SUMMARY
Transcription is a stochastic process occurring mostly in episodic bursts. Although the local chromatin environment is known to influence the bursting behavior on long timescales, the impact of transcription factors (TFs)—especially in rapidly inducible systems—is largely unknown. Using fluorescence in situ hybridization and computational models, we quantified the transcriptional activity of the proto-oncogene c-Fos with single mRNA accuracy at individual endogenous alleles. We showed that, during MAPK induction, the TF concentration modulates the burst frequency of c-Fos, whereas other bursting parameters remain mostly unchanged. By using synthetic TFs with TALE DNA-binding domains, we systematically altered different aspects of these bursts. Specifically, we linked the polymerase initiation frequency to the strength of the transactivation domain and the burst duration to the TF lifetime on the promoter. Our results show how TFs and promoter binding domains collectively act to regulate different bursting parameters, offering a vast, evolutionarily tunable regulatory range for individual genes.
INTRODUCTION
Major leaps in our understanding of transcription have been achieved by studying gene expression at the single-cell level (Raj and van Oudenaarden, 2008). It is now well established that transcription is inherently stochastic and occurs predominantly as an episodic process, characterized by pulsatile bursts (see Supplemental Results for terminology describing bursting) of mRNA production (Sanchez and Golding, 2013). Yet, there is no universal law of gene expression (Larson, 2011), and each gene has its own kinetic parameters (Suter et al., 2011a). The kinetic properties of these bursts can be major regulators of the transcriptional response (Molina et al., 2013). Bursting control has been attributed mainly to chromatin accessibility and modifications (Sanchez and Golding, 2013), and only a few studies establish quantitatively the link between upstream transcription factors (TFs) and downstream transcriptional bursts (Larson, 2011; Larson et al., 2013; Neuert et al., 2013).
To investigate how TFs modulate bursts, we studied the transcriptional response of the early response gene c-Fos. Its promoter is enriched with paused RNA Polymerase II on the transcription start site resulting in a nucleosome-deprived, regulatory-factor-accessible state (Adelman and Lis, 2012). Further, it shows constant level of histone acetylation independently of gene activation (Fowler et al., 2011). Together, this yields a constitutively permissive and open promoter structure (Healy et al., 2013). Therefore, c-Fos is ideally suited to investigate the molecular origin of bursting independently from chromatin accessibility. Like other early response genes, c-Fos reacts to several different stimuli. Two of the major activation pathways acting on the serum response element (SRE) of its promoter are serum induction (Galbraith and Espinosa, 2011) and heavy metal exposure (Murata et al., 1999). Both stimuli activate the MAPK pathway and lead to rapid nuclear accumulation of phosphorylated kinases, i.e., the TF (predominantly ERK for serum and p38 for zinc induction) (Yang et al., 2013). Both TFs induce c-Fos transcription by phosphorylating Elk1 bound to the SRE (Galbraith and Espinosa, 2011). Despite this similarity, vastly different kinetic signatures of c-Fos transcription have been reported for the timing and amount of produced mRNA (Costa et al., 2006; Murata et al., 1999). By studying these activation pathways, we not only investigate the impact of different TFs on c-Fos transcription, but also address potential conserved, fundamental activation mechanisms.
We used single molecule fluorescence in situ hybridization (smFISH) (Femino et al., 1998; Haimovich et al., 2013; Mouaikel et al., 2013; Raj et al., 2008; Zenklusen et al., 2008) to count the nascent and mature c-Fos mRNA with single-transcript sensitivity (Mueller et al., 2013) and modeling to further deepen our understanding of c-Fos transcription dynamics (Munsky et al., 2012; Raj et al., 2006; Zenklusen et al., 2008). We show that c-Fos transcription occurs in bursts in which frequency is modulated by the nuclear TF concentration. Stimulation with synthetic TFs further suggests that other bursting parameters such as burst duration and polymerase initiation frequency can also be altered. Taken together, our study illustrates that TFs can be a major determinant for modulating transcriptional bursts in human cells.
RESULTS
Adapted Transcriptional Response of c-Fos after Different MAPK Activation Stimuli
We used the smFISH approach pioneered by the Singer lab (Femino et al., 1998) to quantify c-Fos mature and nascent mRNA number in individual cells (Mueller et al., 2013) (Figures 1A and S1A; Movie S1). We validated our experimental system by comparison with an alternative smFISH approach (Raj et al., 2008) and quantitative reverse transcription polymerase chain reaction (qRT-PCR) measurements (see Supplemental Results and Figures S1A–S1I).
Figure 1. Quantification of c-Fos Transcriptional Response after Different Stimuli.
(A) smFISH in U2OS 30 min after serum induction. Signal in proximity of the transcription sites (TSs) appears only saturated due to scaling to show individual mature mRNA. Scale bars, 10 μm in all figures. Surface plot (not to scale) for area indicated with red dashed line. Detected mature mRNAs shown as green spots over DAPI image.
(B) Average mature mRNA levels at different time points after serum induction (left) and zinc concentration (right) by smFISH (red line) and qRT-PCR (blue line). Error bars are 95% CI obtained by bootstrap for smFISH and SD for qRT-PCR (three independent experiments).
(C) Selected histograms of smFISH measurement from (B). Cells containing less than 20 mRNAs are show in orange and other cells in green.
(D) Number of active TS after serum (left) or zinc induction (right). Cells containing no active TS are not shown. Inset shows average number of active TS per cell.
(E) Immunofluorescence (IF) against p-ERK or p-p38 (red) for indicated induction condition and nuclei visualized with DAPI (blue). Note that only one cell has elevated p-p38 levels in the 50 μM picture.
(F) Average p-ERK levels (red) and average number of active TS per cell (blue line) after serum induction.
(G) Proportion of cells with elevated p-p38 signal (red line) and average number of active TS per cell (blue line).
We measured c-Fos transcription following different MAPK activation pathways. First, we activated c-Fos in serum-starved human osteosarchoma (U2OS) cells by addition of fetal bovine serum (FBS). In agreement with published results (Shah and Tyagi, 2013), mature mRNA levels increased rapidly to reach a maximum after 30 min and returned to basal expression after 2 hr (Figure 1B). Quantification with qRT-PCR showed similar induction kinetics, further validating our smFISH approach (Figure 1B). The single transcript sensitivity of smFISH allowed us to investigate the cell-to-cell variability (Figure 1C). Noninduced cells expressed on average only four mRNAs, and no cell contained more than 30 mRNAs. After 30 min of serum induction, cells contained an average of 90 mRNAs with large variability (some cells had only a few mRNAs, whereas others contained a few hundred). Our results in Normal Human Dermal Fibroblast (NHDF) cells illustrate similar variability in primary cells (Figures S1J and S1K).
Second, we activated c-Fos by exposing cells for 4 hr to different concentrations of the heavy metal zinc (ZnSO4). In agreement with a previous study (Murata et al., 1999), we observed higher average c-Fos mRNA levels with increasing zinc concentration (Figure 1B). smFISH revealed the presence of two distinct populations: uninduced cells with fewer than 20 mRNAs and strongly induced cells with up to 600 mRNAs (Figure 1C). The transcriptional response appears to be switch-like, in which a few strongly induced cells appear at zinc levels as low as 50 μM, and higher zinc levels increase the number of activated cells. Importantly, these rarely activated cells at low zinc levels cannot be detected from averaged mRNA levels (Figure 1B) (see Supplemental Discussion for potential implications of these rare events).
c-Fos Burst Frequency Correlates with Nuclear TF Concentration
Recent studies showed that key regulatory steps for different genes are the modulation of burst frequency and burst amplitude (Dar et al., 2012; Singh et al., 2010; Suter et al., 2011b). Because we cannot directly measure burst frequency using smFISH, we used the measured number of active TS in the cell population as readout for burst frequency. We detected up to four active c-Fos TSs per cell (Figure 1D), which is caused by the polyploidy of U2OS due to high levels of chromosome fragmentation (Pontén and Saksela, 1967). Serum starved cells had no detectable active TSs (Figure 1D). The number of active sites and average number of mature mRNAs per cell both increased substantially after addition of serum or zinc (Figures 1B and 1D). This suggests that cells adjust mature mRNA levels by activating more TSs (frequency modulation). We also observed large variability in the number of active TSs (Figures 1D and S1L), which likely accounts for the observed variability in mature mRNA levels (Figures 1C and S1J).
We next explored the molecular mechanism behind burst frequency regulation. Addition of serum or zinc led to the nuclear translocation of the phosphorylated kinases ERK1/2 (p-ERK) or p38 (p-p38), respectively. We measured their nuclear concentration by immunofluorescence (IF) and their correlation with c-Fos transcription. First, we quantified p-ERK levels after serum induction (Figure 1E). In agreement with previous reports (Costa et al., 2006; Galbraith and Espinosa, 2011), p-ERK rapidly accumulated in the nucleus and reached a maximum after 10 min (Figure 1F) and then decreased to a near-basal level after 1 hr. Strikingly, the average number of active TSs per cell showed similar kinetics, lagging approximately 10 min behind (Figure 1F). This correlation is consistent with the molecular observation that p-ERK activates c-Fos transcription by interacting on its SRE with ETS-domain family proteins, such as Elk-1 (Galbraith and Espinosa, 2011). To further test this link, we treated cells with tetradecanoylphorbol acetate (TPA), which strongly activates c-Fos via the ERK pathway (Lee et al., 2002). We found that TPA treatment yielded prolonged periods of p-ERK presence in the nucleus as well as continued activation of c-Fos (see Supplemental Results and Figure S1M). Although p-ERK may not be the only regulator of c-Fos, these results validated it as a reliable indicator of c-Fos activation during serum response
After zinc exposure (Figure 1E), only a fraction of the cells displayed elevated p-p38 levels, and the size of that fraction increased with zinc concentration (Figure 1G), as did the number of active TSs per cell. To directly link p-p38 levels with c-Fos bursting, we performed simultaneous IF and smFISH measurements for 100 and 150 μM zinc induction (data not shown; Figure 2A). We found that p-p38 levels were positively correlated with both active TS numbers (Figure 2B) and mature mRNA levels (Figures 2C and 2D, Pearson’s correlation coefficient of 0.51). These results support a model where higher TF levels lead to higher burst frequencies, resulting in more active TSs and higher mature mRNA levels.
Figure 2. MAPK Phosphorylation Level Controls the Burst Frequency.
(A) smFISH against c-Fos (green) combined with IF against p-p38 (red) in U2OS cells 4 hr after induction with 150 μM of zinc. Nuclei are stained with DAPI (blue).
(B) Average number of active TS per cell increases with the average p-p38 level. Cells were pooled in five bins based on their p-p38 levels containing the same number of cells as indicated with black bars in (C) and (E).
(C) Mature mRNA number as a function of p-p38 levels (Pearson’s correlation coefficient of 0.51).
(D) Histogram of mature mRNA data shown with same bins as used in (B).
(E and F) Nascent mRNA number per active TS plotted as function of p-p38 levels (Pearson’s correlation coefficient of −0.02). (F) Same binning as in (B).
All error bars are SEM. See also Figure S2.
Measured Burst Amplitude Is Largely Independent of Activation Condition
Several studies have shown that cells not only modulate the burst frequency but also the burst amplitude (Dar et al., 2012; Skupsky et al., 2010; Suter et al., 2011b). This amplitude depends on the burst duration and the polymerase initiation rate. However, the time an mRNA spends at the TS (retention time, which depends upon the elongation and processing rates and the gene length) determines what we can measure with smFISH. If this time is significantly longer than the burst duration, we can directly observe the burst amplitude. In contrast, if the retention period is shorter than the burst duration, we will only observe the fraction of mRNAs that is currently attached to the TS. We refer to this case as “burst saturation.” We use “measured amplitude” to refer to the amount of nascent mRNAs quantified at the TS, which can be different from the true burst amplitude. Noting that smFISH estimates an equivalent number of fully elongated transcripts, which can be different than the actual polymerase number (Little et al., 2013; Mueller et al., 2013), we analyzed the nascent mRNA data with and without numerical corrections for partial transcripts (for further details, see Supplemental Results and Figure S2).
To test if the measured amplitude depends on the TF concentration, we determined how many nascent mRNAs were present at each active TS. In stark contrast to the above results, this number was independent of the p-p38 levels (Figures 2E and 2F; Pearson’s correlation coefficient of −0.02) suggesting that the measured amplitude at each active TS is independent of the nuclear TF concentration. To further investigate the apparent lack of amplitude regulation, we determined nascent mRNA levels for all activation conditions (serum activation and zinc exposure) and obtained comparable values for all cases (Figures S3A and S3B). A resampling analysis revealed that for most conditions the data share the same underlying mean value close to 5 nascent mRNAs (Figure S3C). The only exceptions occurred at 20 min after serum induction and for 150 μM of zinc, where induction was at the highest levels. NHDF cells showed comparable results (Figure S3D) indicating a similar activation mode.
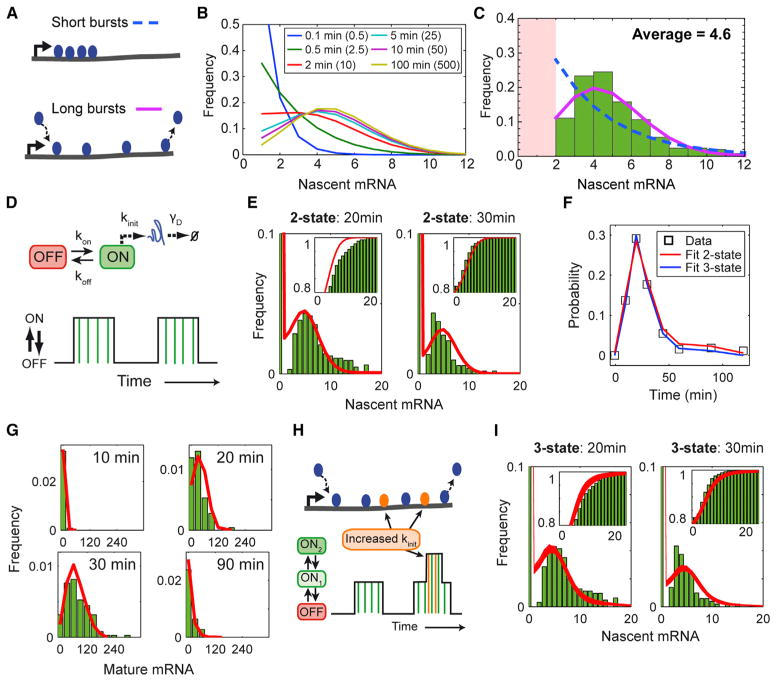
The preceding analysis indicates that the measured amplitude of the c-Fos bursts is largely constant. However, this value could differ from the actual burst amplitude in the case of burst saturation. Kinetic gene expression models predict specific signatures for the nascent mRNA distribution that could distinguish these cases (Golding et al., 2005): below burst saturation, nascent mRNA levels follow a geometrical distribution; but at saturation, the distribution becomes Poissonian (Figures 3A and 3B). To test whether we can discriminate between these two scenarios, we pooled the nascent mRNA data without the identified outliers. The resulting distribution is far more consistent with a Poisson distribution than with a geometric distribution (Figure 3C) indicating burst saturation, where individual bursts can produce larger numbers of mRNAs (Figure 3B). This bursting behavior is also consistent with the observed rapid production of high mature mRNA levels. Even at peak induction, we saw on average only one active c-Fos allele in U2OS cells. For bursts below saturation, we would expect more activation events in order to produce this large amount of mature mRNAs.
Figure 3. Mathematical Modeling of Transcriptional Response of c-Fos after Serum Induction.
(A) Cartoons illustrating concept of burst saturation limit. For short bursts below the saturation limit (upper plot), mRNA attached to all the loaded polymerases can be observed. For burst in the saturation limit (lower plot), only the mRNA produced by the currently loaded polymerases can be detected.
(B) Impact of burst duration on nascent mRNA distribution. Curves share same initiation rate (five mRNA/minute) and burst frequency (0.1 burst/minute) but differ in burst duration as indicated in figure legend. Values in parenthesis indicate average number of mRNAs produced per burst, i.e., the burst amplitude.
(C) Histogram of pooled nascent mRNA numbers from all induction condition (serum and zinc) except identified outliers in Figure S3C. Fit with Poisson distribution (pink solid line; log-likelihood of fit = −536) and truncated geometric distribution (dashed blue line; log-likelihood of fit = −705).
(D) Two-state model of transcription. Gene can switch between inactive (OFF) and active (ON) state. Transitions are described by rate constants kon and koff. Transcripts are produced during ON states as a Poisson process with fixed rate, kinit (vertical green bars in lower plot). Each mRNA undergoes a production period modeled as an irreversible process with fixed completion time, tprod and mature mRNA degrades as a first-order reaction with the constant γD.
(E–G) Fit with two-state model (Parameters from fit L2-8 in Table S1). (E) Fit of nascent mRNA data (green histogram) with two-state model (red line). Insets show cumulative histograms. (F) Probability for one TS to be active (black squares) together with prediction of two-state (red) and three-state model (blue). (G) Fit of mature mRNA data (green histogram) with two-state model (red line).
(H) Three-state model of transcription. A second ON state with a higher initiation frequency can be reached from the first ON state.
(I) Fits of nascent mRNA data (green histogram) with three-state model (red lines). Each line represents an individual fit with parameters defined in Table S2. Insets show cumulative histograms.
See also Figure S3.
Kinetic Model of c-Fos Transcription Reveals that Transcription Occurs in Bursts that Are Longer Than the Retention Time
The interpretation of the smFISH-IF data (Figure 2B) and the correlation of MAPK levels with the number of active TSs (Figure 1F) suggest a regulation mechanism for c-Fos where the nuclear TF concentration controls the burst frequency. To validate our qualitative understanding and determine the c-Fos transcription dynamics, we developed a discrete stochastic model to fit to the mature and nascent mRNA distributions in response to the time varying MAPK signal (Neuert et al., 2013). We focused our analysis on the serum activation data for which we have observed the entire time course. We implemented a two-state random telegraph model (Figure 3D) with a transcriptional active ON and a silent OFF state. Because we observed a correlation between p-ERK level and c-Fos burst frequency, the measured p-ERK concentration affects only the model’s OFF-ON transition; all other model parameters are constant. For further details on the modeling, see Supplemental Experimental Procedures and Figure S3E. The following results are supported by different fitting metrics (see Supplemental Results and Figures S3F–3K). The two-state model fit converged to a satisfactory set of parameters describing the number of active TSs, nascent mRNAs per active TS and mature mRNAs (Table S1 and Figures 3E–3G). The simple model shows that TF modulation of the burst frequency can largely explain the experimental data. The extracted parameters reveal interesting features of the c-Fos transcription kinetics (Table S1): bursts are in the saturation limit and last 3–4 min; even at peak levels of the kinase, an individual allele is only activated every 9–11 min. During a burst, several mRNAs are initiated per minute resulting in the total synthesis of several tens of mRNAs. The estimated time to produce one mRNA, which includes elongation and mRNA processing, is approximately 1 min (see Supplemental Results for more details on the partition between these two processes).
c-Fos Transcription at Peak Induction Is Described by a Second ON State with a Higher Initiation Rate
At 20 min after serum induction, where we measured the highest p-ERK levels, many cells have large numbers of nascent mRNAs represented by an elongated tail in the nascent mRNA distribution (Figure 3E) that is not captured by the two-state model. To better understand this transient behavior, we compared the number of nascent mRNAs to the number of active TS for each cell. These two measurements were uncorrelated at all time points except at 20 min, when cells with more active TS also had more nascent mRNAs (Figure S3I). This suggests that cells with higher levels of activating TF enter another transcription mode beyond that in the two-state model. To confirm the existence of this mode, we treated cells with TPA and observed a similarly strong correlation between nascent mRNAs per active TS and number of active TS per cell (Figure S3I).
From our two-state model fit, we concluded that bursts are in the saturation limit where initiation and termination of polymer-ases are at steady state. Thus, the experimentally observed increase of this steady-state level at 20 min could be achieved by a temporary increase in the initiation rate (Figure 3H). To test this hypothesis, we added a second ON state with an independent initiation rate to our model that can be reached at high p-ERK levels from the first ON state. Upon fitting this expanded model to the data, we could capture both the shape and the tails of the nascent mRNA distribution (Figure 3I). Furthermore, the number of active TS and the mature mRNA levels were still well described (Figures 3F and S3J). The estimated parameters of the three-state model for the first ON state are very similar to those for the two-state model fit (Table S2). However, we found greater parameter uncertainty for the second ON state, and multiple sets of parameters yielded similar distributions and described the data equally well (Figure S3K; Table S2).
Taken together, our model results are consistent with our previous observations and suggest a simple mechanism for c-Fos transcriptional regulation. Cells can use the TF concentration to tune the burst frequency. At peak induction, further increase of mRNA levels occurs by increasing the polymerase initiation frequency.
Synthetic Transcription Factors as a Tool to Dissect c-Fos Bursting Parameters
We next investigated if we could alter the identified burst characteristics more profoundly by changing the properties of the TF. We developed synthetic TFs to induce c-Fos transcription independently of any cellular activation pathway. We engineered four proteins that target different parts of the c-Fos promoter with the transcription activator-like effector (TALE) approach (Perez-Pinera et al., 2013) (Figure 4A). We fused these constructs with activator domains of different strengths (VP16 or VP64) to generate synthetic TFs.
Figure 4. Activation of c-Fos with Synthetic Transcription Factors.
(A) c-Fos promoter with different synthetic TFs (TALE) binding sites indicated by letters A–D. SRE, serum response element. Different activator domain (AD, in red) were fused to the TALE: VP16 or VP64.
(B) Histogram of nascent c-Fos mRNA levels after transfection with one TALE-VP16, one TALE-VP64, or four TALE-VP16. Red lines are model curves (Table S3). Curves for VP64 and 4xVP16 are obtained by changing only one parameter indicated in red in cartoon compared to VP1.
See also Figure S4.
First, we investigated the mature mRNA levels by exposing cells to these synthetic TFs. As a control, we transfected cells with a TALE without an activator domain and found no activation. When transfecting with one TALE-VP16, we observed an increase in the average mature mRNA number from 4 to 28 (Figures S4A and S4B). The mature mRNA levels increased even further after transfecting with one TALE with the stronger VP64 activation domain (52 mature mRNAs), or all four different TALE-VP16 (70 mature mRNAs) (Figure S4B). We also analyzed the nascent mRNA distribution at active TSs. For one TALE-VP16, we detected on average three nascent mRNAs per active TS. The obtained distribution resembled a geometric distribution indicative of a short burst duration compared to the retention time (Figures 4B and 3B). The TALE-VP64 construct yielded increased nascent mRNA levels (average of four) but still resembled a geometric distribution. This change is in agreement with a model where the stronger activator domain leads to higher initiation rate but does not affect the burst duration (Figures 4B and 3B). When transfecting all four different TALE-VP16, the nascent mRNA number per active TS increased further (average of five). Furthermore, the shape of the distribution also changed to resemble a Poisson distribution (Figure 4B). This is in agreement with an increase of the burst duration beyond burst saturation (Figure 3B). Finally, we tested if the observed changes could be reproduced with the two-state model. By changing only one parameter in the model, we could generate the observed distributions and the respective transitions: changing the initiation rate reproduced the difference from one TALE-VP16 to one TALE-VP64, while changing the burst duration yielded the change observed when activating with one TALE-VP16 compared to four TALE-VP16 (Figure 4B and Table S3).
Taken together, these data suggest two additional potential key determinants for transcriptional bursts. The initiation rate could be controlled by the strength of the activator domain, whereas the burst duration could be controlled by the lifetime of the TF on the promoter (Figure 5).
Figure 5. Role for TFs in Shaping c-Fos Transcriptional Bursts.
Model that illustrates how TFs can act on multiple key aspects of transcriptional burst. Increase of TF concentration yields increase of burst frequency (kon, green); duration of TF binding event with DNA binding domain (DBD) affects burst duration (koff, blue); strength of the activator domain (AD) influences initiation rate (kinit, red).
DISCUSSION
Frequency Modulation as a Simple but Versatile Mechanism for c-Fos Transcription
We determined how c-Fos mRNA levels are regulated after serum or zinc induction by smFISH. We found that transcription following MAPK induction occurs in discontinuous bursts where predominately the burst frequency, but not the measured amplitude, is modulated by the TF concentration. To gain a more quantitative understanding of the bursting mechanism, we analyzed the serum induction data with a stochastic gene expression model. This model revealed that c-Fos mRNA production occurs in relatively isolated bursts of several minutes, which is in agreement with a study also suggesting large bursts for c-Fos (Shah and Tyagi, 2013) and another study estimating comparable bursting timescales (Suter et al., 2011b). The model further showed that the measured burst amplitude by smFISH is substantially smaller than the total number of mRNAs produced during one burst. Last, we could determine that during bursts several transcripts are initiated per minute and are produced in approximately 1 min. The latter suggests that elongation and maturation are both fast processes for c-Fos.
Taken together, this suggests a rather simple but effective and versatile system to activate c-Fos under different conditions. Cells control principally the burst frequency—and hence the total mRNA levels—by adjusting the nuclear concentration of TFs. High TF concentrations mean an increased likelihood for activation and high mRNA levels; low TF concentrations give the opposite response. Although this rather simple mechanism explained most of the tested c-Fos activation conditions, additional burst amplitude regulation also occurs at peak induction.
Role of TFs in Modulating c-Fos Bursts
We found that at peak induction—20 min after serum induction—the addition of a second ON state with an increased initiation rate could explain the data better than the simple two-state model. Similar needs for multistate activation processes have been reported in the literature (Dar et al., 2012; Neuert et al., 2013). Initiation frequency modulation could be attributed to several molecular events ranging from preinitiation complex (PIC) formation and stability to promoter escape, and it will be important to investigate this regulation in future studies.
We found that c-Fos burst characteristics can be altered more profoundly by targeting its promoter with synthetic TFs. Using activation domains of different strengths led to different polymerase initiation frequencies. Targeting the promoter with multiple TFs led to longer bursts suggesting a relationship between the TF-promoter binding time and the burst duration. This is compatible with previous observations of synergetic effect of multiple TFs acting on the same promoter (Carey et al., 1990; Perez-Pinera et al., 2013). We therefore propose a possible link between PIC stability and simultaneous binding of multiple TFs.
Taken together, our study establishes that TFs play a major role in affecting c-Fos bursting (Figure 5). TF concentrations can be changed rapidly to change the burst frequency. The duration of TF binding events can affect the burst duration and hence how many transcripts are produced during each burst. Changing the promoter sequence and/or the interaction partners for the promoter could alter this duration, therefore allowing different transcription responses at different promoters. Last, the TF activation domain could play an important role to establish the initiation frequency during active periods. These evolutionarily tunable dynamics provide cells with an extensive toolkit to fine-tune transcriptional responses.
Transcription as a Gene-Specific Multilayered Response
Over the last decade, many of the steps involved in transcription have been shown to affect bursting kinetics, such as the local chromatin environment and chromatin modifications (Dar et al., 2012; Mouaikel et al., 2013; Suter et al., 2011b; Viñuelas et al., 2013), local promoter architecture (Raj et al., 2006; Suter et al., 2011b), nucleosome turnover (Brown et al., 2013), gene looping (Hebenstreit, 2013), and TFs (Larson et al., 2013; Neuert et al., 2013; Raj et al., 2006). Over the recent years, studies in prokaryotic and eukaryotic systems have described a wide range of transcriptional kinetics (Suter et al., 2011a) and have illustrated that there is no universal law of gene expression (Larson, 2011). Accordingly, a variety of regulatory mechanisms have been described for different genes and model organisms where modulation of burst duration (So et al., 2011), burst amplitude (Raj et al., 2006), burst frequency (Brown et al., 2013; Larson et al., 2013), or combinations of these (Dar et al., 2012; Neuert et al., 2013; Suter et al., 2011b) have been reported. Our study identifies burst frequency modulation as the major control point for c-Fos expression and proposes that changing the properties of the TF could further alter burst amplitude and duration—showing that several key aspects of bursts can be affected at the level of initiation. This expands the emerging picture that gene regulation can be seen as a superposition of regulation at different levels that can be tuned to obtain the desired response.
Future studies will further decipher the complex interplay of TFs and their endogenous promoters by combining measurements of transcription in living cells (Annibale and Gratton, 2014) and novel genome editing approaches (Gaj et al., 2013).
EXPERIMENTAL PROCEDURES
More details on the experimental procedures can be found in Supplemental Experimental Procedures.
Cell Culture
NHDF are isolated from the dermis of adult skin (Promocell, C-12302) and maintained at 37°C in DMEM-F12 supplemented with 10% FBS. U2OS cells were cultured at 37°C in DMEM, supplemented with 10% FBS.
smFISH and IF
smFISH was performed according to supplier (Biosearch Technology) and published protocols (Femino et al., 1998).
Gene Expression Analysis by qRT-PCR
Gene expression of c-Fos and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was quantified using the PrimeTime qPCR assay (Integrated DNA Technologies).
Synthetic Transcription Factors
Assembly of a custom TAL effector construct was performed as described previously (Cermak et al., 2011) into custom backbone plasmid. Plasmids were deposited on the Addgene platform (www.addgene.org/Xavier_Darzacq). For sequences and target sites, see Table S4.
smFISH Quantification
Number of mature mRNAs and equivalent full-length transcripts were estimated with FISH-quant (Mueller et al., 2013).
IF Quantification
p-ERK and p-p38 levels were automatically measured in nuclei with homemade programs in MATLAB (MathWorks) and ImageJ (http://rsb.info.nih.gov/ij/).
Mathematical Modeling
Model analyses for mRNA distributions utilized modified finite state projections analyses (Munsky and Khammash, 2006) and the stochastic simulation algorithm (Gillespie, 1977). Parameter estimations were conducted using a combination of local and global optimization routines.
Supplementary Material
Acknowledgments
This work was supported by ANR-10-BLAN-1222, ANR-10-LABX-54, MEMO LIFE/ANR-11-IDEX-0001-02. A.S. was supported by La Ligue nationale contre le cancer and Labex Memo Life. B.M. was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R25GM105608. F.M. was supported by the Région Ile-de-France under C’Nano IdF (Center of Competences in NanoSciences for the Paris region), the Fondation pour la Recherche Médicale (FRM), and Institut Pasteur. We thank Edouard Bertrand, Timothee Lionnet, and Jim McNally for critical reading of the manuscript and Olivier Bensaude, Olivier Collin, Dan Larson, and Robert Singer for helpful discussions.
Footnotes
Supplemental Information includes Supplemental Results, Supplemental Discussion, Supplemental Experimental Procedures, four figures, four tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.05.053.
AUTHOR CONTRIBUTIONS
A.S., C.Z., F.M., and X.D. designed the project, A.S. and X.D. designed experiments, A.S., F.E.B., F.P., and N.L. performed experiments, A.S., B.M., F.M., and X.D. analyzed data, B.M. developed the mathematical modeling, A.S., B.M., F.M., and X.D. interpreted the modeling results, A.S., B.M., F.M., and X.D. wrote the paper. All authors discussed results.
References
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annibale P, Gratton E. Advanced fluorescence microscopy methods for the real-time study of transcription and chromatin dynamics. Transcription. 2014;5:5. doi: 10.4161/trns.28425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Mao C, Falkovskaia E, Jurica MS, Boeger H. Linking stochastic fluctuations in chromatin structure and gene expression. PLoS Biol. 2013;11:e1001621. doi: 10.1371/journal.pbio.1001621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M, Lin YS, Green MR, Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82–e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Marchi M, Cardarelli F, Roy A, Beltram F, Maffei L, Ratto GM. Dynamic regulation of ERK2 nuclear translocation and mobility in living cells. J Cell Sci. 2006;119:4952–4963. doi: 10.1242/jcs.03272. [DOI] [PubMed] [Google Scholar]
- Dar RD, Razooky BS, Singh A, Trimeloni TV, McCollum JM, Cox CD, Simpson ML, Weinberger LS. Transcriptional burst frequency and burst size are equally modulated across the human genome. Proc Natl Acad Sci USA. 2012;109:17454–17459. doi: 10.1073/pnas.1213530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Fowler T, Sen R, Roy AL. Regulation of primary response genes. Mol Cell. 2011;44:348–360. doi: 10.1016/j.molcel.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Espinosa JM. Lessons on transcriptional control from the serum response network. Curr Opin Genet Dev. 2011;21:160–166. doi: 10.1016/j.gde.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–2361. [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín JE, Darzacq X, Choder M. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Healy S, Khan P, Davie JR. Immediate early response genes and cell transformation. Pharmacol Ther. 2013;137:64–77. doi: 10.1016/j.pharmthera.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Hebenstreit D. Are gene loops the cause of transcriptional noise? Trends Genet. 2013;29:333–338. doi: 10.1016/j.tig.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Larson DR. What do expression dynamics tell us about the mechanism of transcription? Curr Opin Genet Dev. 2011;21:591–599. doi: 10.1016/j.gde.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Fritzsch C, Sun L, Meng X, Lawrence DS, Singer RH. Direct observation of frequency modulated transcription in single cells using light activation. eLife. 2013;2:e00750. doi: 10.7554/eLife.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Ahn DH, Crawley SC, Li JD, Gum JR, Jr, Basbaum CB, Fan NQ, Szymkowski DE, Han SY, Lee BH, et al. Phorbol 12-myristate 13-acetate up-regulates the transcription of MUC2 intestinal mucin via Ras, ERK, and NF-kappa B. J Biol Chem. 2002;277:32624–32631. doi: 10.1074/jbc.M200353200. [DOI] [PubMed] [Google Scholar]
- Little SC, Tikhonov M, Gregor T. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell. 2013;154:789–800. doi: 10.1016/j.cell.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina N, Suter DM, Cannavo R, Zoller B, Gotic I, Naef F. Stimulus-induced modulation of transcriptional bursting in a single mammalian gene. Proc Natl Acad Sci USA. 2013;110:20563–20568. doi: 10.1073/pnas.1312310110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel J, Causse SZ, Rougemaille M, Daubenton-Carafa Y, Blugeon C, Lemoine S, Devaux F, Darzacq X, Libri D. High-frequency promoter firing links THO complex function to heavy chromatin formation. Cell Reports. 2013;5:1082–1094. doi: 10.1016/j.celrep.2013.10.013. [DOI] [PubMed] [Google Scholar]
- Mueller F, Senecal A, Tantale K, Marie-Nelly H, Ly N, Collin O, Basyuk E, Bertrand E, Darzacq X, Zimmer C. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat Methods. 2013;10:277–278. doi: 10.1038/nmeth.2406. [DOI] [PubMed] [Google Scholar]
- Munsky B, Khammash M. The finite state projection algorithm for the solution of the chemical master equation. J Chem Phys. 2006;124:044104. doi: 10.1063/1.2145882. [DOI] [PubMed] [Google Scholar]
- Munsky B, Neuert G, van Oudenaarden A. Using gene expression noise to understand gene regulation. Science. 2012;336:183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Gong P, Suzuki K, Koizumi S. Differential metal response and regulation of human heavy metal-inducible genes. J Cell Physiol. 1999;180:105–113. doi: 10.1002/(SICI)1097-4652(199907)180:1<105::AID-JCP12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Neuert G, Munsky B, Tan RZ, Teytelman L, Khammash M, van Oudenaarden A. Systematic identification of signal-activated stochastic gene regulation. Science. 2013;339:584–587. doi: 10.1126/science.1231456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, Guilak F, Crawford GE, Hartemink AJ, Gersbach CA. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Methods. 2013;10:239–242. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontén J, Saksela E. Two established in vitro cell lines from human mesenchymal tumours. Int J Cancer. 1967;2:434–447. doi: 10.1002/ijc.2910020505. [DOI] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A, Golding I. Genetic determinants and cellular constraints in noisy gene expression. Science. 2013;342:1188–1193. doi: 10.1126/science.1242975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, Tyagi S. Barriers to transmission of transcriptional noise in a c-fos c-jun pathway. Mol Syst Biol. 2013;9:687. doi: 10.1038/msb.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Razooky B, Cox CD, Simpson ML, Weinberger LS. Transcriptional bursting from the HIV-1 promoter is a significant source of stochastic noise in HIV-1 gene expression. Biophys J. 2010;98:L32–L34. doi: 10.1016/j.bpj.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skupsky R, Burnett JC, Foley JE, Schaffer DV, Arkin AP. HIV promoter integration site primarily modulates transcriptional burst size rather than frequency. PLoS Comput Biol. 2010;6:6. doi: 10.1371/journal.pcbi.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So LH, Ghosh A, Zong C, Sepúlveda LA, Segev R, Golding I. General properties of transcriptional time series in Escherichia coli. Nat Genet. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter DM, Molina N, Naef F, Schibler U. Origins and consequences of transcriptional discontinuity. Curr Opin Cell Biol. 2011a;23:657–662. doi: 10.1016/j.ceb.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011b;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- Viñuelas J, Kaneko G, Coulon A, Vallin E, Morin V, Mejia-Pous C, Kupiec JJ, Beslon G, Gandrillon O. Quantifying the contribution of chromatin dynamics to stochastic gene expression reveals long, locus-dependent periods between transcriptional bursts. BMC Biol. 2013;11:15. doi: 10.1186/1741-7007-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD, Whitmarsh AJ. MAP kinase signalling cascades and transcriptional regulation. Gene. 2013;513:1–13. doi: 10.1016/j.gene.2012.10.033. [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.