Abstract
Single nucleotide polymorphisms (SNPs) in the let-7 miRNA binding site within the 3’ untranslated region (3’UTR) of KRAS appear related to the risk of cancer. The present case-control study was conducted with 244 BC patients and 204 healthy women to examine whether KRAS polymorphisms (rs61764370 T/G and rs712 G/T) are associated with breast cancer (BC) risk in an Iranian population. The polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) method was used for genotyping of KRAS SNPs. Our results showed that the rs61764370 TG genotype (OR= 3.73; 95% CI =1.38-10.08; P=0.007) as well as the G allele OR= 3.56; 95% CI =1.33-9.53; P=0.008, respectively) increased the risk of BC. However, the KRAS rs712 TT vs GG+GT genotype in a recessive model was associated with a reduced risk of BC (OR= 0.56; 95% CI =0.38-0.84; P=0.006). In addition, the rs712 T allele decreased the risk of BC compared with the G allele (OR=0.75, 95%CI=0.58-0.97, P=0.031). However, we found no relationship among KRAS SNPs and clinicopathological characteristics of BC patients (P>0.05). Taken together, the present study provided evidence of relationships between KRAS polymorphisms and BC risk in a southeast Iranian population. Additional studies using larger sample sizes and diverse ethnicities are now warranted.
Keywords: Let-7, KRAS, polymorphism, PCR-RFLP, breast cancer
Introduction
One of the most common cancers in women globally, breast cancer (BC) influences over 1 million women yearly (Keegan et al., 2007; Babu et al., 2011; Bray et al., 2013). The etiology of BC is unclear, but genetic factors have been suggested to play key functions in the development of BC (Hashemi et al., 2012a; Hashemi et al., 2012b; Hashemi et al., 2013a).
MiRNAs are small, non-coding, sequences of nucleotides that control gene expression through attaching to complementary sites in the 3’-untranslated region (3’UTRs) of target mRNAs (Meltzer, 2005). Till now, nearly 2000 miRNAs have been identified in humans and growing proof highlights their central function in the differentiation, cell propagation, carcinogenesis, tumour progression and response to treatment (Valeri et al., 2010). The lethal-7 (let-7) family members are among the most known miRNAs in human cancers. In general, they function as tumour suppressors by suppressing oncogenes implicated in the regulation of the cell cycle or intracellular signaling cascades (Su et al., 2012). The V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) is an effector molecule that contributes to several important signal transduction pathways such as Ral guanine nucleotide exchange factor and mitogen-activated protein kinase, phosphatidylinositol 3-kinase pathways. KRAS is a recognized target of let-7 and multiple complementary sites for this miRNA were determined in the 3’-untranslated (3’UTR) region of the mRNA.
Recent work has introduced that a germline and functional single-nucleotide polymorphism (SNP) in the KRAS 3’UTR region (rs61764370 T/G) is located in the let-7 complementary site 6 (LCS-6). Current proof indicated that that LCS-6 disrupts the let-7 binding affinity for KRAS, and result in decreased KRAS inhibition and enhanced tumour growth (Johnson et al., 2007; Chung et al., 2014). A number studies have examined the impact of KRAS rs61764370 T/G polymorphism on the risk of various cancers. For example, the LCS6 variant allele was shown to affect non-small cell lung cancer (NSCLC) in moderate smokers (Chin et al., 2008), triple-negative BC in premenopausal women (Paranjape et al., 2011) and ovarian cancer in BRCA negative females from inherited BC and ovarian cancer syndrome families (Ratner et al., 2010).
Another SNP within KRAS gene is in the rs712 region of the let-7 binding site, which is another miRNA target site. A number studies have assessed the impact of rs712 genetic variation in different cancers including BC (Huang et al., 2015), papillary thyroid carcinoma (PTC) (Ning et al., 2016), oral squamous cell carcinoma (OSCC) (Wang et al., 2012) and gastric cancer (Li et al., 2013). Huang et al (Huang et al., 2015) found no relationship between the let-7 rs712 polymorphism and BC risk, but they determined its possible function in BC tumor metastasis that can be used as a new biomarker towards resistant tumor metastasis. In contrast, a meta-analysis by Ying et al (Ying et al., 2014) confirmed that allele T of rs712 enhances genetic risk of cancer in Chinese population.
To the best of our knowledge, no study has examined the influence of KRAS gene polymorphisms on BC susceptibility in an Iranian population. Accordingly, we aimed to investigate the potential relationship between KRAS variants (rs61764370 T/G and rs712 G/T) and BC risk in a sample of southeast Iranian women. Besides, we also evaluated the association of the KRAS variants with clinicopathological features of BC patients.
Materials and Methods
Study population
The present case-control study was done on 244 BC patients and 204 ages adjusted healthy women with no background of any malignancy. The clinicopathological characteristics of BC patients are shown in Table 1. The registration process and study design have been described in our previous investigations (Amininia et al., 2014; Rezaei et al., 2016; Sanaei et al., 2016). Ethical certificate for enrollment was taken from local Ethics Committee of Zahedan University of Medical Sciences (#7082), and informed consent was acquired from all included individuals. We collected blood samples in EDTA-containing tubes from case and control subjects and extracted the DNA by salting out method as explained previously (Hashemi et al., 2013b).
Table 1.
Clinical and Pathological Characteristics of Breast Cancer Patients
| Characteristics | Cases n (%) |
|---|---|
| Tumor Size (cm) | |
| ≤2 | 74 (30.3) |
| >2 | 122 (50.0) |
| Unknown | 48 (19.7) |
| Nodal metastasis | |
| No | 60 (24.6) |
| Yes | 152 (62.3) |
| Unknown | 32 (13.1) |
| Grade | |
| I | 39 (16.0) |
| II | 115 (47.1) |
| III | 40 (16.4) |
| IV | 1 (0.4) |
| Unknown | 49 (20.1) |
| Stage | |
| I | 38 (15.6) |
| II | 85 (34.8) |
| III | 68 (27.9) |
| IV | 39 (16.0) |
| Unknown | 14 (5.7) |
| Histology | |
| Ductal carcinoma | 158 (70.5) |
| Other | 66 (29.5) |
| Estrogen Receptor status | |
| Positive | 137 (56.1) |
| Negative | 81 (33.2) |
| Unknown | 26 (10.7) |
| Progesterone Receptor status | |
| Positive | 133 (54.5) |
| Negative | 84 (34.4) |
| Unknown | 27 (11.1) |
| HER2 status | |
| Positive | 117 (48.0) |
| Negative | 113 (46.3) |
| Unknown | 14 (5.7) |
Genotyping Method
We used polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) method for identification of KRAS SNPs. For KRAS rs61764370, the forward and reverse primers were 5`-GTGTCAGAGTCTCGCTCTTGTC-3` and 5`-AGACCACACTAGCACTACCTAAGGA-3`, respectively. For KRAS rs712 the primers were 5`-AAGGCATACTAGTACAAGTGGTAA-3` and 5`-TGTGTTCCCTCAATGTTTCAGT-3`.
In each 0.20 ml PCR reaction tube, we mixed 1 μl of genomic DNA (~100 ng/ml), 1 μl of each primer and 10 μl of 2X Prime Taq Premix (Genet Bio, Korea) in addition to 7 μl ddH2O. The PCR conditions were standardized as follows: 5 min preheating at 95°C, 30 cycles of 95°C for 30s, 62°C for rs712, and 58°C for rs61764370 for 30s, and 72 °C for 30s followed by a final extension step for 10 min at 72 °C. For rs61764370, the PCR product (10 μl) was digested using HinfI restriction enzyme (Thermo Scientific, Vilnius, Lithuania). The T allele was digested and produced 80, 135, 161 bp fragments, but the G allele produces 296 and 80-bp amplicons (Figure 1).
Figure 1.
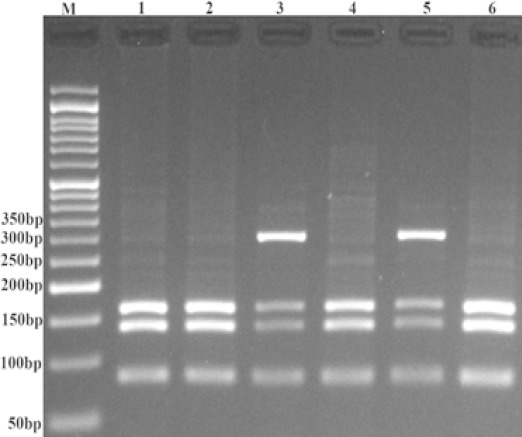
Photograph of the KRAS rs61764370 Polymorphism Using PCR-RFLP. The T allele was digested by HinfI restriction enzyme and produces 80-, 135-, and 161-bp fragments, but the G allele produces 296-and 80-bpfragments. M: DNA marker; Lanes 1, 2, 4, and 6: TT; Lanes 3, and 5:TG.
For KRAS rs712, 10 μl of PCR product was digested using TaqI (Thermo Scientific, Vilnius, Lithuania) restriction enzyme. Product size was 426-bp for T allele (undigested); G allele digested and produces 154-bp and 272-bp (Figure 2). To verify the genotyping quality, we randomly re-genotyped 20% of the samples and the genotypic were 100% concordant.
Figure 2.
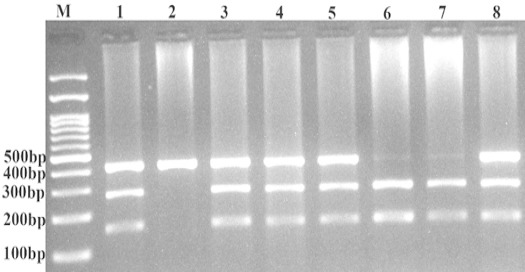
Photograph of the KRASrs712 Polymorphism Using PCR-RFLP. The G allele digested by TaqI restriction enzyme and produces 154-bp and 272-bp, however T allele undigested (426-bp). M: DNA marker; Lanes 1, 3, 4, 5, and 8, GT; Lane 2, TT; Lanes 6, and 7, GG.
Statistical analysis
Statistical analyses were executed by SPSS 19.0 software package. Independent sample t-test and the χ2 test were applied for continuous and categorical data, respectively. The association between genotypes and BC were determined by calculating the odds ratio (OR) and 95% confidence intervals (95% CI) from logistic regression analyses. All reported p values are two-sided and took into account statistically significant if p was below 0.05. Besides, Hardy-Weinberg equilibrium (HWE) was calculated for both cases and controls.
Results
The present study group included 244 BC patients with a mean age of 49.1 ±11.1 years and 204 healthy women with a mean age of 48.9 ±12.1 years. No significant difference was found between the groups regarding age (p=0.838). The HWE calculation indicated that both KRAS SNPs were in HWE in the control group (P>0.05).
The genotype and allelic frequency of KRAS rs712 G>T and rs61764370 T>G polymorphisms are shown in table 2. Our findings indicated that rs61764370 T>G heterozygote TG genotype vs TT was associated with higher risk of developing BC (TT vs GG+GT: OR=3.55, 95% CI=1.31-9.65, P=0.012). Similarly, in allele comparison G vs T, our analysis indicated that the G allele was a risk factor for BC (OR=3.44, 95%CI=1.28-9.29, P=0.013).
Table 2.
Association of KRAS Gene Polymorphisms (Rs712 G/T and Rs61764370 T/G) and the Risk of Breast Cancer
| Polymorphism | Case n (%) | Control n (%) | OR (95%CI) | p-value |
|---|---|---|---|---|
| rs61764370 T>G | ||||
| TT | 224 (91.8) | 199 (97.5) | 1 | - |
| TG | 20 (8.2) | 5 (2.5) | 3.55 (1.31-9.65) | 0.012 |
| GG | 0 (0.0) | 0 (0.0) | - | - |
| Allele | ||||
| T | 468 (95.9) | 403 (98.8) | 1 | - |
| G | 20 (4.1) | 5 (1.2) | 3.44 (1.28-9.29) | 0.013 |
| rs712 G>T | ||||
| Codominant | ||||
| GG | 33 (13.5) | 25 (12.3) | 1 | - |
| GT | 148 (60.7) | 101 (49.5) | 1.11 (0.62-1.98) | 0.768 |
| TT | 63 (25.8) | 78 (38.2) | 0.60 (0.33-1.07) | 0.104 |
| Dominant | ||||
| GG | 33 (13.5) | 25 (12.3) | 1 | - |
| GT+TT | 211 (85.7) | 179 (87.4) | 0.51 (0.51-1.56) | 0.893 |
| Recessive | ||||
| GG+GT | 181 (74.2) | 126 (61.8) | 1 | - |
| TT | 63 (25.8) | 78 (38.2) | 0.56 (0.38-0.84) | 0.006 |
| Allele | ||||
| G | 214 (43.9) | 151 (37.0) | 1 | - |
| T | 274 (56.1) | 257 (63.0) | 0.75 (0.57-0.98) | 0.041 |
With respect to rs712 G>T variation, our results demonstrated that the rs712 G>T variant was associated with a reduced risk of BC in recessive model (TT vs GG+GT: OR=0.56, 95% CI=0.38-0.84, P=0.006) as well as in allele comparison (T vs G: OR=0.75, 95% CI=0.57-0.98, P=0.041).
However, no significant association between clinical characteristics of BC patients including age, histological type, tumor size, stage, grade, ER, PgR and HER2 and the studied KRAS gene polymorphisms was observed (P>0.05, data not shown).
Discussion
The current study confirmed the relationship of KRAS gene polymorphisms and the risk of BC in an Iranian population. According to our results, the rs61764370 TG vs TT and G vs T allele were risk factors for BC. However, the rs712 TT vs GG+GT genotype and T vs G allele reduced the risk of BC with OR of 0.56 and 0.75, respectively.
In accordance with our findings, Pilarski et al., (2012) indicated that KRASrs61764370 polymorphism was associated with the risk of double primary breast and ovarian cancer. Another study Paranjape et al., (2011) showed the KRAS variant is a risk factor for triple negative breast cancer in premenopausal women (OR 2.31, 95% CI 1.26-4.22) highlighting its role as a genetic marker. On the other hand, Uvirova et al., (2015) have found no association between KRAS rs61764370 variant and risk of BC risk, although they suggested that KRAS rs61764370 TG genotype could affect the HER2 gene expression profile. Cerne et al., (2012) also found no proof of relationship of KRAS rs61764370 variant with risk of sporadic and familial breast cancer which supports the findings of a meta-analysis by Zhang et al., (2016) who summarized that KRAS genotype GT/GG of rs61764370 was not related to the risk of breast, ovarian, non.-small cell lung cancer, colorectal, or head-neck carcinoma in Caucasian population.
Let-7 appears to play key roles in the BC advancement, because breast-tumour-initiating cells demonstrated markedly low levels of let-7; therefore, reintroduction of let-7 inhibits these putative BC stem cells by control of their self-renewal and ability to differentiate. Recent proof also suggests a central role for the let-7 miRNA family in progression of BC via altered expression of KRAS, an infrequent target of activating mutations in breast tumors (Kumar and Swanton, 2011). LCS6 in the KRAS 3’-UTR mRNA causes an increase in expression of KRAS in vitro and a reduction in let-7 levels in vivo (Chin et al., 2008). In contrast, Crowley et al., (2014) indicated that targeted knock-in of the polymorphism rs61764370 was not associated with KRAS levels but decreased let-7 expression. The KRAS LCS6 variant is rather infrequent, and extremely rare in East Asians and in Native Americans, and so uncommon in Africans with a minor allele frequency of 7% in the European populations (Chin et al., 2008). Similarly, in our study, the KRAS LCS6 variant allele was 4.1% in BC patients and 1.2% in healthy women.
With respect to rs712, our findings indicated that the rs712 TT vs GG+GT genotype in the recessive model and T vs G allele were protective factors against BC. Compared to our findings, Ning et al., (2016) found a considerable difference in both allelic and genotypic genotypes incidence for rs712 between PTC patients and control subjects. Dai et al., (2016) also demonstrated that TT genotype and T allele of rs712 were increased the risk of CRC. A meta-analysis by Zhao et al., (2015) concluded that allele T, genotype TT and allele T carrier (GT/TT) of rs712 may increase cancer risk in Chinese population, and potentially is capable of being used as a genetic factor for assessment of cancer risk. However, Huang et al., (2015) found no association between the let-7 rs712 polymorphism and BC risk, which supports the results of two studies performed on nasopharyngeal carcinoma (Pan et al., 2014) and PTC (Jin et al., 2014).
There are some limitations in the present study, one of which is investigating only two SNPs of the KRAS gene. Second limitation is lacking the information concerning the known risk factors for BC (e.g., parity, oral contraceptive or hormone therapy use, breast-feeding, smoking and alcohol intake). So, we were unable to determine the interaction between the variants and environmental factors.
Overall, our study provided the evidence regarding the implication of KRAS polymorphisms, rs61764370 and rs712, in developing breast carcinoma with lack of any relationship with clinicopathological features of the patients. The KRAS rs61764370 variant increased the BC risk, but the rs712 variant was a protective factor against BC in the studied Iranian population. More studies on larger sample sizes with different races are warranted to confirm our findings.
Disclosure of Conflicting Interests
The Authors state that there is no conflict of interest to declare.
Acknowledgements
This research was supported financially as a dissertation grant (MSc thesis of SS #7082) by deputy for Research, Zahedan University of Medical Sciences, Zahedan, Iran.
References
- Amininia S, Hashemi M, Ebrahimi M, et al. Association between CCNE1 polymorphisms and the risk of breast cancer in a sample of southeast Iranian population. Med Oncol. 2014;31:189. doi: 10.1007/s12032-014-0189-z. [DOI] [PubMed] [Google Scholar]
- Babu GR, Samari G, Cohen SP, et al. Breast cancer screening among females in Iran and recommendations for improved practice:a review. Asian Pac J Cancer Prev. 2011;12:1647–55. [PubMed] [Google Scholar]
- Bray F, Ren JS, Masuyer E, et al. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–45. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- Cerne JZ, Stegel V, Gersak K, et al. KRAS rs61764370 is associated with HER2-overexpressed and poorly-differentiated breast cancer in hormone replacement therapy users:a case control study. BMC Cancer. 2012;12:105. doi: 10.1186/1471-2407-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS3’untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CH, Lee JW, Slebos RJ, et al. A 3’-UTR KRAS-variant is associated with cisplatin resistance in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann Oncol. 2014;25:2230–6. doi: 10.1093/annonc/mdu367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley EH, Arena S, Lamba S, et al. Targeted knock-in of the polymorphism rs61764370 does not affect KRAS expression but reduces let-7 levels. Hum Mutat. 2014;35:208–14. doi: 10.1002/humu.22487. [DOI] [PubMed] [Google Scholar]
- Dai Q, Wei HL, Huang J, et al. KRAS polymorphisms are associated with survival of CRC in Chinese population. Tumour Biol. 2016;37:4727–34. doi: 10.1007/s13277-015-4314-1. [DOI] [PubMed] [Google Scholar]
- Hashemi M, Eskandari-Nasab E, Fazaeli A, et al. Bi-directional PCR allele-specific amplification (bi-PASA) for detection of caspase-8 -652 6N ins/del promoter polymorphism (rs3834129) in breast cancer. Gene. 2012a;505:176–9. doi: 10.1016/j.gene.2012.05.043. [DOI] [PubMed] [Google Scholar]
- Hashemi M, Eskandari-Nasab E, Fazaeli A, et al. Association between polymorphisms of glutathione S-transferase genes (GSTM1, GSTP1 and GSTT1) and breast cancer risk in a sample Iranian population. Biomarkers Med. 2012b;6:797–803. doi: 10.2217/bmm.12.61. [DOI] [PubMed] [Google Scholar]
- Hashemi M, Fazaeli A, Ghavami S, et al. Functional polymorphisms of FAS and FASL gene and risk of breast cancer -pilot study of 134 cases. PLoS One. 2013a;8:e53075. doi: 10.1371/journal.pone.0053075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi M, Hanafi Bojd H, Eskandari Nasab E, et al. Association of adiponectin rs1501299 and rs266729 gene polymorphisms with nonalcoholic fatty liver disease. Hepat Mon. 2013b;13:e9527. doi: 10.5812/hepatmon.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Yang Y, Guo Y, et al. Association of a let-7KRAS rs712 polymorphism with the risk of breast cancer. Genet Mol Res. 2015;14:16913–20. doi: 10.4238/2015.December.14.19. [DOI] [PubMed] [Google Scholar]
- Jin H, Liang Y, Wang X, et al. Association between a functional polymorphism rs712 within let-7-binding site and risk of papillary thyroid cancer. Med Oncol. 2014;31:221. doi: 10.1007/s12032-014-0221-3. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Keegan TH, Chang ET, John EM, et al. Recent changes in breast cancer incidence and risk factor prevalence in San Francisco Bay area and California women 1988 to 2004. Breast Cancer Res. 2007;9:R62. doi: 10.1186/bcr1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Swanton C. KRAS3’-UTR variants and stratification of breast-cancer risk. Lancet Oncol. 2011;12:318–9. doi: 10.1016/S1470-2045(11)70065-1. [DOI] [PubMed] [Google Scholar]
- Li ZH, Pan XM, Han BW, et al. A let-7 binding site polymorphism rs712 in the KRAS3’UTR is associated with an increased risk of gastric cancer. Tumour Biol. 2013;34:3159–63. doi: 10.1007/s13277-013-0885-x. [DOI] [PubMed] [Google Scholar]
- Meltzer PS. Cancer genomics:small RNAs with big impacts. Nature. 2005;435:745–6. doi: 10.1038/435745a. [DOI] [PubMed] [Google Scholar]
- Ning L, Rao W, Yu Y, et al. Association between the KRAS gene polymorphisms and papillary thyroid carcinoma in a Chinese Han population. J Cancer. 2016;7:2420–6. doi: 10.7150/jca.16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan XM, Jia J, Guo XM, et al. Lack of association between let-7 binding site polymorphism rs712 and risk of nasopharyngeal carcinoma. Fam Cancer. 2014;13:93–7. doi: 10.1007/s10689-013-9681-4. [DOI] [PubMed] [Google Scholar]
- Paranjape T, Heneghan H, Lindner R, et al. A 3’-untranslated region KRAS variant and triple-negative breast cancer:a case-control and genetic analysis. Lancet Oncol. 2011;12:377–86. doi: 10.1016/S1470-2045(11)70044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilarski R, Patel DA, Weitzel J, et al. The KRAS-variant is associated with risk of developing double primary breast and ovarian cancer. PLoS One. 2012;7:e37891. doi: 10.1371/journal.pone.0037891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner E, Lu L, Boeke M, et al. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70:6509–15. doi: 10.1158/0008-5472.CAN-10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaei M, Hashemi M, Sanaei S, et al. Association between vascular endothelial growth factor gene polymorphisms with breast cancer risk in an Iranian population. Breast Cancer (Auckl) 2016;10:85–91. doi: 10.4137/BCBCR.S39649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei S, Hashemi M, Rezaei M, et al. Evaluation of the pri-miR-34b/c rs493∓polymorphism and its association with breast cancer risk. Biomed Rep. 2016;5:125–9. doi: 10.3892/br.2016.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su JL, Chen PS, Johansson G, et al. Function and regulation of let-7 family microRNAs. Microrna. 2012;1:34–9. doi: 10.2174/2211536611201010034. [DOI] [PubMed] [Google Scholar]
- Uvirova M, Simova J, Kubova B, et al. Comparison of the prevalence of KRAS-LCS6 polymorphism (rs61764370) within different tumour types (colorectal, breast, non-small cell lung cancer and brain tumours). A study of the Czech population. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2015;159:466–71. doi: 10.5507/bp.2015.029. [DOI] [PubMed] [Google Scholar]
- Valeri N, Gasparini P, Braconi C, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci U S A. 2010;107:21098–103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WY, Chien YC, Wong YK, et al. Effects of KRAS mutation and polymorphism on the risk and prognosis of oral squamous cell carcinoma. Head Neck. 2012;34:663–6. doi: 10.1002/hed.21792. [DOI] [PubMed] [Google Scholar]
- Ying HQ, Wang F, He BS, et al. The involvement of Kras gene 3’-UTR polymorphisms in risk of cancer and influence on patient response to anti-EGFR therapy in metastatic colorectal cancer:a meta-analysis. Onco Targets Ther. 2014;7:1487–96. doi: 10.2147/OTT.S65496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SY, Shi J. rs61764370 polymorphism of Kras and risk of cancer in Caucasian population:A meta-analysis. J Cancer Res Ther. 2016;12:699–704. doi: 10.4103/0973-1482.147379. [DOI] [PubMed] [Google Scholar]
- Zhao WH, Qu XF, Xing ZG, et al. Association of rs712 polymorphism in Kras gene 3’-luntranslated region and cancer risk:a meta-analysis. J BUON. 2015;20:309–16. [PubMed] [Google Scholar]


