Abstract
Extensive studies have suggested that most miRNA functions are executed through complex miRNA-target interaction networks, and such networks function semiredundantly with other regulatory systems to shape gene expression dynamics for proper physiological functions. We found that knocking down vgln-1, which encodes a conserved RNA-binding protein associated with diverse functions, causes severe larval arrest at the early L1 stage in animals with compromised miRISC functions (an ain-2/GW182 mutant). Through an enhancer screen, we identified five specific miRNAs, and miRNA families, that act semiredundantly with VGLN-1 to regulate larval development. By RIP-Seq analysis, we identified mRNAs that are directly bound by VGLN-1, and highly enriched for miRNA binding sites, leading to a hypothesis that VGLN-1 may share common targets with miRNAs to regulate gene expression dynamics for development.
Keywords: miR-2, miR-45, miR-51, miR-83, miRNA, vgln-1, Vigilin
miRNA-mediated gene silencing is crucial for all aspects of cellular processes, including animal development, metabolism, stress responses, and neuronal behaviors (Ambros 2004; Bartel 2009). Misregulation of miRNAs has been linked to numerous human diseases (Li and Kowdley 2012; Mendell and Olson 2012; Orellana and Kasinski 2015). Although disrupting lin-4 and let-7 produces a robust developmental phenotype (Vella and Slack 2005), the vast majority of miRNAs and miRNA families are not essential for development or viability (Miska et al. 2007; Alvarez-Saavedra and Horvitz 2010). Due to redundancy and complex interactions between multiple miRNAs and multiple targets, single gene genetic methods are not sufficient to elucidate the physiological roles of many miRNAs. Our laboratory, and others, have provided evidence to support the idea that miRNAs function redundantly with other gene regulatory mechanisms to ensure developmental robustness (Hammell et al. 2009a; Brenner et al. 2010; Vidigal and Ventura 2014; Weaver et al. 2014; Ren et al. 2016).
Our previous study demonstrated that compromising overall miRISC function by mutating one of two C. elegans GW182 proteins dramatically enhanced developmental defects associated with RNAi knockdown of a large number of genes (Weaver et al. 2014). In many cases, the phenotypes were synergistic. One such gene identified, C08H9.2, encodes a putative RNA binding protein homologous to mammalian HDLBP or Vigilin (Weber et al. 1997; Rohwedel et al. 2003). We thus named the C08H9.2 gene vgln-1 (ViGiLiN).
Studies in yeast, Drosophila, and humans have shown that Vigilin genes are involved in various cellular processes, including cell division, mRNA transport, RNA editing, translation, heterochromatin formation, and cancer (Weber et al. 1997; Frey et al. 2001; Li et al. 2003; Wang et al. 2005; Hogan et al. 2008; Zhou et al. 2008; Woo et al. 2011; Gelin-Licht et al. 2012; Mitchell et al. 2013; Molyneux et al. 2014; Otsuka et al. 2014; Yang et al. 2014; Mobin et al. 2016). The diversity of functions and phenotypes associated with this RNA binding protein suggests that the Vigilin proteins have diverse RNA targets.
The functional relationship between Vigilin and miRNAs has not been previously explored. However, RNA binding proteins have been shown to directly or indirectly influence miRNA regulation of target mRNAs for specific cellular and physiological functions (Hammell et al. 2009b; van Kouwenhove et al. 2011; Akay et al. 2013; Ciafrè and Galardi 2013). It is not clear whether a more general RNA-binding protein, or domain, is commonly involved in miRNA-mediated silencing. In this study, we applied genetic enhancer analysis to investigate the functional relationship between VGLN-1 and miRNAs during Caenorhabditis elegans larval development. Our study uncovered previously unknown roles of both VGLN-1 and several specific miRNAs in larval development, and the data suggest potential collaborative regulation by VGLN-1 and miRNAs on common mRNA targets.
Materials and Methods
RNA interference (RNAi)
RNAi constructs targeting either the 3′end or 3′UTR of vgln-1 were constructed by amplifying 500 bp of the region of interest, cloning them into the L4440 vector, and transforming into HT115 bacteria. Primer sequences were:
3′ UTR forward (with EagI), ataaCGGCCGGatgattcaaccaacgtcaataac.
3′UTR reverse (with XhoI), atgcCTCGAGTtttgattacaccaattcagtttatta.
5′end forward (with XbaI), ataaTCTAGAAGGACAACTCCAGCGAGGA.
5′end reverse (with HindIII), ataaAAGCTTCTCGACGTTGTCAAGCAAGA.
All other RNAi bacteria were obtained from the ORFeome RNAi feeding library (Reboul et al. 2003) and the C. elegans RNAi library (Kamath et al. 2003). All RNAi was delivered by feeding. An RNAi sensitive strain NL2099 with a rrf-3 mutation was used (Supplemental Material, Figure S1A in File S1). To achieve a strong vgln-1 (RNAi) effect, bacteria were picked from a fresh colony, and grown for ∼8 hr with 100–200 μg/ml ampicillin and 15 μg/ml tetracycline. Alternatively, an overnight culture was diluted and grown for 4 hr. After spotting on nematode growth medium (NGM) plates with 0.4 mM isopropyl-β-d-thiogalactoside (IPTG) and 100 μg/ml ampicillin, plates were allowed to dry for 4 d before adding L4 stage worms. After the worms reached gravid adult stage, they were transferred to new RNAi plates and allowed to lay eggs for 24 hr. The progeny were then scored ∼40 hr later for the L1 arrest phenotype.
For brood size analysis, one L4 stage nematode of the double mutant was randomly selected from each plate (NGM medium) and transferred to a fresh plate. A total of 18 worms was counted. The worms were transferred to fresh plates daily until they stopped laying eggs, and the number of eggs was recorded every day. The nematode brood size was determined based on the sum of total eggs laid by individual hermaphrodites.
Generating transgenic strains
Plasmids were obtained from Addgene. pDD162 (Peft-3::Cas9 + Empty sgRNA) was a gift from B. Goldstein (Addgene plasmid # 47549) (Dickinson et al. 2013). pCFJ350—MCS(ttTi5605, II) and pMA122—peel-1 negative selection were gifts from E. Jorgensen (Addgene plasmid # 34866 and # 34873) (Frøkjær-Jensen et al. 2012). A short guide RNA to target Cas9 to the endogenous vgln-1 locus was designed and cloned into pDD162. A homologous recombination template to replace the vgln-1 gene with unc-119 from pCFJ350 was cloned into pBIISK(+). Selectable markers were used to isolate a null allele based on the published protocol (Dickinson et al. 2013). Loss of the vgln-1 gene in strain MH5004 vgln-1(kuIs104) was further confirmed by PCR.
The transcriptional reporter Pvgln-1::gfp (kuIs103) (strain MH4922) consists of the vgln-1 promoter driving GFP with the vgln-1 3′UTR. The fusion reporter construct Pvgln-1::vgln-1::gfp (kuIs102) (strain MH4920) consists of the vgln-1 promoter driving the vgln-1 cDNA fused to GFP with the vgln-1 3′UTR. These constructs were cloned into the pCFJ350 vector with a functional unc-119 gene from Caenorhabditis briggsae and then introduced into the genome of unc-119 mutant worms by microparticle bombardment (Merritt 2010). Three out of four lines obtained for each reporter exhibited high levels of GFP expression. These strains were also used for CLIP and RIP-Seq.
Crosslinking and immunoprecipitation (CLIP)
Mixed-stage worms containing either Pvgln-1::vgln-1::gfp (kuIs102) (strain MH4920) or Pvgln-1::gfp (kuIs103) (strain MH4922) were grown on NGM plates with HB101 bacteria, collected by washing with M9 buffer, and irradiated by UV (Broughton and Pasquinelli 2013) to crosslink protein to RNA, and then stored at −70°. Worms were lysed by grinding in liquid nitrogen and immediately resuspended in lysis buffer (10 mM Tris-Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, plus addition of complete mini Roche protease inhibitor pills, Thermo Fisher RNaseOut, 100 mM PMSF). Immunoprecipitation of protein crosslinked to RNA was performed with GFP-Trap A fusion proteins coupled to agarose beads according to the vendor’s protocol (Chromotek). RNA was digested, radiolabeled, and visualized as described (Moore et al. 2014).
RNA immunoprecipitation (IP) and sequencing (RIP-Seq)
Worms containing either Pvgln-1::vgln-1::gfp (kuIs102) (strain MH4920) or Pvgln-1::gfp (kuIs103) (strain MH4922) were grown on large peptone-enriched plates spotted with HB101. Worms were collected using S-basal medium, bleached, and synchronized for 30 hr in S-basal without cholesterol. The synchronized L1s were then flash-frozen in liquid nitrogen. Worms were lysed by grinding in liquid nitrogen, and immediately resuspended in lysis buffer (10 mM Tris-Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, plus addition of complete mini Roche protease inhibitor pills, Thermo Fisher RNaseOut, 100 mM PMSF). IP was performed with GFP-Trap A fusion proteins coupled to agarose beads according to the vendor’s protocol (Chromotek). Prior to IP, 100 μl of the lysate was removed and used for total RNA samples (Total). Following IP, RNA from strain MH4920 (IP) or strain MH4922 (CTRL) was digested using Trizol (Molecular Research Center) and 4-bromoanisole (Sigma). IP and CTRL immunoprecipitations were performed synchronously, and repeated for a total of three biological replicates. Library preparation of the resulting RNA samples was performed by the Biofrontiers Institute NGS Core Facility (University of Colorado at Boulder) using the Lexogen SENSE Total RNA-Seq Library Prep Kit. Sequencing was then performed on an Illumina HiSequation 2000 using a read size of 1 × 100.
Three libraries of VGLN-1::GFP (IP), two libraries of GFP (CTRL), and two libraries of total RNA (Total, pre-IP) were sequenced. Analysis was performed using Galaxy (Afgan et al. 2016). Quality controls were analyzed by FastQC from The Babraham Bioinformatics group at https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. The resulting raw reads were processed using the FASTX-Toolkit from the Hannon lab at http://hannonlab.cshl.edu/fastx_toolkit/download.html. First reads were trimmed by removing the first 9 bp (as recommended by Lexogen), followed by removing reads with an overall quality < 30. Reads were then mapped to the genome using TopHat (Trapnell et al. 2009) and the Illumina iGenome WBCel235.
Differential analysis was performed in multiple ways. Mapped reads were run through both the CuffLinks pipeline with CuffDiff (Trapnell et al. 2013) using standard settings, and in parallel through DESeq2 (Love et al. 2014) using HTSeq (Anders et al. 2015) for read counts. In both cases, comparisons were made between replicates of VGLN-1::GFP (IP) vs. GFP (CTRL); fold enrichment represents the ratio of (IP) over (CTRL). Additionally, IP enrichment analysis was performed using HTSeq and DESeq2 according to the following formula:
where “adjusted counts” for any transcript refers to:
Resulting CuffDiff results, DESeq2 results, and IP enrichment results were then compared for enrichment.
3′ UTR analysis
Resulting enriched targets were then analyzed for number of conserved miRNA seed sites per 1 kb of 3′ UTR length. 3′ UTR information, conserved miRNA site predictions (≥7mer), and 3′ UTR length information was all obtained from TargetScanWorm (Jan et al. 2011). For genes containing >1 3′ UTR isoform, the median number of conserved miRNA seed sites between all available isoforms was used. Conserved miRNA sites per 1000 bp were calculated by:
The conserved sites per 1000 bp of enriched targets identified by RIP-Seq were then filtered based on annotated expression during the L1 stage of development, and compared to the conserved sites per 1000 bp of all available 3′ UTRs in genes expressed during the L1 stage of development (Spencer et al. 2011).
Data availability
All strains used are listed in Table S1. Unless otherwise stated, strains were maintained and manipulated, as previously described (Brenner 1974; Stiernagle 2006), on NGM agar with Escherichia coli OP50 as a food source at 20°. Data from miRNA enhance screen, RIP-Seq and DESeq analyses, miRNA seed site analysis of VGLN-1 bound mRNAs, and analysis of VGLN-1-IP enrichment relative to total RNA are shown in Table S2, Table S3, Table S4, and Table S5, respectively.
Results
VGLN-1 is essential for development when miRISC function is compromised
While worms containing a loss of function (lf) ain-2 mutation, or treated with vgln-1(RNAi), are superficially wild type, treating ain-2(lf) with vgln-1(RNAi) led to synthetic developmental arrest of F1 progeny at the first larval stage (Figure S1A in File S1) (Weaver et al. 2014). To eliminate potential off-target effects, we performed RNAi analysis using three constructs targeting two nonoverlapping coding regions, and the 3′UTR region (Materials and Methods). RNAi treatment using each construct caused a partial L1 arrest phenotype in an RNAi-sensitive background, and the phenotype was dramatically enhanced by ain-2(lf), supporting the view that the phenotype is due to silencing of vgln-1 expression (Figure S1A in File S1).
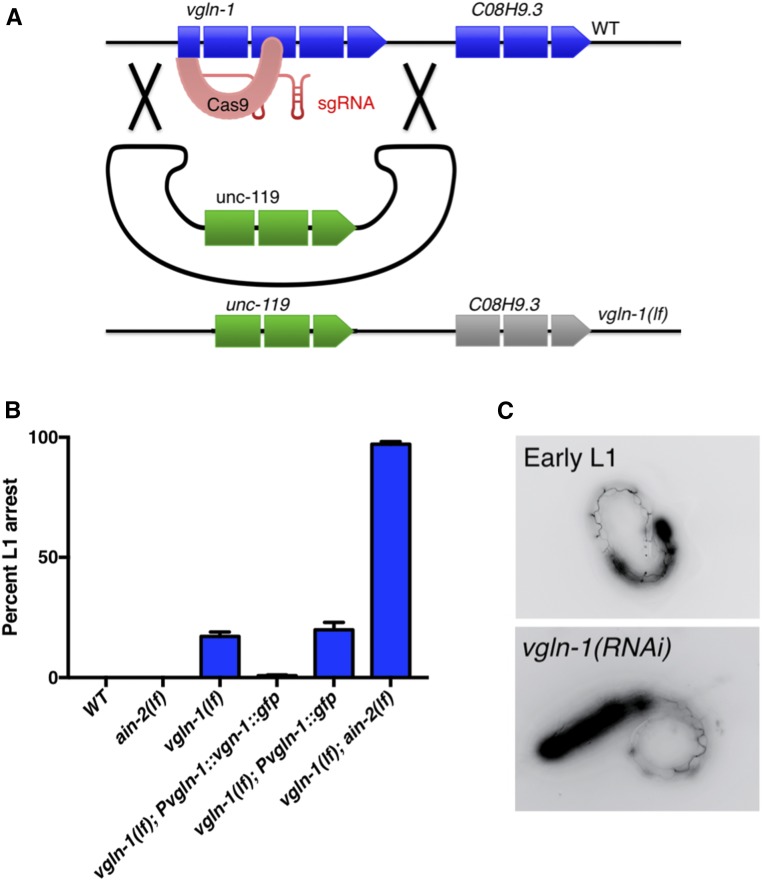
To facilitate further genetic analysis, we generated a vgln-1 null allele by applying the CRISPR gene knockout method (Figure 1A) (Dickinson et al. 2013). This vgln-1(kuIs104, lf) single mutant exhibited 20% L1 arrest, and a slight developmental delay. However, in combination with ain-2(lf), the double mutant exhibited a severe but incomplete sterile phenotype (although average brood size was >100, <75% of eggs hatched after 72 hr) (Figure S1, B and C in File S1). Among the progeny, 97% exhibited L1 arrest (Figure 1B) consistent with vgln-1(RNAi) tests. The partial L1 arrest phenotype of the vgln-1(lf) mutant was suppressed by expression of an integrated, translational fusion reporter Pvgln-1::vgln-1::gfp (kuIs102) (Figure 1B). These results indicate that the putative RNA binding protein VGLN-1 functions in early larval development and reproductive development, and is essential when miRISC function is compromised.
Figure 1.
VGLN-1 is essential for postembryonic development when miRISC is compromised. (A) Cartoon illustration of the construct design of vgln-l(lf) using CRISPR/Cas9. (B) Percentage of hatched animals arrested at the early L1 stage of indicated genotype. Loss of ain-2 dramatically enhances the phenotype of vgln-1(lf). The vgln-1::gfp fusion gene driven by the vgln-1 promoter is functional. Error bars represent SE of the proportion. Number of animals examined = N. vgln-1(lf) N = 410, ain-2(lf) N = 323, vgln-1(lf);ain-2(lf) N = 251, vgln-1(lf);Pvgln-1::vgln-1::gfp N = 824, and vgln-1(lf);Pvgln-1::gfp N = 167. (C) Images of arrested L1 larvae expressing an AJM-1::GFP fusion protein that marks adherens junctions between seam cells. The pattern of seam cells (Knight et al. 2002) indicates that rrf-3;vgln-1(RNAi) animals arrest development within 3 hr of hatching.
To better characterize the developmental arrest that occurs with knockdown of both vgln-1 and ain-2, we observed the pattern of an AJM-1::GFP reporter (Knight et al. 2002). During postembryonic development, seam cells divide in a characteristic pattern, and this reporter marks adherens junctions between seam cells. The pattern observed in arrested vgln-1(RNAi) larvae corresponds to 3 hr after hatching (Figure 1C), visually similar to that observed in larvae under starvation-induced diapause (Baugh and Sternberg 2006). We also found that intestinal tubulogenesis and apical membrane polarity (indicated by ERM-1::GFP), that have been found to be disrupted in some L1 arrested mutants (Zhang et al. 2012; Zhu et al. 2015), were unaffected by loss of vgln-1 (Figure S2 in File S1).
VGLN-1 binds to RNA and is broadly expressed throughout C. elegans development
Based on homology, and the presence of 14 KH domains, VGLN-1 is predicted to function as an RNA binding protein. To test its RNA binding property, proteins and RNAs were identified via UV CLIP from GFP tagged VGLN-1 strains described below and in Materials and Methods. The radiolabeled RNA from IP was detected by electrophoresis, and the RNA-protein complex was found to be sensitive to RNase treatment (Figure S3 in File S1), confirming that VGLN-1 binds RNA.
We found that the translational fusion reporter construct Pvgln-1::vgln-1::gfp (kuIs102) was expressed ubiquitously throughout development, notably in the intestine [consistent with previous observations (McGhee et al. 2007)], spermatheca, and vulva (Figure S4 in File S1). The reporter appears to be mostly excluded from the nucleus suggesting that VGLN-1 functions mainly in the cytoplasm.
Specific miRNAs and miRNA families act with VGLN-1 to regulate early larval development
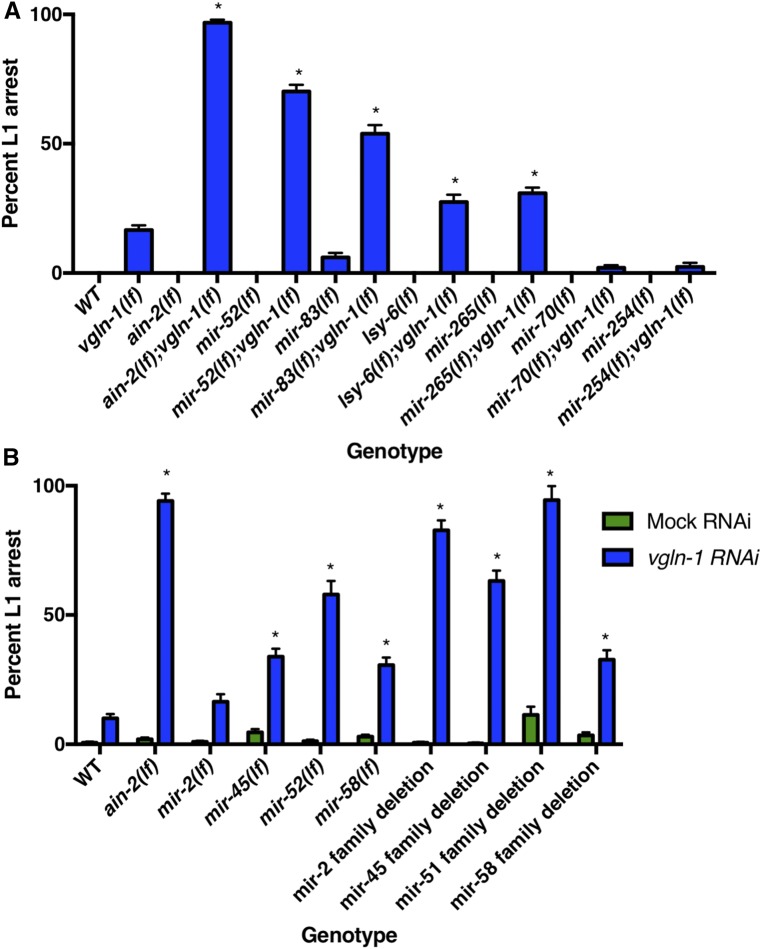
Based on the synergistic effect between vgln-1(lf) and ain-2(lf) on larval lethality, we hypothesized that VGLN-1 collaborates with multiple miRNAs to regulate many mRNA targets to promote larval development. To identify these specific miRNAs, we screened all the available C. elegans miRNA deletion strains for prominent enhancement of the L1 arrest phenotype using vgln-1(RNAi). Our screen identified several miRNAs of which the deletion mutations significantly enhanced the L1 arrest phenotype of vgln-1(RNAi) (Table S2). To exclude the possibility that the enhancement is due to increased sensitivity to RNAi in these miRNA deletion strains (Massirer et al. 2012), we constructed vgln-1(lf);miRNA(lf) double mutants, and confirmed that deletion of mir-52, mir-83, lsy-6, and mir-265 prominently enhanced the L1 arrest phenotype of vgln-1(lf) (Figure 2A).
Figure 2.
Specific miRNAs and miRNA families cooperate with Vigilin to regulate development. (A) Loss-of-function mutations in multiple miRNAs enhance the L1 arrest phenotype of vgln-1(lf). mir-70 and mir-254 were identified in our RNAi screen, but the double mutants did not exhibit a synthetic phenotype. Error bars represent SE of proportion. N = 85–777 worms counted. * P < 0.001 (Fisher’s exact tests comparing the proportion indicated to WT and to vgln-1(lf)). (B) While knockout of single miRNAs miR-2, miR-45, and miR-52 also enhanced the L1 arrest phenotype of vgln-1(RNAi), knockout of the entire miR-2 family, miR-45 family, mir-58 family, and a subset of the miR-51 family further enhanced the L1 arrest phenotype of vgln-1(RNAi). *P < 0.0001 [Fisher’s exact tests comparing the proportion indicated to WT on vgln-1(RNAi)].
miR-52 is part of the miR-51 family in C. elegans, which shares the same seed sequence with the highly conserved miR-99/100 family (Grimson et al. 2008). The miR-51 family is one of the few miRNA families that displays a severe developmental defect (embryonic lethality) when all members are knocked out (Alvarez-Saavedra and Horvitz 2010; Shaw et al. 2010; Brenner et al. 2012). miR-52 is known to be highly expressed at the L1 stage as well as L4, and is expressed significantly higher than other miR-51 family members (Lau et al. 2001; Lee and Ambros 2001; Martinez et al. 2015). The expression difference might explain why loss of miR-52, but not closely related family members, enhances the vgln-1(lf) phenotype (Table S2). miR-83 is expressed throughout larval development in several tissues, including neurons and the intestine, and shares a seed sequence with miR-49 (Martinez et al. 2015). LSY-6 is well characterized for its role in chemosensory receptor expression and neuronal asymmetry, and its expression was detected in <10 neurons (Johnston and Hobert 2003). miR-265 is also expressed in a few head neurons (Martinez et al. 2015). Although seed sequence alone predicted many potential mRNA targets for LSY-6 and miR-265, the specificity of their functional relationship with these mRNAs remains unknown (Didiano and Hobert 2006, 2008; Garcia et al. 2011). Baring the caveats of potential background mutation effect in the miRNA deletion strains (see Discussion below), our genetic analysis likely uncovered previously unknown functions associated with miR-52, miR-83, LSY-6, and miR-265 in early larval development.
To identify interactions between VGLN-1 and miRNAs of multi-member miRNA families, we tested whether knockdown of entire miRNA families could enhance the vgln-1(RNAi) phenotype. We found that miR-2, miR-45, miR-51, and miR-58 family deletions all resulted in significant larval arrest phenotypes on vgln-1(RNAi) (Figure 2B). Both miR-2 and miR-45 family deletions have a slight egg retention phenotype, but no developmental phenotypes (Alvarez-Saavedra and Horvitz 2010). While most of these miRNA families exhibited a more severe phenotype relative to the single miRNA deletions, the miR-58 family phenotype was very similar to that of the single mutant (Figure 2B), so our further analysis focused on the single miRNA miR-58. Therefore, our analysis has also identified novel roles for the miR-2, miR-45, miR-51, and miR-58 families of miRNAs in early larval development. Based on these comprehensive genetic results, we focused further analysis on the miR-2, miR-45, and miR-51 families, as well as miR-58 and miR-83.
We need to point out that it is possible that the synergistic lethality in strains combining a mutation of specific miRNA and vgln-1(lf) [or vgln-1(RNAi)] was due to a background mutation in the miRNA deletion strain created by chemical mutagenesis. The fact that lsy-6 is known to be expressed only in a few neurons invites a more serious consideration of this caveat. However, the possibility that the effects of all or most of these miRNA mutants being the effect of second site mutations is highly unlikely for several circumstantial reasons. First, the enhancement of larval lethality is consistent with the enhancer phenotype associated either ain-2(lf) or ain-1(lf) mutation, both of which are known to specifically compromise overall miRISCs functions (Ding et al. 2005; Zhang et al. 2007; Weaver et al. 2014). Second, in the case of miR-58, similar phenotypes are associated with single miRNA mutant and deletion of corresponding miRNA family. Finally, as shown below the binding sites of the five miRNA families are enriched in VGLN-1 binding RNA at a particularly high level.
VGLN-1 interacts with a set of mRNAs that are enriched for miRNA binding sites
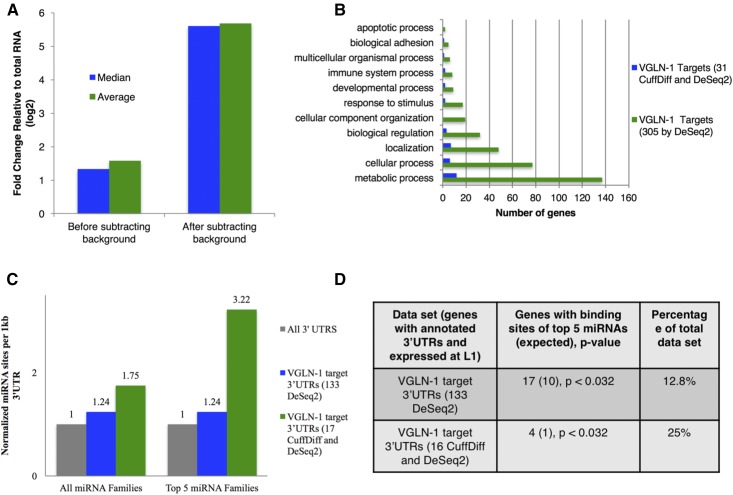
To identify VGLN-1-bound mRNAs, we employed RNA IP and high-throughput sequencing (RIP-Seq) (Zhao et al. 2010), using an anti-GFP antibody on samples from strains expressing either VGLN-1::GFP or control GFP. The resulting reads from three biological replicates were trimmed and mapped to the genome. We then performed differential expression analysis using both the Cufflinks suite and the DESeq2 R software suite to compare the FPKM values or reads of the VGLN-1::GFP IP to that of the GFP control IP. Together, these approaches yielded 304 enriched targets (q < 0.1, Table S3). Targets identified by DESeq2 encompassed the 31 targets identified by CuffDiff. Relative to the median and average fold change of all mRNAs, the median and average fold change of targets in the VGLN-1 IP was highly elevated when accounting for background through use of the GFP IP controls (Figure 3A and Table S5). GOTerm analysis of the 304 targets identified revealed enrichment in genes associated with metabolic processes, localization, and development (Figure 3B). The striking enrichment of genes involved in metabolic processes, which is consistent with the reported roles of human Vigilin (Mobin et al. 2016), may suggest a potential connection between metabolic abnormality and larval arrest.
Figure 3.
Identification of VGLN-1 bound mRNAs and enrichment for miRNA binding sites in these mRNA 3′UTRs. (A) Removing background identified by our GFP control improved identification of VGLN-1 bound RNAs that are enriched in the IP relative to total RNA. Median and average fold change of VGLN-1 bound RNAs from RIP-Seq was highly elevated relative to the median and average fold change of all other mRNAs when accounting for background measured by a control GFP IP. (B) Functional GOTerm analysis of VGLN-1 target mRNAs revealed an enrichment for genes involved in metabolic processes. (C) VGLN-1 bound mRNAs have a high density of all miRNA binding sites, and of miRNAs that we show genetically interact with vgln-1 (miR-2, miR-45, miR-51, miR-83, and miR-58 families). (D) Enrichment and percent of gene sets with miRNA sites of the top five families that we show to interact genetically with vgln-1. P-values were calculated by a hypergeometric distribution. http://nemates.org/MA/progs/overlap_stats.html.
3′UTR analysis of our 133 targets, and 17 high confidence targets known to be expressed at L1 (Spencer et al. 2011), revealed a large increase in the density of conserved (7mer or 8mer) miRNA seed sites relative to annotated UTRs of mRNAs expressed at the L1 stage of larval development. We observed an increase in total seed sites (1.2-fold), and a more dramatic increase in seed sites for the miRNA families that we show interact genetically with vgln-1 (3.2-fold) (Figure 3C). The enrichment of predicted targets of these miRNA families in our target gene sets was determined to be significant by a hypergeometric distribution test (Figure 3D). The five families tested for enrichment in Figure 3, C and D were miR-2, miR-45, miR-51, miR-83, and miR-58 families. mir-265 and lsy-6 were excluded from these analyses due to their limited spatial expression, and extensive list of predicted mRNA targets (Johnston and Hobert 2003; Martinez et al. 2015). These results indicate that many of the 3′UTRs of VGLN-1 targets are likely recognized by miRNAs, and, more specifically, miRNAs that we show genetically interact with vgln-1. Table S4 details the specific seed sites found within each of these target mRNA 3′UTRs.
Discussion
Our interrogation of genetic redundancy has revealed key players in early larval development. Genetic analysis has revealed overlapping, previously unknown roles for VGLN-1 and several individual miRNAs, and miRNA families, during early larval development. Based on the identification of VGLN-1-bound mRNAs and miRNA target predictions, both VGLN-1 and these miRNAs are expected to act on a large number of RNA targets. The diversity of functions associated with these VGLN-1-bound mRNAs implied that VGLN-1 acts in multiple cellular events (Figure 3B). For example, the association of many of these mRNAs with metabolism is consistent with the observation that human Vigilin regulates the expression of many metabolic genes (Graham and Oram 1987; Mobin et al. 2016).
Alone, C. elegans vgln-1 was also reported as one of 25 genes identified in a enhancer screen for a potential role in developmental timing (Hayes and Ruvkun 2006). Phenotypes from genome-wide RNAi analysis also suggested potential functions of vgln-1 in body morphogenesis, locomotion, and larval development (Kamath et al. 2003; Simmer et al. 2003; Ceron et al. 2007; Cantacessi et al. 2009). The sensitized genetic background of ain-2(lf) used in our study, where miRNA function is compromised, critically facilitated the exposure of the role of VGLN-1 in C. elegans larval development. Likewise, the sensitized genetic background of the vgln-1 mutant was critical to expose the roles of many miRNAs and families of miRNAs in regulating the first larval stage of C. elegans development.
Although we were not able to determine a high confidence VGLN-1 binding motif using computational methods [RBPMotif (Kazan et al. 2010)], the enrichment of specific miRNA target sites among VGLN-1 target mRNAs could suggest that alternative methods of functional selectivity may exist. This enrichment and genetic interactions between vgln-1 and miRNA/miRISC genes suggest potential collaboration between RNA binding protein VGLN-1 and miRNAs in regulating gene expressions for developmental functions, but there could be multiple potential models for such an interaction. In one model, this RNA-binding protein could potentially act as a corepressor of miRISCs to regulate the same mRNA targets. It is interesting to note that mammalian GW182 proteins, critical factors involved in miRISC-mediated gene silencing, actually contain RNA binding domains, albeit the roles of such domains have not yet been clearly demonstrated (Braun et al. 2013). Interestingly, the C. elegans AIN-1 (first GW182 protein determined to play an essential role in miRISC) and AIN-2 proteins lack such an RNA binding region (Ding et al. 2005; Rehwinkel et al. 2005; Zhang et al. 2007). Physical association and genetic interaction between RNA binding proteins and miRISCs have been detected in the worm (Zhang et al. 2007; Akay et al. 2013). However, physical interaction between VGLN-1 with miRISCs was not observed in our search for miRISC binding proteins (Zhang et al. 2007), nor in the search for VGLN-1 binding proteins, making the corepressor model unlikely. Lack of interaction between VGLN-1 and DCR-1 may also make a role of VGLN-1 in miRNA biogenesis unlikely (Duchaine et al. 2006). In addition, the genetic redundancy exhibited in vgln-1(lf); miR-x(lf) double mutants argues that the two systems act in parallel (nonindependent) mechanisms. Alternatively, and more consistent with the genetic data, VGLN-1 and miRNA may act in parallel to regulate the expression of many genes, some of which could be common targets, for a specific physiological function. The genetic data does not implicate that the regulations need to be at the same level even for the shared targets, particularly since both miRNAs and VGLN-1 are known to regulate gene expression at levels other than mRNA stability. Specifically, a recent paper indicated that Vigilin regulates gene expression mainly through inhibiting translation (Mobin et al. 2016). mRNA localization has also been suggested to be involved in VGLN-1 function (Gelin-Licht et al. 2012). These regulations might be the key reason that we were not successful in determining the functional relationship between VGLN-1 and miRNAs by analyzing the mRNA levels of selective genes. Growth and genetic background differences between mutant strains may have also presented an obstacle to obtain consistent data at specific developmental stages. Therefore, future mechanistic studies that analyze protein level changes for specific mRNA targets may be required to elucidate the potential regulatory roles of VGLN-1 and miRNAs on these targets at specific developmental stages.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.043414/-/DC1.
Acknowledgments
We thank A. Sewell for critical reading of the paper, A. Sewell, B. Weaver and Han laboratory members for helpful discussions, V. Gobel, R. Goldstein, E. Jorgensen, and the Caenorhabditis Genetics Center (CGC) [funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440)] for strains and materials, the Next Generation Sequencing (NGS) Core Facility at Colorado University (CU) for sequencing and Mass Spec Core Facilities at University of Colorado, and WormBase and Online Mendelian Inheritance in Man® (OMIM) databases for accessible information. Supported by an NIH predoctoral training grant (University of Colorado, 2T32GM008759-11, R.Z.) an NIH R01 grant (GM047869, M.H.), and the Howard Hughes Medical Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions: R.A.Z. and B.M.W. designed research, R.A.Z., B.M.W. and M.C. performed the experiments and analyzed data; R.A.Z. wrote the paper; M.H. supervised the study, and edited the paper.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., et al. , 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44: W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akay A., Craig A., Lehrbach N., Larance M., Pourkarimi E., et al. , 2013. RNA-binding protein GLD-1/quaking genetically interacts with the mir-35 and the let-7 miRNA pathways in Caenorhabditis elegans. Open Biol. 3: 130151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra E., Horvitz H. R., 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., 2004. The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S., 2016 FastQC—a quality control tool for high throughput sequence data. Babraham Bioinformatics. Available at: https://www.bioin-8formatics.babraham.ac.uk/projects/fastqc/.
- Bartel D. P., 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., Sternberg P. W., 2006. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 Arrest. Curr. Biol. 16: 780–785. [DOI] [PubMed] [Google Scholar]
- Braun J. E., Huntzinger E., Izaurralde E., 2013. The role of GW182 proteins in miRNA-mediated gene silencing. Adv. Exp. Med. Biol. 768: 147–163. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner J. L., Jasiewicz K. L., Fahley A. F., Kemp B. J., Abbott A. L., 2010. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr. Biol. 20: 1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner J. L., Kemp B. J., Abbott A. L., 2012. The mir-51 family of microRNAs functions in diverse regulatory pathways in Caenorhabditis elegans. PLoS One 7: e37185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J. P., Pasquinelli A. E., 2013. Identifying argonaute binding sites in Caenorhabditis elegans using iCLIP. Methods 63: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantacessi C., Loukas A., Campbell B. E., Mulvenna J., Ong E. K., et al. , 2009. Exploring transcriptional conservation between Ancylostoma caninum and Haemonchus contortus by oligonucleotide microarray and bioinformatic analyses. Mol. Cell. Probes 23: 1–9. [DOI] [PubMed] [Google Scholar]
- Ceron J., Rual J.-F., Chandra A., Dupuy D., Vidal M., et al. , 2007. Large-scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity. BMC Dev. Biol. 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafrè S. A., Galardi S., 2013. microRNAs and RNA-binding proteins. RNA Biol. 10: 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D., Hobert O., 2006. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 13: 849–851. [DOI] [PubMed] [Google Scholar]
- Didiano D., Hobert O., 2008. Molecular architecture of a miRNA-regulated 3′ UTR. RNA 14: 1297–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M., 2005. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell 19: 437–447. [DOI] [PubMed] [Google Scholar]
- Duchaine T. F., Wohlschlegel J. A., Kennedy S., Bei Y., Conte D., Jr., et al. , 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354. [DOI] [PubMed] [Google Scholar]
- Frey S., Pool M., Seedorf M., 2001. Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem. 276: 15905–15912. [DOI] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D. M., Baek D., Shin C., Bell G. W., Grimson A., et al. , 2011. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 18: 1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelin-Licht R., Paliwal S., Conlon P., Levchenko A., Gerst J. E., 2012. Scp160-dependent mRNA trafficking mediates pheromone gradient sensing and chemotropism in yeast. Cell Reports 1: 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham L., Oram J., 1987. Identification and characterisation of a high density lipoprotein binding protein in cell membranes by ligand blotting. J. Biol. Chem. 262: 7439–7442. [PubMed] [Google Scholar]
- Grimson A., Srivastava M., Fahey B., Woodcroft B. J., Chiang H. R., et al. , 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455: 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell C. M., Karp X., Ambros V., 2009a. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci USA 106: 18668–18673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell C. M., Lubin I., Boag P. R., Blackwell T. K., Ambros V., 2009b. nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell 136: 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes G. D., Ruvkun G., 2006. Misexpression of the Caenorhabditis elegans miRNA let-7 is sufficient to drive developmental programs. Cold Spring Harb. Symp. Quant. Biol. 71: 21–27. [DOI] [PubMed] [Google Scholar]
- Hogan D. J., Riordan D. P., Gerber A. P., Herschlag D., Brown P. O., 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6: e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan C. H., Friedman R. C., Ruby J. G., Bartel D. P., 2011. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. J., Hobert O., 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845–849. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kazan H., Ray D., Chan E. T., Hughes T. R., Morris Q., 2010. RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLOS Comput. Biol. 6: e1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight C. G., Patel M. N., Azevedo R. B. R., Leroi A. M., 2002. A novel mode of ecdysozoan growth in Caenorhabditis elegans. Evol. Dev. 4: 16–27. [DOI] [PubMed] [Google Scholar]
- Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P., 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee R. C., Ambros V., 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864. [DOI] [PubMed] [Google Scholar]
- Li A.-M., Watson A., Fridovich-Keil J. L., 2003. Scp160p associates with specific mRNAs in yeast. Nucleic Acids Res. 31: 1830–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kowdley K. V., 2012. MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics 10: 246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N. J., Ow M. C., Reece-hoyes J. S., Barrasa M. I., Ambros V. R., et al. , 2008. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. 18: 2005–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massirer K. B., Perez S. G., Mondol V., Pasquinelli A. E., 2012. The miR-35–41 family of microRNAs regulates RNAi sensitivity in Caenorhabditis elegans. PLoS Genet. 8: e1002536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Sleumer M. C., Bilenky M., Wong K., McKay S. J., et al. , 2007. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev. Biol. 302: 627–645. [DOI] [PubMed] [Google Scholar]
- Mendell J. T., Olson E. N., 2012. MicroRNAs in stress signaling and human disease. Cell 148: 1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., 2010. Transgenic solutions for the germline. (February 8, 2010), WormBook , ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.148.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E. A., Alvarez-Saavedra E., Abbott A. L., Lau N. C., Hellman A. B., et al. , 2007. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 3: e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S. F., Jain S., She M., Parker R., 2013. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 20: 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobin M. B., Gerstberger S., Teupser D., Campana B., Charisse K., et al. , 2016. The RNA-binding protein vigilin regulates VLDL secretion through modulation of Apob mRNA translation. Nat. Commun. 7: 12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux S. D., Waterhouse P. D., Shelton D., Shao Y. W., Watling C. M., et al. , 2014. Human somatic cell mutagenesis creates genetically tractable sarcomas. Nat. Genet. 46: 964–972. [DOI] [PubMed] [Google Scholar]
- Moore M. J., Zhang C., Gantman E. C., Mele A., Darnell J. C., et al. , 2014. Mapping argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat. Protoc. 9: 263–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana E., Kasinski A., 2015. MicroRNAs in cancer: a historical perspective on the path from discovery to therapy. Cancers (Basel) 7: 1388–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka H., Gotoh Y., Komeno T., Ono T., Kawasaki Y., et al. , 2014. Aralin, a type II ribosome-inactivating protein from Aralia elata, exhibits selective anticancer activity through the processed form of a 110-kDa high-density lipoprotein-binding protein: a promising anticancer drug. Biochem. Biophys. Res. Commun. 453: 117–123. [DOI] [PubMed] [Google Scholar]
- Reboul J., Vaglio P., Rual J.-F., Lamesch P., Martinez M., et al. , 2003. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35–41. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E., 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11: 1640–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z., Veksler-Lublinsky I., Morrissey D., Ambros V., 2016. Staufen negatively modulates microRNA activity in Caenorhabditis elegans. G3 Bethesda 6: 1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwedel J., Kügler S., Engebrecht T., Purschke W., Müller P. K., et al. , 2003. Evidence for posttranscriptional regulation of the multi K homology domain protein vigilin by a small peptide encoded in the 5′ leader sequence. Cell. Mol. life Sci. 60: 1705–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. R., Armisen J., Lehrbach N. J., Miska E. A., 2010. The conserved miR-51 microRNA family is redundantly required for embryonic development and pharynx attachment in Caenorhabditis elegans. Genetics 185: 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Moorman C., van der Linden A. M., Kuijk E., van den Berghe P. V. E., et al. , 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer W. C., Zeller G., Watson J. D., Henz S. R., Watkins K. L., et al. , 2011. A spatial and temporal map of C. elegans gene expression. Genome Res. 21: 325–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T., 2006. Maintenance of C. elegans. (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.101.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., et al. , 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kouwenhove M., Kedde M., Agami R., 2011. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer 11: 644–656. [DOI] [PubMed] [Google Scholar]
- Vella M. C., Slack F. J., 2005. C. elegans microRNAs. (September 21, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.26.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidigal J. A., Ventura A., 2014. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 25: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Z., Blackwell K., Carmichael G. G., 2005. Vigilins bind to promiscuously A-to-I-Edited RNAs and are involved in the formation of heterochromatin. Curr Biol. 15:384–391. [DOI] [PubMed] [Google Scholar]
- Weaver B. P., Zabinsky R., Weaver Y. M., Lee E. S., Xue D., et al. , 2014. CED-3 caspase acts with miRNAs to regulate non-apoptotic gene expression dynamics for robust development in C. elegans. eLife 3: e04265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber V., Wernitznig A., Hager G., Harata M., Frank P., et al. , 1997. Purification and nucleic-acid-binding properties of a Saccharomyces cerevisiae protein involved in the control of ploidy. Eur. J. Biochem. 249: 309–317. [DOI] [PubMed] [Google Scholar]
- Woo H.-H., Yi X., Lamb T., Menzl I., Baker T., et al. , 2011. Posttranscriptional suppression of proto-oncogene c-fms expression by Vigilin in breast cancer. Mol. Cell. Biol. 31: 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. L., Wei L., Huang W. Q., Li R., Shen W. Y., et al. , 2014. Vigilin is overexpressed in hepatocellular carcinoma and is required for HCC cell proliferation and tumor growth. Oncol. Rep. 31: 2328–2334. [DOI] [PubMed] [Google Scholar]
- Zhang H., Kim A., Abraham N., Khan L. A., Hall D. H., et al. , 2012. Clathrin and AP-1 regulate apical polarity and lumen formation during C. elegans tubulogenesis. Development 139: 2071–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ding L., Cheung T. H., Dong M. Q., Chen J., et al. , 2007. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell 28: 598–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Ohsumi T. K., Kung J. T., Ogawa Y., Grau D. J., et al. , 2010. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 40: 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang Q., Chen L.-L., Carmichael G. G., 2008. On the mechanism of induction of heterochromatin by the RNA-binding protein vigilin. RNA 14: 1773–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Sewell A. K., Han M., 2015. Intestinal apical polarity mediates regulation of TORC1 by glucosylceramide in C. elegans. Genes Dev. 29: 1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains used are listed in Table S1. Unless otherwise stated, strains were maintained and manipulated, as previously described (Brenner 1974; Stiernagle 2006), on NGM agar with Escherichia coli OP50 as a food source at 20°. Data from miRNA enhance screen, RIP-Seq and DESeq analyses, miRNA seed site analysis of VGLN-1 bound mRNAs, and analysis of VGLN-1-IP enrichment relative to total RNA are shown in Table S2, Table S3, Table S4, and Table S5, respectively.