Abstract
Ribosome biogenesis requires the intertwined processes of folding, modification, and processing of ribosomal RNA, together with binding of ribosomal proteins. In eukaryotic cells, ribosome assembly begins in the nucleolus, continues in the nucleoplasm, and is not completed until after nascent particles are exported to the cytoplasm. The efficiency and fidelity of ribosome biogenesis are facilitated by >200 assembly factors and ~76 different small nucleolar RNAs. The pathway is driven forward by numerous remodeling events to rearrange the ribonucleoprotein architecture of pre-ribosomes. Here, we describe principles of ribosome assembly that have emerged from recent studies of biogenesis of the large ribosomal subunit in the yeast Saccharomyces cerevisiae. We describe tools that have empowered investigations of ribosome biogenesis, and then summarize recent discoveries about each of the consecutive steps of subunit assembly.
Introduction
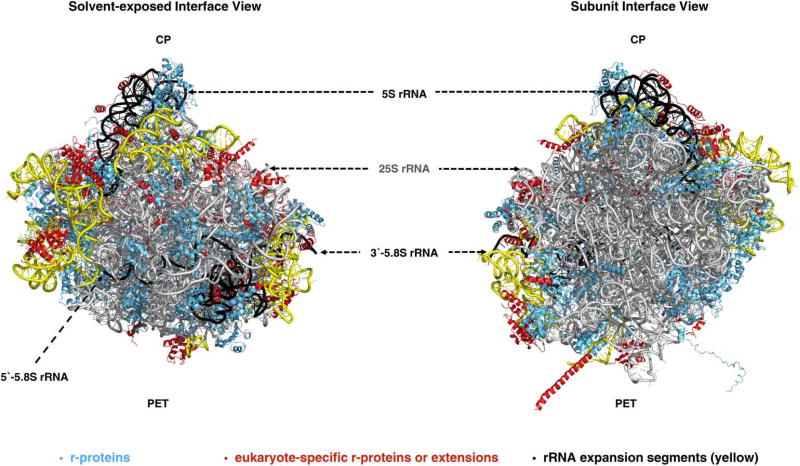
Ribosomes are ribonucleoprotein (RNP) complexes responsible for the synthesis of proteins in all cells in Nature. They contain two asymmetric subunits (large and small subunits), the core structure and function of which are largely conserved across all kingdoms of life [1,2]. Eukaryotic ribosomes differ from their prokaryotic counterparts in size and complexity due to the addition of eukaryote-specific ribosomal proteins (r-proteins), r-protein extensions, and ribosomal RNA (rRNA) expansion segments (ESs) [3–7]. In the yeast Saccharomyces cerevisiae, the large 60S subunit that hosts the catalytic peptidyl transferase center (PTC) contains 25S rRNA (3396 nucleotides), 5.8S rRNA (158 nucleotides), 5S rRNA (121 nucleotides), and 46 r-proteins (Figure 1). The small 40S subunit containing 18S rRNA (1800 nucleotides) and 33 r-proteins functions as the decoding center to bring together messenger RNAs and transfer RNAs (reviewed in ref. [8]).
Figure 1. Structure of the yeast 60S ribosomal subunit.
The crystal structure [5] of the yeast 60S subunit is shown with 25S (gray), 5S and 5.8S rRNAs (black), and r-proteins (blue) viewed from the solvent (left) and subunit interfaces (right). The conserved rRNA core (gray), eukaryote-specific rRNA expansion segments (ESs) in 25S rRNA (yellow), and eukaryote-specific r-proteins/extensions (red) are also indicated (PDB ID: 4V88).
In rapidly growing cells, a vast majority of cellular resources, including the gene expression and nuclear transport machinery, are devoted to the production of these mega-dalton scale nanomachines [9]. Since protein synthesis and translational fidelity are central to cellular homeostasis, major defects in ribosome assembly or function are lethal, observed as embryonic lethality in higher organisms. Partial loss-of-function mutations in ribosomal components or its assembly machinery in humans result in cancers and various other diseases referred to as ribosomopathies (reviewed in refs [10–12]).
The basic framework of ribosome assembly
Ribosome assembly begins with transcription of precursor rRNAs (pre-rRNAs), which then undergo base modification, co-transcriptional folding, pre-rRNA processing, and assembly with r-proteins to form functional ribosomal subunits (reviewed in ref. [13]). These events occur in a co-ordinated manner in a series of events spanning three subcellular compartments: the nucleolus, the nucleoplasm, and the cytoplasm (Figure 2). Studies over several decades have demonstrated that the concerted action of 76 small nucleolar RNPs (snoRNPs) and ~200 trans-acting proteins, referred to as assembly factors (AFs), facilitates production of translation-competent ribosomal subunits in S. cerevisiae. AFs include proteins with a wide range of biochemical activities, such as endonucleases and exonucleases, GTPases, ATP-dependent RNA helicases, AAA-ATPases, kinases, and structural proteins, predicted to function as scaffolds or chaperones. During ribosome assembly, the structural AFs help to stabilize pre-ribosomes, and to establish molecular interaction networks within pre-ribosomes. The enzymatic AFs link maturation events with substantial changes in free energy to remodel molecular interaction networks during assembly (reviewed in refs [14,15]). The structural and functional integrity of pre-ribosomes are tested prior to their entry into the translation pool by AFs (reviewed in ref. [16] and see ‘The Middle Stage: Remodeling Events Required to Trigger Removal of the ITS2 Spacer’ and ‘Cytoplasmic Maturation of Large Ribosomal Subunit’). Finally, the existence of heterogeneous ribosomes and their roles in translational regulation are becoming increasingly evident (reviewed in refs [17–19]), although it remains to be understood if ribosome assembly mechanisms can be altered to construct them.
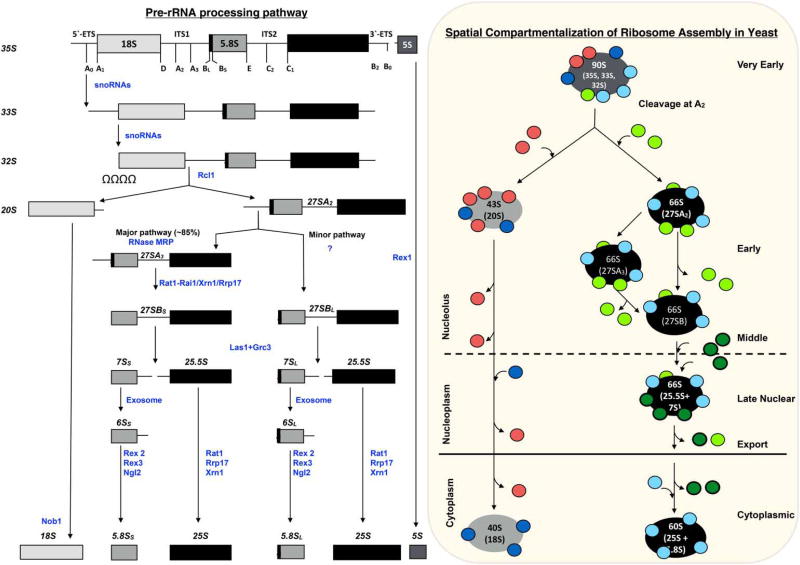
Figure 2. Pathway of ribosome assembly in S. cerevisiae.
Pre-rRNA processing pathway (left). The 35S pre-rRNA transcribed by RNA polymerase I contains sequences for 18S, 5.8S, and 25S rRNAs, separated and flanked by ITS and ETS. The processing sites in pre-rRNA are indicated. The spacer sequences are removed from pre-rRNAs in a series of endonucleolytic and exonucleolytic events (enzymes shown in blue). Pre-5S rRNA is transcribed by RNA polymerase III. It is not yet known where in the cell pre-5S rRNA processing occurs, but assembly with pre-ribosomes occurs in the nucleolus. Spatial compartmentalization of 60S subunit assembly in S. cerevisiae (right). Assembly occurs within RNA–protein complexes called pre-ribosomes. The protein composition of pre-ribosomes is dynamic due to the entry and exit of AFs (red/green) at specific steps of assembly, and due to pre-rRNA processing. Most r-proteins (blue) associate with pre-rRNA during very early steps of assembly.
There are many recent reviews highlighting details of different aspects of eukaryotic ribosome biogenesis (reviewed in refs [13,14–17,20–25]). Here, we focus on principles of eukaryotic ribosome biogenesis emerging from recent studies of 60S subunit assembly in yeast.
Functional sites in the large ribosomal subunit
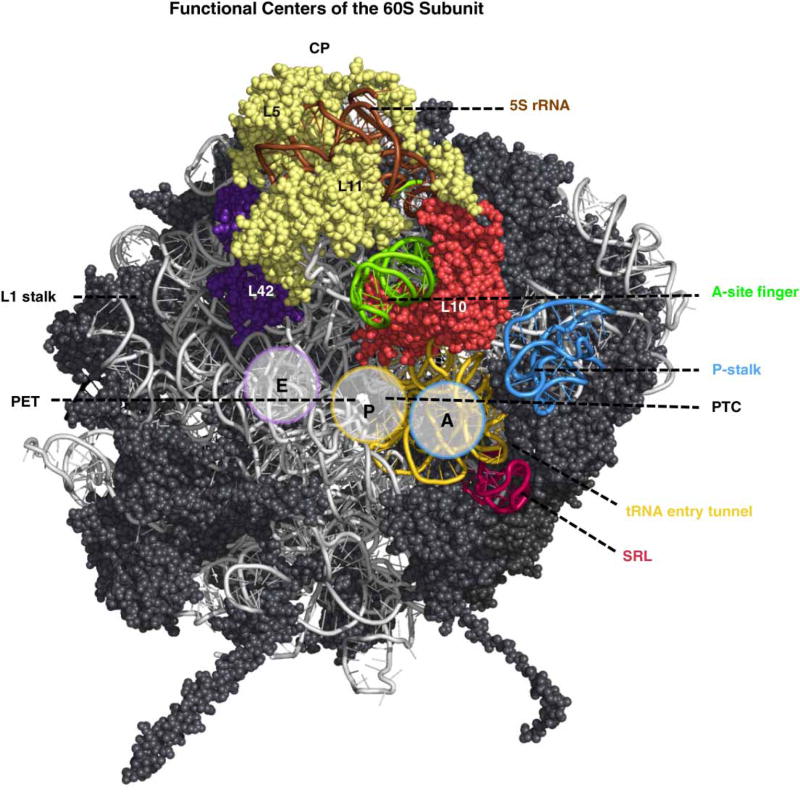
When trying to understand the mechanism of 60S ribosomal subunit biogenesis, it is important to consider that the assembly process must have evolved to inspect the structural and functional integrity of these functional sites at different steps of the assembly, and to protect them from premature binding to 40S subunits and translation factors. Functional sites in the large subunit include: (1) the P-stalk that recruits and activates translation factors; (2) the sarcin ricin loop (SRL) responsible for activation of GTPases involved in translation; (3) the tRNA accommodation corridor that contains the A-, P-, and E-sites to bind tRNAs bearing the substrate amino acid, tRNAs attached to the growing polypeptide chain, and tRNAs from which the 2′-acyl amino acid has been removed, respectively; (4) the PTC that catalyzes peptide bond formation; and (5) the polypeptide exit tunnel (PET) through which all nascent polypeptides exit the 60S subunit (Figure 3). Even though the PET is often considered as a mere passive conduit for nascent polypeptides, many recent studies have shown it to be an active participant in determining the rate of translation and protein folding [26,27]. All of these functional sites, except the PET, are present on the subunit interface of the 60S subunit that interacts with the 40S subunit (reviewed in refs [1,8]).
Figure 3. Functional centers in the large ribosomal subunit.
The P-stalk (blue), SRL (red), tRNA accommodation corridor beginning with helices 89 and 91 (yellow) containing the A- and P-sites, and ending with r-protein L42 (purple) at the end of the E-site, A-site finger (green), and CP containing 5S rRNA (brown) and r-proteins L5 and L11 (pale yellow) are shown. The r-protein L10 (orange-red) is also shown. The subunit interface is oriented to visualize the PET emerging from the active site in the PTC (PDB ID: 4V88).
Breaking down the pathway into steps
Three of the four rRNAs in eukaryotic ribosomes (18S, 5.8S, and 25S rRNAs) are synthesized by RNA polymerase I as a pre-RNA (35S pre-rRNA) that needs to undergo processing to acquire functionality (reviewed in ref. [22]). The 35S pre-rRNA contains mature rRNA sequences separated by two internal transcribed spacers (ITS1 and ITS2), and flanked by two external transcribed spacers (5′-ETS and 3′-ETS) (Figure 2). The fourth rRNA, 5S rRNA, is separately transcribed by RNA polymerase III and loaded onto nascent large subunits. Processing of pre-rRNA is executed in multiple steps by endonucleases and exonucleases. Most r-proteins and a subset of the AFs bind pre-rRNA co-transcriptionally, or very soon after transcription is completed, to generate the earliest pre-ribosomal intermediates. Other AFs enter pre-ribosomes at later times. Unlike r-proteins, association of AFs with pre-ribosomes is transient; they are not present in mature ribosomes. Thus, pre-ribosomes at different stages of ribosome assembly can be distinguished by their protein composition and by the presence of distinct pre-rRNA processing intermediates.
Assembly of 60S subunits can be grouped into at least six steps based on the pre-rRNA content of assembly intermediates (Figure 2). (1) ‘Very early steps’ occur as pre-rRNA is synthesized and initially compacted to form 90S pre-ribosomes, from which the 5′-ETS is removed and ITS1 cleavage occurs. Cleavage at the A2 site in ITS1 separates the 20S pre-rRNA from 27S pre-rRNA. These two pre-rRNAs are packaged into 43S and 66S precursors to mature 40S and 60S subunits, respectively. Thus, after cleavage at the A2 site, assembly of 40S and 60S subunits is largely independent. (2) ‘Early steps’ include removal of the ITS1 spacer sequence from 27SA2 and 27SA3 pre-rRNAs to produce 27SB pre-rRNA in 66S pre-ribosomes. (3) ‘Middle steps’ include cleavage at the C2 site in ITS2 of 27SB pre-rRNA to produce 7S and 25.5S pre-rRNAs, and exit of pre-ribosomes from the nucleolus. (4) ‘Late nuclear steps’ include removal of the ITS2 spacer (processing of 25.5S and 7S pre-rRNAs to 25S and 5.8S rRNAs) and remodeling of the central protuberance (CP). (5) Export of nascent particles through the nuclear pore complex (NPC) into the cytoplasm is enabled by binding of export factors. (6) ‘Final steps’ of subunit maturation occur in the cytoplasm and include the final steps of formation of 5.8S rRNA, assembly of the last few r-proteins, release of remaining AFs, and quality control checks of functional sites.
Evolution of tools to study ribosome assembly
The field of eukaryotic ribosome assembly emerged in the 1960s and 1970s with the discovery of pre-rRNA processing intermediates that were packaged into ribonucleoprotein particles (RNPs) containing r-proteins and unidentified non-r-proteins [28–33]. Owing to the availability of a wide spectrum of genetic, molecular, and proteomic techniques, S. cerevisiae has been the model organism of choice in the ensuing years to investigate ribosome assembly in eukaryotes [34]. Following a quiescent period for eukaryotic ribosome assembly research, the 1990s witnessed the development of genetic screens for ribosome assembly mutants, which led to the discovery of trans-acting AFs (reviewed in ref. [35]). This was followed up in the 2000s with the purification and characterization of pre-ribosomes using epitope-tagged AFs and mass spectrometry, which provided an almost complete catalog of AFs [36–38]. Conditional null mutants of essential AFs were then used to identify the pre-rRNA processing step(s) for which the AFs are required and the role of AFs in large ribosomal subunit biogenesis (reviewed in ref. [18]). Assaying the proteins and pre-rRNAs that co-purified with AFs in wild-type pre-ribosomes revealed their entry and exit points during assembly. These functional genetic and biochemical investigations reached another dimension with the elucidation of low-resolution cryo-EM structures in the 2000s, and high-resolution crystal structures of mature yeast ribosomes about 5–6 years ago, providing us with a detailed blueprint of the final product of the assembly pathway. Most recently, advances in cryo-EM technology have revealed near-atomic-resolution snapshots of assembly intermediates [39,40]. These structures provide valuable tools to begin to understand remodeling events during different steps of ribosome assembly, and help guide the design of experiments to uncover the precise mechanisms of action of AFs. The combination of methods to elucidate binding sites of AFs on rRNAs (CRAC), to determine rRNA structure, and to map protein–protein interaction networks (yeast two-hybrid and pull-down assays), together with advances in mass spectrometry (SILAC, iTRAQ SRM, and SWATH) and structure-based site-directed mutagenesis, is now enabling ribosome biologists to uncover mechanistic details of this complex process at an ever more rapid pace [41–48].
Roles of rRNA in assembly
The 5.8S and 25S rRNAs in the 60S subunit fold into six conserved domains of secondary structure (I–VI), which are then compacted to form the rRNA tertiary structure (Figure 4A,B). The high copy number of rRNA genes in yeast (~200 copies) poses a significant challenge for mutational analysis of rRNA structural elements, although the development of genetic approaches to overcome this challenge has revealed functionally important regions in transcribed spacer sequences (ITSs and ETSs) and rRNA [50–55]. Recently, it has been shown that eukaryotic-specific rRNA sequences, so-called ESs, participate in early, middle, and late nuclear steps of ribosome biogenesis [49]. Their roles in assembly correlate with their position in 60S subunits, identically to the situation for r-proteins (see below). The early-acting ESs localize to 25S rRNA domains I and III, the middle-acting ESs to the neighborhood surrounding the PET, and the late-acting ESs are near the CP and the subunit interface (Figure 4C). Structures of mature and pre-60S subunits revealed that most ESs have co-evolved with the r-protein extensions (‘R-Protein Extensions Are Crucial to Ribosome Assembly’), which potentially aid the folding of the ESs. Some ESs and transcribed spacers also serve as binding sites for AFs [56,57].
Figure 4. Hierarchical assembly of neighborhoods within 60S subunits.
Folding of 60S subunit rRNAs in yeast. The 25S rRNA contains six secondary structure domains (I–VI) (A), which are compacted along with 5.8S and 5S rRNAs to form the tertiary structure of rRNA in the 60S subunit (B). For example, 5.8S rRNA base-pairs with domain I (purple) and is sandwiched between domains I and III (green) of 25S rRNA that are far apart in the primary sequence of 25S rRNA. (C) Correlation between the location and the function of ESs in 60S subunit assembly. ESs cluster on the periphery of the 60S subunit and are required for specific steps of pre-rRNA processing [49]. Deletion of ESs: (1) in the equatorial belt (blue) affects early steps, (2) in the bottom half (orange) affects middle steps, and (3) around the CP and ES19 (green) affects late steps of 60S subunit assembly (green). (D) Correlation between the location and the function of r-proteins. Depletion of r-proteins in the equatorial belt of the solvent interface affects early (blue) steps of 60S subunit assembly. Depletion of r-proteins in the bottom half of 60S subunit (orange) and surrounding the CP and on the subunit interface (green) affects late steps of 60S subunit assembly. Many r-proteins that bind to the subunit interface (purple) associate with pre-ribosomes in the cytoplasm. The secondary structure of large subunit rRNA was obtained from www.ribovision.org (PDB ID: 4V88).
Lessons emerging from studies of r-proteins
In vitro reconstitution of bacterial ribosomal subunits revealed that their assembly is hierarchical and occurs largely 5′ to 3′ with respect to rRNA (reviewed in ref. [58]). Importantly, determination of the crystal structure of yeast ribosomes [3–5], together with the development of methods to purify and characterize ribosome assembly intermediates [36–39], provided the means to investigate how eukaryotic r-proteins assemble into ribosomes in vivo. Systematic examination of the effects of depleting r-proteins demonstrated that the assembly of r-proteins into large subunits in yeast cells is hierarchical [20,59–67]. The solvent-exposed surface of the large subunit, where r-proteins bind domains I, II, and VI of 25S rRNA, is assembled first (Figure 4D). Its construction is required for the earliest pre-rRNA processing steps (removal of ITS1). Subsequently, the neighborhood adjacent to 5.8S rRNA and the rim surrounding the PET are assembled. Depletion of r-proteins located here prevents middle steps of assembly required to initiate removal of ITS2 (cleavage at the C2 site). Finally, the intersubunit surface of large subunits, containing domains IV and V of 25S rRNA, and the neighborhood surrounding the CP are assembled. Depleting r-proteins located here affects the final nuclear steps of pre-rRNA processing, as well as remodeling events that prepare subunits for nuclear export. The few r-proteins that assemble in the cytoplasm are also located in the subunit interface. Thus, an important theme that emerges is that assembly of functional sites of the large subunit is completed last.
Most r-proteins from the large subunit co-immunoprecipitate 27SA2 pre-rRNA and often 35S pre-rRNA [59,68]. Thus, these r-proteins are thought to associate with nascent particles co-transcriptionally, or else very early in the assembly pathway. However, for those r-proteins tested, this initial binding to early pre-ribosomes is relatively weak and becomes stronger as assembly proceeds. Thus in yeast, as observed in bacteria, r-proteins may initially form so-called encounter complexes with pre-ribosomes [69], which then transition to more stable contacts with assembling ribosomes as the pathway continues. These results suggest that the hierarchical assembly of r-proteins in the large subunit may, in fact, represent a hierarchy of rRNP neighborhoods transitioning from encounter complexes to form their more mature configurations. Consistent with this idea, it was found that rRNP intermediates become more stable as their assembly proceeds; blocking early steps in assembly causes rapid turnover of pre-rRNPs, whereas blocking later steps leads to much slower turnover [63,64]. Furthermore, when early-acting r-proteins are depleted, middle- and late-acting r-proteins do not stably associate with pre-ribosomes, whereas when middle- or late-acting r-proteins are depleted, early-acting r-proteins are stably bound to pre-ribosomes.
R-protein extensions are crucial to ribosome assembly
One striking feature revealed by the crystal structure of yeast ribosomes is that a significant fraction of r-proteins contain N- and/or C-terminal extensions or internal loops that protrude from their globular core and reach across the surface or into the interior of the 60S subunit core [2–6,8]. Most of these extensions contain amino acid sequences predicted to be intrinsically disordered [70]. Thus, these extensions have properties ideally suited to enable assembly of ribosomes: (1) they are long and flexible and thus able to access a large surface area and (2) the disordered domains can often become more structured upon binding their ligand and thus might help stabilize assembling ribosomes. An important role of extensions might be to establish networks of protein–protein and protein–rRNA interactions that serve as conduits for information transfer across the surface of pre-ribosomes or mature ribosomes.
Recent studies have begun to reveal specific functions of r-protein extensions [56,66,71–73]. Deletion of r-protein extensions that bind the same domains of rRNA as their globular portion affects ribosome assembly in a manner similar to depletion of that r-protein [71]. This effect probably reflects that the same domains of rRNA are structured by binding to the r-protein globular domains as to the extensions. In contrast, r-protein extensions that span multiple rRNA domains are predicted to play a role in structuring or communicating structural changes across these domains during ribosome assembly. For example, a recent study showed that removing the N-terminal extension of L8 blocks later steps in assembly, than are blocked when the entire protein is depleted [71]. In this case, the globular domain of L8 binds to domain I of 25S rRNA, which assembles early, whereas the extension of L8 threads into domain V of 25S rRNA, which is constructed at later stages.
A burst of recent studies has demonstrated that r-protein extensions facilitate delivery of the r-proteins into pre-ribosomes. The extensions of r-proteins L4, L5 plus L11, and L10 have been found to bind co-translationally to dedicated chaperones (Acl4, Syo1, and Sqt1, respectively), which prevent the aggregation of these r-proteins, transport them into the nucleus, and stably insert them into pre-ribosomes [74–79]. Prior to delivery into pre-ribosomes, the r-protein L4 binds to the karyopherin Kap104 via its C-terminal extension and to the chaperone protein Acl4 via its internal loop [74,75]. In mutants defective in L4–Acl4 interaction, L4 still binds to pre-ribosomes, but there are defects in 60S subunit assembly, suggesting that the Acl4–L4 internal loop interaction might monitor correct insertion of the L4 internal loop into the ribosome. Release of Acl4 from the internal loop of L4 may be triggered by interaction of the C-terminal extension of L4 with r-protein L18, allowing insertion of the loop into the ribosome.
The nuclear import of r-proteins L5 and L11 requires the transport adaptor protein Syo1 that interacts with Kap104 [76]. Syo1 forms a ternary complex with L5 and L11 by its interaction with the N-terminal domain of L5 and the disordered loop of L11 [77]. This Syo1–L5–L11 complex is transported into the nucleus, where it binds to 5S rRNA. Subsequently, AFs, such as Rpf2 and Rrs1, are required to assemble the 5S RNP onto pre-ribosomes [80]. The 5S RNP is bound to the mature 60S subunit via interaction of L11 with helix 84 of 25S rRNA to form the CP [77]. Syo1 is a molecular mimic of helix 84, and thus, the Syo1–L11 interaction might serve as a control mechanism to ensure the entry of properly folded L11 into the pre-ribosome.
Very early steps of 60S subunit assembly
Formation of very early 60S subunit assembly intermediates occurs during and after cleavage at the A2 site by the endonuclease Rcl1 (Figure 2) [81]. A2 cleavage can occur on nascent pre-rRNA transcripts, or else post-transcriptionally following the release of 35S pre-rRNA from rDNA [82–84]. Co- vs. post-transcriptional cleavage of A2 cleavage seems to be closely linked to growth conditions, but the implications for ribosome assembly and translation of these alternative pathways involving cleavage at the A2 site are not yet understood.
Ribosomal subunit assembly at this very early stage includes assembly of proteins, especially subcomplexes of several different AFs, with nascent pre-rRNA, together with chemical modification of pre-rRNA and initial compaction of the pre-rRNP. Cleavage at the A2 site results in the formation of pre-ribosomes containing 20S and 27SA2 pre-rRNAs (Figure 2). Mutations that block 3′-ETS formation prevent cleavage at the A2 and A3 sites [85,86], which suggests that all other steps of pre-rRNA processing specific to large subunits cannot occur until after transcription is completed. In support of this idea, depletion of r-protein L3 that binds to the 3′-end of 25S rRNA has the greatest destabilizing effect on the earliest 60S assembly intermediates containing 27SA2 and 27SA3 pre-rRNAs [64,87], suggesting that the proper configuration of the 3′-end of rRNA is important for early steps of assembly.
A large number of AFs are implicated in the very early steps of 60S subunit biogenesis, because they co-immunoprecipitate only the earliest pre-rRNAs and because their depletion leads to turnover of early pre-rRNAs before later ones can be formed (reviewed in ref. [13]). Included among these very early factors are seven DEAD-box proteins, i.e. ATP-dependent RNA helicases, suggesting that significant remodeling events might occur during this initial stage of 60S subunit assembly. For the most part, though, the roles of very early AFs are poorly defined. The best studied of these are the snoRNPs that enable rRNA modification, and the scaffolding AF Rrp5 that is thought to play a key role in compaction of rRNA in early assembly intermediates (see below).
Approximately 2% of rRNA nucleotides undergo chemical modifications, such as pseudouridylation and 2′-O-ribose methylation, mediated by snoRNA-guided RNPs (snoRNPs) or substrate-specific enzymes (reviewed in refs [88–90]). Interestingly, the modifications cluster at the subunit interface, around the functional sites of the 60S subunit, which is largely devoid of r-proteins. The snoRNPs enter pre-ribosomes co-transcriptionally and are directed to their target sites via snoRNA–pre-rRNA interactions [82,90–92]. SnoRNAs form Watson-Crick base-pairs with their respective target sequences that are located far apart in the primary sequence of 25S rRNA. Thus, together, the different snoRNAs might enable initial compaction of the 35S pre-rRNA during initial stages of ribosome assembly. Although deletion of any single snoRNA gene has little or no effect on assembly, depletion of the proteins common to snoRNPs aborts ribosome assembly at very early stages [93–97]. Chemical modifications also affect the interaction of RNA with water and other molecules, and thus affect rRNA folding and protein binding [98–100]. Consistent with these predictions, misdirecting modifications can have lethal consequences [91].
The methyltransferases Spb1 and Nop2 modify 27S pre-rRNA residues Gm2922 and m5C2870, respectively, in the PTC, suggesting important roles of these methylation events in construction of the active center of the large subunit [101–104]. Mutations affecting the enzymatic activity of Nop2 result in accumulation of 27SA2 pre-rRNA, indicating that the modification occurs during or is important for very early steps of assembly [102]. There is some evidence suggesting that Spb1-mediated methylation occurs during late nuclear steps of 60S subunit assembly [103]. Nevertheless, exactly when, how, and why the action of these two modification enzymes is required for ribosome assembly is largely unclear, and requires further investigation.
The very early AF Rrp5 is a large protein designed to cause the compaction of assembling RNPs; it contains 12 S1 RNA-binding motifs and 7 TPR protein-binding domains. Consistent with this abundance of RNA-binding domains, Rrp5 cross-links to pre-rRNA sequences destined for both subunits [105,106]. Rrp5 also forms a subcomplex with several AFs required for very early steps [106,107]. Depletion of Rrp5 inhibits the earliest steps of ribosome assembly, i.e. cleavage at the A0, A1, and A2 sites, and at the A3 site [108]. When assembling ribosomes (‘Christmas trees’ seen via the Miller spread method) are visualized upon Rrp5 depletion, ‘loose structures’ are observed, suggesting that protein–RNA subcomplexes can form but are not compacted [106].
Early steps of 60S subunit assembly
Following the initial compaction of pre-ribosomes, plus cleavage at the A2 site and completion of transcription to form 27SA2 pre-rRNA, the spacer sequence ITS1 is removed. In rapidly growing yeast cells, ~85–90% of pre-rRNA is cleaved at the A3 site by the endonuclease MRP [109–112], followed by processing of the remaining sequences in ITS1 by the exonucleases Rat1, Xrn1, and Rrp17 to form 27SBs pre-rRNA [51,112–115].
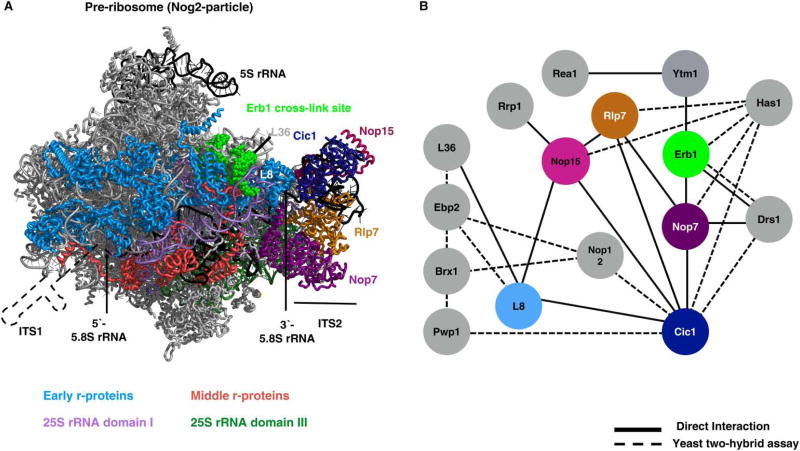
In contrast, a smaller fraction, ~10–15%, is cleaved by an unknown endonuclease at the BL site to form 27SBL pre-rRNA [51,115]. Twelve AFs (Ytm1, Erb1, Nop7, Rlp7, Cic1, Nop15, Has1, Drs1, Rpf1, Pwp1, Nop12, and Rrp1) and 11 r-proteins were initially implicated in 27SA3 pre-rRNA processing, because their depletion caused accumulation of this RNA (Figure 5) [13,37,84,116–128]. The r-proteins involved in this step bind domains I, II, and IV of 25S rRNA (reviewed in ref. [20]). The three AFs Erb1, Ytm1, and Nop7 bind to each other and to domains I and III of 25S rRNA, potentially helping to bring together these rRNA domains during early stages of 60S subunit assembly (Figure 5A) [41,57]. The remaining AFs bind to ITS2 or are thought to be located near or at the proximal stem [41,57,62,117,127]. Thus, most of these proteins required for 27SA3 pre-rRNA processing are not located immediately adjacent to ITS1, where the processing occurs (Figure 5A). Upon depletion of any of these r-proteins or AFs, assembly of r-proteins L17, L26, L35, and L37 that bind to 5.8S rRNA is greatly diminished [59,63,64,68,127], indicating that this stage of assembly includes base-pairing between the 5′- and 3′-ends of 5.8S rRNA and the 5′-end of 25S rRNA, and association of r-proteins with these rRNA domains. Importantly, depletion of these proteins also causes rapid turnover of pre-ribosomes mediated by the 5′ →3′ exonuclease Rat1, before 27SA3 can be processed into 27SB pre-rRNA [68]. Therefore, the most parsimonious explanation for the apparent block in 27SA3 pre-rRNA processing when each of these 23 AFs or r-proteins is depleted is that they serve structural roles to create an early 66S pre-rRNP stable enough and properly configured to undergo further maturation, including processing of 27SA3 pre-rRNA, rather than these proteins being directly involved in ITS1 removal.
Figure 5. ‘A3’ AFs localize close to the ITS2 spacer.
(A) The ‘A3’ AFs bind in or close to ITS2 in pre-ribosomes (shown in the cryo-EM structure of the Nog2-particle) [57]. A cartoon representation of RNA is included at the 5′-end of 5.8S rRNA from which ITS1 emerges. The location of early-acting r-proteins (light blue) relative to 25S rRNA domains I (light violet) and III (dark green). Colors of AFs correspond to their colors in the interaction network (right). The ‘A3’ AFs bind on or close to the ITS2 spacer. Depletion of A3 AFs strongly diminishes the association of r-proteins L17, L26, L35, and L37 (red) with 5.8S rRNA (black). (B) The protein–protein interaction network of A3 AFs as inferred from refs [5,48,57,116–119] (PDB ID: 3JCT).
The middle stage: remodeling events required to trigger removal of the ITS2 spacer
The middle stage of large ribosomal subunit maturation includes remodeling events that trigger removal of the ITS2 spacer from 27SB pre-rRNA and the exit of pre-ribosomes from the nucleolus. ITS2 removal is initiated by cleavage at the C2 site in ITS2 by the endonuclease Las1 [130–132] and requires the association of many AFs with pre-ribosomes (reviewed in ref. [13]). Pre-ribosomes transit from the nucleolus to the nucleoplasm following ITS2 cleavage [123], although it is unclear what enables this movement. During this period, many early-acting AFs exit from pre-ribosomes [57]. The mechanisms underlying their release, and the consequences of doing so, have only been investigated for three factors, such as Nsa1, Ytm1, and Erb1, which are released by the AAA-ATPases Rix7 and Rea1 [133–135].
Interestingly, analysis of a large number of r-protein and AF mutants defective in C2 cleavage and ITS2 removal suggests that checkpoints exist to prevent this irreversible step if other neighborhoods distal to ITS2, including several functional sites of the large subunit, have not properly assembled (Konikkat S. and Woolford J.L., unpublished work). These neighborhoods include the CP situated on top of the subunit, as well as the PET and the PTC.
At early steps of large subunit assembly, the AFs Rpf2 and Rrs1 deliver the 5S rRNP complex, containing 5S rRNA and r-proteins L5 and L11, to the top of nascent particles to form the CP (Figure 6A) [80]. However, mutations in these four proteins or 5S rRNA that block this step block middle steps of assembly, resulting in accumulation of 27SB and 7S pre-rRNAs. Thus, docking of the CP onto early assembly intermediates is necessary for later steps of subunit maturation [80].
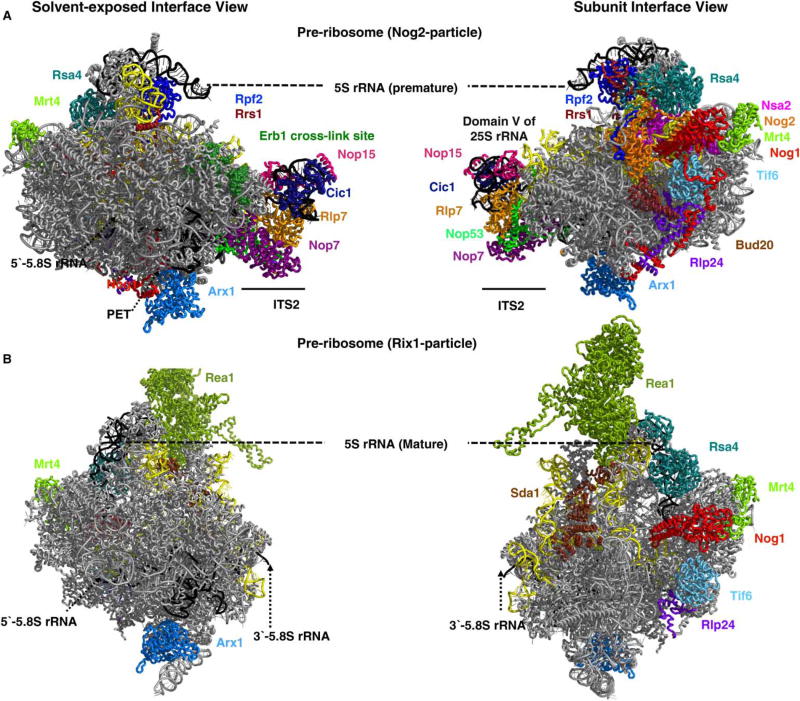
Figure 6. Localization of AFs on the particles undergoing ITS2 spacer removal.
Cryo-EM structure of Nog2 (A) and Rix1 (B) particles representing two late nuclear intermediates. The Nog2 particles represent the earlier assembly intermediate in which the ITS2 spacer has not yet been removed and the 5S rRNP is still in a premature conformation. In wild-type Rix1 particles, ITS2 is removed and rotation of 5S rRNP is completed. Domain V of 25S rRNA containing the PTC (yellow), the subunit interface, and the PET of the pre-60S particles are bound by many AFs that help to construct and inspect the functional centers [57,129] (PDB IDs: 5FL8, 3JCT).
The GTPase Nog1 also assembles into pre-ribosomes early in the biogenesis pathway [136]. The N-terminal domain of Nog1 is positioned at the entrance of the PET, whereas the extremely long C-terminal tail extends from the P-stalk base to protrude deep into the PET from its exit almost back to the PTC (Figure 6A) [57,137]. This location suggests several functions for Nog1: as a scaffold for the assembly of the PET, as a tunnel inspector to ensure its proper construction, and/or as a means to prevent premature or inappropriate entry of other proteins into the tunnel. Depletion or inactivation of Nog1 prevents cleavage at the C2 site; thus, proper assembly of the PET is probably a prerequisite for removal of ITS2 [137,138]. Consistent with this idea, depletion of any one of six r-proteins that bind the rRNA surrounding the tunnel exit prevents cleavage of ITS2 [64–66].
Construction of the PTC also appears to be necessary to initiate removal of ITS2. Five AFs (Nsa2, Nog2, Dbp10, Nug1, and Rsa4) bind at or adjacent to the PTC [57,129,139–142] immediately before C2 cleavage (Figure 6) [38,59,143]. Depletion of any one of these AFs results in accumulation of uncleaved 27SB pre-rRNA-containing ITS2 [38,143–147].
Before most of these AFs bind to the PTC, the AAA-ATPase Rea1 enables the release of the AFs Ytm1 and Erb1 from pre-ribosomes [135]. This remodeling event is required for stable association of r-proteins around the PET exit and assembly of the AFs with the PTC, as well as for the removal of ITS2 (Konikkat S. and Woolford J. L., unpublished work). Mutations that prevent release of Ytm1 and Erb1 block each of these three events.
Taken together, the effects of these assembly mutants suggest that five remodeling events involved in construction of essential functional sites of the large subunit must occur during early and middle stages of large subunit biogenesis, to commit the assembly pathway to the irreversible step of ITS2 removal. Early in the pathway, the 5S rRNP docks onto the top of the subunit and Nog1 assembles to enable proper formation of the PET. Just before C2 cleavage, Rea1 releases AFs Ytm1 and Erb1. This structural transition may trigger both proper construction of the rim around the PET exit, including the ‘bowtie structure’ formed by 5.8S rRNA base-pairing with 25S rRNA, and binding of AFs to the PTC. Failure to carry out any one of these remodeling events may activate a checkpoint to prevent C2 cleavage.
It is not yet known how the endonuclease Las1 recognizes the C2 site, nor whether any of the mutants described above alter accessibility of Las1 to its substrate, although studies in mammalian systems have demonstrated direct interactions between Las1 and homologs of the Rix1 (Ipi) sub-complex that recruit Rea1 to pre-ribosomes [148].
Late nuclear stages
Late nuclear steps of 60S subunit maturation include several major remodeling events: processing of 7S and 25.5S pre-rRNAs [132,149], a 180° rotation of the CP [140], and further assembly of 25S rRNA domain V containing the PTC (discussed below). Successful completion of these structural transitions is coupled to subsequent export of nascent 60S subunits to the cytoplasm.
The 5′-end of 25.5S pre-rRNA is trimmed by the Rat1 exonuclease, which is a component of the Las1 complex [120,132,149]. The 3′-end of 7S pre-rRNA is processed to form 6S pre-rRNA by the exosome ([150], reviewed in ref. [22]). The exosome is recruited to pre-ribosomes by interaction with the AF Nop53 [151] and potentially by interaction with other AFs [152–154]. Following removal of Ytm1 and Erb1 by Rea1, Nop53 binds to 25S rRNA near ITS2 [57] (Figure 6), and to the exosome-associated helicase, Mtr4 [151]. In mammalian cells, interactions have been reported between Nvl2 (the homolog of Rix7) and the exosome complex [152–154]. Exonucleolytic trimming of the ITS2 spacer must require removal of the AFs Nop15, Cic1, and Rlp7 that are bound to ITS2 to render it accessible to the exosome [57,155]. How and when these three AFs exit pre-ribosomes remain to be understood.
It was surprising to find that the 5S rRNP (r-proteins L5 and L11 bound to 5S rRNA) is initially docked onto pre-60S subunits ~180° rotated from its mature conformation (Figure 6A) [140]. The AFs Rpf2 and Rrs1 are necessary to deliver the 5S RNP to early assembly intermediates [80], but it is unclear how they do so. Consistent with the role of Rpf2 and Rrs1 in CP assembly, cryo-EM structures of late nuclear pre-ribosomes show that these two AFs are located adjacent to the CP, bridging the CP with the body of the large subunit [57]. However, we do not know whether Rpf2 and Rrs1 assemble into pre-ribosomes before the 5S rRNP or enter preribosomes bound the 5S rRNP.
During late nuclear stages of 60S subunit assembly, the 5S rRNP undergoes a dramatic 180° rotation, most probably mediated by removal of Rpf2 and Rrs1, and action of the AAA-ATPase Rea1 (reviewed in ref. [156]). Concurrent with rotation of the CP, the A-site finger also undergoes rearrangement, as well as helices surrounding the PTC in domain V of 25S rRNA [57,140]. Rea1 is recruited to late pre-ribosomes by the Rix1 sub-complex [129,157]. Following rotation of the CP, Rea1 removes the WD40 protein Rsa4 from pre-ribosomes [57]. An interaction partner of Rsa4 is the AF Nsa2 bound to helix 89 at the PTC [57,141]. Although Nsa2 is not released from pre-ribosomes along with Rsa4 [46], Nsa2 is proposed to have a crucial role in transmitting the mechano-chemical energy generated by the action of Rea1 on Rsa4, to remodel neighboring rRNA helices [141]. The K+-dependent GTPase Nog2 bound at the PTC plays a crucial step in this remodeling event [139]. A GTP-binding mutant of Nog2 fails to initiate the removal of Rsa4 by Rea1. The removal of Nog2, in turn, facilitated by Rea1, requires GTP hydrolysis by Nog2. This scenario illustrates how the activity of two AFs can be coupled to drive assembly forward. The exit of Nog2 opens up the binding site for the essential export factor Nmd3, thus preparing pre-ribosomes for nuclear export. [139,158]. The neighborhood, including the CP and the PTC, is the binding site for other NTPases such as Dbp10, Nug1, and Nog1 [57,142]. Hence, we are only beginning to understand the dynamic chain of potentially coupled events leading to CP rotation, PTC maturation, and acquisition of export competence.
In addition, the recent determination of cryo-EM structures and localization of AF-binding sites by cross-linking with pre-rRNA has provided glimpses into rRNA structural rearrangement events that occur proximal to ITS2 during late nuclear steps of ribosome assembly [57,129,140,157]. An overlap between pre-rRNA structures in the Nog2- and Rix1-assembly intermediates with those for mature rRNA shows that domains I, II, and VI of 25S rRNA are largely formed prior to late nuclear stages [5,57,129] (data not shown). However, 25S rRNA domain III, which surrounds the exit of the PET, and domain V of 25S rRNA, which contains the PTC, are not yet in their final conformations in these nucleoplasmic pre-ribosomes. The exit of AFs and activity of NTPases, including AAA-ATPases, facilitate the reorientation of helices to restructure the rRNP. For example, ES19 undergoes a structural transition close to 5.8S rRNA during late nuclear steps, potentially coinciding with the exit of Nop7, which is positioned between these two structures (Figure 7). In addition, the 5S rRNP is rotated ~180° during late nuclear stages of assembly [140]. These structural changes are thought to be facilitated by the exit of AFs such as Rpf2, Rrs1, and Rsa4, via the action of the AAA-ATPase Rea1 and the GTPase Nog2 (Figure 6A) [57,129,139–141].
Figure 7. Removal of AFs facilitates structural transitions of RNA helices.
The exit of Nop7 repositions 25S rRNA ES19 to its mature conformation. Nop7 is positioned between helices that constitute ES3 and ES19 in Nog2 particles [57]. The exit of Nop7 allows mature 60S-like ES3–ES19 contacts [5,57,129] (PDB IDs: 3JCT, 5FL8, 4V88).
Nuclear export of pre-60S particles
Pre-60S particles are exported into the cytoplasm through the hydrophobic NPC in a RAN-GTP-dependent fashion [159,160]. This irreversible step requires the action of multiple AFs, including Arx1, Alb1, Rrp12, Bud20, Nmd3, Mtr2, Mex67, Npl3, Ecm1, and Gle2, that bind to different sites on pre-60S subunits (reviewed in detail in ref. [25]). Interestingly, Nmd3 is the only essential protein among these 10 export factors in yeast. Although deletion of any single one of the other nine export factors is not lethal, many double mutant combinations are lethal. This potentially redundant nature of export factors might be necessary to ensure efficient export of ~25 pre-ribosomal particles/minute through each NPC in log-phase yeast cells [9]. The concerted action of these AFs is crucial to prevent misassembled pre-ribosomes from poisoning the pool of translating 60S ribosomes in the cytoplasm.
For export of pre-ribosomes through the hydrophobic NPC to occur, three things must happen: pre-ribosomes must be compacted so that they fit through the NPC, the negatively charged surface of pre-ribosomes must be shielded, and the pre-ribosomes must productively dock onto the NPC.
To fit large asymmetric pre-ribosomes into and through the NPC, pre-rRNP composition is simplified by release of many late nuclear AFs before export [46]. Release of these factors can also serve as timers to ensure completion of nuclear steps in subunit assembly before committing to export. For example, the binding site of the AF Nog2 in the PTC neighborhood overlaps considerably with that of the export factor Nmd3. Thus, release of Nog2, which is thought to be signaled by completion of late nuclear remodeling events, allows binding of Nmd3 and timely export [139]. It is possible that rRNA helices that protrude from pre-60S particles can also block the transit of pre-60S particles through the NPC. Interaction between Arx1 and the dynamic ES27 is thought to lock ES27 into a more compact form during late nuclear stages of 60S subunit biogenesis [161].
Compared with the solvent interface, the subunit interface of mature 60S subunits is relatively devoid of r-proteins (Figure 4D). This is even more true for late nuclear pre-60S particles, since they lack r-proteins L10, L24, L29, L40, L42, P0, and P1, which assemble on the subunit interface after export of pre-ribosomes to the cytoplasm [16]. The absence of these cytoplasmic r-proteins in nuclear pre-rRNPs further accentuates the electrostatic barrier presented for export of pre-ribosomes through the NPC. Therefore, multiple AFs that bind the pre-ribosomes were predicted to facilitate transport of nascent 60S particles out of the nucleus by shielding the negative charges on the exposed RNA. Indeed, recent cryo-EM structures demonstrate that export factors, such as Bud20 and Arx1, and AFs, such as Nog1, Nug1, Nsa2, Rlp24, Tif6, and Mrt4, are distributed across the intersubunit surface (Figure 8) [57,139,140,161]. Export-competent pre-60S particles are then presented to the NPC by AFs that specifically interact with export adaptors or FG-rich nucleoporins [162–167].
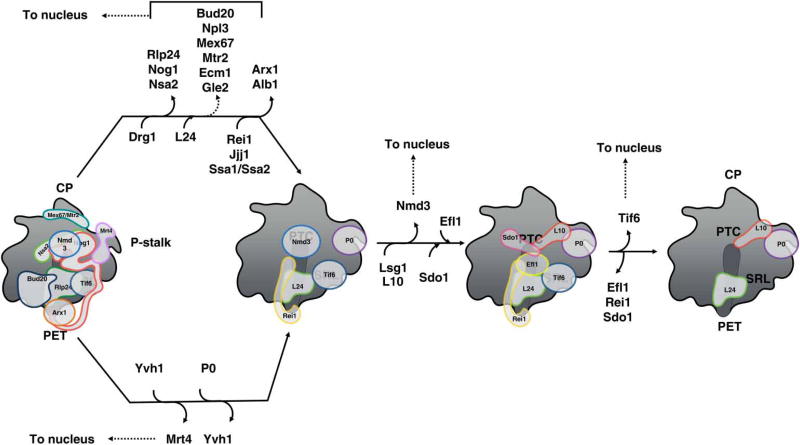
Figure 8. Nuclear export and cytoplasmic maturation of 60S subunits.
66S pre-ribosomes that are exported from the nucleoplasm to the cytoplasm are bound to multiple AFs, which facilitate their transport through the NPC. The cytoplasmic maturation pathway of 60S subunit maturation prior to entering translation is shown. AFs with known binding sites on pre-ribosomes are indicated. AFs that function in the nucleus are shuttled back after release from cytoplasmic assembly intermediates.
Cytoplasmic maturation of large ribosomal subunit
The last steps of maturation occur in the cytoplasm and are among the best-defined events in 60S subunit biogenesis. These include formation of the 3′-end of 5.8S rRNA by processing of 6S pre-rRNA [149,168], release of the few remaining AFs, assembly of several r-proteins [169], and final proofreading of functional sites (reviewed in ref. [16,24,25]).
Release and recycling of eight different AFs from cytoplasmic pre-ribosomes occur by the recruitment and activation of the four NTPases Drg1, Ssa1, Efl1, and Lsg1, and the phosphatase Yvh1, coupled with the removal of their respective substrates (Figure 8). Most notably, the pathway proceeds in an ordered, stepwise fashion, as each step is coupled to the next. For example, the AAA-ATPase Drg1 is recruited and activated by its substrate Rlp24, whose release is coupled to the subsequent insertion of its homolog r-protein L24 [169–171]. Because L24 lacks the Drg1-recruiting motif found in the Rlp24 ‘placeholder’, it remains locked into mature ribosomes as a bona fide r-protein. The presence of L24 then enables Rei1 to bind and initiate removal of Arx1. Nog1 is released together with Rlp24, perhaps resulting from the interaction of its extended C-terminal tail with Rlp24 [57]. Many other AFs, e.g. Nug1 and Nsa2, appear to be present in cytoplasmic pre-60S particles [46], but their mechanism of release has not yet been determined.
Another important theme that emerges from investigations of cytoplasmic maturation is that functional regions in pre-ribosomes are test-driven or proofread for their proper structure and function by binding with AFs or r-proteins before the nascent subunits are allowed to enter the pool of functional ribosomes (reviewed in ref. [16]). Binding of AFs to functional sites also prevents premature association with the translational machinery. Drg1-mediated release of Nog1 opens up the binding sites for Rei1 in the PET and Sdo1 in the P-site [57,172–174]. Rei1 is thought to probe the conformational space in the PET, as the addition of bulky groups to its C-terminal domain affects 60S subunit assembly [172]. The protein phosphatase Yvh1 enables release of Mtr4 from the P-stalk [175,176]. Mtr4 is a homolog of r-protein P0. Thus, its removal opens up the binding site for P0 and the neighboring r-protein L10, the entry of which facilitates folding of the P-stalk into its mature conformation. Formation of the mature P-stalk enables recruitment of the GTPase Efl1, resulting in release of Tif6 and Nmd3 from the GAC (GTPase-activating center) and PTC, respectively. Sdo1 binds the P-site with its N-terminal domain inserted into the PTC and acts as a cofactor for Efl1 [173,174]. The C-terminal portion of Sdo1 contains dynamic domains II–III, which potentially scan the conformational maturation of the RNP neighborhood surrounding it. Mutations in a flexible loop of the r-protein L10 that extends toward the P-site and the PTC block activation of Efl1 and subsequent release of Nmd3 and Tif6 [69]. Thus, it is concluded that Efl1 is activated by the P-site loop of L10 and the SRL, which is responsible for the activation of other GTPases involved in translation. Consequently, Efl1, Tif6, and the SRL act in concert to test-drive translation–initiation competency before Nmd3 is finally released from the PTC.
In the cytoplasm, pre-60S subunits are prevented from entering the translation pool by AFs that provide steric hindrance to the association of translation factors. For example, Arx1 prevents premature association of methionine aminopeptidase, which binds nascent polypeptides emerging from the ribosome tunnel [161,171]. Release of AFs that bind to the PET, CP, tRNA accommodation corridor, SRL, P-stalk, and PTC, respectively, are required to create the binding sites for tRNAs, translation factors, and the 40S subunit.
Prospective
It seems likely that the near future will include many more high-resolution cryo-EM structures of pre-ribosomes during different stages of assembly. These structures will provide much more information about RNP interaction networks and how they change during assembly. Hence, it is likely that these pre-ribosome structures will inspire the creation of numerous targeted mutations to test the importance of molecular interaction networks, leading to more extensive and refined models for the dynamic events occurring during ribosome biogenesis. Although highly impactful, cryo-EM is a complex and an expensive tool to study the structure of pre-ribosomes. Therefore, an important gap to be filled is the development of additional facile tools to probe rRNA and rRNP structural changes during assembly.
Acknowledgments
The authors wish to thank members of the Woolford Laboratory, especially Stephanie Biedka and Jelena Jakovljevic, for their critical reading of this manuscript.
Abbreviations
- AF
assembly factor
- CP
central protuberance
- CRAC
cross-linking and analysis of cDNAs
- ES
expansion segment
- ETS
external transcribed spacer
- FG repeats
phenylanine (F) glycine (G) repeats
- GAC
GTPase-activating center
- ITS
internal transcribed spacer
- iTRAQ
isobaric tag for relative and absolute quantitation
- MRP
RNase MRP
- NPC
nuclear pore complex
- PET
polypeptide exit tunnel
- pre-rRNA
precursor rRNA
- PTC
peptidyl transferase center
- RNP
ribonucleoprotein
- r-protein
ribosomal protein
- rRNA
ribosomal RNA
- SILAC
stable isotope labeling with amino acids in cell culture
- snoRNA
small nucleolar RNA
- snoRNPs
small nucleolar ribonucleoproteins
- SRL
sarcin ricin loop
- SWATH
sequential window acquisition of all theoretical mass spectra
- TPR
tetratricopeptide repeat (a structural motif present in proteins)
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Wilson DN, Doudna Cate JH. The structure and function of the eukaryotic ribosome. Cold Spring Harb. Perspect. Biol. 2012;4:a011536. doi: 10.1101/cshperspect.a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melnikov S, Ben-Shem A, de Loubresse NG, Jenner L, Yusupova G, Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012;19:560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 4.Armache J-P, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. Proc. Natl Acad. Sci. U.S.A. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 6.Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N. Atomic structures of the eukaryotic ribosome. Trends Biochem. Sci. 2012;37:189–198. doi: 10.1016/j.tibs.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Anger AM, Armache J-P, Berninghausen O, Habeck M, Subklewe M, Wilson DN, et al. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 8.Yusupova G, Yusupov M. High-resolution structure of the eukaryotic 80S ribosome. Annu. Rev. Biochem. 2014;83:467–486. doi: 10.1146/annurev-biochem-060713-035445. [DOI] [PubMed] [Google Scholar]
- 9.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/S0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 10.Armistead J, Triggs-Raine B. Diverse diseases from a ubiquitous process: the ribosomopathy paradox. FEBS Lett. 2014;588:1491–1500. doi: 10.1016/j.febslet.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 11.Danilova N, Gazda HT. Ribosomopathies: how a common root can cause a tree of pathologies. Dis. Model Mech. 2015;8:1013–1026. doi: 10.1242/dmm.020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brighenti E, Treré D, Derenzini M. Targeted cancer therapy with ribosome biogenesis inhibitors: a real possibility? Oncotarget. 2015;6:38617–38627. doi: 10.18632/oncotarget.5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woolford JL, Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kressler D, Hurt E, Baßler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 16.Karbstein K. Quality control mechanisms during ribosome maturation. Trends Cell Biol. 2013;23:242–250. doi: 10.1016/j.tcb.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert WV. Functional specialization of ribosomes? Trends Biochem. Sci. 2011;36:127–132. doi: 10.1016/j.tibs.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byrgazov K, Vesper O, Moll I. Ribosome heterogeneity: another level of complexity in bacterial translation regulation. Curr. Opin. Microbiol. 2013;16:133–139. doi: 10.1016/j.mib.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Cruz J, Karbstein K, Woolford JL. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 2015;84:93–129. doi: 10.1146/annurev-biochem-060614-033917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gamalinda M, Woolford JL. Paradigms of ribosome synthesis: lessons learned from ribosomal proteins. Translation. 2015;3:e975018. doi: 10.4161/21690731.2014.975018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández-Pevida A, Kressler D, de la Cruz J. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdiscip. Rev. RNA. 2015;6:191–209. doi: 10.1002/wrna.1267. [DOI] [PubMed] [Google Scholar]
- 23.Lafontaine DLJ. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 2015;22:11–19. doi: 10.1038/nsmb.2939. [DOI] [PubMed] [Google Scholar]
- 24.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 2010;35:260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nerurkar P, Altvater M, Gerhardy S, Schütz S, Fischer U, Weirich C, et al. Eukaryotic ribosome assembly and nuclear export. Int. Rev. Cell Mol. Biol. 2015;319:107–140. doi: 10.1016/bs.ircmb.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Wilson DN, Bhushan S, Becker T, Beckmann R. Nascent Polypeptide Chains Within the Ribosomal Tunnel Analyzed by Cryo-EM. The Ribosome: Structure, Function, & Evolution. Springer; New York: 2011. pp. 387–398. [Google Scholar]
- 27.Ramu H, Vázquez-Laslop N, Klepacki D, Dai Q, Piccirilli J, Micura R, et al. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol. Cell. 2011;41:321–330. doi: 10.1016/j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 28.Warner JR, Soeiro R. Nascent ribosomes from HeLa cells. Proc. Natl Acad. Sci. U.S.A. 1967;58:1984–1990. doi: 10.1073/pnas.58.5.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warner JR. The assembly of ribosomes in yeast. J. Biol. Chem. 1971;246:447–454. [PubMed] [Google Scholar]
- 30.Warner JR, Udem SA. Temperature sensitive mutations affecting ribosome synthesis in Saccharomyces cerevisiae. J. Mol. Biol. 1972;65:243–257. doi: 10.1016/0022-2836(72)90280-X. [DOI] [PubMed] [Google Scholar]
- 31.Udem SA, Warner JR. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J. Mol. Biol. 1972;65:227–242. doi: 10.1016/0022-2836(72)90279-3. [DOI] [PubMed] [Google Scholar]
- 32.Trapman J, Planta RJ. Detailed analysis of the ribosomal RNA synthesis in yeast. Biochim. Biophys. Acta. 1975;414:115–125. doi: 10.1016/0005-2787(75)90214-2. [DOI] [PubMed] [Google Scholar]
- 33.Vaughan MH, Warner JR, Darnell JE. Ribosomal precursor particles in the HeLa cell nucleus. J. Mol. Biol. 1967;25:235–251. doi: 10.1016/0022-2836(67)90140-4. [DOI] [PubMed] [Google Scholar]
- 34.Altvater M, Schütz S, Chang Y, Panse VG. Dissecting ribosome assembly and transport in budding yeast. Methods Cell Biol. 2014;122:437–461. doi: 10.1016/B978-0-12-417160-2.00020-5. [DOI] [PubMed] [Google Scholar]
- 35.Ripmaster TL, Vaughn GP, Woolford JL. DRS1 to DRS7, novel genes required for ribosome assembly and function in Saccharomyces cerevisiae. Mol. Cell Biol. 1993;13:7901–7912. doi: 10.1128/MCB.13.12.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/S1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 37.Wehner KA, Baserga SJ. The σ70-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell. 2002;9:329–339. doi: 10.1016/S1097-2765(02)00438-0. [DOI] [PubMed] [Google Scholar]
- 38.Baßler J, Grandi P, Gadal O, Leßmann T, Petfalski E, Tollervey D, et al. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/S1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 39.Henderson R. Overview and future of single particle electron cryomicroscopy. Arch. Biochem. Biophys. 2015;581:19–24. doi: 10.1016/j.abb.2015.02.036. [DOI] [PubMed] [Google Scholar]
- 40.Cheng Y. Single-particle cryo-EM at crystallographic resolution. Cell. 2015;161:450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Granneman S, Petfalski E, Swiatkowska A, Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA–protein cross-linking. EMBO J. 2010;29:2026–2036. doi: 10.1038/emboj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubota M, Tran C, Spitale RC. Progress and challenges for chemical probing of RNA structure inside living cells. Nat. Chem. Biol. 2015;11:933–941. doi: 10.1038/nchembio.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Leitner A, Faini M, Stengel F, Aebersold R. Crosslinking and mass spectrometry: an integrated technology to understand the structure and function of molecular machines. Trends Biochem. Sci. 2016;41:20–32. doi: 10.1016/j.tibs.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Altvater M, Chang Y, Melnik A, Occhipinti L, Schütz S, Rothenbusch U, et al. Targeted proteomics reveals compositional dynamics of 60S pre-ribosomes after nuclear export. Mol. Syst. Biol. 2012;8:628. doi: 10.1038/msb.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.O111.016717. O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCann KL, Charette JM, Vincent NG, Baserga SJ. A protein interaction map of the LSU processome. Genes Dev. 2015;29:862–875. doi: 10.1101/gad.256370.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramesh M, Woolford JL. Eukaryote-specific rRNA expansion segments function in ribosome biogenesis. RNA. 2016;22:1153–1162. doi: 10.1261/rna.056705.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cole SE, LaRiviere FJ. Analysis of nonfunctional ribosomal RNA decay in Saccharomyces cerevisiae. Methods Enzymol. 2008;449:239–259. doi: 10.1016/S0076-6879(08)02412-9. [DOI] [PubMed] [Google Scholar]
- 51.Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allmang C, Tollervey D. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J. Mol. Biol. 1998;278:67–78. doi: 10.1006/jmbi.1998.1693. [DOI] [PubMed] [Google Scholar]
- 53.Peculis BA, Greer CL. The structure of the ITS2-proximal stem is required for pre-rRNA processing in yeast. RNA. 1998;4:1610–1622. doi: 10.1017/S1355838298981420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Côté CA, Peculis BA. Role of the ITS2-proximal stem and evidence for indirect recognition of processing sites in pre-rRNA processing in yeast. Nucleic Acids Res. 2001;29:2106–2116. doi: 10.1093/nar/29.10.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Côté CA, Greer CL, Peculis BA. Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA. 2002;8:786–797. doi: 10.1017/S1355838202023063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Granneman S, Petfalski E, Tollervey D. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 2011;30:4006–4019. doi: 10.1038/emboj.2011.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature. 2016;534:133–137. doi: 10.1038/nature17942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 59.Ohmayer U, Gamalinda M, Sauert M, Ossowski J, Pöll G, Linnemann J, et al. Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae. PLoS ONE. 2013;8:e68412. doi: 10.1371/journal.pone.0068412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Babiano R, de la Cruz J. Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2010;38:5177–5192. doi: 10.1093/nar/gkq260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Babiano R, Gamalinda M, Woolford JL, de la Cruz J. Saccharomyces cerevisiae ribosomal protein L26 is not essential for ribosome assembly and function. Mol. Cell. Biol. 2012;32:3228–3241. doi: 10.1128/MCB.00539-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Babiano R, Badis G, Saveanu C, Namane A, Doyen A, Díaz-Quintana A, et al. Yeast ribosomal protein L7 and its homologue Rlp7 are simultaneously present at distinct sites on pre-60S ribosomal particles. Nucleic Acids Res. 2013;41:9461–9470. doi: 10.1093/nar/gkt726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jakovljevic J, Ohmayer U, Gamalinda M, Talkish J, Alexander L, Linnemann J, et al. Ribosomal proteins L7 and L8 function in concert with six A3 assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. RNA. 2012;18:1805–1822. doi: 10.1261/rna.032540.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gamalinda M, Ohmayer U, Jakovljevic J, Kumcuoglu B, Woolford J, Mbom B, et al. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev. 2014;28:198–210. doi: 10.1101/gad.228825.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gamalinda M, Jakovljevic J, Babiano R, Talkish J, de la Cruz J, Woolford JL. Yeast polypeptide exit tunnel ribosomal proteins L17, L35 and L37 are necessary to recruit late-assembling factors required for 27SB pre-rRNA processing. Nucleic Acids Res. 2013;41:1965–1983. doi: 10.1093/nar/gks1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Beekvelt CA, de Graaff-Vincent M, Faber AW, van’t Riet J, Venema J, Raué HA. All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:5001–5008. doi: 10.1093/nar/29.24.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohmayer U, Gil-Hernández Á, Sauert M, Martín-Marcos P, Tamame M, Tschochner H, et al. Studies on the coordination of ribosomal protein assembly events involved in processing and stabilization of yeast early large ribosomal subunit precursors. PLoS ONE. 2015;10:e0143768. doi: 10.1371/journal.pone.0143768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J. 2011;30:4020–4032. doi: 10.1038/emboj.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peng Z, Oldfield CJ, Xue B, Mizianty MJ, Dunker AK, Kurgan L, et al. A creature with a hundred waggly tails: intrinsically disordered proteins in the ribosome. Cell. Mol. Life Sci. 2014;71:1477–1504. doi: 10.1007/s00018-013-1446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tutuncuoglu B, Jakovljevic J, Wu S, Gao N, Woolford JL. The N-terminal extension of yeast ribosomal protein L8 is involved in two major remodeling events during late nuclear stages of 60S ribosomal subunit assembly. RNA. 2016;22:1386–1399. doi: 10.1261/rna.055798.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Espinar-Marchena FJ, Fernández-Fernández J, Rodríguez-Galán O, Fernández-Pevida A, Babiano R, de la Cruz J. Role of the yeast ribosomal protein L16 in ribosome biogenesis. FEBS J. 2016;283:2968–2985. doi: 10.1111/febs.13797. [DOI] [PubMed] [Google Scholar]
- 73.Bussiere C, Hashem Y, Arora S, Frank J, Johnson AW. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J. Cell Biol. 2012;197:747–759. doi: 10.1083/jcb.201112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stelter P, Huber FM, Kunze R, Flemming D, Hoelz A, Hurt E. Coordinated ribosomal L4 protein assembly into the pre-ribosome is regulated by its eukaryote-specific extension. Mol. Cell. 2015;58:854–862. doi: 10.1016/j.molcel.2015.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pillet B, García-Gómez JJ, Pausch P, Falquet L, Bange G, de la Cruz J, et al. The dedicated chaperone Acl4 escorts ribosomal protein Rpl4 to its nuclear pre-60S assembly site. PLoS Genet. 2015;11:e1005565. doi: 10.1371/journal.pgen.1005565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kressler D, Bange G, Ogawa Y, Stjepanovic G, Bradatsch B, Pratte D, et al. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science. 2012;338:666–671. doi: 10.1126/science.1226960. [DOI] [PubMed] [Google Scholar]
- 77.Calviño FR, Kharde S, Ori A, Hendricks A, Wild K, Kressler D, et al. Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA-binding site. Nat. Commun. 2015;6:6510. doi: 10.1038/ncomms7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Madru C, Lebaron S, Blaud M, Delbos L, Pipoli J, Pasmant E, et al. Chaperoning 5S RNA assembly. Genes Dev. 2015;29:1432–1446. doi: 10.1101/gad.260349.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pausch P, Singh U, Ahmed YL, Pillet B, Murat G, Altegoer F, et al. Co-translational capturing of nascent ribosomal proteins by their dedicated chaperones. Nat. Commun. 2015;6:7494. doi: 10.1038/ncomms8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, et al. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Horn DM, Mason SL, Karbstein K. Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. J. Biol. Chem. 2011;286:34082–34087. doi: 10.1074/jbc.M111.268649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koš M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, et al. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 84.Talkish J, Biedka S, Jakovljevic J, Zhang J, Tang L, Strahler JR, et al. Disruption of ribosome assembly in yeast blocks cotranscriptional pre-rRNA processing and affects the global hierarchy of ribosome biogenesis. RNA. 2016;22:852–866. doi: 10.1261/rna.055780.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kufel J, Dichtl B, Tollervey D. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA. 1999;5:909–917. doi: 10.1017/S135583829999026X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hitchen J, Ivakine E, Melekhovets YF, Lalev A, Nazar RN. Structural features in the 3′ external transcribed spacer affecting intragenic processing of yeast rRNA. J. Mol. Biol. 1997;274:481–490. doi: 10.1006/jmbi.1997.1376. [DOI] [PubMed] [Google Scholar]
- 87.Rosado IV, Kressler D, de la Cruz J. Functional analysis of Saccharomyces cerevisiae ribosomal protein Rpl3p in ribosome synthesis. Nucleic Acids Res. 2007;35:4203–4213. doi: 10.1093/nar/gkm388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sharma S, Lafontaine DLJ. ‘View from a bridge’: a new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 2015;40:560–575. doi: 10.1016/j.tibs.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 89.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/S0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 90.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 91.van Nues RW, Granneman S, Kudla G, Sloan KE, Chicken M, Tollervey D, et al. Box C/D snoRNP catalysed methylation is aided by additional pre-rRNA base-pairing. EMBO J. 2011;30:2420–2430. doi: 10.1038/emboj.2011.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schimmang T, Tollervey D, Kern H, Frank R, Hurt EC. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt EC. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-F. [DOI] [PubMed] [Google Scholar]
- 96.Gautier T, Bergès T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell Biol. 1997;17:7088–7098. doi: 10.1128/MCB.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lafontaine DL, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5:455–467. doi: 10.1017/S135583829998192X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 99.Arnez JG, Steitz TA. Crystal structure of unmodified tRNAGln complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 100.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA. 2011;2:611–631. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- 101.Kressler D, Rojo M, Linder P, de la Cruz J. Spb1p is a putative methyltransferase required for 60S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:4598–4608. doi: 10.1093/nar/27.23.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hong B, Brockenbrough JS, Wu P, Aris JP. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell Biol. 1997;17:378–388. doi: 10.1128/MCB.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lapeyre B, Purushothaman SK. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell. 2004;16:663–669. doi: 10.1016/j.molcel.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 104.Sharma S, Yang J, Watzinger P, Kötter P, Entian K-D. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013;41:9062–9076. doi: 10.1093/nar/gkt679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Eppens NA, Rensen S, Granneman S, Raué HA, Venema JAAP. The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA. 1999;5:779–793. doi: 10.1017/S1355838299990313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lebaron S, Segerstolpe Å, French SL, Dudnakova T, de lima Alves F, Granneman S, et al. Rrp5 binding at multiple sites coordinates pre-rRNA processing and assembly. Mol. Cell. 2013;52:707–719. doi: 10.1016/j.molcel.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hierlmeier T, Merl J, Sauert M, Perez-Fernandez J, Schultz P, Bruckmann A, et al. Rrp5p, Noc1p and Noc2p form a protein module which is part of early large ribosomal subunit precursors in S. cerevisiae. Nucleic Acids Res. 2013;41:1191–1210. doi: 10.1093/nar/gks1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Venema J, Tollervey D. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 1996;15:5701–5714. [PMC free article] [PubMed] [Google Scholar]
- 109.Lindahl L, Archer RH, Zengel JM. A new rRNA processing mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:295–301. doi: 10.1093/nar/20.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell Biol. 1993;13:7935–7941. doi: 10.1128/MCB.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chu S, Archer RH, Zengel JM, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc. Natl Acad. SciU.SA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lygerou Z, Allmang C, Tollervey D, Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 113.Johnson AW. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell Biol. 1997;17:6122–6130. doi: 10.1128/MCB.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xue Y, Bai X, Lee I, Kallstrom G, Ho J, Brown J, et al. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell Biol. 2000;20:4006–4015. doi: 10.1128/MCB.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Oeffinger M, Zenklusen D, Ferguson A, Wei KE, Hage EA, Tollervey D, et al. Rrp17p is a eukaryotic exonuclease required for 5′ end processing of pre-60S ribosomal RNA. Mol. Cell. 2009;36:768–781. doi: 10.1016/j.molcel.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Miles TD, Jakovljevic J, Horsey EW, Harnpicharnchai P, Tang L, Woolford JL. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell Biol. 2005;25:10419–10432. doi: 10.1128/MCB.25.23.10419-10432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dembowski JA, Ramesh M, McManus CJ, Woolford JL. Identification of the binding site of Rlp7 on assembling 60S ribosomal subunits in Saccharomyces cerevisiae. RNA. 2013;19:1639–1647. doi: 10.1261/rna.041194.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shimoji K, Jakovljevic J, Tsuchihashi K, Umeki Y, Wan K, Kawasaki S, et al. Ebp2 and Brx1 function cooperatively in 60S ribosomal subunit assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:4574–4588. doi: 10.1093/nar/gks057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Talkish J, Campbell IW, Sahasranaman A, Jakovljevic J, Woolford JL. Ribosome assembly factors Pwp1 and Nop12 are important for folding of 5.8S rRNA during ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell Biol. 2014;34:1863–1877. doi: 10.1128/MCB.01322-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Strezoska Ž, Pestov DG, Lau LF. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol. Cell Biol. 2000;20:5516–5528. doi: 10.1128/MCB.20.15.5516-5528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Adams CC, Jakovljevic J, Roman J, Harnpicharnchai P, Woolford JL. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA. 2002;8:150–165. doi: 10.1017/S1355838202010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang L, Sahasranaman A, Jakovljevic J, Schleifman E, Woolford JL. Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes. Mol. Biol. Cell. 2008;19:2844–2856. doi: 10.1091/mbc.E07-12-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gadal O, Strauss D, Petfalski E, Gleizes P-E, Gas N, Tollervey D, et al. Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J. Cell Biol. 2002;157:941–951. doi: 10.1083/jcb.200111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jäger S, Strayle J, Heinemeyer W, Wolf DH. Cic1, an adaptor protein specifically linking the 26S proteasome to its substrate, the SCF component Cdc4. EMBO J. 2001;20:4423–4431. doi: 10.1093/emboj/20.16.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fatica A, Oeffinger M, Tollervey D, Bozzoni I. Cic1p/Nsa3p is required for synthesis and nuclear export of 60S ribosomal subunits. RNA. 2003;9:1431–1436. doi: 10.1261/rna.5130503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Oeffinger M, Tollervey D. Yeast Nop15p is an RNA-binding protein required for pre-rRNA processing and cytokinesis. EMBO J. 2003;22:6573–6583. doi: 10.1093/emboj/cdg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dembowski JA, Kuo B, Woolford JL. Has1 regulates consecutive maturation and processing steps for assembly of 60S ribosomal subunits. Nucleic Acids Res. 2013;41:7889–7904. doi: 10.1093/nar/gkt545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fabian GR, Hopper AK. RRP1, a Saccharomyces cerevisiae gene affecting rRNA processing and production of mature ribosomal subunits. J. Bacteriol. 1987;169:1571–1578. doi: 10.1128/jb.169.4.1571-1578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Barrio-Garcia C, Thoms M, Flemming D, Kater L, Berninghausen O, Baßler J, et al. Architecture of the Rix1–Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nat. Struct. Mol. Biol. 2015;23:37–44. doi: 10.1038/nsmb.3132. [DOI] [PubMed] [Google Scholar]
- 130.Schillewaert S, Wacheul L, Lhomme F, Lafontaine DLJ. The evolutionarily conserved protein Las1 is required for pre-rRNA processing at both ends of ITS2. Mol. Cell Biol. 2012;32:430–444. doi: 10.1128/MCB.06019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Castle CD, Sardana R, Dandekar V, Borgianini V, Johnson AW, Denicourt C. Las1 interacts with Grc3 polynucleotide kinase and is required for ribosome synthesis in Saccharomyces cerevisiae. Nucleic Acids Res. 2013;41:1135–1150. doi: 10.1093/nar/gks1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gasse L, Flemming D, Hurt E. Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol. Cell. 2015;60:808–815. doi: 10.1016/j.molcel.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 133.Gadal O, Strauß D, Braspenning J, Hoepfner D, Petfalski E, Philippsen P, et al. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 2001;20:3695–3704. doi: 10.1093/emboj/20.14.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thoms M, Ahmed YL, Maddi K, Hurt E, Sinning I. Concerted removal of the Erb1–Ytm1 complex in ribosome biogenesis relies on an elaborate interface. Nucleic Acids Res. 2016;44:926–939. doi: 10.1093/nar/gkv1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Baßler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, Hurt E. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol. Cell. 2010;38:712–721. doi: 10.1016/j.molcel.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Saveanu C, Namane A, Gleizes P-E, Lebreton A, Rousselle J-C, Noaillac-Depeyre J, et al. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fuentes JL, Datta K, Sullivan SM, Walker A, Maddock JR. In vivo functional characterization of the Saccharomyces cerevisiae 60S biogenesis GTPase Nog1. Mol. Genet. Genomics. 2007;278:105–123. doi: 10.1007/s00438-007-0233-1. [DOI] [PubMed] [Google Scholar]
- 138.Lapik YR, Misra JM, Lau LF, Pestov DG. Restricting conformational flexibility of the switch II region creates a dominant-inhibitory phenotype in Obg GTPase Nog1. Mol. Cell Biol. 2007;27:7735–7744. doi: 10.1128/MCB.01161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Matsuo Y, Granneman S, Thoms M, Manikas R-G, Tollervey D, Hurt E. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature. 2014;505:112–116. doi: 10.1038/nature12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Leidig C, Thoms M, Holdermann I, Bradatsch B, Berninghausen O, Bange G, et al. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat. Commun. 2014;5:3491. doi: 10.1038/ncomms4491. [DOI] [PubMed] [Google Scholar]
- 141.Baßler J, Paternoga H, Holdermann I, Thoms M, Granneman S, Barrio-Garcia C, et al. A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation. J. Cell Biol. 2014;207:481–498. doi: 10.1083/jcb.201408111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Manikas R-G, Thomson E, Thoms M, Hurt E. The K+-dependent GTPase Nug1 is implicated in the association of the helicase Dbp10 to the immature peptidyl transferase centre during ribosome maturation. Nucleic Acids Res. 2016;44:1800–1812. doi: 10.1093/nar/gkw045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lebreton A, Saveanu C, Decourty L, Jacquier A, Fromont-Racine M. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J. Biol. Chem. 2006;281:27099–27108. doi: 10.1074/jbc.M602199200. [DOI] [PubMed] [Google Scholar]
- 144.Saveanu C, Bienvenu D, Namane A, Gleizes P-E, Gas N, Jacquier A, et al. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. doi: 10.1093/emboj/20.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.de la Cruz J, Sanz-Martínez E, Remacha M. The essential WD-repeat protein Rsa4p is required for rRNA processing and intranuclear transport of 60S ribosomal subunits. Nucleic Acids Res. 2005;33:5728–5739. doi: 10.1093/nar/gki887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Baßler J, Kallas M, Hurt E. The NUG1 GTPase reveals an N-terminal RNA-binding domain that is essential for association with 60S pre-ribosomal particles. J. Biol. Chem. 2006;281:24737–24744. doi: 10.1074/jbc.M604261200. [DOI] [PubMed] [Google Scholar]
- 147.Talkish J, Zhang J, Jakovljevic J, Horsey EW, Woolford JL. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:8646–8661. doi: 10.1093/nar/gks609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Castle CD, Cassimere EK, Denicourt C. LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis. Mol. Biol. Cell. 2012;23:716–728. doi: 10.1091/mbc.E11-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Geerlings TH, Vos JC, Raué HA. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′→3′ exonucleases. RNA. 2000;6:1698–1703. doi: 10.1017/S1355838200001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/S0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 151.Thoms M, Thomson E, Baßler J, Gnädig M, Griesel S, Hurt E. The exosome is recruited to RNA substrates through specific adaptor proteins. Cell. 2015;162:1029–1038. doi: 10.1016/j.cell.2015.07.060. [DOI] [PubMed] [Google Scholar]
- 152.Nagahama M, Yamazoe T, Hara Y, Tani K, Tsuji A, Tagaya M. The AAA-ATPase NVL2 is a component of pre-ribosomal particles that interacts with the DExD/H-box RNA helicase DOB1. Biochem. Biophys. Res. Commun. 2006;346:1075–1082. doi: 10.1016/j.bbrc.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 153.Hiraishi N, Ishida Y-i, Nagahama M. AAA-ATPase NVL2 acts on MTR4-exosome complex to dissociate the nucleolar protein WDR74. Biochem. Biophys. Res. Commun. 2015;467:534–540. doi: 10.1016/j.bbrc.2015.09.160. [DOI] [PubMed] [Google Scholar]
- 154.Yoshikatsu Y, Ishida Y-i, Sudo H, Yuasa K, Tsuji A, Nagahama M. NVL2, a nucleolar AAA-ATPase, is associated with the nuclear exosome and is involved in pre-rRNA processing. Biochem. Biophys. Res. Commun. 2015;464:780–786. doi: 10.1016/j.bbrc.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 155.Makino DL, Baumgärtner M, Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013;495:70–75. doi: 10.1038/nature11870. [DOI] [PubMed] [Google Scholar]
- 156.Shchepachev V, Tollervey D. Motoring toward pre-60S-ribosome export. Nat. Struct. Mol. Biol. 2016;23:3–4. doi: 10.1038/nsmb.3154. [DOI] [PubMed] [Google Scholar]
- 157.Galani K, Nissan TA, Petfalski E, Tollervey D, Hurt E. Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J. Biol. Chem. 2004;279:55411–55418. doi: 10.1074/jbc.M406876200. [DOI] [PubMed] [Google Scholar]
- 158.Johnson AW. Ribosomes: lifting the nuclear export ban. Curr. Biol. 2014;24:R127–R129. doi: 10.1016/j.cub.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 159.Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell Biol. 1999;144:389–401. doi: 10.1083/jcb.144.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Stage-Zimmermann T, Schmidt U, Silver PA. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell. 2000;11:3777–3789. doi: 10.1091/mbc.11.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bradatsch B, Leidig C, Granneman S, Gnädig M, Tollervey D, Böttcher B, et al. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat. Struct. Mol. Biol. 2012;19:1234–1241. doi: 10.1038/nsmb.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bradatsch B, Katahira J, Kowalinski E, Bange G, Yao W, Sekimoto T, et al. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell. 2007;27:767–779. doi: 10.1016/j.molcel.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 163.Oeffinger M, Dlakić M, Tollervey D. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev. 2004;18:196–209. doi: 10.1101/gad.285604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hackmann A, Gross T, Baierlein C, Krebber H. The mRNA export factor Npl3 mediates the nuclear export of large ribosomal subunits. EMBO Rep. 2011;12:1024–1031. doi: 10.1038/embor.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Askjaer P, Bachi A, Wilm M, Bischoff FR, Weeks DL, Ogniewski V, et al. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell Biol. 1999;19:6276–6285. doi: 10.1128/MCB.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Occhipinti L, Chang Y, Altvater M, Menet AM, Kemmler S, Panse VG. Non-FG mediated transport of the large pre-ribosomal subunit through the nuclear pore complex by the mRNA export factor Gle2. Nucleic Acids Res. 2013;41:8266–8279. doi: 10.1093/nar/gkt675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yao Y, Demoinet E, Saveanu C, Lenormand P, Jacquier A, Fromont-Racine M. Ecm1 is a new pre-ribosomal factor involved in pre-60S particle export. RNA. 2010;16:1007–1017. doi: 10.1261/rna.2012310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Thomson E, Tollervey D. The final step in 5.8S rRNA processing is cytoplasmic in Saccharomyces cerevisiae. Mol. Cell Biol. 2010;30:976–984. doi: 10.1128/MCB.01359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lo K-Y, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kappel L, Loibl M, Zisser G, Klein I, Fruhmann G, Gruber C, et al. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J. Cell Biol. 2012;199:771–782. doi: 10.1083/jcb.201205021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Greber BJ, Boehringer D, Montellese C, Ban N. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2012;19:1228–1233. doi: 10.1038/nsmb.2425. [DOI] [PubMed] [Google Scholar]
- 172.Greber BJ, Gerhardy S, Leitner A, Leibundgut M, Salem M, Boehringer D, et al. Insertion of the biogenesis factor Rei1 probes the ribosomal tunnel during 60S maturation. Cell. 2016;164:91–102. doi: 10.1016/j.cell.2015.11.027. [DOI] [PubMed] [Google Scholar]
- 173.Ma C, Yan K, Tan D, Li N, Zhang Y, Yuan Y, et al. Structural dynamics of the yeast Shwachman-Diamond syndrome protein (Sdo1) on the ribosome and its implication in the 60S subunit maturation. Protein Cell. 2016;7:187–200. doi: 10.1007/s13238-015-0242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Weis F, Giudice E, Churcher M, Jin L, Hilcenko C, Wong CC, et al. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2015;22:914–919. doi: 10.1038/nsmb.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lo K-Y, Li Z, Wang F, Marcotte EM, Johnson AW. Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J. Cell Biol. 2009;186:849–862. doi: 10.1083/jcb.200904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kemmler S, Occhipinti L, Veisu M, Panse VG. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J. Cell Biol. 2009;186:863–880. doi: 10.1083/jcb.200904111. [DOI] [PMC free article] [PubMed] [Google Scholar]