1978) Association kinetics with coupled diffusion III. Ionic-strength dependence of the lac repressor-operator association. Biophys. Chem. 8, 271–280 [DOI] [PubMed] [Google Scholar]
- 21. Coyne H. J. 3rd, De S., Okon M., Green S. M., Bhachech N., Graves B. J., and McIntosh L. P. (2012) Autoinhibition of ETV6 (TEL) DNA binding: appended helices sterically block the ETS domain. J. Mol. Biol. 421, 67–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De S., Chan A. C., Coyne H. J. 3rd, Bhachech N., Hermsdorf U., Okon M., Murphy M. E., Graves B. J., and McIntosh L. P. (2014) Steric mechanism of auto-inhibitory regulation of specific and non-specific DNA binding by the ETS transcriptional repressor ETV6. J. Mol. Biol. 426, 1390–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De S., Okon M., Graves B. J., and McIntosh L. P. (2016) Autoinhibition of ETV6 DNA binding is established by the stability of its inhibitory helix. J. Mol. Biol. 428, 1515–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Green S. M., Coyne H. J. 3rd, McIntosh L. P., and Graves B. J. (2010) DNA binding by the ETS protein TEL (ETV6) is regulated by autoinhibition and self-association. J. Biol. Chem. 285, 18496–18504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davis T. M., and Wilson W. D. (2000) Determination of the refractive index increments of small molecules for correction of surface plasmon resonance data. Anal. Biochem. 284, 348–353 [DOI] [PubMed] [Google Scholar]
- 26. Record M. T. Jr., Anderson C. F., and Lohman T. M. (1978) Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q. Rev. Biophys. 11, 103–178 [DOI] [PubMed] [Google Scholar]
- 27. Kowalczykowski S. C., Paul L. S., Lonberg N., Newport J. W., McSwiggen J. A., and von Hippel P. H. (1986) Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry 25, 1226–1240 [DOI] [PubMed] [Google Scholar]
- 28. Alma N. C., Harmsen B. J., de Jong E. A., Ven J., and Hilbers C. W. (1983) Fluorescence studies of the complex formation between the gene 5 protein of bacteriophage M13 and polynucleotides. J. Mol. Biol. 163, 47–62 [DOI] [PubMed] [Google Scholar]
- 29. Bresloff J. L., and Crothers D. M. (1975) DNA-ethidium reaction kinetics: demonstration of direct ligand transfer between DNA binding sites. J. Mol. Biol. 95, 103–123 [DOI] [PubMed] [Google Scholar]
- 30. Lohman T. M., DeHaseth P. L., and Record M. T. Jr. (1978) Analysis of ion concentration effects of the kinetics of protein-nucleic acid interactions. Application to lac repressor-operator interactions. Biophys. Chem. 8, 281–294 [DOI] [PubMed] [Google Scholar]
- 31. Lohman T. M. (1986) Kinetics of protein-nucleic acid interactions: use of salt effects to probe mechanisms of interaction. CRC Crit. Rev. Biochem. 19, 191–245 [DOI] [PubMed] [Google Scholar]
- 32. Kowalczykowski S. C., Lonberg N., Newport J. W., and von Hippel P. H. (1981) Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. J. Mol. Biol. 145, 75–104 [DOI] [PubMed] [Google Scholar]
- 33. Wang S., Linde M. H., Munde M., Carvalho V. D., Wilson W. D., and Poon G. M. (2014) Mechanistic heterogeneity in site recognition by the structurally homologous DNA-binding domains of the ETS family transcription factors Ets-1 and PU.1. J. Biol. Chem. 289, 21605–21616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stephens D. C., Kim H. M., Kumar A., Farahat A. A., Boykin D. W., and Poon G. M. K. (2016) Pharmacologic efficacy of PU.1 inhibition by heterocyclic dications: a mechanistic analysis. Nucleic Acids Res. 44, 4005–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stephens D. C., and Poon G. M. (2016) Differential sensitivity to methylated DNA by ETS-family transcription factors is intrinsically encoded in their DNA-binding domains. Nucleic Acids Res. 44, 8671–8681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang S., Poon G. M. K., and Wilson W. D. (2015) Quantitative investigation of protein–nucleic acid interactions by biosensor surface plasmon resonance. in DNA-Protein Interactions (Leblanc B. P., and Rodrigue S., eds) pp. 313–332, Springer, New York: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsodikov O. V., Holbrook J. A., Shkel I. A., and Record M. T. Jr. (2001) Analytic binding isotherms describing competitive interactions of a protein ligand with specific and nonspecific sites on the same DNA oligomer. Biophys. J. 81, 1960–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poon G. M., Abu-Ghazalah R. M., and Macgregor R. B. Jr. (2004) Ionic mobilities of duplex and frayed wire DNA in discontinuous buffer electrophoresis: evidence of interactions with amino acids. Biochemistry 43, 16337–16347 [DOI] [PubMed] [Google Scholar]
- 39. Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.