Abstract
Improving water-use efficiency by incorporating drought avoidance traits into new wheat varieties is an important objective for wheat breeding in water-limited environments. This study uses genome wide association studies (GWAS) to identify candidate loci for water-soluble carbohydrate accumulation—an important drought-avoidance characteristic in wheat. Phenotypes from a multi-environment trial with experiments differing in water availability and separate single nucleotide polymorphism (SNP) and diversity arrays technology (DArT) marker sets were used to perform the analyses. Significant associations for water-soluble carbohydrate accumulation were identified on chromosomes 1A, 1B, 1D, 2D, and 4A. Notably, these loci did not collocate with the major loci identified for relative maturity. Loci on chromosome 1D collocated with markers previously associated with the high molecular weight glutenin Glu-D1 locus. Genetic × environmental interactions impacted the results strongly, with significant associations for carbohydrate accumulation identified only in the water-deficit experiments. The markers associated with carbohydrate accumulation may be useful for marker-assisted selection of drought tolerance in wheat.
Keywords: water-soluble carbohydrates, nonstructural carbohydrates, association analysis, genotype-by-environment interaction, molecular marker
Reduction in grain yield and quality due to drought decrease the sustainability of farming systems, and threatens global food security (Ray et al. 2012; Reynolds et al. 2016). Incorporating traits that improve water-use efficiency (WUE) in water-limited environments into elite breeding germplasm is an important aim for wheat genetic improvement (Rebetzke et al. 2009; Reynolds et al. 2015). Water soluble carbohydrate (WSC) accumulation and remobilization are promising traits that could contribute to improved grain-filling under water-limited conditions, and, consequently, improved WUE (Bidinger et al. 1977; Pheloung and Siddique 1991; Gebbing and Schnyder 1999; Foulkes et al. 2007; Piaskowski et al. 2016). Carbohydrate accumulation occurs when the crop synthesizes assimilate at a rate greater than sink requirement. In wheat, most of the carbohydrate is stored in the form of fructans, with a minor component of sucrose and hexose (Schnyder 1993; Wardlaw and Willenbrink 1994). Both the accumulation and remobilization of WSC is modified by environmental conditions that alter the balance between sources and sinks of assimilate. In particular, the availability of source carbon (as sucrose) affects accumulation (Xue et al. 2013). The WSC can be remobilized for use in growth or respiration (Kiniry 1993). However, the main sink for remobilization is the developing grain (Schnyder 1993; van Herwaarden et al. 1998; Takahashi et al. 2001), with remobilized WSC contributing as much as 30–50% of grain yield under terminal drought conditions, and 10–20% under well-watered conditions (Bidinger et al. 1977; Pheloung and Siddique 1991; Schnyder 1993; Gebbing and Schnyder 1999; Piaskowski et al. 2016).
Flowering time is a key trait associated with WSC accumulation owing to the nature of WSC accumulation across crop growth stages (Passioura 1996; Rebetzke et al. 2008). Accumulation of WSC increases from before anthesis to a peak at 7–20 d after anthesis (Gebbing 2003; Ehdaie et al. 2008; Zhu et al. 2010) where WSC concentration (WSCC) can reach as much as 40% of total stem weight (Schnyder 1993). After anthesis, WSC levels decline due to remobilization to other sinks. Under water deficit conditions, this peak can sometimes occur before anthesis (Goggin and Setter 2004), and remobilization is earlier and proportionally greater (Bidinger et al. 1977; Virgona and Barlow 1991).
A number of studies have reported on genetic control and quantitative trait loci (QTL) for WSC accumulation and related characters in wheat (Snape et al. 2007; Yang et al. 2007; Rebetzke et al. 2008; McIntyre et al. 2010; Pinto et al. 2010; Bennett et al. 2012). Mapping populations typically varied for the major developmental genes for photoperiod sensitivity and reduced plant height, which can indirectly cause much of the observed phenotypic variability for grain yield and other traits (Rattey et al. 2009; Bennett et al. 2012; Edae et al. 2014). The biparental populations assessed in Rebetzke et al. (2008) were segregating for the photoperiod sensitivity locus Ppd-D1 and the semi-dwarfing loci Rht-B1 and Rht-D1, and these loci collocated with QTL for WSCC, WSC total amount per square meter, and WSC per tiller.
Genomic strategies show significant promise for the improvement and understanding of drought tolerance traits (Langridge and Reynolds 2015). The primary objectives of this study were to identify markers associated with WSCC by genome-wide association studies (GWAS) and characterize the dependency of marker associations on environments. We conducted GWAS separately for two molecular marker sets (SNP and DArT markers) and for each experiment to assess the variability of marker-trait associations due to genotype × environment (G × E) interaction (Oldmeadow et al. 2011; Zila et al. 2013). We also conducted GWAS for relative maturity to ascertain if loci with significant associations with WSCC were due to the indirect effects of relative maturity on WSCC.
Materials and Methods
Genotypes used in this study
The genotypes for the GWAS analyses were selected from evaluation trials conducted in multiple environments in 2009 and 2010. Each field trial contained 990 genotypes. Some genotypes were not repeated at every experiment, with a total of 1314 genotypes tested. Thus, for relative maturity GWAS, all 990 genotypes were used in 2009 experiments, and 972 genotypes were used in 2010 experiments. The accumulation of WSC varies with plant development (Ehdaie et al. 2008), so the subset used for WSCC GWAS consisted of 312 breeding lines from the 2009 experiments constrained to a 3–5 d difference in anthesis date as well as 46 commercially grown varieties. For the second year of this study in 2010, except for 11 breeding lines excluded from the overall experiments, the same breeding lines and varieties were evaluated for WSCC.
Experimental design and site locations
Experiments with contrasting irrigation and rainfed treatments were grown at Yanco Agricultural Institute (Yanco, Australia) and Coleambally Community Experimental Demonstration Farm (Coleambally, Australia) in 2009 and 2010. A split-plot design was used, in which the main-plot factor was irrigation treatment (irrigated or rainfed), and the 990 genotype entries (including the subset of genotypes for WSC measurement) were the subplot factor. There were two replicates of each treatment at each location. Genotype placement was optimized with the spatial design package DiGGer (Coombes 2002). For the laboratory phase measuring WSC using near-infrared spectroscopy (NIRS), an experimental design structured by day of measurement and NIRS instrument carousel and well was implemented to account for extraneous variation originating from laboratory processes. Samples from both field locations were pooled into one laboratory phase experimental design for each year, and the placement of genotypes within the laboratory experimental phase was also optimized with DiGGer (Coombes 2002), with partial replication of 20% of experiment field plots sampled (i.e., a replication level of 1.2), following the methods in Cullis et al. (2006) and Smith et al. (2006).
All experiments were sown on a full soil profile of moisture, achieved by flood irrigating each site 4–6 wk before sowing, so that the focus on water deficit conditions would be in the later stages of crop growth. Sowing dates were targeted for the first 2 wk of May, and sowing rates were 115 kg ha−1 in the irrigated and 70 kg ha−1 in the rainfed treatments, respectively. Presowing nitrogen was targeted to be 120 kg N ha−1 from the combination of deep soil nitrogen (following soil testing—data not shown) and fertilizer applied at sowing. Irrigated experiments were fertilized supplementally through the growing season to a total of ∼300 kg N ha−1 consistent with predicted N demand by the crop. Experiments were subject to a strict weed, pest, and disease control regime to maximize yield potential. Soil moisture at each experiment was monitored using gypsum block AM400 soil moisture data loggers (Hansen, Wenatchee, WA). Onsite weather stations (Davis Instruments, Hayward, CA) were used to record rainfall and air temperature. Irrigation treatments were flood irrigated when soil water potential fell below −75 kPa. Both sites had below average rainfall and above average temperatures in 2009, while conditions at both sites in 2010 had higher rainfall and lower temperatures than average.
Measurements and observations
Relative maturity at a common date around the median flowering date for all entries within each experiment was determined using the Zadoks decimal score for plant development (Zadoks et al. 1974). Scores for each field experiment were taken when most lines were in the range Z50–Z69 (head emergence to completion of anthesis).
Lines selected for WSCC analysis were sampled from a 50-cm long section of row (0.09 m2) when the irrigated treatments at each site were ∼180° d postanthesis, following the sampling method of Rebetzke et al. (2008). For WSC analysis, ∼5–10 stalks (including leaves, leaf sheaths, and heads, but not senesced plant material) were subsampled from each biomass sample, and ground to pass through a 2 mm-sized screen. Ground biomass samples were homogenized, desiccated, and scanned by NIRS with a Bruker Multi-purpose Analyzer (Bruker Optik GmbH, Ettlingen, Germany) and OPUS software (version 5.1). Scanned spectra were transformed using the first derivative and multiplicative scatter correction. Calibrations to obtain predicted WSCC values from spectra measurements were constructed using the “Quant 2 Method” component of the OPUS software with a randomly selected 10% subset of samples. WSCC for the calibration samples was determined using the alkaline ferricyanide method (Piltz and Law 2007).
Statistical methods for phenotype values
A multiplicative mixed linear model was used to analyze the multi-experiment phenotype data for both traits following Gilmour et al. (1997) and Beeck et al. (2010). The linear mixed model is given by
where y is the data vector of the response variable across p experiments with N plots per experiment; is a vector of fixed effects (including linear trends across range and row) with associated design matrix The term is a random component with associated design matrix and contains experimental blocking structures used to capture extraneous variation (including field range and row for both traits, and laboratory day of measurement, NIRS carousel and well for WSCC only).
The residual error is which, at the jth experiment, was assumed to have distribution where is the residual variance for the jth experiment and is a matrix that contains a parameterization for a separable autoregressive process to model potential spatial correlation of the observations for the relative maturity analysis. For WSCC analysis, unique residual variances for each year were modeled.
The term is a random component with associated design matrix used to model the genotype within experiment effects, which combine the genotype and G × E interaction effects. Organizing the genotype within environment effects as a matrix of rows corresponding to genotypes and columns corresponding to environments facilitates modeling as a multiplicative k-factor analytic (FA) model (Smith et al. 2001):
where is a matrix with jth column containing the jth factor loadings for the p experiments, is a vector of genotype scores across the p experiments, and is a residual genetic term, where, at the jth experiment, and is the residual genetic variance for the jth experiment. The term represents an identity matrix.
The variance model for the combined genotype and G × E effects is given by
where is a diagonal matrix of the p environment specific variances.
For each analysis, the most parsimonious FA model was identified using the Akaike Information Criterion (AIC) (Akaike 1974). The nongenetic random effects were maintained in the model if they were significant according to log likelihood ratio tests relative to the full model with all nongenetic random effects (Stram and Lee 1994). Fixed effects were tested for significance using Wald F-statistics (Kenward and Roger 1997).
Empirical best linear unbiased predictors (E-BLUPs) for phenotypic values were obtained from the FA models for each individual experiment (Kelly et al. 2007; Cullis et al. 2010). For both relative maturity and WSCC, experiments were clustered using the matrix of genetic correlations between experiments (Cullis et al. 2010). All data were analyzed using the software package ASReml-R (Butler et al. 2009), in the R statistical software environment (R Development Core Team 2012).
Genotyping methods
Two separate marker sets were used: 985 lines from the overall experiment were genotyped using the Illumina 9k Infinum iSelect beadchip array (Cavanagh et al. 2013), resulting in 4883 polymorphic SNPs across the population. Similarly, 955 lines were genotyped with Diversity Arrays technology (DArT) (Akbari et al. 2006) resulting in 2013 polymorphic markers across the population. Genotyping included all 358 lines phenotyped for WSCC. Genotype information for SNP and DArT marker datasets were prepared separately for analysis using the R software package Synbreed (Wimmer et al. 2012). Imputation of missing values (3.5% for SNPs and 15% for DArTs) was performed using the software package Beagle (Browning and Browning 2009). Each marker dataset was filtered for duplicated and monomorphic markers, as well as markers with minor allele frequency of <5%. The resulting 4162 SNP markers and 1773 DArT markers were used to compute a separate scaled identity by descent relationship matrix () after Endelman and Jannink (2012) for each marker dataset.
Consensus maps were used for marker physical positions. For the DArT dataset this study used the Wheat Interpolated Maps (version 6) as a reference to locate the positions of DArT markers (Dr Andrzej Kilian, Diversity Arrays Pty Limited, personal communication), and for the SNP dataset the 9K Consensus Map (version 4) was used (Dr Matthew Hayden, DEPI Victoria, personal communication.).
Linkage disequilibrium analysis
Patterns of linkage disequilibrium (LD) in the SNP and DArT marker sets were estimated using the methods of Breseghello and Sorrells (2006). Pairwise LD estimates (r2) were calculated with the software package PLINK (Purcell et al. 2007) for unlinked loci pairs and for syntenic loci separately. Syntenic r2 was plotted against pairwise genetic distance from the consensus maps for all chromosomes on each genome with a second degree locally weighted polynomial regression (LOESS) curve fitted to each scatter plot (Cleveland 1979). All of the unlinked r2 estimates were square-root-transformed to approximate a normal distribution, and the 5% quantile of that distribution was determined following Breseghello and Sorrells (2006). The intersection of the LOESS curve and the 5% quantile for unlinked marker pairs was taken as an estimate of the extent of LD decay within each genome following Laidò et al. (2014).
GWAS methods
Separate association analyses for each trait at each experiment were performed using the phenotype E-BLUPs described above. Associations using SNP and DArT marker sets were performed separately. The compressed mixed linear model approach (Zhang et al. 2010) was implemented in the R software package Genome Association and Prediction Integrated Tool (GAPIT) (Lipka et al. 2012) as follows:
where is the vector of E-BLUPs for one trait measured in one experiment, is a vector of fixed effects for the corresponding design matrix (), including a molecular marker. The vector of overall genetic line effects (with associated design matrix ) is modeled as where is the relationship matrix and is the estimated additive genetic variance. is the vector of random residuals. False discovery rates (FDR) were estimated separately for each experiment following Benjamini and Hochberg (1995) with a nominal threshold of 10% to declare significant associations.
Data availability
Supplemental Material, File S1 contains a detailed description of all Supplemental files. File S2 contains phenotype information for WSCC. File S3 contains phenotype information for relative maturity at flowering time. File S4 contains SNP genotypes for each individual. File S5 contains DArT genotypes for each individual.
Results
Genotype × environment interactions
Consistent with experimental weather conditions, genetic correlations for WSCC between experiments showed two distinct environment groups, with the Yanco and Coleambally 2009 rainfed experiments, which experienced terminal water deficit, forming one cluster, and the other experiments collectively representing a well-watered cluster. Within these clusters, the genetic correlations were maintained at rG = 0.87 between the two rainfed experiments that make up the water deficit environment cluster, and ranged from rG = 0.74–0.98 in the well-watered environment cluster. Between the two clusters, genetic correlations ranged from rG = 0.02–0.35. For relative maturity, no environmental clustering was evident, and genetic correlations between all experiments were very high, ranging from rG = 0.92 to rG = 0.99.
LD and minor allele frequency
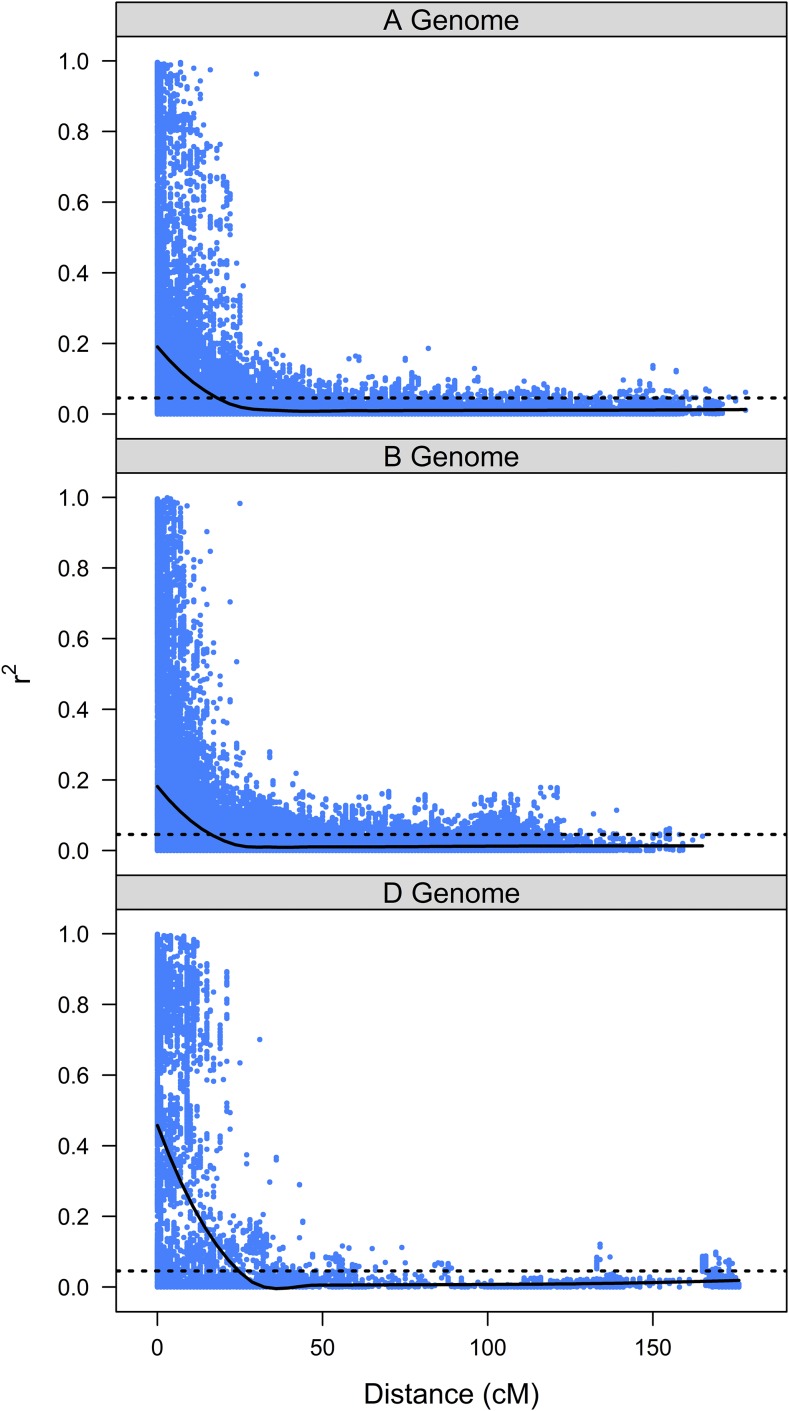
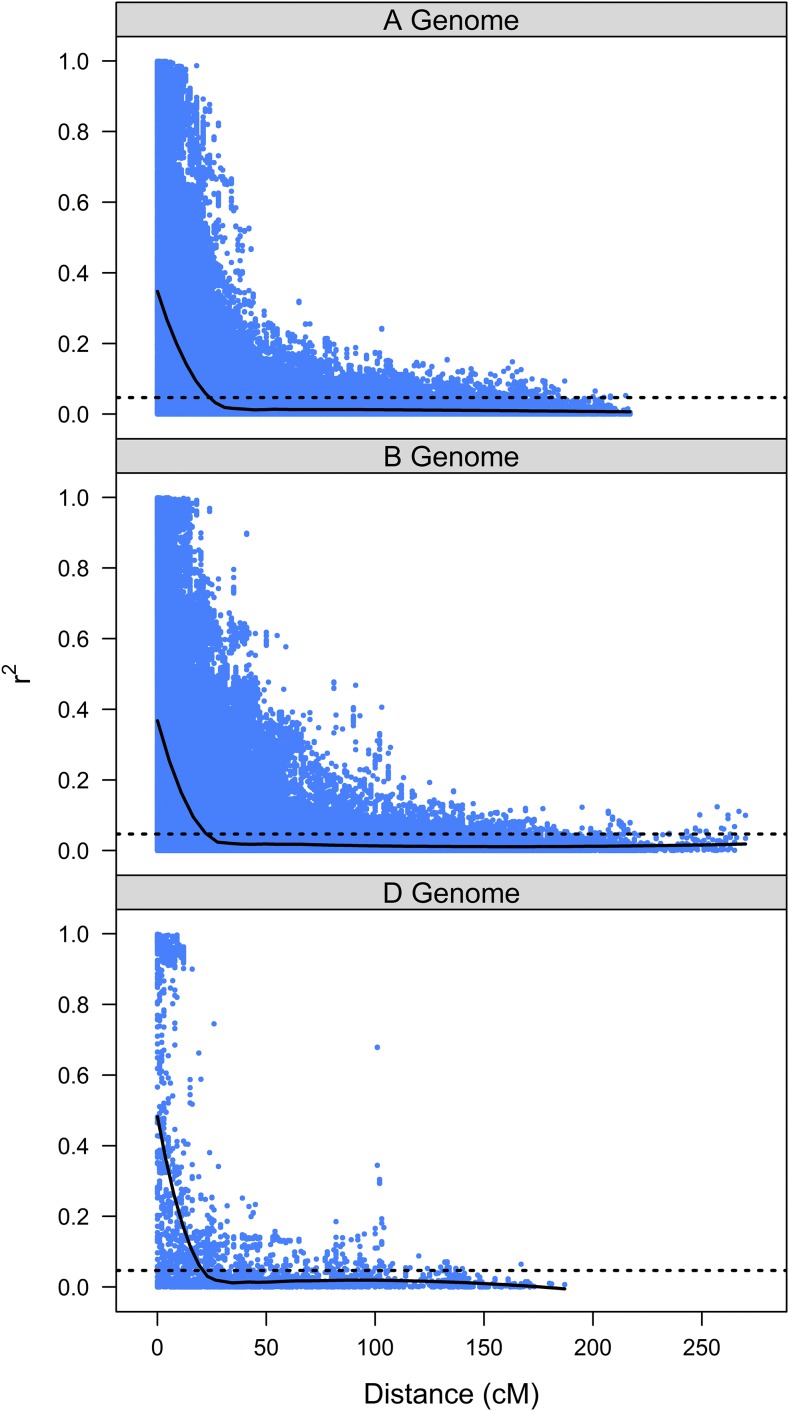
We evaluated the distribution of LD within chromosomes separately for each marker set and for each of the three wheat genomes. LD was more extensive with respect to linkage distances within the D genome for the DArT marker set, as the average DArT marker LD did not decrease below the 5% quantile for unlinked marker pairs (r2 = 0.0456) until the distance between markers was ≥25 cM (Figure 1). By comparison, the average LD was <0.0456 at distances of 16–18 cM for the A and B genomes respectively. In contrast, LD decreased below the 5% quantile (r2 = 0.0470) for the SNP marker set at 21 cM for both the D and B genomes, while LD for the A genome was higher at 24 cM (Figure 2).
Figure 1.
Pairwise LD estimates (r2) plotted against Euclidian pairwise marker distances for markers on the same consensus chromosome for the DArT marker set.
Figure 2.
Pairwise LD estimates (r2) plotted against Euclidian pairwise marker distances for markers on the same consensus chromosome for the SNP marker set.
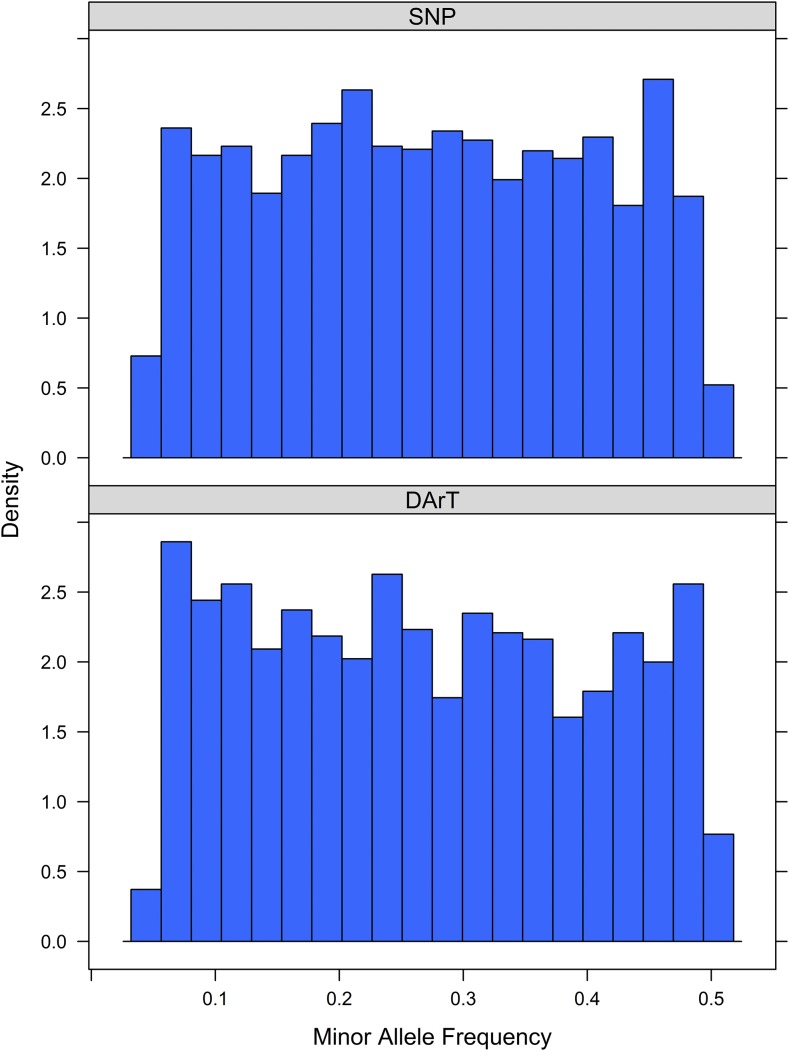
The minor allele frequency distribution for SNP markers was similar to that for the DArT markers, although the DArT markers had a lower proportion of the rarest allele class (MAF = 5–7.25%; Figure 3). SNPs with MAF below 5% were not included in the analysis because of their reduced power for GWAS.
Figure 3.
Minor allele frequency distribution for SNP and DArT marker datasets used for associations.
GWAS
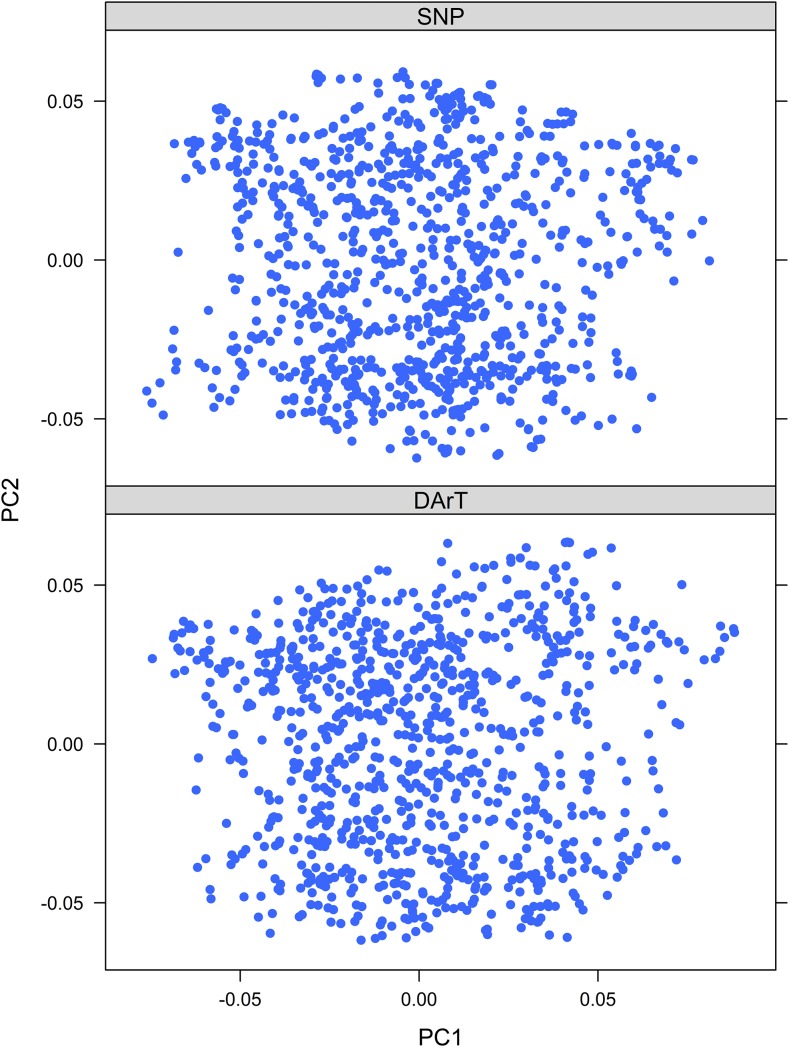
Both marker sets displayed a low degree of population structure (Figure 4) with no obvious patterns among genotypes. For genomic relationship matrices computed from either SNP or DArT markers separately, the first two eigenvectors collectively explained only 15% of the variation in genomic relationships, indicating a lack of strong subpopulation structure in the association population. Only markers identified as statistically significant (with P-values below the 10% FDR threshold) in more than one experiment in the association studies were considered reliable associations (Table 1 and Table 2).
Figure 4.
PCA plot of the first two eigenvectors from the relationship matrix of each marker set. For both marker types, the first two principle components account for ∼15% of the observed variation in genomic relationships.
Table 1. Markers significant for water-soluble carbohydrate concentration at >1 experiment for the DArT marker set.
| Experiments | Marker | Chromosome | Distance (cM) | P-Value Range | Minor Allele Frequency | FDR Value Range | Nearby Genes |
|---|---|---|---|---|---|---|---|
| 09COLE_RFD, 09YANA_RFD | wPt-9592 | 1A | 68.3 | 0.000520–0.000789 | 0.491 | 0.0725–0.0999 | |
| 09COLE_RFD, 09YANA_RFD | wPt-665784 | 1A | 69.2 | 0.000557–0.0006277 | 0.474 | 0.0725–0.0923 | |
| 09COLE_RFD, 09YANA_RFD | wPt-7359 | 1B | 11.3 | 6.97 × 10−5 – 9.73 × 10−5 | 0.107 | 0.0228–0.0323 | |
| 09COLE_RFD, 09YANA_RFD | wPt-666719 | 1D | 83.1 | 1.56 × 10−5 – 2.97 × 10−5 | 0.278 | 0.00784–0.0193 | Glu-D1 |
| 09COLE_RFD, 09YANA_RFD | wPt-3743 | 1D | 83.3 | 1.58 × 10−5 – 3.88 × 10−5 | 0.275 | 0.00784–0.0193 | Glu-D1 |
| 09COLE_RFD, 09YANA_RFD | wPt-733835 | 1D | 86.5 | 0.000205–9.16 × 10−5 | 0.285 | 0.0228–0.0510 | Glu-D1 |
| 09COLE_RFD, 09YANA_RFD | wPt-797974 | 2D | 41.1 | 0.000519–0.000607 | 0.378 | 0.0725–0.0923 | Ppd-D1 |
| 09COLE_RFD, 09YANA_RFD | wPt-800147 | 4A | 62.7 | 0.000804–0.000527 | 0.327 | 0.0725–0.0999 |
No SNP markers showed significantly associations. Experiment is given as year-site-irrigation treatment. Chromosome and position are from the consensus map. Nearby genes are from CMAP GrainGene database (http://wheat.pw.usda.gov/cmap/) searches within the LD blocks estimated for each genome. Association analyses returned significant markers for the two water deficit environments (2009 Coleambally and Yanco Rainfed), and not any of the well-watered environment experiments.
Table 2. Markers significant for relative maturity at >1 experiment for the DArT and SNP marker datasets.
| Experiments | Marker | Chromosome | Distance (cM) | P-Value Range | Minor Allele Frequency | FDR Value Range | Nearby Genes |
|---|---|---|---|---|---|---|---|
| 10COLE_IRR, 10COLE_RFD, 10YANA_IRR | wPt-730744 | 2D | 73.0 | 4.85 × 10−5 – 7.91 × 10‑5 | 0.161 | 0.0482–0.0787 | Ppd-D1 |
| 09COLE_IRR, 09COLE_RFD, 09YANA_IRR, 09YANA_RFD, 10COLE_IRR, 10COLE_RFD, 10YANA_IRR, 10YANA_RFD | wsnp_CAP12_c812_428290 (IWA989) | 2D | 57.9 | 3.91 × 10−16 – 1.414 × 10−14 | 0.134 | 1.50 × 10−12 – 5.41 × 10−11 | Ppd-D1 |
| 09YANA_IRR, 09YANA_RFD | wsnp_BE422566B_Ta_1_2 (IWA76) | 4B | 43.3 | 0.000169–0.000171 | 0.0582 | 0.0498–0.0507 | |
| 09COLE_IR, 09COLE_RFD, 09YANA_IRR, 09YANA_RFD, 10COLE_IRR, 10COLE_RFD, 10YANA_IRR, 10YANA_RFD | wsnp_AJ612027A_Ta_2_1 (IWA1) | 5A | 66.2 | 6.30 × 10−9 – 3.74 × 10−6 | 0.360 | 1.20 × 10−5 – 0.00410 | Vrn-A1 |
| 09COLE_IRR, 09COLE_RFD, 09YANA_IRR, 09YANA_RFD, 10COLE_IRR | wsnp_AJ612027A_Ta_2_5 (IWA2) | 5A | 66.2 | 1.29 × 10−5 – 0.000219 | 0.151 | 0.00619–0.0764 | Vrn-A1 |
| 09COLE_RFD, 09YANA_IRR, 09YANA_RFD | wsnp_BE404341A_Ta_2_3 (IWA46) | 5A | 66.7 | 6.07 × 10−5 – 0.000105 | 0.136 | 0.0258–0.0446 | Vrn-A1 |
| 09COLE_IRR, 09COLE_RFD, 09YANA_IRR, 09YANA_RFD, 10COLE_IRR, 10COLE_RFD, 10YANA_IRR, 10YANA_RFD | wsnp_BF293620A_Ta_2_1 (IWA454) | 5A | 66.2 | 1.19 × 10−6 – 0.000215 | 0.150 | 0.000909–0.0937 | Vrn-A1 |
| 09COLE_IRR, 09COLE_RFD, 09YANA_IRR, 09YANA_RFD, 10COLE_IRR, 10COLE_RFD, 10YANA_IRR, 10YANA_RFD | wsnp_BJ224975A_Ta_2_1 (IWA589) | 5A | 66.2 | 1.53 × 10−8 – 4.29 × 10−6 | 0.357 | 1.47 × 10−5 – 0.00410 | Vrn-A1 |
| 09COLE_RFD, 09YANA_IRR, 09YANA_RFD | wsnp_BJ224975A_Ta_2_2 (IWA590) | 5A | 66.2 | 7.20 × 10−5 – 0.000118 | 0.140 | 0.0276–0.0450 | Vrn-A1 |
| 09COLE_IRR, 09COLE_RFD, 09YANA_IRR, 09YANA_RFD, 10COLE_IRR, 10COLE_RFD, 10YANA_IRR, 10YANA_RFD | wsnp_Ex_c22727_31934296 (IWA2743) | 5A | 66.7 | 9.26 × 10−10 – 6.31 × 10−7 | 0.359 | 1.77 × 10−6 – 0.00121 | Vrn-A1 |
| 09COLE_RFD, 09YANA_IRR, 09YANA_RFD | wsnp_Ex_c31799_40545376 (IWA3362) | 5A | 69.1 | 0.000154–0.000243 | 0.369 | 0.0490–0.0581 | Vrn-A1 |
| 09COLE_IRR, 09COLE_RFD, 09YANA_IRR, 09YANA_RFD, 10COLE_IRR, 10COLE_RFD | wsnp_Ex_c31799_40545478 (IWA3363) | 5A | 69.1 | 1.74 × 10−6 – 0.000195 | 0.379 | 0.0009498–0.0747 | Vrn-A1 |
| 09COLE_IRR, 09COLE_RFD, 09YANA_IRR, 09YANA_RFD, 10COLE_IRR, 10COLE_RFD | wsnp_Ex_c7729_13177883 (IWA4719) | 5A | 69.1 | 1.43 × 10−6 – 0.000152 | 0.379 | 0.000911–0.0647 | Vrn-A1 |
| 09COLE_RFD, 09YANA_IRR, 09YANA_RFD | wsnp_Ex_rep_c66689_65010988 (IWA5287) | 5A | 66.7 | 0.000126–0.000203 | 0.400 | 0.0438–0.0518 | Vrn-A1 |
| 09YANA_IRR, 09YANA_RFD, 10COLE_IRR, 10COLE_RFD, 10YANA_IRR, 10YANA_RFD | wsnp_Ex_c12048_19288999 (IWA1577) | 5B | 71.1 | 3.83 × 10−5 – 0.000223 | 0.266 | 0.0291–0.0533 | Vrn-B1 |
| 09YANA_IRR, 09YANA_RFD | wsnp_Ra_c20970_30293078 (IWA7732) | 5B | 71.1 | 0.000332–0.000433 | 0.272 | 0.0747–0.0976 | Vrn-B1 |
| 10YANA_IRR, 10YANA_RFD | wsnp_Ra_c20970_30293227 (IWA7733) | 5B | 71.1 | 6.16 × 10−5 – 0.000183 | 0.262 | 0.0383–0.0499 | Vrn-B1 |
| 10YANA_IRR, 10YANA_RFD | wsnp_Ex_c508_1008029 (IWA4087) | 5D | 61.0 | 3.90 × 10−5 – 0.000332 | 0.202 | 0.0258–0.0978 | Vrn-D1 |
Experiment is given as year-site-irrigation treatment. Chromosome and position are from the consensus maps. Nearby genes are from CMAP GrainGene database (http://wheat.pw.usda.gov/cmap/) searches within the LD blocks estimated for each genome.
The GWAS for relative maturity using the DArT markers identified only one marker (on chromosome 2D), which was detected as significant in three experiments, all of them in the well-watered environment cluster (10COLE_IRR, 10COLE_RFD, and 10YANA_IRR; Table 2). This marker is located >30 cM from the marker associated with WSCC on the same chromosome. In contrast, GWAS for relative maturity using the SNP marker set identified 17 markers significant in more than one experiment (Table 2). SNP associations with relative maturity within specific experiments did not follow the pattern of environmental clustering observed for WSCC. Rather, five of 17 SNP associations with relative maturity were observed in all experiments. Some environmental-specificity was observed for relative maturity associations, but this did not reflect differences between well-watered and water-limited conditions. For example, four of 17 SNP associations were detected at both treatments within the same year-location combination (Table 2), suggesting G × E patterns for relative maturity due to local weather patterns rather than water availability.
Among the SNPs associated with relative maturity, 11 markers were located within 3 cM of each other on the consensus map on chromosome 5A, and three markers collocated on chromosome 5B. Additionally, one marker was identified on each of chromosomes 2D, 4B, and 5D. Four of the significant markers on 5A and the marker on 2D were detected in all eight experiments.
The range in MAF of trait-associated loci ranged from 0.107 to 0.491 for the DArT marker set, and at least one relatively rare SNP allele was detected for relative maturity (MAF = 0.058 on chromosome 4B). The highest MAF for associated loci in the SNP marker set was 0.400.
Discussion
Comparison of analysis at individual experiments
GWAS results for both relative maturity and WSCC show that significant associations can be experiment-specific, and relatable to the overall G × E relationships between experiments for each trait. Genetic correlations between all experiments were very high for relative maturity (indicating limited G × E interaction), and significant loci were detected in all experiments. In contrast, G × E was strong for WSCC, with factor analysis revealing two distinct environment types, corresponding to well-watered and water-deficit environments. Reflecting these differences in overall G × E patterns between relative maturity and WSCC, several markers were associated with relative maturity across all experiments, whereas the significant associations for WSCC were detected only in two experiments in the water deficit environment cluster. No significant associations for WSCC were detected in the well-watered environment cluster of experiments.
Combinations of well-watered and water deficit environments have been used for WSCC QTL detection previously, and, in some studies, such as Yang et al. (2007) and Pinto et al. (2010), QTL for WSCC were detected in well-watered, or water deficit environments but not in both. This discrepancy illustrates the importance of environmental characterization in QTL analysis, and the value in understanding the target population of environments that each QTL analysis is performed in. Once established, GWAS can be conducted separately for each experiment, or separately for traits values averaged over environments within well-defined clusters. Experiment-by-experiment GWAS should allow a means to replicate QTL detection in comparable environments, and as both Oldmeadow et al. (2011) and Zila et al. (2013) indicate, to understand possible QTL × environment interactions.
Loci associated with WSCC
GWAS detected associations between markers on chromosomes 1A, 1B, 1D, 2D, and 4A with WSCC measured in water-limited conditions. No markers were associated with both WSCC and relative maturity, in contrast to Rebetzke et al. (2008), where flowering time loci explained large proportions of variation for WSCC. The results herein may reflect the sampling methods used for phenotyping WSCC, or because the association population lines for WSCC were selected to be constrained for development.
Among the markers significantly associated with WSCC in the two water deficit environment experiments, wPt-7359 on 1B has not been previously reported in trait associations, but wPt-800147 on 4A was associated with plant height (Yu et al. 2014) and seedling shoot dry weight under normal and saline conditions (Masoudi et al. 2015). Marker wPt-3743 on 1D was associated with a range of other traits, including grain yield and resistance to yellow rust, powdery mildew, and leaf rust (Crossa et al. 2007), grain yield and spike length under salt stress conditions (Azadi et al. 2015), and spike number (tiller number) per plant (Cui et al. 2014). Marker wPt-9592 on 1A was previously associated with grain yield under water deficit conditions (in particular, terminal drought; Crossa et al. 2007), heading date after vernalization (Le Gouis et al. 2012), and seed dormancy (Singh et al. (2010).
Marker wPt-3743 on chromosome 1D was previously reported to be located near the high molecular weight glutenin Glu-D1 locus and the storage protein activator gene locus SPA-D (Plessis et al. 2013; Deng et al. 2015; Jin et al. 2015). Marker wPt-733835 is also in this region (Jin et al. 2015). The Glu-D1 locus is important for selection as, along with the Glu-A1 and Glu-B1 loci, it is responsible for a large percentage of the phenotypic variation for dough quality. The combination of glutenin alleles present at the Glu-1D locus will largely determine the end use and grain quality class of wheat varieties (Payne 1987; Whiting 2004). Glutenin protein complexes play an important role in conferring elasticity and strength in wheat dough (Plessis et al. 2013), and the Glu loci have been shown to collocate with QTL for nitrogen and dry matter accumulation in grain (Charmet et al. 2005).
Rebetzke et al. (2008) identified QTL for WSC per tiller that collocated with the glutenin loci Glu-A1, and Glu-B1. The Glu-D1 and SPA-D loci contribute to phenotypic variation for grain yield and grain number through the plant response to nitrogen (Bordes et al. 2013). Potentially the Glu loci could be involved with the inheritance of WSCC through an interaction between nitrogen use, tiller number, and grain weight. WSCC is influenced by nitrogen content, as higher nitrogen availability in the plant drives sink demand for assimilate (van Herwaarden et al. 1998; Ruuska et al. 2008), and WSC tends to accumulate in the absence of sink demand (Gebbing 2003).
Loci associated with relative maturity
Significant associations were identified in the vicinity of a number of the known major flowering time loci, including the main photoperiod and vernalization loci under selection in wheat breeding germplasm pool globally (Yan et al. 2004; Eagles et al. 2009, 2010; Cane et al. 2013; Slafer et al. 2015). Both DArT and SNP markers were identified close to the photoperiod-sensitivity locus Ppd-D1 on chromosome 2D on the consensus map (Table 1). Given the importance of the Ppd-D1 locus to selection of growth duration and adaptability (Kamran et al. 2014; Slafer et al. 2015), the SNP and DArT markers identified here may prove useful to supplement other markers for this locus, such as those outlined in Cane et al. (2013).
The analyses were able to detect significant associations near the Vrn-A1, Vrn-B1, and Vrn-D1 loci across multiple experiments, although only SNP markers were identified as statistically significant, including 11 markers near Vrn-A1, three markers near Vrn-B1, and one marker was detected near Vrn-D1 (Table 1). For the genotypes in this study, variation in at these loci would be expected to include alleles for both spring and winter alleles, as well as winter alleles that confer different vernalization requirements (Eagles et al. 2010, 2014; Harris et al. 2017). One marker identified (wsnp_AJ612027A_Ta_2_1) was also reported to be associated with the Vrn-A1 locus by Lopes et al. (2015). Three markers identified on 5B are within 10 cM of the Vrn-B1 locus reported by Guedira et al. (2014). The single marker on chromosome 5D associated with relative maturity (wsnp_Ex_c508_1008029) at both 10YANA_IRR and 10YANA_experiments corresponds to the vicinity of the Vrn-D1 locus (Eagles et al. 2009).
An additional marker associated with relative maturity at both the 09YANA_IRR and 09YANA_RFD experiments was located on chromosome 4B (wsnp_BE422566B_Ta_1_2). This marker was significant in both the water deficit environments as well as the well-watered environments in this study. QTL for heading date on this chromosome have been previously reported by Hanocq et al. (2007), Griffiths et al. (2009), and Le Gouis et al. (2012), and photoperiod sensitivity QTL on this chromosome have been reported by Shindo et al. (2003) and Sourdille et al. (2003).
Comparison of DArT and SNP molecular marker sets
DArT markers were developed using methylation-sensitive restriction enzymes, and, as such, can represent methylation polymorphisms that provide both genetic and epigenetic information (Wenzl et al. 2004; Akbari et al. 2006). It is also possible for DArT markers to be located in insertion/deletion sites (indels), although ∼80% are SNPs (Kilian et al. 2005). The minor allele frequency distribution for both marker sets was close to uniform across most of the allele frequency range, in contrast to the expected inflation of rare alleles expected for markers under drift–mutation equilibrium (Hamblin et al. 2011). This frequency spectrum in the SNP marker set was noted previously by Cavanagh et al. (2013), who concluded “The observed MAF is the consequence of intentional bias in SNP selection, where common alleles were favored by choosing more broadly distributed SNPs.”
Our genotype set had more than twice as many polymorphic SNP markers than DArT markers, and the DArT marker set had a higher proportion of missing data. Across the A and B genomes, DArT markers exhibit a more rapid breakdown of LD than SNP markers, but the pattern was reversed in the D genome. The longer linkage blocks in the D genome for both SNP and DArT markers is consistent with previous reports (Wang et al. 2014). The differences in marker density, distribution and LD structure between the two markers sets are the most likely causes of the observed differences in association analyses.
Conclusions
This study highlights the need to characterize G × E interactions in multi-environment datasets, and to define target populations of environments for marker-trait associations. These populations of environments define the scope of inference for interpreting GWAS results. In this study, we identified two clusters of experiments based on their genotypic correlations for the expression of WSCC. Marker associations for WSCC were identified only in the water deficit experiments, which represented a minority of the experiments; these associations would have been missed if the trait values were averaged across all experiments.
The loci identified for WSCC have both previously been associated with performance under water limited conditions, but did not reflect linkage to major effect relative maturity loci. The marker on 1D colocates with the Glu-D1 locus, which may have some pleiotropic effect on WSCC. These reported associations may be useful for marker-assisted selection of WSCC in water-limited environments, independent of relative maturity.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.039842/-/DC1.
Acknowledgments
We thank Kerry Schirmer and Aaron Hutchison for their expert technical contributions and data collection. We also thank the referees for their improvements to the manuscript. The breeding lines used in this study were kindly provided by Australian Grains Technologies, InterGrain, HRZ Wheats (now Dow Seeds), Sydney University, and LongReach Plant Breeders. The authors gratefully acknowledge the Grains Research and Development Corporation (GRDC) of Australia funding for this project (ICF00007), and a Grains Industry Research Scholarship for B.O. The American Australian Association is gratefully acknowledged for the ConocoPhillips Education Fellowship awarded to B.O.
Footnotes
Communicating editor: E. Akhunov
Literature Cited
- Akaike H., 1974. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19(6): 716–723. [Google Scholar]
- Akbari M., Wenzl P., Caig V., Carling J., Xia L., et al. , 2006. Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor. Appl. Genet. 113(8): 1409–1420. [DOI] [PubMed] [Google Scholar]
- Azadi A., Mardi M., Hervan E., Mohammadi S., Moradi F., et al. , 2015. QTL mapping of yield and yield components under normal and salt-stress conditions in bread wheat (Triticum aestivum L.). Plant Mol. Biol. Rep. 33(1): 102–120. [Google Scholar]
- Beeck C. P., Cowling W. A., Smith A. B., Cullis B. R., 2010. Analysis of yield and oil from a series of canola breeding trials. Part I. Fitting factor analytic mixed models with pedigree information. Genome 53(11): 992–1001. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57: 289–300. [Google Scholar]
- Bennett D., Izanloo A., Reynolds M., Kuchel H., Langridge P., et al. , 2012. Genetic dissection of grain yield and physical grain quality in bread wheat (Triticum aestivum L.) under water-limited environments. Theor. Appl. Genet. 125(2): 255–271. [DOI] [PubMed] [Google Scholar]
- Bidinger F., Musgrave R. B., Fischer R. A., 1977. Contribution of stored pre-anthesis assimilate to grain yield in wheat and barley. Nature 270(5636): 431–433. [Google Scholar]
- Bordes J., Ravel C., Jaubertie J. P., Duperrier B., Gardet O., et al. , 2013. Genomic regions associated with the nitrogen limitation response revealed in a global wheat core collection. Theor. Appl. Genet. 126(3): 805–822. [DOI] [PubMed] [Google Scholar]
- Breseghello F., Sorrells M. E., 2006. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172: 1165–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning B. L., Browning S. R., 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84(2): 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D., Cullis B. R., Gilmour A. R., Gogel B. J., 2009. ASReml-R Reference Manual, Queensland Department of Primary Industries and Fisheries, Brisbane, QLD. [Google Scholar]
- Cane K., Eagles H. A., Laurie D. A., Trevaskis B., Vallance N., et al. , 2013. Ppd-B1 and Ppd-D1 and their effects in southern Australian wheat. Crop Pasture Sci. 64: 100–114 [Google Scholar]
- Cavanagh C. R., Chao S., Wang S., Huang B. E., Stephen S., et al. , 2013. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 110(20): 8057–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmet G., Robert N., Branlard G., Linossier L., Martre P., et al. , 2005. Genetic analysis of dry matter and nitrogen accumulation and protein composition in wheat kernels. Theor. Appl. Genet. 111(3): 540–550. [DOI] [PubMed] [Google Scholar]
- Cleveland W. S., 1979. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 74(368): 829–836. [Google Scholar]
- Coombes, N. E., 2002 The reactive tabu search for efficient correlated experimental designs. Ph.D. Thesis, Liverpool John Moores University, Liverpool, UK. [Google Scholar]
- Crossa J., Burgueño J., Dreisigacker S., Vargas M., Herrera-Foessel S. A., et al. , 2007. Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 177: 1889–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F., Zhao C., Ding A., Li J., Wang L., et al. , 2014. Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor. Appl. Genet. 127(3): 659–675. [DOI] [PubMed] [Google Scholar]
- Cullis B. R., Smith A. B., Coombes N. E., 2006. On the design of early generation variety trials with correlated data. J. Agric. Biol. Environ. Stat. 11(4): 381–393. [Google Scholar]
- Cullis B. R., Smith A. B., Beeck C. P., Cowling W. A., 2010. Analysis of yield and oil from a series of canola breeding trials. Part II. Exploring variety by environment interaction using factor analysis. Genome 53(11): 1002–1016. [DOI] [PubMed] [Google Scholar]
- Deng Z., Tian J., Chen F., Li W., Zheng F., et al. , 2015. Genetic dissection on wheat flour quality traits in two related populations. Euphytica 203(1): 221–235. [Google Scholar]
- Eagles H. A., Cane K., Vallance N., 2009. The flow of alleles of important photoperiod and vernalisation genes through Australian wheat. Crop Pasture Sci. 60(7): 646–657. [Google Scholar]
- Eagles H. A., Cane K., Kuchel H., Hollamby G. J., Vallance N., et al. , 2010. Photoperiod and vernalization gene effects in southern Australian wheat. Crop Pasture Sci. 61(9): 721–730. [Google Scholar]
- Eagles H. A., Cane K., Trevaskis B., Vallance N., Eastwood R. F., et al. , 2014. Ppd1, Vrn1, ALMT1 and Rht genes and their effects on grain yield in lower rainfall environments in southern Australia. Crop Pasture Sci. 65(2): 159–170. [Google Scholar]
- Edae E. A., Byrne P. F., Haley S. D., Lopes M. S., Reynolds M. P., 2014. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor. Appl. Genet. 127(4): 791–807. [DOI] [PubMed] [Google Scholar]
- Ehdaie B., Alloush G. A., Waines J. G., 2008. Genotypic variation in linear rate of grain growth and contribution of stem reserves to grain yield in wheat. Field Crops Res. 106(1): 34–43. [Google Scholar]
- Endelman J. B., Jannink J.-L., 2012. Shrinkage estimation of the realized relationship matrix. G3 2: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes M. J., Sylvester-Bradley R., Weightman R., Snape J. W., 2007. Identifying physiological traits associated with improved drought resistance in winter wheat. Field Crops Res. 103(1): 11–24. [Google Scholar]
- Gebbing T., 2003. The enclosed and exposed part of the peduncle of wheat (Triticum aestivum): spatial separation of fructan storage. New Phytol. 159(1): 245–252. [DOI] [PubMed] [Google Scholar]
- Gebbing T., Schnyder H., 1999. Pre-anthesis reserve utilization for protein and carbohydrate synthesis in grains of wheat. Plant Physiol. 121(3): 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour A. R., Cullis B. R., Verbyla A. P., 1997. Accounting for natural and extraneous variation in the analysis of field experiments. J. Agric. Biol. Environ. Stat. 2(3): 269–293. [Google Scholar]
- Goggin D. E., Setter T. L., 2004. Fructosyltransferase activity and fructan accumulation during development in wheat exposed to terminal drought. Funct. Plant Biol. 31(1): 11–21. [DOI] [PubMed] [Google Scholar]
- Griffiths S., Simmonds J., Leverington M., Wang Y., Fish L., et al. , 2009. Meta-QTL analysis of the genetic control of ear emergence in elite European winter wheat germplasm. Theor. Appl. Genet. 119(3): 383–395. [DOI] [PubMed] [Google Scholar]
- Guedira M., Maloney P., Xiong M., Petersen S., Murphy J. P., et al. , 2014. Vernalization duration requirement in soft winter wheat is associated with variation at the Vrn-B1 locus. Crop Sci. 54(5): 1960–1971. [Google Scholar]
- Hamblin M. T., Buckler E. S., Jannink J.-L., 2011. Population genetics of genomics-based crop improvement methods. Trends Genet. 27(3): 98–106. [DOI] [PubMed] [Google Scholar]
- Hanocq E., Laperche A., Jaminon O., Lainé A. L., Le Gouis J., 2007. Most significant genome regions involved in the control of earliness traits in bread wheat, as revealed by QTL meta-analysis. Theor. Appl. Genet. 114(3): 569–584. [DOI] [PubMed] [Google Scholar]
- Harris F. A. J., Eagles H. A., Virgona J. M., Martin P. J., Condon J. R., et al. , 2017. Effect of VRN1 and PPD1 genes on anthesis date and wheat growth. Crop Pasture Sci. 68(3): 195–201. [Google Scholar]
- Jin H., Wang Z., Li D., Wu P., Dong Z., et al. , 2015. Genetic analysis of chromosomal loci affecting the content of insoluble glutenin in common wheat. J. Genet. Genomics 42(9): 495–505. [DOI] [PubMed] [Google Scholar]
- Kamran A., Iqbal M., Spaner D., 2014. Flowering time in wheat (Triticum aestivum L.): a key factor for global adaptability. Euphytica 197(1): 1–26. [Google Scholar]
- Kelly A. M., Smith A. B., Eccleston J. A., Cullis B. R., 2007. The accuracy of varietal selection using factor analytic models for multi-environment plant breeding trials. Crop Sci. 47(3): 1063–1070. [Google Scholar]
- Kenward M. G., Roger J. H., 1997. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics 53(3): 983–997. [PubMed] [Google Scholar]
- Kilian, A., E. Huttner, P. Wenzl, D. Jaccoud, J. Carling et al., 2005 The fast and the cheap: SNP and DArT-based whole genome profiling for crop improvement, pp. 443–461 in Proceedings of the International Congress ’In the Wake of the Double Helix: from the Green Revolution to the Gene Revolution,’, edited by R. Tuberosa, R. L. Phillips, and M. Gale. Avenue Media, Bologna, Italy. [Google Scholar]
- Kiniry J. R., 1993. Nonstructural carbohydrate utilization by wheat shaded during grain growth. Agron. J. 85(4): 844–849. [Google Scholar]
- Laidò G., Marone D., Russo M. A., Colecchia S. A., Mastrangelo A. M., et al. , 2014. Linkage disequilibrium and genome-wide association mapping in tetraploid wheat (Triticum turgidum L.). PLoS One 9(4): e95211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge P., Reynolds M. P., 2015. Genomic tools to assist breeding for drought tolerance. Curr. Opin. Biotechnol. 32: 130–135. [DOI] [PubMed] [Google Scholar]
- Le Gouis J., Bordes J., Ravel C., Heumez E., Faure S., et al. , 2012. Genome-wide association analysis to identify chromosomal regions determining components of earliness in wheat. Theor. Appl. Genet. 124(3): 597–611. [DOI] [PubMed] [Google Scholar]
- Lipka A. E., Tian F., Wang Q., Peiffer J., Li M., et al. , 2012. GAPIT: genome association and prediction integrated tool. Bioinformatics 28(18): 2397–2399. [DOI] [PubMed] [Google Scholar]
- Lopes M. S., Dreisigacker S., Peña R. J., Sukumaran S., Reynolds M. P., 2015. Genetic characterization of the wheat association mapping initiative (WAMI) panel for dissection of complex traits in spring wheat. Theor. Appl. Genet. 128(3): 453–464. [DOI] [PubMed] [Google Scholar]
- Masoudi B., Mardi M., Hervan E. M., Bihamta M., Naghavi M., et al. , 2015. QTL mapping of salt tolerance traits with different effects at the seedling stage of bread wheat. Plant Mol. Biol. Rep. 33: 1790–1803. [Google Scholar]
- McIntyre C. L., Mathews K. L., Rattey A., Chapman S. C., Drenth J., et al. , 2010. Molecular detection of genomic regions associated with grain yield and yield-related components in an elite bread wheat cross evaluated under irrigated and rainfed conditions. Theor. Appl. Genet. 120(3): 527–541. [DOI] [PubMed] [Google Scholar]
- Oldmeadow C., Riveros C., Holliday E. G., Scott R., Moscato P., et al. , 2011. Sifting the wheat from the chaff: prioritizing GWAS results by identifying consistency across analytical methods. Genet. Epidemiol. 35(8): 745–754. [DOI] [PubMed] [Google Scholar]
- Passioura J. B., 1996. Drought and drought tolerance. Plant Growth Regul. 20(2): 79–83. [Google Scholar]
- Payne P. I., 1987. Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Annu. Rev. Plant Physiol. 38(1): 141–153. [Google Scholar]
- Pheloung P., Siddique K., 1991. Contribution of stem dry matter to grain yield in wheat cultivars. Funct. Plant Biol. 18(1): 53–64. [Google Scholar]
- Piaskowski J. L., Brown D., Garland Campbell K., 2016. Near-infrared calibration of soluble stem carbohydrates for predicting drought tolerance in spring wheat. Agron. J. 108(1): 285–293. [Google Scholar]
- Piltz J., Law D., 2007. AFIA-Laboratory Methods Manual Australian Fodder Industry Association Inc, Balwyn, VIC. [Google Scholar]
- Pinto R. S., Reynolds M., Mathews K., McIntyre C. L., Olivares-Villegas J.-J., et al. , 2010. Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Theor. Appl. Genet. 121(6): 1001–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessis A., Ravel C., Bordes J., Balfourier F., Martre P., 2013. Association study of wheat grain protein composition reveals that gliadin and glutenin composition are trans-regulated by different chromosome regions. J. Exp. Bot. 64(12): 3627–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81(3): 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2012 R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna. Available at: http://www.R-project.org/
- Rattey A., Shorter R., Chapman S., Dreccer F., van Herwaarden A., 2009. Variation for and relationships among biomass and grain yield component traits conferring improved yield and grain weight in an elite wheat population grown in variable yield environments. Crop Pasture Sci. 60(8): 717–729. [Google Scholar]
- Ray D. K., Ramankutty N., Mueller N. D., West P. C., Foley J. A., 2012. Recent patterns of crop yield growth and stagnation. Nat. Commun. 3: 1293. [DOI] [PubMed] [Google Scholar]
- Rebetzke G. J., van Herwaarden A. F., Jenkins C., Weiss M., Lewis D., et al. , 2008. Quantitative trait loci for water-soluble carbohydrates and associations with agronomic traits in wheat. Aust. J. Agric. Res. 59(10): 891–905. [Google Scholar]
- Rebetzke G. J., Chapman S. C., McIntyre C. L., Richards R. A., Condon A. G., et al. , 2009. Grain yield improvement in water-limited environments, pp. 215–249 in Wheat: Science and Trade, edited by Carver B. F. Wiley-Blackwell, Ames, IA. [Google Scholar]
- Reynolds M., Tattaris M., Cossani C. M., Ellis M., Yamaguchi-Shinozaki K., et al. , 2015. Exploring genetic resources to increase adaptation of wheat to climate change, pp. 355–368 in Advances in Wheat Genetics: From Genome to Field: Proceedings of the 12th International Wheat Genetics Symposium, edited by Ogihara Y., Takumi S., Handa H. Springer, Tokyo, Japan. [Google Scholar]
- Reynolds M. P., Quilligan E., Aggarwal P. K., Bansal K. C., Cavalieri A. J., et al. , 2016. An integrated approach to maintaining cereal productivity under climate change. Glob. Food Secur. 8: 9–18. [Google Scholar]
- Ruuska S., Lewis D., Kennedy G., Furbank R., Jenkins C., et al. , 2008. Large scale transcriptome analysis of the effects of nitrogen nutrition on accumulation of stem carbohydrate reserves in reproductive stage wheat. Plant Mol. Biol. 66(1): 15–32. [DOI] [PubMed] [Google Scholar]
- Schnyder H., 1993. The role of carbohydrate storage and redistribution in the source-sink relations of wheat and barley during grain filling—a review. New Phytol. 123(2): 233–245. [Google Scholar]
- Shindo C., Tsujimoto H., Sasakuma T., 2003. Segregation analysis of heading traits in hexaploid wheat utilizing recombinant inbred lines. Heredity 90(1): 56–63. [DOI] [PubMed] [Google Scholar]
- Singh R., Matus-Cádiz M., Båga M., Hucl P., Chibbar R., 2010. Identification of genomic regions associated with seed dormancy in white-grained wheat. Euphytica 174(3): 391–408. [Google Scholar]
- Slafer G. A., Kantolic A. G., Appendino M. L., Tranquilli G., Miralles D. J., et al. , 2015. Genetic and environmental effects on crop development determining adaptation and yield, pp. 285–319 in Crop Physiology, Ed. 2, chap. 12, edited by Sadras V. O., Calderini D. F. Academic Press, San Diego. [Google Scholar]
- Smith A., Cullis B., Thompson R., 2001. Analyzing variety by environment data using multiplicative mixed models and adjustments for spatial field trend. Biometrics 57(4): 1138–1147. [DOI] [PubMed] [Google Scholar]
- Smith A., Lim P., Cullis B. R., 2006. The design and analysis of multi-phase plant breeding experiments. J. Agric. Sci. 144(5): 393. [Google Scholar]
- Snape J. W., Foulkes M. J., Simmonds J., Leverington M., Fish L. J., et al. , 2007. Dissecting gene x environmental effects on wheat yields via QTL and physiological analysis. Euphytica 154(3): 401–408. [Google Scholar]
- Sourdille P., Cadalen T., Guyomarc’h H., Snape J., Perretant M., et al. , 2003. An update of the Courtot × Chinese Spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theor. Appl. Genet. 106(3): 530–538. [DOI] [PubMed] [Google Scholar]
- Stram D. O., Lee J. W., 1994. Variance components testing in the longitudinal mixed effects model. Biometrics 50: 1171–1177. [PubMed] [Google Scholar]
- Takahashi T., Chevalier P. M., Rupp R. A., 2001. Storage and remobilization of soluble carbohydrates after heading in different plant parts of a winter wheat cultivar. Plant Prod. Sci. 4(3): 160–165. [Google Scholar]
- van Herwaarden A. F., Angus J. F., Richards R. A., Farquhar G. D., 1998. ‘Haying-off’, the negative grain yield response of dryland wheat to nitrogen fertiliser II. Carbohydrate and protein dynamics. Aust. J. Agric. Res. 49(7): 1083–1094. [Google Scholar]
- Virgona J. M., Barlow E. W. R., 1991. Drought stress induces changes in the non-structural carbohydrate composition of wheat stems. Funct. Plant Biol. 18(3): 239–247. [Google Scholar]
- Wang S., Wong D., Forrest K., Allen A., Chao S., et al. , 2014. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12(6): 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw I. F., Willenbrink J., 1994. Carbohydrate storage and mobilisation by the culm of wheat between heading and grain maturity: the relation to sucrose synthase and sucrose-phosphate synthase. Funct. Plant Biol. 21(3): 255–271. [Google Scholar]
- Wenzl P., Carling J., Kudrna D., Jaccoud D., Huttner E., et al. , 2004. Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc. Natl. Acad. Sci. USA 101(26): 9915–9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting D., 2004. Wheat Varieties in Australia, 1968–2001. D. Whiting, Snowtown, SA. [Google Scholar]
- Wimmer V., Albrecht T., Auinger H.-J., Schön C.-C., 2012. Synbreed: a framework for the analysis of genomic prediction data using R. Bioinformatics 28(15): 2086–2087. [DOI] [PubMed] [Google Scholar]
- Xue G.-P., Drenth J., Glassop D., Kooiker M., McIntyre C. L., 2013. Dissecting the molecular basis of the contribution of source strength to high fructan accumulation in wheat. Plant Mol. Biol. 81(1–2): 71–92. [DOI] [PubMed] [Google Scholar]
- Yan L., Helguera M., Kato K., Fukuyama S., Sherman J., et al. , 2004. Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor. Appl. Genet. 109(8): 1677–1686. [DOI] [PubMed] [Google Scholar]
- Yang D.-L., Jing R.-L., Chang X.-P., Li W., 2007. Identification of quantitative trait loci and environmental interactions for accumulation and remobilization of water-soluble carbohydrates in wheat (Triticum aestivum L.) stems. Genetics 176: 571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Mao S.-L., Chen G.-Y., Pu Z.-E., Wei Y.-M., et al. , 2014. QTLs for uppermost internode and spike length in two wheat RIL populations and their affect upon plant height at an individual QTL level. Euphytica 200(1): 95–108. [Google Scholar]
- Zadoks J. C., Chang T. T., Konzak C. F., 1974. A decimal code for the growth stages of cereals. Weed Res. 14(6): 415–421. [Google Scholar]
- Zhang Z., Ersoz E., Lai C.-Q., Todhunter R. J., Tiwari H. K., et al. , 2010. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42(4): 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Li S., Liang Z., Zhang Z., Xu X., 2010. Relationship between yield, carbon isotope discrimination and stem carbohydrate concentration in spring wheat grown in Ningxia Irrigation Region (North-west China). Crop Pasture Sci. 61(9): 731–742. [Google Scholar]
- Zila C. T., Samayoa L. F., Santiago R., Butrón A., Holland J. B., 2013. A genome-wide association study reveals genes associated with fusarium ear rot resistance in a maize core diversity panel. G3 3: 2095–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplemental Material, File S1 contains a detailed description of all Supplemental files. File S2 contains phenotype information for WSCC. File S3 contains phenotype information for relative maturity at flowering time. File S4 contains SNP genotypes for each individual. File S5 contains DArT genotypes for each individual.