Abstract
Background & Aims
Nearly 20% of the global cancer burden can be linked to infectious agents. Fusobacterium nucleatum promotes tumor formation by epithelial cells via unclear mechanisms. We aimed to identify microRNAs (miRNAs) induced by F nucleatum and evaluate their ability to promote colorectal carcinogenesis in mice.
Methods
Colorectal cancer (CRC) cell lines were incubated with F nucleatum or control reagents and analyzed in proliferation and would healing assays. HCT116, HT29, LoVo, and SW480 CRC cell lines were incubated with F nucleatum or phosphate buffer saline (PBS control) and analyzed for miRNA expression patterns and in chromatin immunoprecipitation assays. Cells were incubated with miRNAs mimics, control sequences, or small interfering (si) RNAs; expression of reporter constructs was measured in luciferase assays. CRC cells were incubated with F nucleatum or PBS and injected into BALB/C nude mice; growth of xenograft tumors was measured. C57BL APCmin/+, C57BL miR21a−/−, and C57BL mice with full-length miR21a (controls) were given F nucleatum by gavage; some mice were given azoxymethane (AOM) and dextran sodium sulfate (DSS) to induce colitis and colon tumors. Intestinal tissues were collected and tumors were counted. Serum samples from mice were analyzed for cytokine levels by ELISAs. We performed in situ hybridization analyses to detect enrichment of F nucleatum in CRC cells. F nucleatum DNA in 90 tumor and matched non-tumor tissues from patients in China were explored for the expression correlation analysis; levels in 125 tumor tissues from patients in Japan were compared with their survival times.
Results
F nucleatum increased proliferation and invasive activities of CRC cell lines, compared with control cells. CRC cell lines infected with F nucleatum formed larger tumors, more rapidly, in nude mice than uninfected cells. APCmin/+ mice gavaged with F nucleatum developed significantly more colorectal tumors than mice given PBS and had shorter survival times. We found several inflammatory factors to be significantly increased in serum from mice given F nucleatum (interleukin 17F [IL17F], IL21, IL22, and MIP3A). We found 50 miRNAs to be significantly upregulated and 52 miRNAs to be significantly downregulated in CRCs incubated with F nucleatum vs PBS; levels of miR21 increased by the greatest amount (more than 4-fold). Inhibitors of miR21 prevented F nucleatum from inducing cell proliferation and invasion in culture. miR21a−/− mice had a later appearance of fecal blood and diarrhea after administration of AOM and DSS, and had longer survival times, compared with control mice. The colorectum of miR21a−/− mice had fewer tumors, of smaller size, and the miR21a−/− mice survived longer than control mice. We found RASA1, which encodes a RAS GTPase, to be one of the target genes consistently downregulated in cells that overexpressed miR21 and upregulated in cells exposed to miR21 inhibitors. Infection of cells with F nucleatum increased expression of miR21 by activating TLR4 signaling to MYD88, leading to activation of the nuclear factor NFκB. Levels of F nucleatum DNA and miR21 were increased in tumor tissues (and even more so in advanced tumor tissues), compared with non-tumor colon tissues from patients. Patients whose tumors had high amounts of F nucleatum DNA and miR21 had shorter survival times than patients whose tumors had lower amounts.
Conclusions
We found infection of CRC cells with F nucleatum to increase their proliferation, invasive activity, and ability to form xenograft tumors in mice. F nucleatum activates TLR4 signaling to MYD88, leading to activation of the nuclear factor NFκB and increased expression of miR21; this miRNA reduces levels of the RAS GTPase RASA1. Patients with both high amount of tissue F nucleatum DNA and miR21 demonstrated a higher risk for poor outcomes.
Keywords: microbe, signal transduction, gene regulation, carcinogenesis
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer worldwide and the fourth most frequent cause of death following lung, liver and stomach cancer1. Surprisingly, the incidence of CRC in Asian countries, which were previously considered to have lower rates, has increased dramatically during past decade2. Unfortunately, the underlying mechanism(s) for the initiation and development of this global malignancy have not been yet fully elucidated.
It has been estimated that approximately 20% of the global cancer burden can be linked to infectious agents3. Human papillomaviruses and helicobacter pylori, for example, are well-known pathogens that cause cervical and gastric cancer. Considering almost 1.5 kilograms of microbiota inhabit the human gut, it is not surprising that certain viruses and/or bacteria might be related to human cancers4. However, the carcinogenic pathogens which cause CRC remain incompletely explored5.
A growing body of evidence suggests a potential link between the Fusobacterium nucleatum (F nucleatum) and colorectal carcinogenesis. Several studies have demonstrated that F nucleatum is enriched in human colorectal adenomas and carcinomas compared with adjacent normal tissue6, 7. Furthermore, a higher amount of tissue F nucleatum DNA has been indicated to be associated with advanced tumor stage and poor prognosis in CRC8–10. Recent studies showed F nucleatum adheres to and invades endothelial and epithelial cells via its virulence factors such as adhesin A (FadA), fusobacterium autotransporter protein 2 (Fap2) and fusobacterial outer membrane protein A (FomA)11–14. Nevertheless, few studies have by far revealed downstream events of F nucleatum infection that can trigger colonic inflammation and carcinogenesis.
Our previous study demonstrated that microRNA21 (miR21) plays a pathogenic role in chronic inflammatory processes and the development of colitis-associated colon cancer15. Furthermore, data from our group and others have highlighted that specific miRNAs contribute to colorectal carcinogenesis, and several of these can serve as biomarkers for diagnosis, prognosis, and metastasis prediction in CRC patients15–20. Therefore, we hypothesized that F nucleatum infection may regulate several cancer-specific miRNAs to promote the development of CRC. To the best of our knowledge, the impact of F nucleatum-infection and its impact on downstream effects on miRNA regulation have not been interrogated to date.
In the present study, we aimed to identify F nucleatum-induced miRNAs, decipher the mechanism(s) for the regulation of miRNAs by F nucleatum, and demonstrate its biological and clinical significance in CRC. Our study attempts to use miRNA regulation as novel angle to clarify the carcinogenic role of F nucleatum in CRC, and provide a novel evidence that F nucleatum could serve as a potential prognostic and therapeutic target in patients with this malignancy.
MATERIALS AND METHODS
Experimental methods (including statistical analysis) are described in detail in the Supplementary Information.
RESULTS
F nucleatum promotes CRC cell proliferation and invasion in cell lines and a xenograft animal model
Compared to the untreated cells or those treated with E coli DH5α, F nucleatum significantly promoted cell growth in HCT116 and LoVo cells after treatment at 48 hr (P < 0.05 and P < 0.001 respectively, Figure 1A and Supplementary Figure 1A). Consistently, we observed F nucleatum treatment significantly induced S-phase accumulation in LoVo cells (P < 0.01) but less marked increase in HCT116 cells (Figure 1B and Supplementary Figure 1B). Furthermore, F nucleatum treatment remarkably enhanced cell invasion in both HCT116 and LoVo cells (P < 0.01 in both cell lines after F nucleatum treatment at 48 hr, Figure 1C and Supplementary Figure 2).
Figure 1. F nucleatum promotes CRC cell proliferation and invasion in vitro and in vivo.
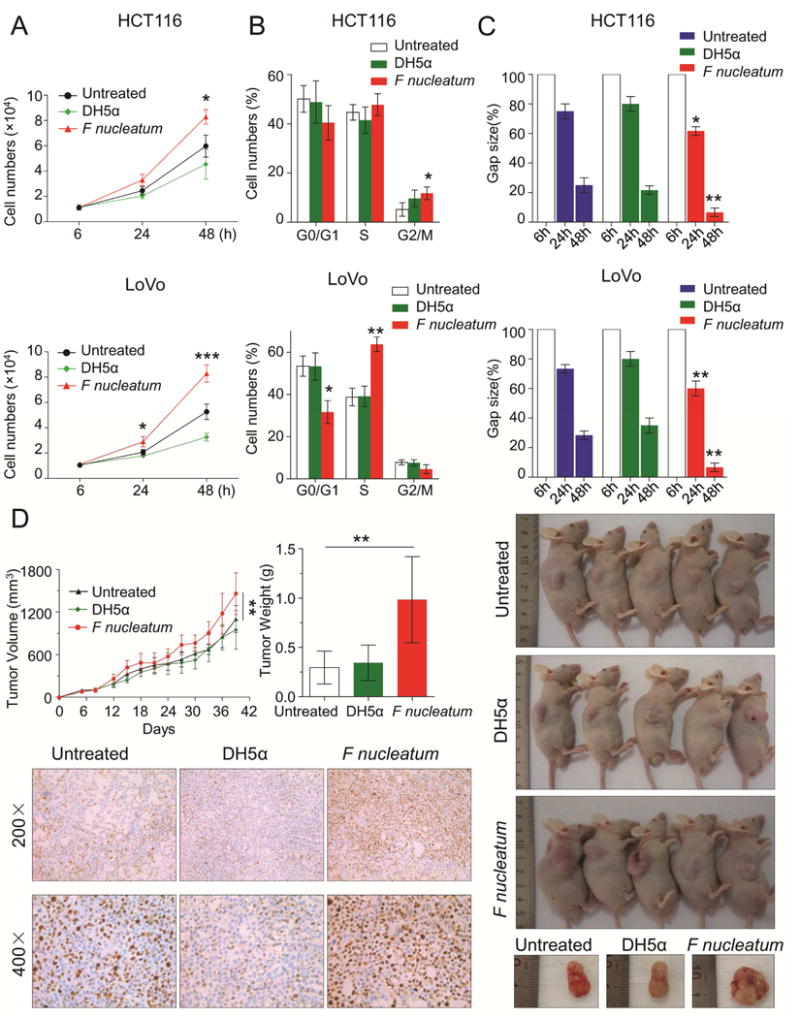
A) HCT116 and LoVo cells were incubated with PBS, DH5α E coli or F nucleatum. The cell proliferation rates were evaluated by cell number counting after treatment at 6, 24 and 48 hr (*P < 0.05, ***P < 0.001, unpaired Student’s t test). B) The cell cycle distribution of treated or control cells was determined by flow cytometry-based assay after 48 hr (*P < 0.05, **P < 0.01, unpaired Student’s t test). C) The scratch wound healing assay was performed to evaluate the invasive capability of treated or control cells (F nucleatum treated cells vs control cells, *P < 0.05, **P < 0.01, unpaired Student’s t test). D) HCT116 cells treated with PBS, DH5α, or F nucleatum were subcutaneously injected into male BALB/C nude mice to produce xenograft tumors in animals (n=5 per group). The upper left panel of figures illustrate tumor growth curves (**P < 0.01 by one-way analysis of variance [ANOVA] and Bonferroni’s multiple comparison test) and tumor weight (**P < 0.01 by one-way ANOVA,). The right panel of figure shows the representative images of xenograft mice. The bottom left figures depict representative immunostaining (200× and 400×) for Ki-67 in xenograft tumor tissues. These results are representative of at least three independent experiments. Bars represent standard deviation (SD).
To confirm these in vitro results, we subcutaneously injected F nucleatum-treated HCT116 cells into male BALB/C nude mice. In concordance with our previous findings, F nucleatum treated cells reported an accelerated tumor growth and heavier tumor weight compared to untreated or DH5α treated cells (P < 0.01). Moreover, we observed enhanced staining for the proliferation marker Ki-67 in F nucleatum-treated xenograft tissues (Figure 1D). Similarly, we generated a xenograft model derived from F nucleatum-treated LoVo cells and successfully validated above results (Supplementary Figure 3). However, F nucleatum treatment did not affect cell apoptosis in both HCT116 and LoVo cells (Supplementary Figure 4). Together, our results highlight that F nucleatum exerts oncogenic function in CRC.
F nucleatum promotes tumorigenicity in APCMin/+ mouse
The classic Vogelgram multistep model for CRC indicates tumor suppressor gene, adenomatous polyposis coli (APC) gene acts as a gatekeeper and alterations in the APC/Wnt-signaling trigger adenoma–carcinoma sequence21. The APCMin/+ mice carry APC mutations, which demonstrates a consequent predisposition to multiple intestinal neoplasia22. Therefore, we hypothesized that chronic F nucleatum infection in APCMin/+ mice may induce synergistically oncogenic effect by enhancing the aberrant epithelial cell growth, and consequently leading to the initiation of CRC.
To confirm our hypothesis, we administrated F nucleatum or phosphate buffer saline (PBS as negative control, NC) into 5–6 weeks old APCMin/+ mice, which received antibiotics by gavage administration for 3 days beforehand (Figure 2A). Surprisingly, 20 weeks after F nucleatum treatment, APCMin/+ mice presented serious complications including ascites (60% versus 30%, P = 0.178), bloody diarrhea (80% versus 20%, P = 0.0246), gut dilatation (60% versus 30%, P = 0.178) and splenomegaly (80% versus 40%, P = 0.0679) when compared with the PBS group (Figure 2B). Notably, we further found that the colorectum showed significant increase in tumor numbers (3.10 ± 3.38 versus 0.67 ± 1.00, P = 0.042), tumor size (7% versus 0% for tumors with size > 3mm) and tumor load (6.40 ± 6.64 versus 1.27 ± 1.90, P = 0.0161; Figure 2C). The small intestine demonstrated mild increase in the tumor numbers, size and tumor load as well (Sumpplementary Figure 5). In particular, F nucleatum-treated APCMin/+ mice had a shorter survival period compared to control group (median survival of 164.5 days for F nucleatum-treated mice and 200 days for NC mice, P=0.0129, Figure 2D). In addition, we found colorectal tumors from F nucleatum treated mice showed high expression levels of proliferative marker PCNA compared to negative control, suggesting that F nucleatum could effectively stimulate cell proliferation (Figure 2E).
Figure 2. F nucleatum promotes tumorigenicity and CRC progression in ApcMin/+ mice.
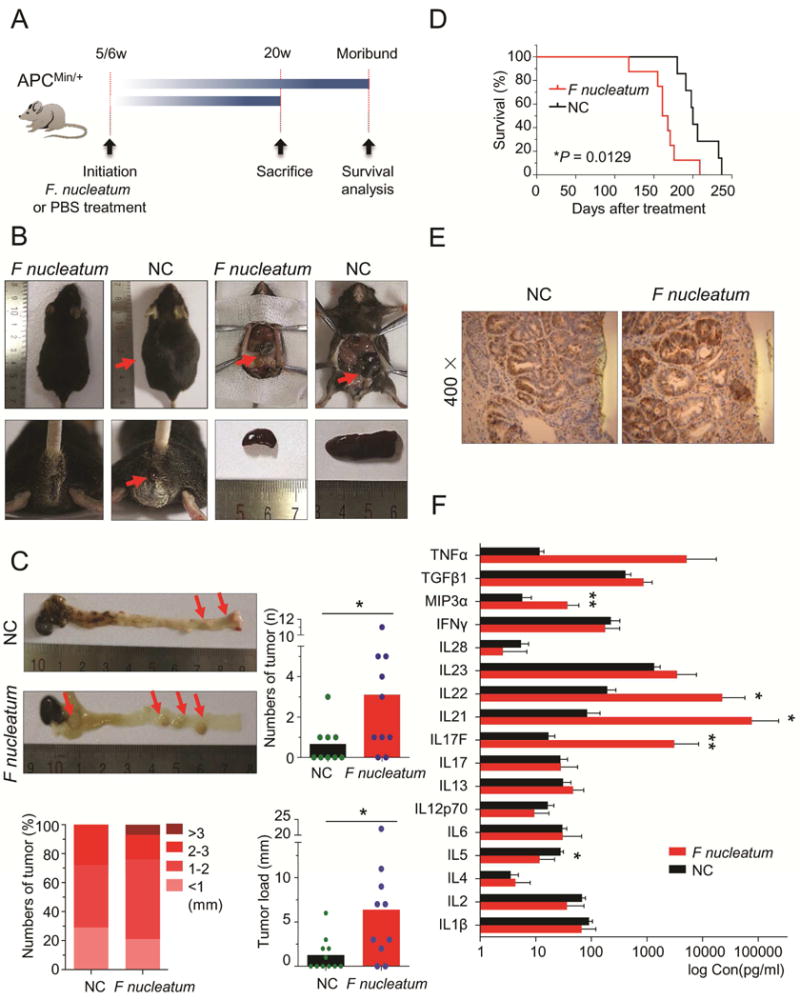
A) APCMin/+ mice were administrated with F nucleatum or PBS (negative control, NC) and sacrificed after treatment at 20 weeks. For the survival analysis, we compared survival time between F nucleatum treated group and PBS group. B) On 20 weeks, F nucleatum-treated APCMin/+ mice presented with ascites, bloody diarrhea, gut dilatation and splenomegaly. C) The left upper figure illustrates a representative image of a colon of APCMin/+ mice with or without F nucleatum treatment. The upper right figure and the bottom figure shows the administration of F nucleatum for 20 weeks which resulted in significant increase in tumor numbers, size and tumor load in the colorectum (n = 10 per group,*P<0.05, Mann-Whitney U test). D) The Kaplan-Meier survival curves show F nucleatum treatment reduced the survival rates of APCmin/+ mice (n = 8 per group, Log-rank (Mantel-Cox) test). E) Representative image of an immunostaining for PCNA in F nucleatum-treated or F nucleatum-untreated CRC tissues. F) The serum level of inflammatory factors in APCmin/+ mice with or without treatment of F nucleatum. (n = 10 per group, *P < 0.05, **P < 0.01, Mann-Whitney U test). Results represent means ± SD. The red arrows indicated the positive location.
Accumulating evidence demonstrates that inflammatory cells secrete a variety of pro-inflammatory or growth factors during tumorigenesis and these factors contribute to cancer develpoment23. To examine whether these factors could also be induced by F nucleatum infection, we measured serum levels of a panel of inflammatory factors. Interestingly, we found several inflammatory factors were significantly up-regulated in F nucleatum-treated mice including IL17F (P = 0.0022), IL21 (P = 0.0152), IL22 (P = 0.0411) and MIP3a (P = 0.0087, Figure 2F). Collectively, our results suggest that F nucleatum could stimulate immune response, increase serum inflammatory factors and promote tumorigenicity in APCMin/+ mice.
MiR21 is a downstream target of F nucleatum
Accumulating evidence have shown deregulation of miRNA expression in CRC24. Given the importance role of miRNAs in cell homeostasis and colorectal carcinogenesis, it is no surprise that F nucleatum might take advantage of miRNA-mediated regulation of gene expression to promote tumorigenesis.
To determine whether F nucleatum affected miRNA expression, we first treated HCT116, HT29, SW480 and LoVo cells with F nucleatum or PBS and subsequently performed microarray analysis to identify differentially expressed miRNAs. Impressively, among the 657 miRNAs, 50 miRNAs were significantly up-regulated (P < 0.05) while 52 miRNAs were significantly down-regulated (P < 0.05, Figure 3A and Supplementary Table 1). To obtain more accurate results, we used several criteria (signal intensity ≥ 500, P < 0.001 and log2 Fold change ≥ 2) to screen robust miRNA candidates. Only 6 miRNAs (let-7f-5p, let-7i-5p, miR-23a-3p, let-7a-5p, miR-23b-3p, and miR-21-5p) were found to be dramatically over-expressed. We further performed independent experiments, in which we treated multiple CRC cell lines with F nucleatum, to validate our microarray results (Figure 3A and Supplementary Figure 6). Strikingly, miR21 consistently showed to be the most up-regulated miRNA induced by F nucleatum infection, with a nearly ≥ 4-fold increase, compared to other miRNAs, suggesting miR21 is probably a direct target of F nucleatum.
Figure 3. F nucleatum regulates miR21 expression in CRC.
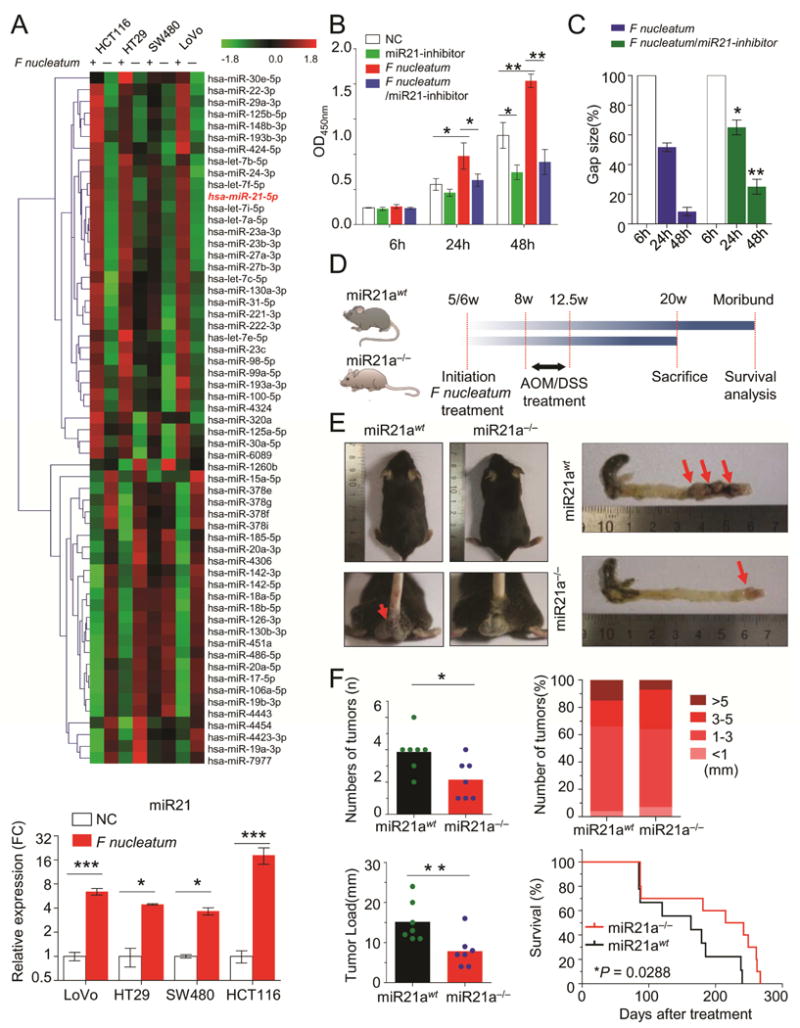
A) Microarrays were performed to identify the differentially expressed miRNA between F nucleatum-treated and control cells. The miR21 expression was consistently up-regulated in different CRC cells after F nucleatum treatment (*P < 0.05, ***P < 0.001, unpaired Student’s t test). B and C) CCK-8 cell viability assay and wound healing assays illustrated that inhibition of miR21 completely abolished F nucleatum’s effect on cell growth and invasion(*P < 0.05, **P < 0.01 by unpaired Student’s t test). D) Both miR21 WT and knockout (KO) mice were initially administrated F nucleatum and then subjected to AOM/DSS treatment. At 20 weeks, mice were sacrificed for various experiments. E) The left representative image showed miR21 KO decreased the bloody diarrhea rates during the F nucleatum infection (n = 7 per group), and the right image showed a representative colon of the miR21 KO or wild type mice. F) The colorectum in miR21 KO mice had fewer tumor numbers, smaller size and less tumor load compared to WT mice (n = 7 per group,*P<0.05, Mann-Whitney U test). Furthermore, miR21 KO mice had longer survival rates than WT mice (n = 10 per group by log-rank (Mantel-Cox) test). These results are representative of at least three independent experiments. Results represent means values ± SD. The red arrows indicated the positively stained cells.
To determine whether F nucleatum exerts oncogenic function in a miR21 dependent manner, we performed miR21 loss of function assays. Since HCT116 has the highest level of miR21 (Supplementary Figure 7A), we treated HCT116 with miR21 inhibitors alone or together with F nucleatum. As expected, F nucleatum treatment significantly promoted tumor growth and cell invasion, while inhibition of miR21 showed remarkable tumor suppressive effects (Figure 3B–3C). However, when we treated cell lines with both F nucleatum and miR21 inhibitors, inhibition of miR21 completely abolished F nucleatum’s effect on cell growth and invasion, highlighting the important role of miR21 in sustaining F nucleatum’s functions (Figure 3B–3C).
To further confirm our assumption that F nucleatum promotes colorectal progression through miR21, we next employed the miR21a knockout (KO, miR21a−/−) mice (Figure 3D). Azoxymethane (AOM) is a recognized procarcinogen with organotropism for the colon and AOM induced tumorigenesis is significantly enhanced by the inflammatory agent dextran sodium sulfate (DSS). Thus, both miR21a wild-type (WT) and KO mice were initially administrated F nucleatum and then subjected to AOM/DSS treatment. WT mice showed an earlier appearance of fecal blood and diarrhea than miR21a−/− mice (Figure 3E). As shown in Figures 3F, the colorectum in miR21a−/− mice had fewer tumor numbers (2.14 ± 1.21 versus 3.86 ± 1.21, P = 0.0309), smaller tumor size (7% versus 15% for > 5mm tumors) and less tumor load (7.86 ± 4.06 versus 15.14 ± 5.01, P = 0.0093) compared to WT mice. Likewise, but a non-significant trend was observed in small intestine in miR21a−/− mice (Supplementary Figure 7B–C). Furthermore, miR21a−/− mice had longer survival than WT mice (median survival of 228.5 days for miR21a−/− mice and 163 days for WT mice, P = 0.0288, Figure 3F).
Taken together, we for the first time report that miR21 is a direct target of F nucleatum and miR21 knockdown/knockout can alleviate the oncogenic function of F nucleatum during colorectal carcinogenesis.
F nucleatum regulates expression of miR21 novel target RASA1 and activates MAPK signaling pathway
Since F nucleatum promoted tumorigenesis through miR21, we therefore questioned whether F nucleatum is able to regulate miR21 targets as well. Several already established targets of miR21 (PTEN, RECK, SPRY1 and RHOB) were tested in HCT116 and LoVo cells. After F nucleatum treatment, all of these miR21 downstream targets were downregulated in F nucleatum-treated cells when compared to F nucleatum-untreated group (Supplementary Figure 8), suggesting that F nucleatum infection affects CRC development likely through miR21. To identify novel miR21 target genes, we began to query the different public prediction database including DIANA-microT-CDSv5.0, miRanda-rel2010, miRDB4.0, miRWalk, PicTar5, PITA, RNA22v2, RNAhybrid2.1 and Targetscan6.225. Interestingly, we identified several novel potential candidates (KCNMB2, LUM, MSH2, TRPM7, CHD7, RASA1, and C17orf39), which were predicted in at least 8 databases (Figure 4A and Supplementary Table 2). We next overexpressed or inhibited miR21 in HCT116 and LoVo cells by using miR21 mimics or inhibitors. We found RASA1 was the only gene that was consistently downregulated in miR21 overexpressing cells while up-regulated in miR21 inhibition cells, suggesting RASA1 was a putative target gene of miR21 (Supplementary Figures 9–10 and Figure 4B). Moreover, we found RASA1 expression was downregulated in cancer vs. normal tissues in CRC patients, indicating the up-regulation of miR21 may lead to the reduced expression of RASA1 in CRC (Figure 4C).
Figure 4. F nucleatum regulates expression of miR21 target gene RASA1 and activates MAPK signaling pathway.
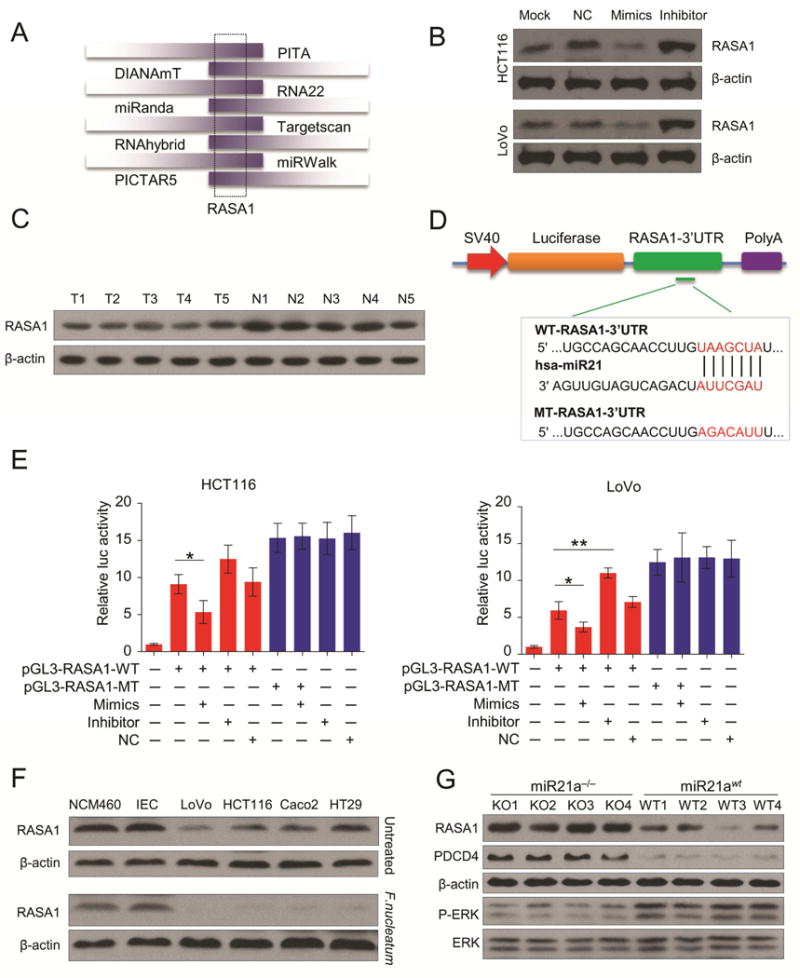
A) RASA1 was predicted as a putative miR21 target through analysis of 8 public prediction databases. B) The representative images of Western blot show overexpression or inhibiton of miR21 downregulated or upregulated RASA1 expression in HCT116 cells (n = 3 per group) and LoVo cells (n = 3 per group). C) Western blot showed RASA1 expression was lower in cancer tissues compared to matched normal tissues from CRC patients (n = 5 per group). D) We generated luciferase reporter plasmids which harbor either wild type (WT) or mutant (MT) miR21 binding sites in 3′-URT of RASA1. E) HCT116 and LoVo were transiently transfected with luciferase constructs along with miR21 mimics, inhibitors or negative controls. After 48hr, the luciferase activity was measured. All bars represent the mean values ± s.d. of three independent experiments. (*P < 0.05, **P < 0.01 by unpaired Student’s t test) F) The protein expression levels of RASA1 were significantly down-regulated in F nucleatum-infected CRC cells, while F nucleatum infections failed to increase RASA1 expression in normal intestinal epithelial cells NCM460 and IEC cells (n = 3). Compared to miR21 KO mice, the expression of RASA1 and another known miR21 target PDCD4 was significantly reduced, but P-ERK overexpressed in F nucleatum-infected miR21 WT mice tissues (n = 4 mice per group).
To support our hypothesis that miR21 directly regulates RASA1 expression through its 3′-UTR, we generated luciferase reporter plasmids which harbored either wild type (WT) or mutated (MT) miR21 binding sites within the 3′-UTR of RASA1 (Figure 4D). HCT116 and LoVo were transiently transfected with luciferase constructs along with miR21 mimics, inhibitors or negative controls. In line with our previous results, miR21 overexpression or inhibition significantly suppressed or increased luciferase activity of the reporter genes containing WT 3′-UTR of RASA1, but no inhibitory effects were observed in mutated cell lines (Figure 4E). Collectively, these results indicated that miR21 suppressed expression of RASA1 through direct binding within the putative 3′-UTR binding sites of RASA1.
RASA1 is a member of RAS GTPase activating proteins (RAS-GAP) family. The well-known oncoprotein RAS can be inactivated through binding to the RAS-GAP members26. Several studies have shown that RASA1 mutation or loss of function in CRC results in the activation of RAS-MAPK cascade27–29. Therefore, we assumed that F nucleatum infection may lead to the reduced expression of RASA1 expression through miR21, and result in the activation of MAPK signaling pathway. As shown in Figure 4F, the expression of RASA1 was significantly down-regulated in F nucleatum-infected CRC cells. On the contrary, F nucleatum infections failed to increase RASA1 expression in normal intestinal epithelial cells NCM460 and IEC cells probably due to their low expression of miR21. Likewise, compared to miR21a−/− mice, the expression of RASA1 and another known miR21 target PDCD4 were significantly reduced, but the phosphorylation of extracellular signal-regulated kinase (P-ERK) was overexpressed in F nucleatum-infected miR21 WT mice tissues (Figure 4F). Taken together, we identified novel miR21 target RASA1 in CRC, and that F nucleatum can regulate RASA1 expression and activate MAPK pathway in a miR21 dependent manner.
TLR4/MYD88/NFκB pathway is activated by F nucleatum infection
Although F nucleatum infection can induce miR21 expression in CRC, the exact mechanisms for regulation of miR21 upon F nucleatum infection remain unclear. Hence, we aimed to find certain pathways that are affected by F nucleatum and identify the upstream regulators that could potentially control expression levels of miR21. We treated HCT116 cells with F nucleatum or PBS and then performed gene expression microarray analysis. The pathway analysis (KEGG and Reactome pathways) implicated F nucleatum infection can significantly stimulate TLR4/MYD88/NFκB pathway in CRC cells (Figure 5A). To validate this result, we incubated HCT116 cells with F nucleatum, DH5α or PBS for 6h, 12h and 24h. The phosphorylation levels of NFκB subunit p65 and p50 were substantially up-regulated, while NFκB inhibitor IκB-α was down-regulated by F nucleatum or DH5α treatment compared to PBS treatment, suggesting that activation of the NFκB pathway is a frequent event that occurs following bacterial infection (Figure 5B). Furthermore, silencing of NFκB significantly impaired the F nucleatum’s oncogenic effect on cell proliferation and cell invasion (Figure 5C and Supplementary Figure 11). Of note, we found CRC tissues with high amount of F nucleatum showed hyperactivity of NFκB, suggesting that F nucleatum requires NF- κB activation to affect CRC cells phenotype (Figure 5D).
Figure 5. TLR4/MYD88/NFκB pathway is activated by F nucleatum infection.
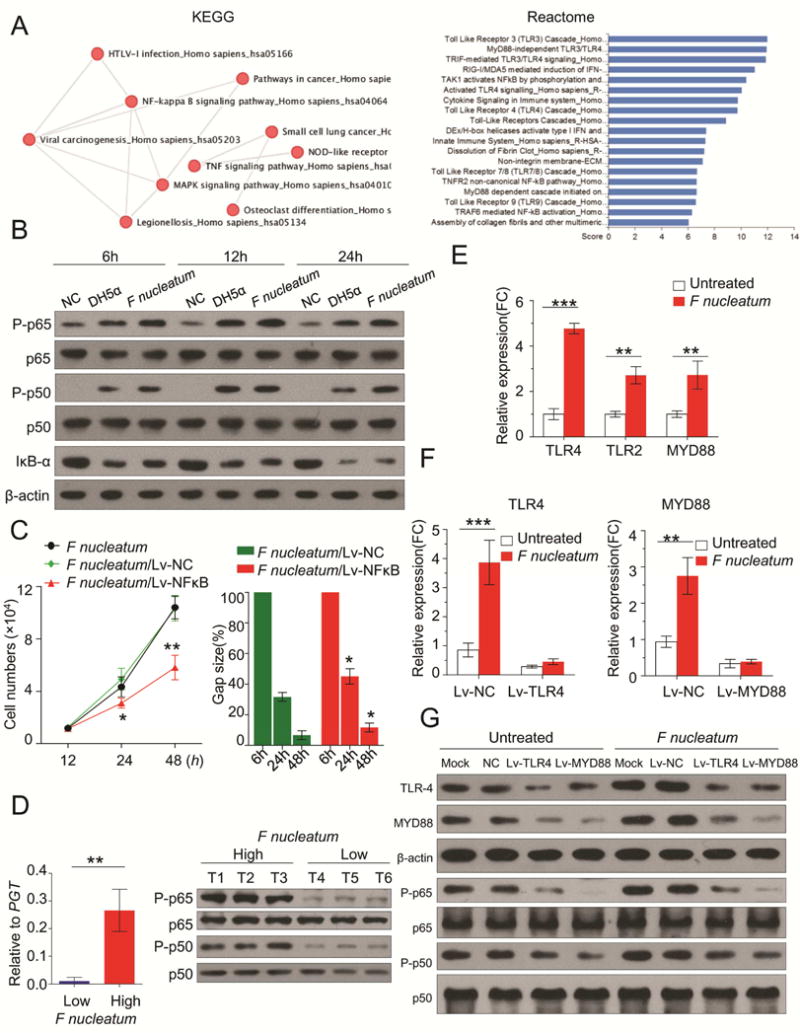
A) We treated HCT116 cells with F nucleatum or PBS (n = 3 per group) and then performed gene expression microarray analysis. The pathway analysis (KEGG and Reactome pathways) implicates F nucleatum infection can significantly stimulate TLR4/MYD88/NFκB pathway in CRC cells. B) HCT116 cells were incubated with F nucleatum, DH5α or PBS for 6h, 12h and 24h. NFκB subunit p65 and p50, phosphorylation level of p65 and p50 (P-p65 and P-p50), and inhibitor IκB-α were measured by Western blot. C) Silencing of NFκB significantly impaired the F nucleatum’s oncogenic effect on cell proliferation (proliferation assay) and cell invasion (wound healing assay) (*P < 0.05, **P < 0.01 by unpaired Student’s t-test. Bars represent SD of three experiments). D) qPCR results of F nucleatum DNA level in the low or high group (left figure, n = 6). Western blots analysis indicated that CRC tissues with high burden of F nucleatum showed activation of NFκB (right figure, n = 3). E) F nucleatum treatment induced the TLR4, TLR2, MYD88 expression in HCT116 (**P < 0.01, ***P < 0.001 by unpaired Student’s t-test. Bars represent SD of three experiments). F) qRT-PCR showed successfully silencing of TLR4 or MYD88 by Lv-TLR4 or Lv-MYD88 (**P < 0.01, ***P < 0.001 by unpaired Student’s t-test. Bars represent SD of three experiments). Western blots illustrated either TLR4 or MYD88 knockdown prevented NFκB activation from F nucleatum infection.
We next investigated the specific surface receptors which can bind to F nucleatum and transmit signaling to NFκB pathway. As shown in Figure 5A, our microarray data showed that toll-like receptor (TLRs)/MYD88 cascade was substantially activated upon F nucleatum infection. We further validated that F nucleatum treatment induced the TLR4, TLR2, MYD88 expression in HCT116 cells (Figure 5E). Recently, several studies have reported that lipopolysaccharide (LPS) of F nucleatum can be recognized by TLR430, 31. Also, it is widely acknowledged that NFκB is the key downstream effector of TLR4 signaling32, 33. Thus, we conclude that F nucleatum stimulates NFκB pathway through activation of TLR4/MYD88 cascade. We performed knockdown of either TLR4 or MYD88 in HCT116 cells and subsequently incubated HCT116 with F nucleatum or PBS. The results showed that silencing of TLR4/MYD88 cascade prevents NFκB activation from F nucleatum infection (Figures 5F). Collectively, we herein identified TLR4/MYD88/NFκB as a direct signaling pathway that is stimulated by F nucleatum in CRC cells.
F nucleatum regulates miR21 expression through TLR4/MYD88/NFκB pathway
Since F nucleatum is able to activate TLR4/MYD88/NFκB pathway, we wondered whether this pathway could be involved in the regulation of miR21 by F nucleatum. Because NFκB is a transcription factor, we assumed TLR4/MYD88/NFκB pathway may regulate miR21 expression in a promoter dependent manner. Therefore, we exploited bioinformatic analysis to decipher potential NFκB binding sites in upstream of transcription start site (TSS) of miR21. Interestingly, we identified the consensus binding sequence (5′GGAGGACTCC3′) for p65 in the promoter region of miR21. To determine whether NFκB can bind to this region and regulate miR21 expression, we first knocked down p65 in HCT116 and LoVo and observed significant down-regulation of miR21 expression (Figure 6A and 6B). Further, we generated luciferase reporter plasmids containing either wild type (WT) or mutant (MT) p65 binding sites in miR21 promoter region to examine the direct interaction between NFκB and miR21 promoter (Figure 6C). We transfected p65 shRNA or luciferase reporter constructs (pGL3-WT and pGL3-MT) in HEK293T cells. The luciferase reporter assay showed that co-transfection of pGL3-WT and p65 shRNA resulted in a significant decrease in the promoter activity of miR21, while p65 knockdown had no effect on mutant miR21 promoter activity (Figure 6C), supporting that NFκB regulates miR21 promoter activity in CRC. To further support our hypothesis that F nucleatum enhanced the binding of NFκB to miR21 promoter, we treated HCT116 and LoVo cells with F nucleatum or PBS following Chromatin immunoprecipitation (ChIP) assay. Consistently, qPCR results showed a more than 5 fold enrichment of miR21 promoter in p65 pulled-down DNA samples compared to IgG samples (Figure 6D and 6E). Our results clearly showed that F nucleatum induce miR21 expression through NFκB.
Figure 6. F nucleatum regulates miR21 expression through TLR4/MYD88/NFκB pathway.
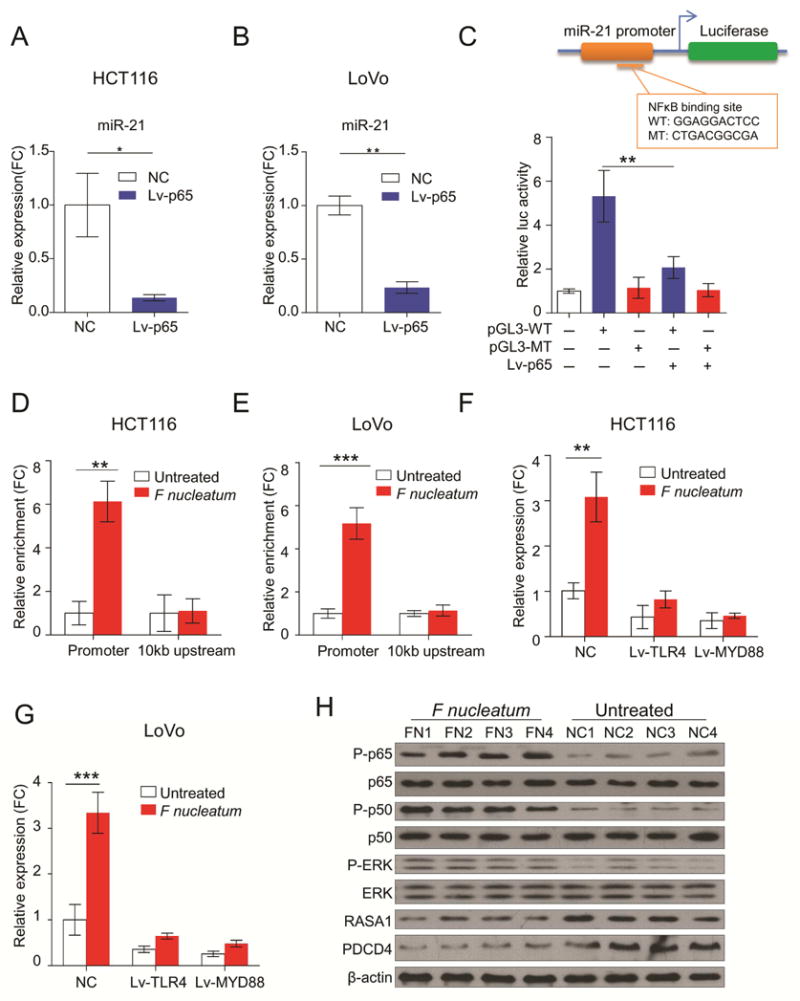
A and B) Knockdown of p65 in HCT116 or LoVo significantly down-regulated miR21 expression (**P < 0.01 by unpaired Student’s t test. Bars represent SD of three experiments). C) We generated luciferase reporter plasmids containing either wild type (WT) or mutant type (MT) of p65 binding sites in miR21 promoter. HEK293T cells were transfected with p65 shRNA or luciferase constructs (pGL3-WT and pGL3-MT) (**P < 0.01 by unpaired Student’s t test). D and E) HCT116 cells and LoVo were treated with F nucleatum or PBS following ChIP assay. qPCR results showed a more than 5 fold enrichment of miR21 promoter in p65 pulled-down DNA samples compared to IgG samples (**P < 0.01 by unpaired Student’s t test). F and G) The qRT-PCR assay indicated that F nucleatum failed to up-regulate miR21 expression when TLR4 or MYD88 was silenced in HCT116 and LoVo (**P < 0.01 by unpaired Student’s t test). H) Western blot analysis showed that CRC tissues from F nucleatum-treated APCmin/+ mice showed activation of NFκB, as well as other members including RASA1, PDCD4 and MAPK pathway (n = 4 mice per group).
More interestingly, we observed that F nucleatum failed to up-regulate miR21 expression when TLR4 or MYD88 were silenced in HCT116 and LoVo cells, highlighting the importance of TLR4/MYD88/NFκB pathway in the regulation of miR21 by F nucleatum (Figure 6F and 6G). Furthermore, CRC tissues from F nucleatum treated APCmin/+ mice showed hyperactivity of miR21 upstream regulator: NFκB, as well as activation of miR21 downstream targets: RASA1, PDCD4 and MAPK pathway (Figure 6H). Together, our results strongly suggest that F nucleatum regulates miR21 expression through TLR4/MYD88/NFκB pathway in CRC.
Overabundance of F nucleatum correlates with high expression of miR21 in human CRC tissues and indicates poor clinical outcome
Although we have interrogated the molecular regulation of miR21 by F nucleatum, the clinical significance of this regulation remains unknown. We used fluorescence in situ hybridization (FISH) assays to image the F nucleatum infection as well as miR21 expression in CRC tissues and adjacent normal tissues. Compared to normal mucosa, F nucleatum was found to invade CRC tissues, often accompanied by high levels of miR21 (Figure 7A). To further demonstrate the expression correlation between F nucleatum and miR21, we measured F nucleatum at both DNA level and miR21 transcript expression in 90 matched fresh frozen CRC and normal tissues. Strikingly, both F nucleatum DNA and miR21 levels were higher in cancer tissues (P = 0.0083 for F nucleatum, P < 0.001 for miR21, Figure 7B). Furthermore, the F nucleatum DNA and miR21 expression was specifically much higher in advanced tumors (Stage III/IV, Figure 7C). Notably, the overabundance of F nucleatum was associated with advanced T stage (P = 0.027), proliferation marker Ki-67 expression (P = 0.017) and lymphatic invasion (P = 0.031). The up-regulation of miR21 was associated with distant metastasis (P=0.006), and TNM stage (P=0.008), suggesting that F nucleatum and miR21 have the same oncogenic role in cell proliferation and invasion, and promotion of CRC development (Supplementary Table 3). Consistently, we found high F nucleatum levels were robustly associated with high expression of miR21 in cancer tissues, suggesting that F nucleatum infection might have led to the induction of miR21 in invaded tumor tissues (r = 0.4537, P < 0.001, Figure 7D).
Figure 7. Overabundance of F nucleatum correlates with high expression of miR21 in CRC and indicates poor clinical outcome.
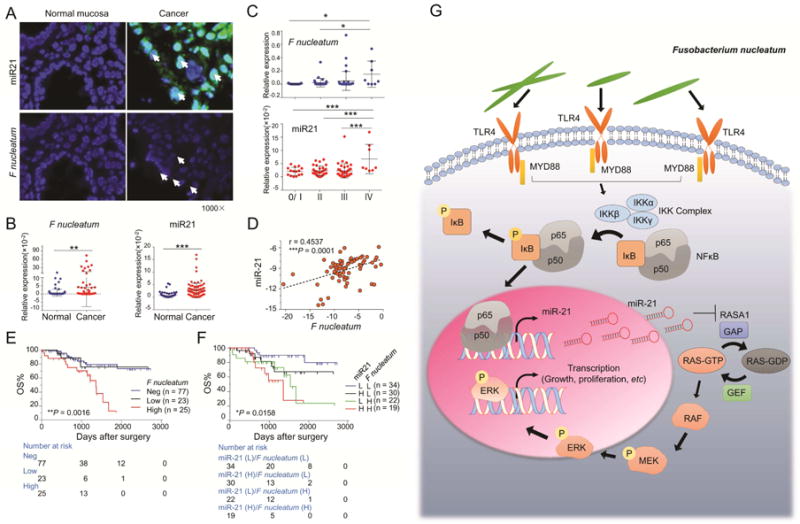
A) FISH assay showed F nucleatum (1000×) was enriched in the mucosa of CRC tissues and accompanied by high level of miR21 (1000×). The white arrows indicate positive staining. B) The qRT-PCR analysis showed that both the expression levels of F nucleatum and miR21 were higher in the cancer tissues than in the adjacent normal tissues (n = 90. **P < 0.01, ***P < 0.001 by unpaired Student’s t test. Bars represent SD). C) The qRT-PCR analysis showed F nucleatum and miR21 was overexpressed in cancer tissues in a stage dependent manner (n = 90. *P < 0.05, ***P < 0.001 by one way ANOVA and Bonferroni’s multiple comparison test. Bars represent SD). D) The amount of F nucleatum DNA was positively associated with miR21 expression in cancer tissues (n = 90. ***P < 0.001 by two-tailed nonparametric Spearman correlation.). E) Kaplan–Meier survival curve for 125 clinical specimens showed high amount of F nucleatum were associated with remarkably poor overall survival (OS) in CRC patients (log-rank (Mantel-Cox) test). Furthermore, subgroup of CRC patients with both high F nucleatum DNA level and miR21 expression has a higher risk clinical outcome (log-rank (Mantel-Cox) test). Neg, negative; L, low; H, high. F) The illustration of the hypothetical mechanism by which F nucleatum regulates miR21 expression through TLR4/MYD88/NFκB pathway in CRC.
To investigate the prognostic impact of regulation of miR21 by F nucleatum in CRC patients, we used another independent cohort of CRC patients, with complete follow-up information (fresh frozen tissues, n = 125). As shown in Figure 7E, high amount of F nucleatum correlated with remarkably poor overall survival in CRC patients, compared to the patients with low or undetectable amount of F nucleatum. More importantly, when we segregated patients into 4 groups based on F nucleatum levels and miR21 expression, we discovered that the group with both high F nucleatum and high miR21 expression showed worst prognosis compared to other 3 groups (Figure 7E).
Taken together, our results showed that F nucleatum is heavily enriched in cancer tissues and more so in the advanced tumors, leading to the high expression of miR21 in locally-advanced tumors. Furthermore, a subgroup of CRC patients with both high F nucleatum levels and miR21 expression has a higher risk for poor clinical outcomes, which has important implications for their clinical management.
DISCUSSION
Identification of specific carcinogenic microorganisms remain an important area of research for many cancers, particularly CRC34. Among the recently identified and published CRC-related microbes, the recently-discovered F nucleatum has garnered most attention. Herein, we for the first time have identified that miR21 is a downstream target of F nucleatum in the colon. Furthermore we have unraveled the underlying molecular mechanism(s) that TLR4/MYD88/NFκB signaling pathway is activated upon the F nucleatum infection, and hyperactive NFκB subsequently binds to the promoter of miR21 and initiates an oncogenic cascade in CRC (Figure 7F). Consistent with these results, expression levels of F nucleatum and miR21 are heavily enriched in cancer tissues and particularly in advanced tumors. The F nucleatum levels were positively associated with miR21 in cancer tissues, suggesting that F nucleatum infection leads to the induction of miR21 in tumor tissues, and such patients often have higher risk and worse clinical outcomes.
Although recent studies have implicated overabundance of F nucleatum in association with colorectal adenomas and cancer, it remains uncertain whether F nucleatum is a causative pathogen for CRC or only an opportunistic pathogen or simply a commensal bacteria associated with the CRC microenvironment35, 36. Our data reveal that F nucleatum infection increase tumor burden in APCMin/+ mice and stimulate the increase of serum inflammatory factors, suggesting that F nucleatum plays a driver role, instead of being simple a passenger, in intestinal tumorigenesis. Furthermore, we also found that F nucleatum infection promotes the proliferation and invasion both in vitro and in vivo, and F nucleatum infection is a more frequent phenomenon in advanced tumors; suggesting F nucleatum infections not only contribute to the initiation of CRC but also to the development of CRC.
To understand how F nucleatum promotes CRC, several groups have previously characterized the interaction between F nucleatum and intratumoral immune cells, and have shown that F nucleatum promotes inflammation and tumorigenesis by modulating the tumor immune microenvironment via expansion of myeloid derived immune cells6, 37. Interestingly, we noticed that a recent study provided novel mechanistic insights that F nucleatum can invade epithelial cells via its virulence factors FadA and activate pro-oncogenic signals to promote CRC, suggesting that F nucleatum may not only regulate immune cells but also targets tumor cell itself12. Since we have previously reported the role of several miRNAs in CRC, we hypothesized that F nucleatum infection may have a potential impact on miRNA expression during the progression of CRC. Surprisingly, we discovered that a large number of miRNAs were differentially expressed upon F nucleatum infection and miR21 was among the most robustly induced by F nucleatum infection. Our previous studies have demonstrated that miR21 plays a pathogenic role in chronic inflammatory processes and the development of colitis-associated colon cancer15. It is conceivable that the presence of F nucleatum drives oncogenic signaling through induction of oncogenic miR21. Nevertheless, our results for the first time have established a crosstalk between F nucleatum infection and epithelial cells miRNA genes. Interestingly, a recent review published by Nosho et al. in 201638 predicted the underlying association of F nucleatum with miR21 in CRC, which happens to be consistent with our present results. It is worth noting that a recent study showed that host fecal miRNAs modulated genes expression in F nucleatum and therefore affected bacterial growth, suggesting that miRNAs may bridge the communication gap between F nucleatum and its host39.
Our data also firstly illustrated that F nucleatum can downregulate the expression of miR21 downstream targets such as RASA1. The MAPK signaling pathway is a major miR21 regulated network that empowers cells with an aggressive phenotype, and is often inappropriately activated in CRC40, 41. Despite the presence of KRAS mutations, recent studies indicate that CRC are deficient in GAP due to its collaborative action with onco-miRs29. To this end, F nucleatum could be a promising therapeutic target which can help alleviate inhibition of RASA1 from miR21 and prevent activation of the MAPK pathway.
In this study, we attempted to reveal the mechanism for the regulation of miR21 by F nucleatum. We found F nucleatum-mediated infection leads to a strong induction of TLR4/MYD88 and results in the sustained activation of NFκB. The hyperactive NFκB binds to the promoter of miR21 and increases its transcription level. However, the modulation of miR21 by F nucleatum seems to be more complicated. Despite the fact that F nucleatum activated TLR4/MYD88/NFκB, we also noticed that DH5α could activate this pathway. However, we did not notice up-regulation of miR21 following DH5α infection, suggesting that F nucleatum adopts other collaborative mechanisms to enhance the effect of TLR4/MYD88/NFκB on miR21 expression. Notwithstanding the above, TLR4/MYD88/NFκB could, at least in part, explain how F nucleatum infection stimulates the overexpression of miR21 in CRC.
From a clinical viewpoint, we further propose that in addition to its functional and therapeutic role, using F nucleatum and miR21 levels offer an added value as integrated biomarkers for better prognosis assessment of CRC, considering that a subgroup of CRC patients with their high levels associate with a higher risk and worse clinical outcome.
In conclusion, our current findings provide critical insights into the molecular mechanisms for the regulation of miR21 by F nucleatum. The clinical results presented here also highlight the emergence of both F nucleatum and miR21 as risk factors for survival outcomes in CRC patients. Our data provides previously unrecognized and novel evidence for a pathogenic role of F nucleatum in CRC, which opens new horizons for targeting microbiota as well as miRNAs alterations for colorectal cancer prevention and treatment.
Supplementary Material
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (No. 81372615, No.81230057, No.81472262, and No.81302066), the National High Technology Research and Development Program (863 Program; Grant No.2014AA020803), the National Science and Technology Major Projects (No.2013ZX09103003-16 and No.2012YQ18013209), the Shanghai Health System Outstanding Young Talent Training Plan (No. XYQ2013118), and the Emerging cutting-edge technology joint research projects of Shanghai (No.SHDC12012106). In addition, the present work was supported by the grants R01 CA72851, CA 181572, CA184792 and U01 CA187956 from the National Cancer Institute, National Institute of Health, pilot grants from the Baylor Sammons Cancer Center and Foundation, as well as funds from the Baylor Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors have no competing financial, personal or professional interests to disclose.
Author contribution:
Study concept and design: HQ, YY, SC, WW, YM, AG; Specimen provider: LH, LY, RG, ML, MY, CP, HL, BG, QZ, QW; NCM460 provider: MPM; Acquisition of clinical data: YY, RG, CP, HL, QW, WW; Analysis and interpretation of data and statistical analysis: YM, HQ, YY, WW, PW, SC, JP, and AG; Animal experiments: YY, LH, CP, HL; Drafting of the manuscript: YM, YY, HQ, SC, WW, AG.
References
- 1.Song M, Garrett WS, Chan AT. Nutrients, foods, and colorectal cancer prevention. Gastroenterology. 2015;148:1244–60 e16. doi: 10.1053/j.gastro.2014.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Zackular JP, Baxter NT, Iverson KD, et al. The gut microbiome modulates colon tumorigenesis. MBio. 2013;4:e00692–13. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sommer F, Backhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–38. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 5.Marchesi JR, Adams DH, Fava F, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–9. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–15. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito M, Kanno S, Nosho K, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137:1258–68. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 8.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2015 doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahara T, Yamamoto E, Suzuki H, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–8. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–55. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakagaki H, Sekine S, Terao Y, et al. Fusobacterium nucleatum envelope protein FomA is immunogenic and binds to the salivary statherin-derived peptide. Infect Immun. 2010;78:1185–92. doi: 10.1128/IAI.01224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abed J, Emgard JE, Zamir G, et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–25. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi C, Yang Y, Xia Y, et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut. 2015 doi: 10.1136/gutjnl-2014-308455. [DOI] [PubMed] [Google Scholar]
- 16.Hur K, Toiyama Y, Okugawa Y, et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2015 doi: 10.1136/gutjnl-2014-308737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada A, Horimatsu T, Okugawa Y, et al. Serum miR-21, miR-29a, and miR-125b Are Promising Biomarkers for the Early Detection of Colorectal Neoplasia. Clin Cancer Res. 2015;21:4234–42. doi: 10.1158/1078-0432.CCR-14-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hur K, Toiyama Y, Schetter AJ, et al. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/dju492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y, Zhang P, Wang F, et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61:1447–53. doi: 10.1136/gutjnl-2011-301122. [DOI] [PubMed] [Google Scholar]
- 20.Ma Y, Zhang P, Wang F, et al. Elevated oncofoetal miR-17–5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun. 2012;3:1291. doi: 10.1038/ncomms2276. [DOI] [PubMed] [Google Scholar]
- 21.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 22.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–4. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 23.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cantini L, Isella C, Petti C, et al. MicroRNA-mRNA interactions underlying colorectal cancer molecular subtypes. Nat Commun. 2015;6:8878. doi: 10.1038/ncomms9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 26.Ohta M, Seto M, Ijichi H, et al. Decreased expression of the RAS-GTPase activating protein RASAL1 is associated with colorectal tumor progression. Gastroenterology. 2009;136:206–16. doi: 10.1053/j.gastro.2008.09.063. [DOI] [PubMed] [Google Scholar]
- 27.Kent OA, Mendell JT, Rottapel R. Transcriptional Regulation of miR-31 by Oncogenic KRAS Mediates Metastatic Phenotypes by Repressing RASA1. Mol Cancer Res. 2016;14:267–77. doi: 10.1158/1541-7786.MCR-15-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun D, Wang C, Long S, et al. C/EBP-beta-activated microRNA-223 promotes tumour growth through targeting RASA1 in human colorectal cancer. Br J Cancer. 2015;112:1491–500. doi: 10.1038/bjc.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun D, Yu F, Ma Y, et al. MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1) J Biol Chem. 2013;288:9508–18. doi: 10.1074/jbc.M112.367763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.To TT, Gumus P, Nizam N, et al. Subgingival Plaque in Periodontal Health Antagonizes at Toll-Like Receptor 4 and Inhibits E-Selectin Expression on Endothelial Cells. Infect Immun. 2016;84:120–6. doi: 10.1128/IAI.00693-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H, Redline RW, Han YW. Fusobacterium nucleatum induces fetal death in mice via stimulation of TLR4-mediated placental inflammatory response. J Immunol. 2007;179:2501–8. doi: 10.4049/jimmunol.179.4.2501. [DOI] [PubMed] [Google Scholar]
- 32.Mukherji A, Kobiita A, Ye T, et al. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–27. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa S, Lozach J, Benner C, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–21. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hold GL, Garrett WS. Gut microbiota. Microbiota organization–a key to understanding CRC development. Nat Rev Gastroenterol Hepatol. 2015;12:128–9. doi: 10.1038/nrgastro.2015.25. [DOI] [PubMed] [Google Scholar]
- 35.Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141–7. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colucci F. An oral commensal associates with disease: chicken, egg, or red herring? Immunity. 2015;42:208–10. doi: 10.1016/j.immuni.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 37.Mima K, Sukawa Y, Nishihara R, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1:653–61. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nosho K, Sukawa Y, Adachi Y, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22:557–66. doi: 10.3748/wjg.v22.i2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S, da Cunha AP, Rezende RM, et al. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe. 2016;19:32–43. doi: 10.1016/j.chom.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma SB, Lin CC, Farrugia MK, et al. MicroRNAs 206 and 21 cooperate to promote RAS-extracellular signal-regulated kinase signaling by suppressing the translation of RASA1 and SPRED1. Mol Cell Biol. 2014;34:4143–64. doi: 10.1128/MCB.00480-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung EJ, Calin GA. The Meaning of 21 in the MicroRNA world: perfection rather than destruction? Cancer Cell. 2010;18:203–5. doi: 10.1016/j.ccr.2010.08.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


