Abstract
A key step in ribosome biogenesis is the nuclear export of pre-ribosomal particles. Nmd3, a highly conserved protein in eukaryotes, is a specific adaptor required for the export of pre-60S particles. Here we used cryo-electron microscopy (cryo-EM) to characterize Saccharomyces cerevisiae pre-60S particles purified with epitope-tagged Nmd3. Our structural analysis indicates that these particles belong to a specific late stage of cytoplasmic pre-60S maturation in which ribosomal proteins uL16, uL10, uL1111, eL40 and eL41 1 are deficient, but ribosome assembly factors Nmd3, Lsg1, Tif6 and Reh1 1 are present. Nmd3 and Lsg1 1 are located near the peptidyl-transferase center (PTC). In particular, Nmd3 recognizes the PTC in its near-mature conformation. In contrast, Reh1 1 is anchored to the exit of the polypeptide tunnel, with its C terminus inserted into the tunnel. These findings pinpoint a structural checkpoint role for Nmd3 in PTC assembly, and provide information about functional and mechanistic roles of these assembly factors in the maturation of the 60S ribosomal subunit.
The assembly of eukaryotic ribosomes is a complex process that requires several hundred assembly factors located in different cellular compartments1–3. One key step is the export of nascent premature subunits through the nuclear pore complex to the cytoplasm for the final steps of maturation3,4. The nuclear export of both pre-40S5 and pre-60S particles6,7 is dependent on the Crm1 (Xpo1) pathway and is further regulated by the Ran GTPase cycle8. Nmd3 is a highly conserved protein in all eukaryotes that functions as a specific adaptor protein for pre-60S export9,10. Nmd3 contains a nuclear-export-signal sequence at its C terminus that is responsible for the recruitment of Crm1 to pre-60S particles10–12. Besides Nmd3, a few other adaptor proteins, including Arx1 (refs. 13,14), Rrp12 (ref. 15), Ecm1 (ref. 16), Bud20 (ref. 17) and the Mex67–Mtr2 complex18,19, are also part of the nuclear export machinery for pre-60S particles, but they function in a Crm1-independent manner.
The nuclear recruitment of Nmd3 to pre-60S particles is dependent on the departure of Nog2, an assembly factor whose binding site overlaps with that of Nmd3 (refs. 20–22). The cytoplasmic release of Nmd3 from pre-60S particles requires the loading of the late ribosomal protein uL16 and is promoted by the GTPase Lsg1 (refs. 23,24). Previous low-resolution cryo-EM22 and UV cross-linking20 data indicated that Nmd3 binds at the intersubunit face of the 60S subunits adjacent to the binding site of uL16, consistent with reported genetic interactions between Nmd3 and uL16 (refs. 24,25). Given the highly specific role of Nmd3 in pre-60S export, it was suggested that the binding of Nmd3 might be the last checkpoint for nuclear assembly4. Furthermore, Nmd3 remains associated with pre-60S particles for nearly all cytoplasmic maturation events26, which further supports the idea of a critical role for Nmd3 in quality control of 60S subunit production.
Although the functional role of Nmd3 is well recognized, its structure, its atomic contacts with 60S subunits, and the underlying molecular mechanism for its removal have remained unclear. In the present study, we purified pre-60S ribosomal particles from S. cerevisiae through epitope-tagged Nmd3 and used cryo-EM to obtain a 3.07-Å structure of the Nmd3 particles. Our structural analysis indicates that four assembly factors—Nmd3, Lsg1, Tif6 and Reh1—are present in a pre-60S complex that typifies a certain cytoplasmic stage lacking a few late-binding proteins. These structural observations highlight the final steps of the cytoplasmic maturation of pre-60S particles, and also provide mechanistic insights into the molecular roles of these factors.
RESULTS
Structural overview of the pre-60S Nmd3 particles
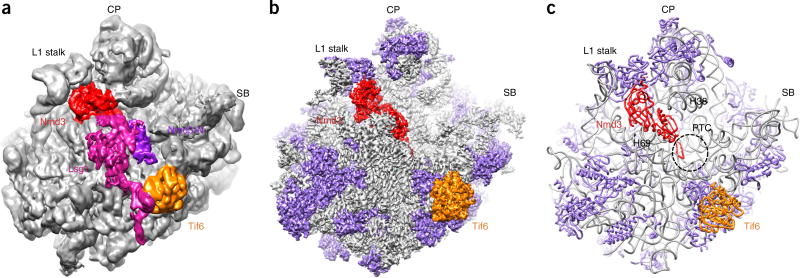
We purified pre-60S particles using epitope-tagged Nmd3 and then subjected them to mass spectrometry (Supplementary Table 1) and single-particle cryo-EM analysis (Supplementary Fig. 1). Classification of particles revealed only one predominant structure for the Nmd3 particles (Supplementary Fig. 2), which was solved at a nominal resolution of 3.07 Å (Fig. 1 and Supplementary Fig. 1). Starting from the crystal structure of the yeast ribosome27, we were able to build an atomic model for the pre-60S structure (Table 1). The overall conformation of 25S rRNA in the Nmd3 particles is very close to its mature form, but four regions differ considerably from the crystal structure27. These include two long, flexible helices (the L1 stalk and H38), a central helix (H69) and the base of the P0 stalk (H43–H44) (Supplementary Fig. 3). The L1 stalk, together with the already assembled ribosomal protein L1, is in an inward position (Fig. 1 and Supplementary Fig. 3a,c), in contrast to the one seen in Nog2 particles21. H38 bends toward H69, with its terminal loop ending right below the central protuberance (CP) (Supplementary Fig. 3c,e). The conformational difference exhibited by H43–H44 is likely due to the fact that the P0 stalk is not fully assembled in Nmd3 particles19,26, because the density at the expected position of P0 is nearly absent. This is consistent with our mass spectrometry data, which showed that both P0 and Yvh1 were present in low abundance in the Nmd3 particles (Supplementary Table 1).
Figure 1.
The structure of the pre-60S Nmd3-TAP ribosomal particle. (a,b) Low-pass-filtered (a) and B-factor-sharpened (b) cryo-EM density maps of the pre-60S particle, shown as surface representations. Lsg1, Nmd3, the N-terminal domain of Nmd3 (Nmd3-N; violet in a) and Tif6 are indicated by color-coding. Ribosomal proteins are shown in purple in b and c. (c) A representation of the final atomic model. The dashed circle indicates the PTC. SB, P0 stalk base.
Table 1.
Statistics of structural determination and model refinement
| Pre-60S ribosomal particle (EMD-9569, PDB 5H4P) |
|
|---|---|
| Data collection | |
| Electron microscope | Titan Krios |
| Voltage (kV) | 300 |
| Electron detector | Falcon II camera |
| Electron dose (e−/Å2) | ~20 |
| Pixel size (Å) | 1.08 |
| Defocus range (µm) | 1.5–2.5 |
| Reconstruction and model refinement | |
| Particles for final refinement | 84,240 |
| Resolution of unmasked map (Å) | 3.4 |
| Resolution of masked map (Å) | 3.07 |
| Map-sharpening B factor (Å2) | –30 |
| R factor | 0.3198 |
| Fourier shell correlation | 0.7245 |
| Model composition | |
| Peptide chains | 44 |
| Residues | 9,809 |
| RNA chains | 3 |
| RNA bases | 3,377 |
| R.m.s. deviation | |
| Bond length (Å) | 0.0053 |
| Bond angle (°) | 0.9611 |
| Ramachandran plot | |
| Favored (%) | 88.80 |
| Outliers (%) | 2.47 |
| Validation (protein) | |
| MolProbity score | 2.01 (99th percentile) |
| Clashscore | 1.74 (100th percentile) |
| Good rotamers (%) | 85.91 |
| Validation (RNA) | |
| Correct sugar puckers (%) | 98.76 |
| Good backbone conformations (%) | 73.85 |
In terms of ribosomal proteins, uL16, uL10, uL11, eL40 and eL41 were clearly missing from the Nmd3 particles (Supplementary Fig. 3a,b). eL41, as a single α-helical protein, binds at the subunit interface, with a stronger association with the 40S subunit27. Therefore, its absence is not surprising. The deficiency of uL16, uL11 and eL40, which are all late ribosomal proteins assembled in the cytoplasm1, correlates well with observed structural differences at H38 and the P0 stalk base, because they are located on the 60S subunit next to H38 and H44, respectively. Notably, the absence of uL16 in the Nmd3 particle complex confirms the previously determined binding order of uL16 and Nmd3 during the assembly of pre-60S particles23. Therefore, these observations indicate that the Nmd3 particles purified in this study were a collection of late-stage cytoplasmic pre-60S particles26.
Assembly factors in Nmd3 particles
Mass spectrometry data suggested the presence of over a dozen factors in the Nmd3 particles21 (Supplementary Table 1). However, only four of them could be identified in the structure: Lsg1, Nmd3, Tif6 and Reh1. In the unsharpened map, a large piece of additional density is seen at the intersubunit face, occupying the position immediately above H69 (Fig. 1a). This density mass could be readily attributed to Lsg1, as its size matches that of a typical GTPase domain. However, because of the substoichiometric occupancy and the structural flexibility of Lsg1, this mass is highly fragmented in the sharpened map. We therefore reclassified the particles according to the local density of Lsg1 using a ‘signal-subtraction’ method28. As a result, we started to see slightly better resolution of some secondary features (Supplementary Fig. 4a), but we still could not build a model to quantitatively analyze Lsg1. Nevertheless, the map suggests that Lsg1 contains a sequence extension that reaches out to contact Tif6 (Supplementary Fig. 4d and Supplementary Video 1). Notably, the binary interaction between Lsg1 and Tif6 was reported in a previous proteomics study29.
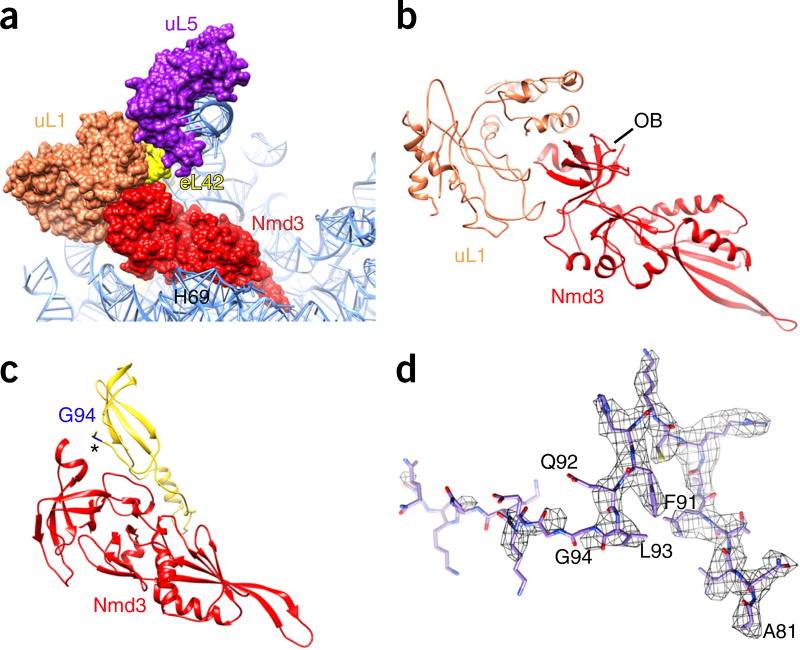
In contrast to that of Lsg1, the sequence assignment of Nmd3 and Tif6 in the map is straightforward, as both of them are well resolved (Fig. 1b and Supplementary Fig. 1g,i). Unexpectedly, we found that the polypeptide exit tunnel (PET) is occluded by an α-helix (Supplementary Fig. 5a). Interestingly, this fragment belongs to the C-terminal helix of Reh1, rather than to either of the two previously discovered tunnel-probing factors, Nog1 (ref. 21) and Rei1 (ref. 30), as inferred from the agreement between the side chains of Reh1 and the map density (Supplementary Fig. 5b–i). This observation is consistent with previous findings that Reh1 and Rei1 have partially redundant roles30,31, and that Arx1 and Alb1 are not present in Reh1 particles31. However, only the tunnel portion of Reh1 (residues 377–432) is resolved in the map; the remaining sequences are untraceable. Nevertheless, this indicates that the PET in pre-60S particles is continuously blocked during the maturation steps from the nucleoplasm to the cytoplasm by Nog1, Rei1 and Reh1 in sequential order.
Structure of Nmd3
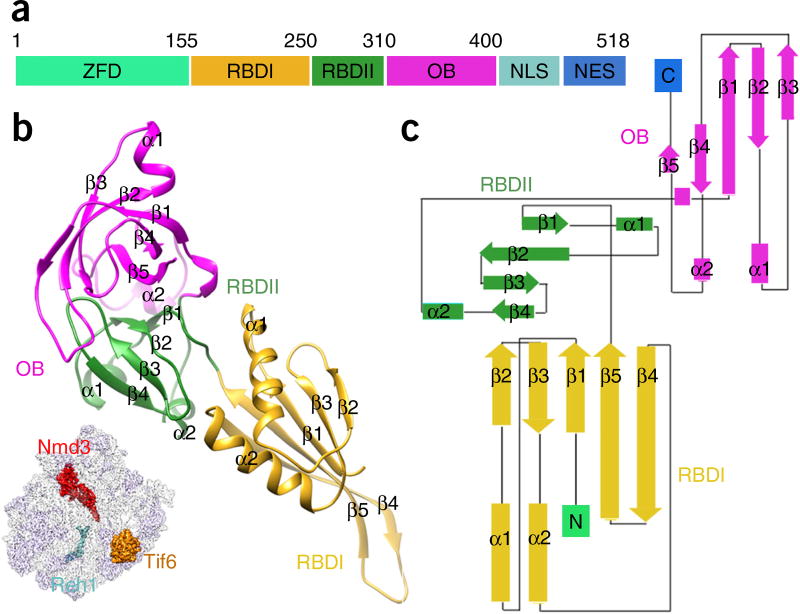
The resolved density of Nmd3 allowed us to build an atomic model for residues 155–402. Most of the residues in this range are resolved at side chain resolution; at a minimum, backbone atoms could be reliably assigned, except for an α-helix (residues 167–182) (Supplementary Fig. 1g). This structure makes it clear that Nmd3 can be divided into four domains (Fig. 2). The N-terminal ~150 residues, although not well ordered in the map, show clear separation from the remaining density, indicating that they probably fold into a separate zinc-finger-containing domain32 and apparently interact with the GTPase domain of Lsg1 (Supplementary Fig. 4c and Supplementary Video 1). Residues 155–310 constitute the major RNA-binding domain (RBD) and can be further divided into two subdomains (RBDI and RBDII). RBDI is composed of a five-stranded β-sheet and two α-helices in the order β1-α1-β2-β3-α2-β4-β5 (Fig. 2b,c). In particular, β4 and β5 form a long β-hairpin with the terminal loop situated in the PTC (Fig. 1c). RBDII is organized in a symmetric arrangement (β-α-β-β-β-α-β) of two short α-helices on a β-sheet backbone (Fig. 2c). The third domain (residues 310–400) is a typical oligonucleotide-binding domain (OB) (Fig. 2b), like those commonly seen in ribosomal proteins and translation factors33. The C-terminal ~100 residues, which include both the nuclear localization sequence and the nuclear export signal, are less ordered, which could be partially due to the presence of the affinity tag at the C terminus. Nevertheless, on the basis of the unsharpened map, it can be assumed that the C-terminal domain also binds to 25S rRNA and is responsible for pulling the terminal loop of H38 toward the CP (Supplementary Fig. 4a,b).
Figure 2.
The structure of Nmd3. (a) Schematic illustration of Nmd3 domain organization. ZFD, zinc-finger-containing domain; NLS, nuclear localization sequence; NES, nuclear export signal. (b) The atomic model of Nmd3, with individual domains (RBDI, RBDII and OB) indicated by color-coding. The orientation of Nmd3 on the pre-60S particle is shown in the illustration on the lower left. (c) Domain topology of Nmd3.
Interactions of Nmd3 with the 2 25S rRNA
Nmd3 is deeply embedded inside the tRNA corridor on the 60S subunit and completely fills the space from the PTC to the tRNA exit site (Fig. 1 and Supplementary Fig. 6). This location of Nmd3 is reminiscent of the unrelated bacterial assembly GTPase EngA, which binds to an equivalent position on the 50S subunit34, thus implying a certain extent of functional conservation of ribosome assembly in prokaryotes and eukaryotes.
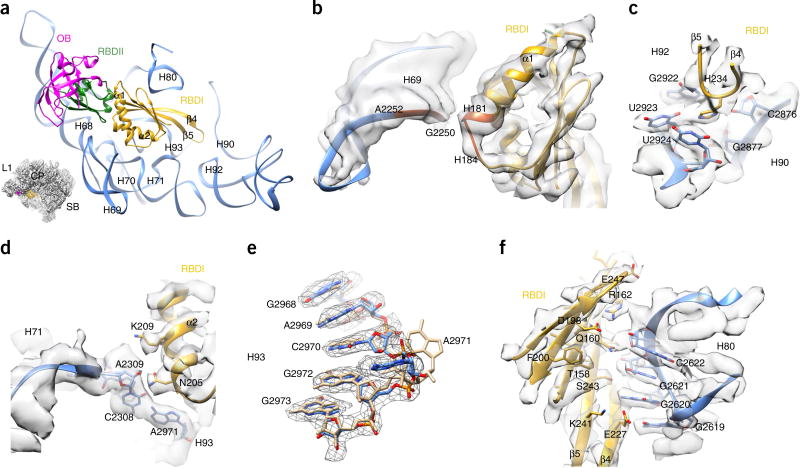
RBDI is surrounded by helices in the proximity of the PTC, including H69, H70, H71, H80, H90, H92 and H93 (Fig. 3). Three contact sites (C1–C3) are responsible for mediating the interaction between RBDI and 25S rRNA. The C1 contact is formed between the C-terminal end of the α1 helix (His181–His184) and the backbone phosphates of H69 (G2250–A2252) (Fig. 3b). Notably, this interface is not resolved in side chain resolution, but apparently this local interaction accounts for the observed conformational difference of H69 relative to the mature 60S subunit (Supplementary Fig. 3c,f). Contact C2 is formed between the α2 helix and the helical junction of H69–H71 and H93 (Fig. 3d). Specifically, Asn205 stabilizes a potential noncanonical base pair between A2971 (H93) and C2308 (H71) (Fig. 3d). A large conformational change of A2971 compared with the mature 25S rRNA (Fig. 3e) could be attributed to the interaction from the C2 site. Contact C3, in contrast, is formed by multiple interactions between surface residues of the β-sheet and the backbone of H80 (Fig. 3f). The most intriguing observation was that the β4–β5 hairpin loop exactly covers the nascent peptide entrance of the PTC (Fig. 3a). His234 from the tip of this hairpin loop could potentially form specific hydrogen bonds with the bases of G2922, U2923, U2924 and C2876 (Fig. 3c).
Figure 3.
Interaction of Nmd3 RBDI with 25S rRNA. (a) Binding position of Nmd3 on the pre-60S particle. Different Nmd3 domains are indicated by color-coding. The orientation is represented in the illustration on the lower left. (b) Zoomed-in view of the interaction between the α1 helix of RBDI and H69. (c) H234 is situated in the PTC and interacts with G2922, U2923, U2924 and C2876 of 25S rRNA. (d) Zoomed-in view of the interaction between the α2 helix of RBDI and H71–H93. It is likely that A2971 of H93 forms a noncanonical base pair with C2308 of H71. (e) Conformational difference between A2971 in the pre-60S map and in its mature state. Coordinates of the mature state (tan) are from the crystal structure of the yeast 80S ribosome (PDB 3U5E)27. (f) Extensive interactions between the β-sheet surface of RBDI and H80.
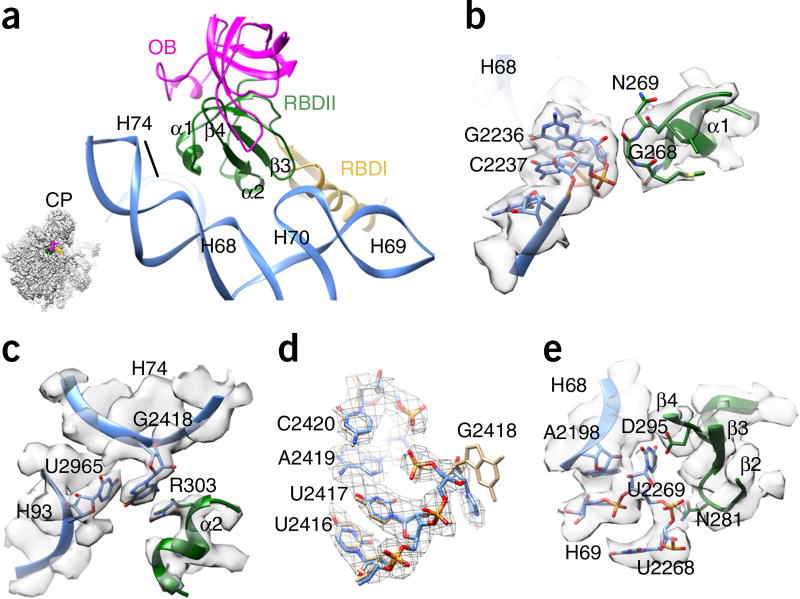
Similarly to what was observed for RBDI, the two short α-helices in RBDII participate in rRNA recognition (Fig. 4a). The backbone of Gly268 (α1-RBDII) probably interacts with the sugar backbone of G2236 (H68) (Fig. 4b). The side chain of Arg303 (α2-RBDII) stacks with the base of G2418 (H74) and U2965 (H93) (Fig. 4c), resulting in a large conformational difference for the base of G2418 (Fig. 4d). In addition, a β-hairpin of RBDII stabilizes the junction of H69 and H68, resulting in a base flip of U2269 (Fig. 4e).
Figure 4.
Interaction of Nmd3 RBDII with 25S rRNA. (a) Overview of the interaction between RBDII and 25S rRNA. The orientation is represented in the illustration on the lower left. (b) The α1 helix of RBDII interacts with H68. (c) R303 from the α2 helix of RBDII stacks with G2418 of H74 and U2965 of H93. (d) Conformational difference between G2418 in the pre-60S map and in its mature state. Coordinates of the mature state (tan) are from the crystal structure of the yeast 80S ribosome (PDB 3U5E)27. (e) Interactions of the β-strand elements of RBDII with the junction of H68–H69.
Interactions of Nmd3 with L1 1 and L42
In contrast to the OBs in ribosomal proteins, the OB of Nmd3 does not recognize 25S rRNA. Instead it interacts with two ribosomal proteins, eL42 and uL1 (Fig. 5). The uL1 stalk is in an inward position; this conformation of the L1 stalk was also observed in nucleoplasmic pre-60S particles bound with Sda1 (ref. 21,35). Because it is an intrinsically flexible component of the 60S subunit, uL1 is not resolved at side chain resolution, but the map is good enough for reliable fitting of a previous atomic model of uL1 (PDB 4V91)36. It is apparent that one side of the OB has a very strong interaction with uL1 (Fig. 5b). A β-strand in the insertion sequence between two neighboring OB strands parallels the C-terminal strand of eL42 (Fig. 5c), which is a late binder that assembles in the cytoplasm. Therefore, Nmd3 might facilitate the incorporation of eL42 into cytoplasmic pre-60S particles. It should be noted that the C-terminal residues of eL42 beyond Gly94 are flexible (full length: 106 residues), which suggests that eL42 has yet to be fully accommodated (Fig. 5d).
Figure 5.
Interaction of the Nmd3 OB with uL1 and eL42. (a) Overall orientation of Nmd3 relative to ribosomal proteins uL1 and eL42. (b) Schematic of the extensive interaction between uL1 and the Nmd3 OB, highlighting a perfect shape match at the interface. (c) An insertion loop on the Nmd3 OB interacts with eL42 (marked with an asterisk). (d) The C-terminal residues of eL42 beyond Gly94 are flexible (full length: 106 residues).
The C-terminal tail of Reh1 1 is in the PET
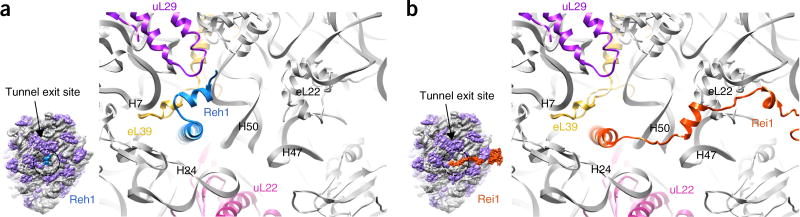
The presence of the Reh1 C-terminal helix in the PET suggests that Reh1 and Rei1 do not coexist in pre-60S particles31 (Supplementary Fig. 5a,b). The carboxy-terminal halves of these two factors are highly homologous, especially their very C termini (Supplementary Fig. 5c). Although the orientation of the C-terminal helix of Reh1 is very similar to that of Rei1 (Supplementary Fig. 5b), the side chains of selected positions on Reh1 provide unambiguous assignments of Reh1 residues in the density within the tunnel (Supplementary Fig. 5d–i). Inside the PET, the C terminus of Reh1 makes contact with a number of ribosomal proteins, including uL29, uL22, eL39 and uL4 (Supplementary Fig. 7). A major difference between Reh1 and Rei1 is found at the tunnel exit (Fig. 6): Reh1 interacts with H7 and extends toward uL29, whereas Rei1 extends away from H7 and toward eL22.
Figure 6.
The C terminus of Reh1 inserts into the PET. (a) The atomic model of the Nmd3 particle viewed from the tunnel exit. The orientation of Reh1 is shown in the illustration on the left (surface representation). (b) Same as a, except that here the atomic model is superimposed on the aligned model of Rei1 (PDB 5APN)30.
DISCUSSION
Role of Nmd3 in quality control of PTC assembly and pre-60S nuclear export
In the structure of Nmd3 particles, nearly all of the 25S rRNA elements are in their mature state, which suggests that, as in pre-rRNA processing, conformational maturation of rRNA occurs almost exclusively in the nucleus. Nmd3 binds to late nucleoplasmic pre-60S particles and directly recruits the export receptor Crm1. Therefore, it is conceivable that association with Nmd3 marks the final nuclear step of assembly of pre-60S particles, and that Nmd3 might have a role in structural proofreading4,37. In pre-60S particles bound with Nog2, the PTC, H69–H71 and rRNA helices from the CP are dramatically different from those in the mature 60S subunit21. As suggested by biochemical data20, the binding site of Nmd3 indeed largely overlaps with that of Nog2 on pre-60S particles (Supplementary Fig. 8). Because of the specific recognition of the nearly mature 25S rRNA helices around the PTC by the RBD of Nmd3, complete remodeling of the PTC (including the neighboring H68–H71) after the release of Nog2 has to take place to prepare the binding sites for Nmd3. In this context, the binding of Nmd3 to the functional center of the 60S subunit is not trivial, as it could be conveniently used as a means of quality control before nuclear export.
Therefore, our structural observations provide direct evidence supporting the proposal that Nmd3 is involved in the quality control of nascent ribosomal subunits4. The persistent association of Nmd3 with cytoplasmic pre-60S particles26 further strengthens this view. Blockage of the tRNA passageway and the entrance of the polypeptide tunnel by Nmd3 may help prevent the undesired association of premature 60S subunits with other cytoplasmic factors in vivo. In our structure, Lsg1 binds at the intersubunit face of H69, which could impede premature association with the 40S subunit. Together with the anti-association role of Tif6 (ref. 38), this suggests that multiple mechanisms might exist to keep defective subunits from engaging in translation.
Another observation from the structure of the 60S–Nmd3 complex is that Nmd3 directly interacts with two flexible rRNA helices (H38 and the L1 stalk) (Fig. 1a), which leads to centripetal rearrangements of those helices. Notably, this sequestering of the peripheral components might facilitate the passage of pre-60S particles across the hydrophobic ‘FG network’39.
Functional implication of the C termini of Nog1, Rei1 1 and Reh1 in 60S-subunit maturation
An unexpected observation from our structural data is the insertion of the C terminus of Reh1 into the PET, which makes for an interesting comparison with Nog1 and Rei1 (refs. 21,30). In late nuclear pre-60S particles, the C-terminal extension of Nog1 also completely blocks the tunnel. Given the early entry of Nog1 into pre-ribosomal particles, it is likely that the C-terminal extension of Nog1 is involved in the assembly of the tunnel itself. Notably, Nog1 remains on pre-60S particles until the early stages of cytoplasmic maturation, at which point its departure is promoted by the ATPase remodeling enzyme Drg1 (ref. 40). The release of Nog1 is required for binding of Rei1 (ref. 40), which again entirely blocks the tunnel. Therefore, it seems that the PET is completely blocked throughout the nucleoplasmic and cytoplasmic stages of pre-60S maturation by three consecutive tunnel-probing factors, Nog1, Rei1 and Reh1. The physiological importance of these observations could be two-fold. First, these factors might test-drive the tunnel as a form of quality control. Second, the presence of their C termini along the entire path of the tunnel might help prevent potential translation inhibitors—for example, the tunnel-targeting antimicrobial peptides produced by insects and mammals41,42—from accessing the tunnel.
Furthermore, the binding site of Reh1 that overlaps with that of Rei1 in the tunnel suggests a role for Reh1 in releasing Rei1 from late cytoplasmic pre-60S particles. Although Rei1 was thought to release Arx1 (refs. 43,44), pre-60S particles immunoprecipitated with N-terminally Flag-tagged Rei1 were found to contain a high level of Arx1 (ref. 31). It is possible that another cytoplasmic factor, in conjunction with Jjj1and Ssa26,45,46, is required for Arx1 and Rei1 to be released simultaneously30. Our structural data suggest that Reh1 might be that factor. Consistent with this hypothesis, pre-60S particles pulled down by N-terminally Flag-tagged Reh1 from both wild-type and Δrei1 cells did not contain either Rei1 or Arx1 (ref. 31).
Model for cytoplasmic maturation of pre-60S particles
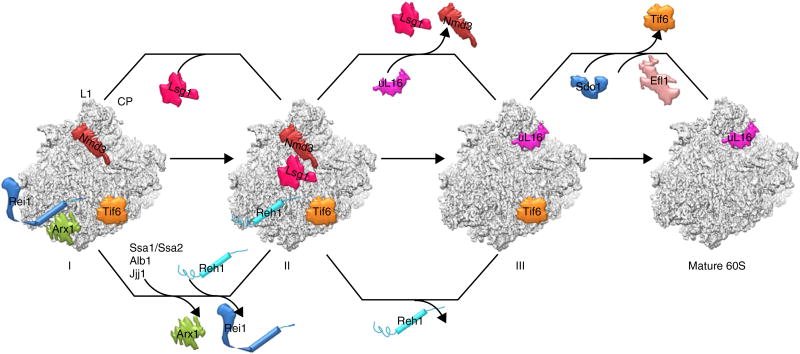
Pre-60S particles immediately after nuclear export contain, in addition to Nmd3, a set of other assembly factors including Nog1, Rlp24, Arx1, Alb1, Tif6, Mrt4, Nug1 and Nsa2, as well as export factors Mex67, Bud20 and Mtr2 (refs. 1,47). In our structure, all of these factors are absent except for Tif6. This indicates that the majority of particles in our sample had already undergone many of the steps of cytoplasmic maturation (Fig. 7). Specifically, Nog1 and Rlp24 had been released by the AAA+ ATPase Drg1 (ref. 40), and the subsequent release of Arx1 and Alb1 by Rei1 (refs. 30,43,44) had also occurred. Although the stalk proteins P0, P1 and P2 are not yet fully assembled in our structure, Mrt4 is absent, suggesting that Yvh1 had already carried out its function to release Mrt4 in our sample (refs. 19,48,49).
Figure 7.
The model for the final cytoplasmic stages of pre-60S maturation. The map features two parallel pathways for the maturation of two functional centers of the 60S subunit, the PTC-CP region and the tunnel exit region. Three states of pre-60S particles are shown (I, II and III). State I represents a stage following the release of Nog1/Rlp24 and the binding of Rei1. State II is the one reported in the present work.
Our structure represents a snapshot of cytoplasmic 60S subunits just before the assembly of uL16 (state II in Fig. 7). The fact that only a predominant population was present in our cryo-EM particles indicates that the incorporation of uL16 is likely to be a rate-limiting step for late cytoplasmic maturation. This could be a conserved mechanism across species, as structures of bacterial late-stage 45S assembly intermediates are deficient in uL16 as well50,51. uL16 has a loop that resides in the ribosomal P site. It was shown that the integrity of this P loop is required for the release of Tif6 and Nmd3 (ref. 52). Interestingly, superposition of uL16 with Nmd3 on the 60S subunit showed that the P-site loop of uL16 is very close to the tunnel-covering loop of the Nmd3 RBD (Supplementary Fig. 8g). This suggests that the loading of uL16 could potentially destabilize the binding of Nmd3 or that a certain rearrangement of these two loops could take place, because Nmd3 also binds to uL16-containing mature 60S subunits22. More intriguingly, when we overlaid Sdo1 (refs. 53,54), the factor that functions together with Efl1 to release Tif6 (refs. 26,55–57), we noted an apparent incompatibility with Nmd3 on the 60S subunit (Supplementary Fig. 8f), indicating that the binding of Sdo1 and the subsequent release of Tif6 would require the prior departure of Nmd3. However, this is in conflict with previous genetic data showing that the release of Tif6 probably occurs before the departure of Nmd3 (ref. 26). Nevertheless, two lines of evidence also support our structural observation. First, the timing of Sdo1 loading onto the late cytoplasmic pre-60S particles is currently unknown, but it appears that Sdo1 binds after uL16 incorporation, because mutations in the P-site loop of uL16 decrease the affinity of Sdo1 for the ribosome in vitro58 and the release of Tif6 in vivo59. Second, a recent cryo-EM study of 60S subunits bound with the ribosome-maturation protein SBDS (the higher eukaryotic homolog of Sdo1) and eIF6 (the higher eukaryotic homolog of Tif6) revealed a specific interaction between SBDS (domain I) and the P-site loop of uL16 (Supplementary Fig. 8h)53.
In summary, our structural data enabled us to reexamine the order of the two last cytoplasmic maturation events after uL16 loading, the release of Nmd3 (by Lsg1) and of Tif6 (by Sdo1 and Efl1) (Fig. 7). In particular, the revised order, which places Tif6 as the last remaining factor on nascent 60S subunit, is in accordance with the hypothesis that Tif6 mediates a continuum between ribosome assembly and translation initiation60.
METHODS
Methods, including statements of data availability and any associated accession codes and references, are available in the online version of the paper.
ONLINE METHODS
Purification of pre-60S Nmd3 particles
Pre-60S particles were purified by tandem affinity purification61. The C-terminally tagged Nmd3-TAP strain SC1121 (SC0000; MATa, ura3-52, leu2-3, 112, YHR170w::TAP-KlURA3) was obtained from EUROSCARF (http://www.euroscarf.de). Cells were cultured in 3 L of YEPD medium at 30 °C to an OD600 of 1.0, collected by centrifugation and washed with lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.075% (v/v) NP-40). Resuspended cells were disrupted ten times with glass beads in lysis buffer containing complete protease inhibitor cocktail (1 pill per 50 ml; Roche). Cell lysates were centrifuged at 13,000 r.p.m. for 1 h at 4 °C with a JA 25.50 motor (Beckman Coulter). Supernatants were collected and applied to a column containing IgG Sepharose beads (GE Healthcare) and incubated for 2 h at 4 °C. The beads were then washed several times with lysis buffer, and proteins were eluted via the addition of 1 ml of TEV cleavage buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2) containing the TEV protease. The cleavage reaction continued for 2 h at 16 °C. The elution was collected and concentrated. The composition of the pre-60S particles was analyzed by mass spectrometry (Supplementary Table 1).
Cryo-EM specimen preparation
The pre-60S sample was diluted to a concentration of ~100 nM. Prior to sample freezing, holey carbon grids (Quantifoil R2/2) were coated with a thin layer of freshly prepared carbon and glow-discharged with a plasma cleaner. Cryo-grids were prepared with an FEI Vitrobot Mark IV at 4 °C and 100% humidity, and inspected with a 300-kV FEI Titan Krios transmission electron microscope (FEI Falcon II direct electron detector). Micrographs were collected at a magnification of 75,000×, which rendered a pixel size of 1.08 Å at the object scale, and with the defocus ranging from −1.5 to −2.5 µm. Data collection was done under low-dose conditions by the semiautomatic software AutoEMation II (written by J.L.). For each micrograph stack, 20 frames were collected, with a total dose of 40 electrons per pixel.
Image processing
Motion correction at the micrograph level was done with MotionCorr (http://cryoem.ucsf.edu/software/driftcorr.html). Micrograph selection, automatic particle picking and normalization were done with SPIDER62. The contrast transfer function parameter was estimated with the CTFFIND3 program63 in the RELION package64. Further image processing, including 2D and 3D classification, refinement and post-processing, was done with RELION 1.3. A total of 2,285 micrographs were collected and 278,881 particles were picked for cascade 2D and 3D classification with a binning factor of two. About 50% of particles were removed during two rounds of 2D classification, and 127,787 particles were subjected to three rounds of 3D classification (Supplementary Fig. 2). After the final round of 3D classification, a total of 84,240 particles were applied for high-resolution refinement (without binning). Final refinement was done with a dose-reduced data set, using only a total dose of 20 electrons per pixel. During the final refinement, a soft-edged mask was applied from the 1.8° step, resulting in a 3.4-Å map (gold-standard FSC 0.143 criteria). After refinement, the map was sharpened using the post-processing options of RELION with a soft mask and B factor of −30 Å2 applied, which resulted in an overall resolution of 3.07 Å. The local resolution was estimated using ResMap65 in RELION.
To improve the local density of Lsg1, we further classified the final data set (84,240 particles) using a signal-subtraction method28. Specifically, we first created an artificial ‘Lsg1-free’ pre-60S map using the refined density map of the Nmd3 particles by masking out the Lsg1 density from the final map. Then, we prepared a data set of modified particles by subtracting the 2D projections of the Lsg1-free pre-60S structure from the respective raw Nmd3 particles. These modified particles, which supposedly should be dominated by density from Lsg1, were subjected to further rounds of 3D classification (with a large soft mask of Lsg1 and also with local search restrictions applied). Nevertheless, the final structure from a subset of the original particles (around 40,000 particles) only marginally improved the density appearance of Lsg1 (Supplementary Fig. 4a), with the consequence of decreased overall resolution (3.7 Å).
Atomic model building and refinement
The amino acid sequence of Nmd3 (UniProt P38861) was subjected to secondary-structure prediction using PSIPRED66. We first built a polyalanine model using Coot67 and then performed sequence assignment using the information from predicted secondary structures and side chain densities in the map. For model building of the pre-60S subunit, the 3.0-Å crystal structure of the yeast ribosome (PDB 3U5D and PDB 3U5E)27 was fitted into the density map using Chimera68. For the rRNAs, the structure of the 5.8S, 5S and 25S rRNAs (PDB 3U5D) were adjusted manually with Coot. Modeling of the L1 stalk was also facilitated by the coordinates of L1-stalk RNA (G2440–U2509) from a crystallography study69 (PDB 3O5H). For ribosomal proteins, the crystal structures were first fitted as rigid bodies and optimized with Coot. The coordinate of protein uL1 was taken from a previous cryo-EM study36 (PDB 4V91). The atomic-model refinement was done via real-space refinement (phenix.real_space_refine)70 in Phenix71 and also by Fourier space refinement in REFMAC72. The atomic model was cross-validated according to previously described procedures73, and model statistics are summarized in Table 1. In the final model, we were able to build residues 155–402 for Nmd3, residues 377–432 for Reh1, and residues 1–227 for Tif6. Notably, a few flexible components of the 25S rRNA, including H38 (1,000–1,050) and the L1 stalk (2,443–2,506), were removed from the final model because they were highly fragmented in the final sharpened map. Structural analysis and figure preparation were done with PyMOL74 and UCSF Chimera.
Supplementary Material
Acknowledgments
We thank the National Center for Protein Sciences (Beijing, China) for providing resources for data collection and computation. Part of the computation was done on the Computing Platform of the Center for Life Science, Peking University. This work was supported by the National Natural Science Foundation of China (grants 31422016, 31470722 and 31630087 to N.G.), the Ministry of Science and Technology of China (grants 2016YFA0500700 and 2013CB910404 to N.G.) and the NIH (grant R01GM028301 to J.L.W.). N.L. is supported by a postdoctoral fellowship from the Peking-Tsinghua Center for Life Sciences.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.L.W. and N.G. conceived the study; C.M., S.W., N.L., Y.C., K.Y., Z.L., L.Z. and J.L. performed the study; C.M., J.L.W. and N.G. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Woolford JL, Jr, Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Gerhardy S, Menet AM, Peña C, Petkowski JJ, Panse VG. Assembly and nuclear export of pre-ribosomal particles in budding yeast. Chromosoma. 2014;123:327–344. doi: 10.1007/s00412-014-0463-z. [DOI] [PubMed] [Google Scholar]
- 4.Johnson AW, Lund E, Dahlberg J. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 2002;27:580–585. doi: 10.1016/s0968-0004(02)02208-9. [DOI] [PubMed] [Google Scholar]
- 5.Moy TI, Silver PA. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13:2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stage-Zimmermann T, Schmidt U, Silver PA. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell. 2000;11:3777–3789. doi: 10.1091/mbc.11.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurt E, et al. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell Biol. 1999;144:389–401. doi: 10.1083/jcb.144.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fung HY, Chook YM. Atomic basis of CRM1-cargo recognition, release and inhibition. Semin. Cancer Biol. 2014;27:52–61. doi: 10.1016/j.semcancer.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadal O, et al. Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho JH, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas F, Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 2003;116:2409–2419. doi: 10.1242/jcs.00464. [DOI] [PubMed] [Google Scholar]
- 12.Trotta CR, Lund E, Kahan L, Johnson AW, Dahlberg JE. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 2003;22:2841–2851. doi: 10.1093/emboj/cdg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hung NJ, Lo KY, Patel SS, Helmke K, Johnson AW. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol. Biol. Cell. 2008;19:735–744. doi: 10.1091/mbc.E07-09-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradatsch B, et al. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell. 2007;27:767–779. doi: 10.1016/j.molcel.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 15.Oeffinger M, Dlakic M, Tollervey D. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev. 2004;18:196–209. doi: 10.1101/gad.285604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao Y, et al. Ecm1 is a new pre-ribosomal factor involved in pre-60S particle export. RNA. 2010;16:1007–1017. doi: 10.1261/rna.2012310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassler J, et al. The conserved Bud20 zinc finger protein is a new component of the ribosomal 60S subunit export machinery. Mol. Cell. Biol. 2012;32:4898–4912. doi: 10.1128/MCB.00910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao W, et al. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell. 2007;26:51–62. doi: 10.1016/j.molcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 19.Sarkar A, Pech M, Thoms M, Beckmann R, Hurt E. Ribosome-stalk biogenesis is coupled with recruitment of nuclear-export factor to the nascent 60S subunit. Nat. Struct. Mol. Biol. 2016;23:1074–1082. doi: 10.1038/nsmb.3312. [DOI] [PubMed] [Google Scholar]
- 20.Matsuo Y, et al. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature. 2014;505:112–116. doi: 10.1038/nature12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu S, et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature. 2016;534:133–137. doi: 10.1038/nature17942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sengupta J, et al. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J. Cell Biol. 2010;189:1079–1086. doi: 10.1083/jcb.201001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West M, Hedges JB, Chen A, Johnson AW. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol. Cell. Biol. 2005;25:3802–3813. doi: 10.1128/MCB.25.9.3802-3813.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedges J, West M, Johnson AW. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005;24:567–579. doi: 10.1038/sj.emboj.7600547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofer A, Bussiere C, Johnson AW. Mutational analysis of the ribosomal protein Rpl10 from yeast. J. Biol. Chem. 2007;282:32630–32639. doi: 10.1074/jbc.M705057200. [DOI] [PubMed] [Google Scholar]
- 26.Lo KY, et al. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 28.Scheres SH. Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 2016;579:125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 29.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 30.Greber BJ, et al. Insertion of the biogenesis factor Rei1 probes the ribosomal tunnel during 60S maturation. Cell. 2016;164:91–102. doi: 10.1016/j.cell.2015.11.027. [DOI] [PubMed] [Google Scholar]
- 31.Parnell KM, Bass BL. Functional redundancy of yeast proteins Reh1 and Rei1 in cytoplasmic 60S subunit maturation. Mol. Cell. Biol. 2009;29:4014–4023. doi: 10.1128/MCB.01582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hedges J, Chen YI, West M, Bussiere C, Johnson AW. Mapping the functional domains of yeast NMD3, the nuclear export adapter for the 60S ribosomal subunit. J. Biol. Chem. 2006;281:36579–36587. doi: 10.1074/jbc.M606798200. [DOI] [PubMed] [Google Scholar]
- 33.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, et al. Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly. Nucleic Acids Res. 2014;42:13430–13439. doi: 10.1093/nar/gku1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrio-Garcia C, et al. Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nat. Struct. Mol. Biol. 2016;23:37–44. doi: 10.1038/nsmb.3132. [DOI] [PubMed] [Google Scholar]
- 36.Fernández IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014;157:823–831. doi: 10.1016/j.cell.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson AW. Ribosomes: lifting the nuclear export ban. Curr. Biol. 2014;24:R127–R129. doi: 10.1016/j.cub.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 38.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 39.Knockenhauer KE, Schwartz TU. The nuclear pore complex as a flexible and dynamic gate. Cell. 2016;164:1162–1171. doi: 10.1016/j.cell.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pertschy B, et al. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol. 2007;27:6581–6592. doi: 10.1128/MCB.00668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seefeldt AC, et al. Structure of the mammalian antimicrobial peptide Bac7(1–16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 2016;44:2429–2438. doi: 10.1093/nar/gkv1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gagnon MG, et al. Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res. 2016;44:2439–2450. doi: 10.1093/nar/gkw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hung NJ, Johnson AW. Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:3718–3727. doi: 10.1128/MCB.26.10.3718-3727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lebreton A, et al. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell Biol. 2006;173:349–360. doi: 10.1083/jcb.200510080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer AE, Hung NJ, Yang P, Johnson AW, Craig EA. The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. USA. 2007;104:1558–1563. doi: 10.1073/pnas.0610704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer AE, Hoover LA, Craig EA. The cytosolic J-protein, Jjj1, and Rei1 function in the removal of the pre-60 S subunit factor Arx1. J. Biol. Chem. 2010;285:961–968. doi: 10.1074/jbc.M109.038349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 2010;35:260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kemmler S, Occhipinti L, Veisu M, Panse VG. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J. Cell Biol. 2009;186:863–880. doi: 10.1083/jcb.200904111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lo KY, Li Z, Wang F, Marcotte EM, Johnson AW. Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J. Cell Biol. 2009;186:849–862. doi: 10.1083/jcb.200904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jomaa A, et al. Functional domains of the 50S subunit mature late in the assembly process. Nucleic Acids Res. 2014;42:3419–3435. doi: 10.1093/nar/gkt1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li N, et al. Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit. Nucleic Acids Res. 2013;41:7073–7083. doi: 10.1093/nar/gkt423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bussiere C, Hashem Y, Arora S, Frank J, Johnson AW. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J. Cell Biol. 2012;197:747–759. doi: 10.1083/jcb.201112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weis F, et al. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2015;22:914–919. doi: 10.1038/nsmb.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma C, et al. Structural dynamics of the yeast Shwachman-Diamond syndrome protein (Sdo1) on the ribosome and its implication in the 60S subunit maturation. Protein Cell. 2016;7:187–200. doi: 10.1007/s13238-015-0242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Menne TF, et al. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat. Genet. 2007;39:486–495. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 56.Finch AJ, et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011;25:917–929. doi: 10.1101/gad.623011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senger B, et al. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- 58.Sulima SO, et al. Eukaryotic rpL10 drives ribosomal rotation. Nucleic Acids Res. 2014;42:2049–2063. doi: 10.1093/nar/gkt1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Keersmaecker K, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat. Genet. 2013;45:186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miluzio A, Beugnet A, Volta V, Biffo S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep. 2009;10:459–465. doi: 10.1038/embor.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 62.Shaikh TR, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 64.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 69.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 70.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 73.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Version 1.3r1. Schrodinger; 2010. The PyMOL Molecular Graphics System. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.