Abstract
The accurate classification of non-small cell lung carcinoma (NSCLC) into lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) is essential for both clinical practice and lung cancer research. Although the standard WHO diagnosis of NSCLC on biopsy material is rapid and economic, more than 13% of NSCLC tumors in the USA are not further classified. The purpose of this study was to analyze the genome-wide pattern differences in copy number variations (CNVs) and to develop a CNV signature as an adjunct test for the routine histopathologic classification of NSCLCs. We investigated the genome-wide CNV differences between these two tumor types using three independent patient datasets. Approximately half of the genes examined exhibited significant differences between LUAD and LUSC tumors and the corresponding non-malignant tissues. A new classifier was developed to identify signature genes out of 20 000 genes. Thirty-three genes were identified as a CNV signature of NSCLC. Using only their CNV values, the classification model separated the LUADs from the LUSCs with an accuracy of 0.88 and 0.84, respectively, in the training and validation datasets. The same signature also classified NSCLC tumors from their corresponding non-malignant samples with an accuracy of 0.96 and 0.98, respectively. We also compared the CNV patterns of NSCLC tumors with those of histologically similar tumors arising at other sites, such as the breast, head, and neck, and four additional tumors. Of greater importance, the significant differences between these tumors may offer the possibility of identifying the origin of tumors whose origin is unknown.
1 | INTRODUCTION
Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) are the two major histological types of non-small cell lung cancer (NSCLC), constituting 54% and 28% of NSCLC, respectively (http://seer.cancer.gov/). Considering that large cell cancer (LCC) samples are undifferentiated NSCLCs that do not show morphologic or immunostaining evidence of glandular or squamous differentiation,1,2 these two subtypes cover over 90% of NSCLC cases in the USA.
One of the first clinical diagnostic tasks is to distinguish between NSCLC and SCLC.3 However, recent advances, especially in personalized medicine and inclusion in clinical trials, have made it imperative to distinguish LUAD from LUSC.4–6 The current recommended WHO methodology for this separation, which is based on routine H&E examination combined with immunostaining, is rapid, accurate, and economic.7
However, most NSCLC tumors (~70%) are currently diagnosed from small biopsies or cytology specimens, which greatly increases the difficulties of accurate classification, even with the use of immunostaining.6–9 More importantly, the agreement between expert lung cancer pathologists on the diagnosis of poorly differentiated NSCLC is modest.8,9 According to the SEER database, a National Cancer Institute initiative that provides information on cancer statistics in the USA, more than 13% of NSCLC tumors (amounting to an estimated 23 971 cases per year in the USA) are not further classified (https://seer.cancer.gov/csr/1975_2013/browse_csr.php?sectionSEL=15&pageSEL=sect_15_table.28.html) and are usually referred to as NSCLC-NOS (not otherwise specified)9,10. Such patients may not receive optimal therapy or become included in a clinical trial for specific types of NSCLC. All of these facts point to the value of molecular classifications as nonsubjective adjuvant methods. These methods include digital nuclear imaging, mutation analysis, mRNA expression values and various other molecular methods, either alone or in combination.10–16
As several papers have addressed copy number variation (CNV) patterns in NSCLC, a gain in corresponding knowledge is incremental. 17,18 However, a comprehensive analysis across independent datasets and associations with other parameters, such as known driver genes in lung adenocarcinoma (e.g., mutEGFR), is warranted. The CNVs between LUADs and LUSCs are genome-wide and of wide deflection. 19 Some CNVs are present in both types, whereas other CNVs are tumor-specific.1 Previous studies have shown that CNVs play important roles in histologic classifications of NSCLC.20,21 More importantly, DNA-based tests are more robust when applied to formalin-fixed paraffin-embedded tissues. Therefore, we aimed to use the global patterns of CNVs as a method to subclassify NSCLC, especially the poorly differentiated cases of NSCLC.
The purposes of our study were as follows: (1) to investigate and document the genome-wide CNV differences between these two tumor types, (2) to develop a molecular classifier and to identify signature genes of NSCLC based on the differences in CNVs, and (3) to compare the CNV patterns of LUADs and LUSCs to those of tumors with similar histology arising in other major organs.
2 | MATERIALS AND METHODS
Three independent datasets were used in this study. The Cancer Genome Atlas (TCGA) dataset was downloaded through the public TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp). The level 3 CNV data of LUAD and LUSC patients measured by the Affymetrix Genome-Wide Human SNP Array 6.0 were used in our study. The CNV value of a gene was defined as the average value of all of the segments’ CNV values corresponding to the gene. There were 496 LUAD samples and 490 LUSC samples. Additionally, paired normal tissues from 556 LUAD and 490 LUSC cases were used to test the classification performance of the identified signature genes. The University of Texas Lung Specialized Program of Research Excellence (UT Lung SPORE) patient dataset is an in-house dataset that includes 248 samples for aCGH analysis using the Agilent 244K Chip by Myriad Genetics (Salt Lake City, UT). The CNV information was available for 151 tumors (105 LUAD samples and 46 LUSC samples) and can be downloaded from the GEO database (GSE74948). A set of 79 LUADs was obtained from British Columbia by Dr. Stephen Lam in collaboration with the Early Detection Research Network (EDRN) and the Canary Foundation EDRN/Canary Project (http://edrn.jpl.nasa.gov/ecas/data/dataset/urn:edrn:UTSW_CopyNumberData). The CNVs of the EDRN/Canary patient dataset were also measured by the Affymetrix Genome-Wide Human SNP Array 6.0. Clinical information on these three datasets is summarized in Supporting Information Table S1.
Data analysis was restricted to autosomes. Genes with CNV values missing in all samples were removed. In total, CNV information was available for 23 494, 21 581, and 22 574 genes in the TCGA, SPORE, and EDRN/Canary datasets, respectively. There were 18 819 genes present in all three datasets for signature gene identification. Supporting Information Table S2 summarizes these three datasets. As the TCGA dataset has the largest sample size and includes many nonmalignant samples, this dataset was used to train the classification (supervised pattern recognition) model and to identify the signature genes. The SPORE and EDRN/Canary datasets were used for validation. Because level 3 TCGA CNV values were log2 (copy number/2)-transformed, the SPORE and EDRN/Canary CNV values were processed in the same manner. All data were normalized by Z-score transformation.
We first investigated the global differences of CNV patterns between LUAD and LUSC tumor samples. The CNVs were further compared between LUSC and LUAD samples by a two-tailed t-test. Bonferroni correction was used as a way to control for the family-wise error rate. Since this analysis was limited only to the TCGA dataset, all 23 494 genes were considered.
Next, we compared the CNV differences between LUAD or LUSC and the following related major cancer types: head and neck carcinoma (HNSC), esophageal squamous cell carcinoma (ESSC), colorectal carcinoma (CRCA), breast carcinoma (BRCA), ovarian serous cystadenocarcinoma (OV), and prostate adenocarcinoma (PRAD). All CNV data regarding these cancer types were downloaded from the public TCGA dataset and were preprocessed in the same manner as the LUAD and LUSC data.
To overcome the inherent noise of the microarray data and the complicated multi-relationships among genes and to select potential signature genes that distinguish LUADs from LUSCs out of approximately 20 000 genes, an algorithm integrating elastic net (EN),22 partial least squares (PLS),23 and naive bayes (NB)24 was applied to complement the advantages of each method. Each method is briefly summarized in the Supporting Information. The integrated EN-PLS-NB algorithm is described in detail in the Supporting Information with the corresponding flowchart in Supporting Information Figure S1A.
The procedure used to identify signature genes classifying LUADs and LUSCs is described below with the corresponding flowchart in Supporting Information Figure S1B. We first applied the EN-PLS-NB algorithm to the TCGA lung cancer dataset consisting of 496 LUAD samples and 490 LUSC samples with 18 819 overlapping genes to obtain a raw list of signature genes, which we termed the “statistically selected gene list.”
To avoid the known important genes from being buried with thousands of whole-genome genes, 11 previously proven tumor-associated genes of NSCLC were identified by a literature review (Supporting Information Table S2). These were then combined with the statistically selected gene list already generated by the EN-PLS-NB algorithm (105 out of 18 819 genes) and were used as candidates for steps (3)–(6) of the EN-PLS-NB algorithm to identify the final set of signature genes.
Sensitivity, specificity and accuracy measurements were used to validate the classification performance. The corresponding definitions are available in the Supporting Information.
A score for each sample was calculated as the confidence of prediction. The cutoff value was set as the standard deviation of the absolute score values (±18.28, see Supporting Information) of the training dataset and can be viewed as a prediction threshold: values above 18.28 are predicted to be LUAD, while values below −18.28 are predicted to be LUSC. Intermediate values are considered poorly differentiated.
After the training procedure using LUADs and LUSCs of the TCGA dataset, the final classification model with the signature gene set was validated on three independent datasets (see Supporting Information Table S2). Using only CNV values of the same signature genes, we first validated whether the classifier could separate tumors from corresponding non-malignant tissues in the TCGA dataset; second, we validated whether it could separate the tumor types in the SPORE dataset; and third, we examined its sensitivity in the EDRN/Canary dataset consisting of only LUAD samples. The SPORE dataset was measured by a completely different platform from those of the TCGA and EDRN/Canary datasets.
All analyses were performed using MATLAB codes, which are available upon request. We are currently working on the online running edition (https://clickgwas.org).
3 | RESULTS
3.1 | Genome-wide CNV differences between LUAD and LUSC
Figure 1 shows the genome-wide median CNV values of all LUAD, LUSC, and non-malignant tissues in different colors. Over 47% of the genes (more than 11 000 genes) showed significant differences in terms of CNVs between LUADs and LUSCs, with losses (28.8% compared with corresponding non-malignant samples) being greater than gains (18.5%) after applying Bonferroni correction for family-wise error control (Figures 1 and 2, Supporting Information Table S3).
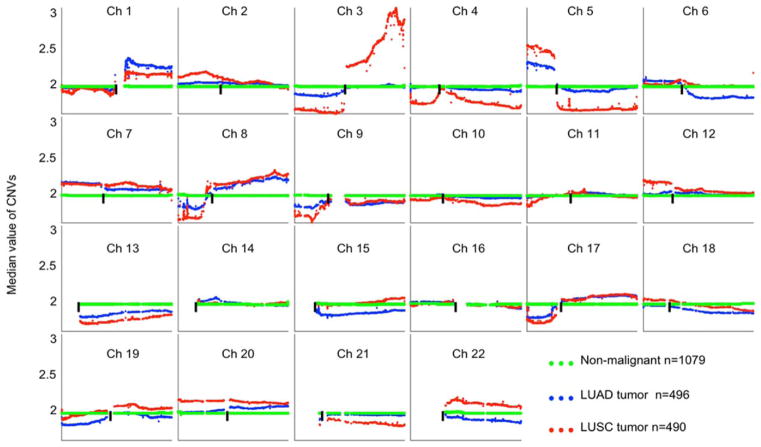
FIGURE 1.
Comparison of CNV patterns between LUAD and LUSC tumors for each chromosome, TCGA only. Each spot is the median value of copy numbers of each gene in the corresponding group. The genes are sorted according to their locations. The space between two arms of each chromosome is the location of the corresponding centromere. [Color figure can be viewed at wileyonlinelibrary.com]
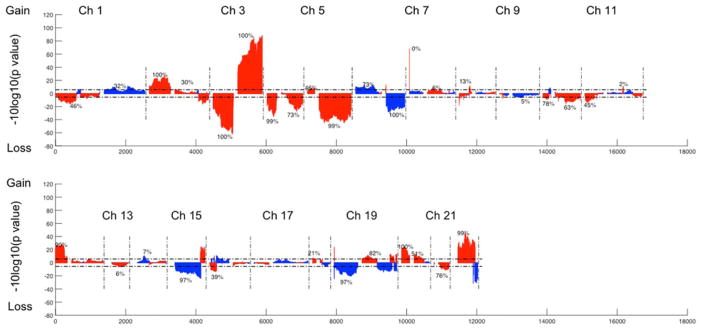
FIGURE 2.
Genome-wide CNVs t-test between LUAD and LUSC tumor samples in the TCGA dataset. Blue color indicates that the deflection (tumor vs. non-malignant samples) was greater for LUAD, whereas red color indicates that the deflection was greater for LUSC. The dashed horizontal lines are the cutoff lines according to the Bonferonni correction (2.1 × 10−6). The vertical dashed lines separate the data of each chromosome. A gap within the individual chromosome data indicates the location of the centrosome. For chromosomes 13, 14, 15, 21, and 22 only genes on the q arm were represented on the microarray. [Color figure can be viewed at wileyonlinelibrary.com]
According to the TCGA annotation (Figure 1 and Table 1), four p arms (chromosomes 13, 14, 15, and 22) contain no listed genes, and 21p contains only six genes. Of the remaining 39 somatic chromosomal arms, 33 (84.6%) had significant CNV differences between LUAD and LUSC, ranging from 1% to 100% (mean = 45.6%). Of the two NSCLC types, LUSC tumors showed more deflections (either gains or losses) compared with the corresponding non-malignant samples, involving 32.1% of the genes, while LUAD only showed 15.2% of genes deflected. LUSC tumors also showed the greatest deflections for gain (chromosome arms 2p, 12p, and part of 22q) and loss (3p, 4p, and 5q). For LUADs, the greatest long-segment deflections involved losses on parts of 6q, 15q, 19p, and 22q.
TABLE 1.
The significance analysis of the two tumor types with Bonferroni correction of each arm (TCGA dataset)
| Chrom | P arm
|
Q arm
|
||||
|---|---|---|---|---|---|---|
| Number of genesa | LUAD vs NMb | LUSC vs NM | Number of genesa | LUAD vs NM | LUSC vs NM | |
| 1 | 1263 | 8% | 74% | 1162 | 100% | 100% |
| 2 | 602 | 100% | 100% | 965 | 78% | 80% |
| 3 | 580 | 99% | 100% | 700 | 6% | 100% |
| 4 | 287 | 2% | 96% | 617 | 67% | 76% |
| 5 | 210 | 100% | 100% | 915 | 53% | 100% |
| 6 | 737 | 100% | 9% | 543 | 99% | 2% |
| 7 | 393 | 100% | 100% | 778 | 98% | 98% |
| 8 | 350 | 83% | 97% | 545 | 100% | 100% |
| 9 | 261 | 100% | 52% | 730 | 100% | 14% |
| 10 | 224 | 6% | 52% | 717 | 34% | 91% |
| 11 | 522 | 4% | 52% | 991 | 43% | 11% |
| 12 | 335 | 16% | 100% | 897 | 30% | 62% |
| 13 | 0 | /c | / | 479 | 87% | 92% |
| 14 | 0 | / | / | 819 | 26% | Noned |
| 15 | 0 | / | / | 861 | 97% | 20% |
| 16 | 552 | 70% | 26% | 461 | 30% | 29% |
| 17 | 427 | 91% | 97% | 991 | 100% | 67% |
| 18 | 99 | None | 97% | 268 | 63% | 35% |
| 19 | 673 | 99% | 23% | 994 | 57% | 36% |
| 20 | 237 | 13% | 100% | 443 | 100% | 100% |
| 21 | 6 | 100% | 100% | 301 | None | 88% |
| 22 | None | None | None | 559 | 71% | 95% |
Number of genes: total number of genes with known gene symbols in each arm;
NM: non-malignant;
/: Not available;
None: None of genes is significantly different;

The most striking changes (for both percentages of involved genes and amplitude of the changes) were observed on chromosome 3, with 100% gains on 3q in LUSC, confirming previously reported findings.18,25–27 In contrast to 3q gains in LUSC, a loss of most or all of 3p characterized both LUAD and LUSC, being more prominent in LUSC (Figure 1). Losses of 3p were the first cytogenetic alteration found in lung cancers, occurring early during lung cancer pathogenesis, and multiple tumor suppressor genes are located in this region.28–30
Long segmental gains at 5p were characteristic of both LUAD and LUSC (more so in LUAD) and involved the telomerase reverse transcriptase gene TERT (5p15). Chromosome 7, the location of Met (7q.31), is the only chromosome that showed gains of almost all contained genes on both arms for both LUSCs and LUADs (Figure 1). Chromosome 22 showed gains of almost all contained genes for LUSCs and a small loss for LUADs (Figure 1).
In addition to large segment gains, short segment gains (amplicons) were also present. LUADs were characterized by narrow region gains in the EGFR pathway, including EGFR (7p12), ERBB2 (17q12), KRAS, and NKX2-1. Supporting Information Figure S2 shows their chromosome locations and distribution in the TCGA dataset. While mutations in KRAS are characteristic of LUADs, gains of KRAS (12p12.1) were more common in LUSC (33.3%) than in LUAD (21.4%) (P=4.3E-7). KRAS is located in a region of long segment gain involving all of 12p that is characteristic of LUSC.
Narrow band regions of loss (deletions) were fewer than narrow band regions of gain, and the most prominent one was at 9p21 (Supporting Information Figure S3). This deleted region contains CDKN2A (also known as p16) and related genes, namely CDKN2B and MTAP. CDKN2A inactivation is more common in LUSC,31 and homozygous deletions are more common in LUSC (20.2%) than in LUAD (11.9%).
Both LUSC and LUAD tumors had approximately equal frequencies of gains for the MDM2 gene (16.5% and 19.2%, respectively), consistent with previous reports32,33 (see Supporting Information Figure S4).
Supporting Information Table S2 summarizes 11 known important lung tumor-associated genes that were identified by a literature review. Four of them—PIK3CA, SOX2, FGFR1 (for LUSCs), and NKX2-1 (for LUADs)—are significantly different in LUAD and LUSC samples according to the t-test with Bonferroni correction (P-value <2.10E-6). The top two genes (PIK3CA and SOX2) were selected for inclusion in the final 33 signature genes (see Table 2).
TABLE 2.
The location and average copy numbers the 33 signature genes
| Rank | Gene | Location | Category | TCGA Tumors
|
TCGA on-malignant
|
Spore Tumors
|
EDRN/Canary Tumors | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LUAD | LUSC | P-value | LUAD | LUSC | LUAD | LUSC | |||||
| 1 | SPATA16 | 3q26.31 | A | 2.09 | 3.31 | 3.1E-77 | 2.01 | 2.01 | 2.08 | 3.38 | 1.98 |
|
| |||||||||||
| 2 | USP13 | 3q26.2-q26.3 | A | 2.07 | 3.36 | 6.32E-76 | 2.01 | 2.01 | 2.20 | 3.54 | 1.99 |
|
| |||||||||||
| 3 | TPRG1 | 3q28 | A | 2.05 | 3.09 | 7.29E-85 | 2.01 | 2.01 | 2.17 | 3.51 | 1.96 |
|
| |||||||||||
| 4 | ZMAT3 | 3q26.32 | A | 2.07 | 3.39 | 6.76E-73 | 2.00 | 2.01 | 2.20 | 3.54 | 2.00 |
|
| |||||||||||
| 5 | MFN1 | 3q26.33 | A | 2.07 | 3.36 | 9.98E-74 | 2.01 | 2.01 | 2.20 | 3.54 | 2.00 |
|
| |||||||||||
| 6 | HTR3D | 3q27.1 | A | 2.07 | 3.45 | 8.75E-67 | 2.01 | 2.01 | 2.14 | 3.36 | 2.01 |
|
| |||||||||||
| 7 | ABCF3 | 3q27.1 | A | 2.06 | 3.40 | 1.34E-71 | 2.01 | 2.01 | 2.14 | 3.36 | 2.03 |
|
| |||||||||||
| 8 | ABCC5 | 3q27 | A | 2.06 | 3.45 | 1.86E-66 | 2.01 | 2.01 | 2.14 | 3.36 | 2.01 |
|
| |||||||||||
| 9 | VWA5B2 | 3q27.1 | A | 2.06 | 3.38 | 1.19E-72 | 2.01 | 2.01 | 2.18 | 3.53 | 2.11 |
|
| |||||||||||
| 10 | FLJ42393 | 3q27.3 | A | 2.05 | 3.14 | 1.58E-81 | 2.01 | 2.02 | 2.17 | 3.51 | 2.02 |
|
| |||||||||||
| 11 | ADIPOQ | 3q27 | A | 2.05 | 3.17 | 4.87E-80 | 2.01 | 2.01 | 2.17 | 3.51 | 2.00 |
|
| |||||||||||
| 12 | RTP2 | 3q27.3 | A | 2.05 | 3.14 | 4.58E-81 | 2.01 | 2.01 | 2.17 | 3.51 | 1.99 |
|
| |||||||||||
| 13 | SST | 3q28 | A | 2.05 | 3.14 | 4.58E-81 | 2.01 | 2.01 | 2.17 | 3.51 | 1.99 |
|
| |||||||||||
| 14 | YEATS2 | 3q27.1 | A | 2.06 | 3.49 | 5.64E-59 | 2.01 | 2.01 | 2.18 | 3.53 | 2.01 |
|
| |||||||||||
| 15 | LOC100131635 | 3q27.3 | A | 2.05 | 3.14 | 6.37E-81 | 2.01 | 2.01 | 2.17 | 3.51 | 1.97 |
|
| |||||||||||
| 16 | PPP1R2 | 3q29 | A | 2.06 | 3.08 | 2.35E-85 | 2.01 | 2.01 | 1.93 | 2.90 | 2.06 |
|
| |||||||||||
| 17 | OSTN | 3q28 | A | 2.05 | 3.09 | 2.14E-83 | 2.01 | 2.02 | 2.17 | 3.51 | 1.99 |
|
| |||||||||||
| 18 | SERPINI2 | 3q26.1 | A | 2.11 | 3.20 | 8.5E-78 | 2.01 | 2.01 | 2.08 | 3.49 | 1.98 |
|
| |||||||||||
| 19 | PDCD10 | 3q26.1 | A | 2.11 | 3.20 | 3.22E-77 | 2.01 | 2.01 | 2.08 | 3.49 | 1.98 |
|
| |||||||||||
| 20 | PIK3CA | 3q26.3 | A&B | 2.07 | 3.17 | 1.55E-78 | 2.01 | 2.01 | 2.20 | 3.54 | 1.98 |
|
| |||||||||||
| 21 | SOX2 | 3q26.3-q27 | B | 2.07 | 3.45 | 7.48E-57 | 2.01 | 2.01 | 2.24 | 3.65 | 2.02 |
|
| |||||||||||
| 22 | STIM2 | 4p15.2 | A | 2.04 | 1.79 | 3.12E-25 | 2.01 | 2.01 | 1.96 | 1.66 | 2.08 |
|
| |||||||||||
| 23 | TBC1D19 | 4p15.2 | A | 2.04 | 1.79 | 3.83E-25 | 2.01 | 2.01 | 2.0 | 1.85 | 2.08 |
|
| |||||||||||
| 24 | RBPJ | 4p15.2 | A | 2.04 | 1.80 | 4.34E-18 | 2.01 | 2.02 | 1.98 | 1.75 | 2.08 |
| 25 | C6orf203 | 6q21 | A | 1.83 | 2.07 | 1.15E-06 | 2.01 | 2.01 | 1.88 | 2.12 | 1.87 |
| 26 | HACE1 | 6q16.3 | A | 1.82 | 2.06 | 1.32E-06 | 2.01 | 2.02 | 1.86 | 2.10 | 1.86 |
| 27 | BVES | 6q21 | A | 1.82 | 2.07 | 1.24E-06 | 2.01 | 2.01 | 1.88 | 2.12 | 1.86 |
| 28 | ALKBH7 | 19p13.3 | A | 1.81 | 1.95 | 4.88E-18 | 2.01 | 2.01 | 1.87 | 1.95 | 2.18 |
|
| |||||||||||
| 29 | C9orf53 | 9p21.3 | A | 1.75 | 1.59 | 7.27E-08 | 2.01 | 2.01 | 1.79 | 1.67 | 2.04 |
| 30 | CLPP | 19p13.3 | A | 1.81 | 1.95 | 4.88E-18 | 2.01 | 2.01 | 1.87 | 1.95 | 2.18 |
| 31 | MIR1233-2 | 15q14 | A | 1.80 | 1.95 | 5.25E-12 | 1.94 | 1.97 | 2.33 | 2.30 | 1.88 |
|
| |||||||||||
| 32 | NKX2-6 | 8p21.2 | A | 1.81 | 1.74 | 0.10E-03 | 2.01 | 2.01 | 1.85 | 1.69 | 1.92 |
| 33 | SIGLEC14 | 19q13.32 | A | 1.80 | 1.94 | 5.04E-06 | 1.88 | 1.90 | 1.91 | 2.13 | 2.19 |
• Rank: ranked by the coefficients to the classification model. P-value is obtained by the two-tailed t-test between copy numbers of LUADs and LUSCs.
• There are no LUSC samples in EDRN/Canary dataset.
• For convenience, CNVs of TCGA dataset were transformed back to the regular copy number format (2 × 2CNV).
• Category A: Genes selected by multivariate classification methods; Category B: Proved tumor associated genes for lung cancer;
3.2 | Identifying CNV signature genes to distinguish LUAD from LUSC and to distinguish tumors from their corresponding non-malignant samples
To overcome the challenge of identifying molecular signatures from microarray data, which contains a huge difference between the number of variables (approximately 20 000) and the number of samples (up to several hundreds), a novel EN-PLS-NB algorithm was developed to identify important genes iteratively using sophisticated machine learning algorithms (details are shown in the Materials and Methods and Supporting Information). As a result, 33 genes were identified as the final set of CNV signatures for the histologic classification model of NSCLC (ranked by their contribution (coefficients) to the classification model in descending order in Table 2). Among them, SOX2 and PIK3CA are well-known key lung tumor-associated genes. Adiponectin (ADIPOQ), ABCF3, ABCC5, SERPINI2, SST, and RBPJ have also been proven to play important roles in NSCLC (Boelens, et al. 2009; Cui, et al. 2011; Kang, et al. 2009; Lv, et al. 2015; Umekawa, et al. 2013).34–38
Twenty-six of the signature genes have greater deflections for LUSC tumors, whereas seven have greater deflections for LUAD tumors (highlighted in yellow in Table 2). There are 21 genes located on 3q, the site of the greatest deflection for LUSCs. Others are located on 4p, 6q, 8p, 9p, 15q, and 19.
The 33-signature gene classifier separated tumors from the nonmalignant samples with an accuracy of 0.96 and 0.98 for LUADs and LUSCs, respectively (Table 3). It classified the two tumor types with an accuracy of 0.88 in the TCGA NSCLC tumors (training data) and 0.84 in the SPORE dataset. In the EDRN/Canary dataset, it classified LUADs with a sensitivity of 0.96 (Table 3). The SPORE data were achieved with platforms that are different from the one used for the TCGA dataset. These results strongly confirm that our classifier possesses high classification performance regardless of the data platform used.
TABLE 3.
The classification results among LUAD, LUSC and non-malignant samples obtained by CNV
| Comparison | Training/Validation | Dataset | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|---|
| LUAD vs non-malignant | Both | TCGA | 0.95 | 0.97 | 0.96 |
| LUSC vs non-malignant | Both | TCGA | 0.97 | 0.99 | 0.98 |
| LUAD vs LUSC | Training | TCGA | 0.94 | 0.81 | 0.88 |
| Validation | SPORE | 0.83 | 0.85 | 0.84 | |
| EDRN/Canary | 0.96 | NA | NA |
There are only LUAD samples in EDRN/Canary dataset, so only Sensitivity measurement can be calculated.
NA: not available.
We further investigated whether we could reduce the number of signature genes without considerably decreasing the accuracy of our classification model. From the original 33 genes, we removed genes one by one, starting with the gene with the least absolute coefficient value. We generated a 7-gene model, which corresponds to the top seven genes in Table 2. The classification results are summarized in Supporting Information Table S5. The 7-gene model exhibited similar accuracy for both TCGA and SPORE datasets (only a modest decrease in accuracy of 0.04). The sensitivity decreased from 0.94 to 0.81 for the TCGA training dataset, but the specificity increased from 0.81 to 0.94. For the independent validation datasets, there was a decrease in specificity in SPORE (0.74 compared to 0.85) and a modest increase in sensitivity in the EDRN/Canary dataset (from 0.96 to 0.97). All seven genes are located on 3q. Any further decrease in the number of genes in the model resulted in much greater decreases in these measurements. The 7-gene model separated tumors from the non-malignant samples with an accuracy of 0.91 and 0.97 for LUADs and LUSCs, respectively.
3.3 | Comparison of CNV patterns for LUAD and LUSC with histologically similar tumors arising at other sites
Because the identification of tissue origin of tumors is often of critical clinical importance, we compared the CNV patterns of LUADs and LUSCs with those of histologically similar tumors arising at other sites. We focused on tumors that frequently cause clinical and pathological difficulties with regard to the identification of their tissue of origin. Therefore, the genome-wide CNV patterns of HNSC, ESSC, CRCA, BRCA, OV, and PRAD were explored for further analysis.
Figures 3 and 4 show the global CNV patterns for these five adenocarcinomas (ADCs) and three squamous cell carcinomas (SCCs). Supporting Information Figure S5 shows the genome-wide CNV t-test results between LUAD and CRCA/BRCA/PRAD/OV tumor samples in the TCGA dataset. Supporting Information Figure S6 shows the genome-wide CNV t-test between LUSC and HNSC/ESSC tumor samples in the TCGA dataset. As shown in these figures and Supporting Information Table S6, in all comparisons except for LUSC from ESSC (only 2.9%), there were significant differences ranging from 42.5% to 55% of the genes after using Bonferroni-correction as the family-wise control.
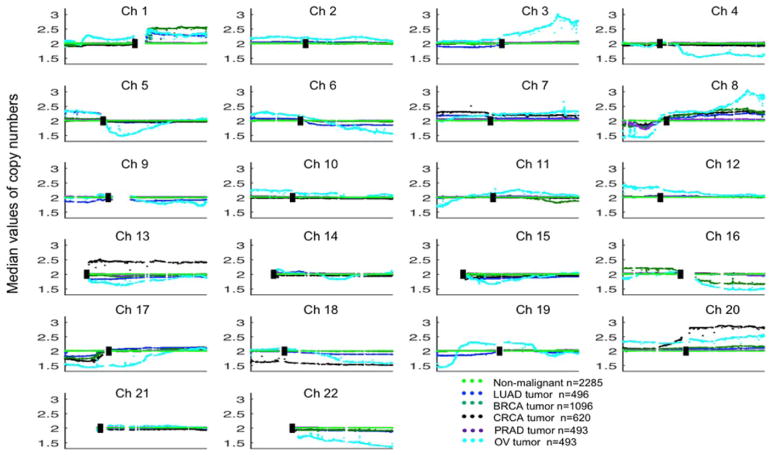
FIGURE 3.
Comparison of CNV patterns for LUAD, BRCA, CRCA, OV, PRAD, and non-malignant tissues. Each spot is the median value of copy numbers of each gene in the corresponding group. The genes are sorted according to their locations. The space between two arms of each chromosome is the location of the corresponding centromere. [Color figure can be viewed at wileyonlinelibrary.com]
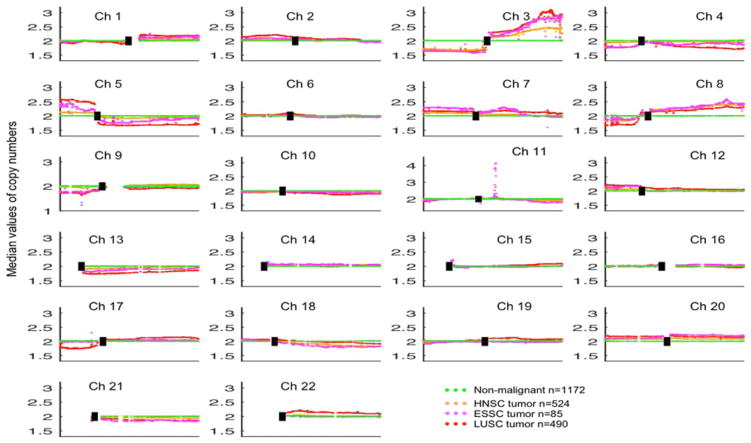
FIGURE 4.
Comparison of CNV patterns for LUSC, HNSC, ESSC, and non-malignant tissues. Each spot is the median value of copy numbers of each gene in the corresponding group. The genes are sorted according to their locations. The space between two arms of each chromosome is the location of the corresponding centromere. Note: Chromsome 11 has different axis limits. [Color figure can be viewed at wileyonlinelibrary.com]
As shown in Figure 3, it is clear that, except for chromosome 21, the CNV pattern for OV differs completely from those for other ADCs across all other chromosomes. BRCA is characterized by 100% losses on 16q and gains on 16p, which distinguishes it from other primary ADCs, including LUAD. Additionally, the whole arm losses of 5p, 3p, 9p, and 9q can easily distinguish LUAD from BRCA. The losses of almost whole arms of 3p, 6q, 9q, 9p, and 19p can easily separate LUAD from BRCA, CRCA, and PRAD. Chromosomes 13, 18, and 20 suggest that CRCA has a different CNV pattern from LUAD, BRCA, and PRAD. Except for the narrow loss in 8p, there is no other loss or gain segment compared with the corresponding non-malignant samples for PRAD.
From the genome-wide point of view, the most striking changes (for both percentages of involved genes and amplitude of the changes) were observed on chromosome 3, with 100% losses on 3p and gains on 3q in all three SCCs (Figure 4). The telomeric peak on 3q contains three genes that are crucial for squamous cell differentiation, including TP63, SOX2, and PIK3CA. This region also contains the telomerase RNA component gene TERC. Supporting Information Figure S7 shows the chromosomal location of these genes in all SCCs. Many other known or putative oncogenes are localized in this arm,2 and gains are an early event during the multistage pathogenesis of LUSC.
Chromosome 8 showed marked changes for both arms that were nearly identical for all ADCs and SCCs (except for 8p of PRAD). Both tumor types had losses of 8p and gains of 8q, with OV possessing the greatest variations on both arms (see Figure 3). The telomeric narrow segment amplicon was present in all tumor types (except for PRAD), which contains MYC (8q24). Of interest, it may present pan-cancer patterns of somatic copy number alterations.39 Amplification of the MYC family genes MYC and MYCL (1p34.2) is common in lung cancers,40 and narrow band gains of both genes were noted in LUADs and LUSCs.
Large peaks on 11q13.3 are quite obvious for all SCCs. ESSC shows the largest amplicon, while LUSC and HNSC show smaller ones. All genes in this narrow amplicon are shown in Supporting Information Figure S7, including FADD, FGF19, FGF4, and FGF3. The median CNV value of FADD is the highest compared with local genes in this area.
4 | DISCUSSION
Our results confirmed global CNV differences that distinguish the two major subtypes of NSCLC. The uneven deflections along the arms indicate that the CNVs do not simply reflect uniform duplications of entire arms or chromosomes. In particular, major gains along the entire 3q arm characterized LUSC tumors, compared to minimal gains involving a minor part of the 3q arm for LUADs.19,25,41 These observations formed the basis of developing a CNV-based classification model for NSCLC.
While the examination of CNV differences between LUAD and LUSC cancers may have limited application in diagnostic pathology, it has provided many important molecular changes between the two major forms of NSCLC. For further applications, we focused on the CNV differences among tumors that frequently cause clinical and pathological difficulties as to the identification of their tissue of origin. The accurate identification of the site of origin may play an important role in clinical management.
Figures 3 and 4 show that SCCs share many common CNV changes involved in their pathogenesis. Adenocarcinomas from different sites, in general, showed more changes between tumor sites, indicating that adenocarcinomas as a whole show more heterogeneity than SCCs.
In Supporting Information Figure S7, FADD shows a very impressive narrow amplicon on 11q13.3 that is only in ESSC, while no published research mentions the relationship between them, even though it has already been proven to play an important role in LUSC and HNSC.41,42
Of the FGF members, FGF19 is located at 11q13.3. FGF19 overexpression has been implicated in the pathogenesis of some cancers, especially in murine models of hepatocellular carcinoma.43 We found only one reference to the role of FGF19 in lung cancer.44 The 11q13 amplicon also contains the associated FGF genes FGF3 and FGF4 and has been reported to be present in other squamous carcinomas, including head and neck and esophageal carcinomas,45 as well as breast cancers. 46 Of great interest, the 8p and 11q amplicons containing FGFR1 and the FGF ligands 3, 4, and 19 are also co-amplified in breast cancers, 46 suggesting a link between the amplification of these FGF pathway members. Moreover, FGF12 is located at 3q29, a large region of gain that is characteristic of SCC tumors. FGF6 and FGF23 are located at 12p13.32, a broad region of gain that is characteristic of SCC tumors. Thus, in addition to the well-described FGFR1 gene, multiple members of the FGF family show copy number gains in SCC tumors. While the occurrence of FGFR1 amplification in SCCs is well known, the roles of other FGFR receptors and their ligands in lung cancer are poorly documented.47,48
As a result of the reasons mentioned above, 13% of NSCLCs are still referred to as NSCLC-NOS, indicating that auxiliary methods are required for the full classification of poorly graded or undifferentiated lung cancers. Accurate classification would benefit from adjuvant tests such as molecular classifiers.
A list of 33 signature genes was generated. Because the magnitude of deflections was greater in LUSC tumors and because the most notable difference between the two cancer types was selective amplification of 3q in LUSCs, the 33-gene list is biased in favor of LUSCs (26 genes, 79%), as well as the chromosomal location on 3q (21 genes, 64%). When the gene list was further reduced to the top 7 contributing genes, there was only a modest decrease in accuracy by 0.04. We decided to stay with the 33-gene classification model, in part because the 7-gene model was unbalanced with all genes located on 3q and all of the genes being LUSC related.
The new WHO classification recommends using immunostaining to classify poorly differentiated lung cancers including NSCLC-NOS.49 Unfortunately, immunostaining data were not available for many of the poorly differentiated lung cancers in the three datasets examined herein. For this reason, we compared the results of the CNV-based classification with the expression of genes frequently used for the immunostaining-based histologic diagnosis of poorly differentiated NSCLC tumors using the TCGA dataset. For LUAD, we used NKX2-1 (TTF-1) and Napsin A (NAPSA), and for LUSC, we used TP63 and keratin 5 (KRT5), the most robust of the high-molecular-weight keratins. The mRNA expression values of these four markers in the TCGA dataset are plotted in Supporting Information Figure S8. Their RNAseqV2 level 3 data were downloaded from the public TCGA portal, which included 515 LUAD samples (490 of them with CNV data available), 502 LUSC samples (487 with CNV data available), and 110 lung nonmalignant samples.
NKX2-1 (TTF-1) is the primary marker used to distinguish LUAD from LUSC. However, from Supporting Information Figure S8, we can clearly see a large overlap between the expression value distributions in these two subgroups. Normally, the sensitivity of NKX2-1 (TTF-1) is approximately 80%.50,51 Supporting Information Figure S8 shows that the lower cutoff value will result in higher sensitivity but a higher FP (false positive) rate. We chose the cutoff value as 10.3 (log2-transformed). The corresponding sensitivity is 81%, and the corresponding FP is 5%, which means that 5% of LUSC cases would be falsely diagnosed as LUAD. Under the same considerations, we chose the cutoff value of NAPSA as 12.5 (log2-transformed). The corresponding sensitivity of LUAD is 81%, and the FP rate for LUSC is 16%. Supporting Information Figure S8 also proves that it is impossible to distinguish LUAD from non-malignant samples using these two markers. However, our signatures classified LUAD from non-malignant samples with an accuracy as high as 96%. For the two LUSC markers, namely TP63 and KRT5, we chose the mean value ± standard deviation of the expression values of the corresponding non-malignant samples as the cutoffs. If the expression values of the corresponding markers were larger than or equal to the cutoff, the sample was classified as the corresponding tumor type. For example, if a sample’s expression value of NKX2-1 was ≥10.3, the sample would be considered an LUAD tissue.
In total, eight TCGA LUAD samples were considered double-negative by the two LUAD markers but double-positive by the two LUSC markers. Additionally, seven TCGA LUSC samples were considered double-negative by the two LUSC markers but double-positive by the two LUAD markers, indicating that these 15 samples may not be classified correctly by TCGA.
Supporting Information Table S7 shows the expression values of these four markers and the predicted scores. The scores of seven so-called LUSC samples were all positive but in the gray area (between −18.28 and 18.28) and therefore considered “NSCLC-NOS, favor ADC.” Two of the eight suspicious LUAD samples were classified as LUSC samples by our method. Four of the remaining six samples had positive scores in the gray area and were therefore also considered “NSCLC-NOS, favor ADC.”
Considering that a fraction of the TCGA diagnostic materials were of less-than-optimal quality (e.g., frozen sections instead of permanently fixed H&E slides)9,10, and in spite of the partially subjective nature of pathologic diagnosis, these 15 samples require a further diagnosis. It is highly possible that these so-called LUAD and LUSC cases were miss-classified by the TCGA database.
The immunostaining method is not limited to the expression values of markers. As previously shown, the sensitivity of NKX2-1 is only approximately 80%, and that of TP63 is even lower.50,51 The inconsistency between TCGA diagnosis and the expression values of these four makers proves the necessity of a third objective classification method or new markers. One of the main purposes of this article was to provide an adjuvant classification method. Our signature has the additional advantage of being of prognostic importance and may be useful in selecting new markers.
Because tumor tissue consists of mixtures of malignant and nonmalignant cells in varying proportions, it is important for any classifier to separate malignant from non-malignant tissue. The classification model using only the CNVs of the 33 signature genes separated both types of NSCLC tumors from the corresponding non-malignant samples with great sensitivity, specificity and accuracy (>0.95), which increases the usefulness of the model.
For the two most frequently mutated oncogenes in LUADs of the lung, that is, KRAS and EGFR,3,52–55 we examined whether these mutations were associated with specific CNV alterations. Tumors with either one of these oncogenic changes failed to exhibit copy number changes that were significantly different from those of wild type tumors. There was no significant difference in CNVs for these 33 genes according to either gender or smoking history (data not shown). We also compared the performance of our 33-gene classifier with that of another CNV classifier proposed by Li et al.14 In their study, the classifier consisting of 266 probes was not tested by an independent validation dataset. According to their results, it had a comparable accuracy to our classifier; however, its sensitivity was much lower (0.65).
Because different platforms were used to measure the CNV, only 26 of our 33 signature genes were included in Li’s data, indicating that more than 20% of our signature genes were missing. Therefore, we could not use our signature genes and model to classify the data used by them.
In summary, we developed and validated an accurate CNV classifier for NSCLC that distinguishes LUAD from LUSC and lung cancer from normal lung tissue. Three different datasets with different platforms confirmed that this classifier was largely independent of the major CNV platforms in common usage. Several of the genes in the classifier are relevant to lung cancer. Thus, this classifier has the additional advantage of being of prognostic importance and may be useful in selecting the subpopulation of curative resected lung cancer patients that will benefit from adjuvant therapy. An especially relevant use would be for large multinational clinical trials where no central pathology review is available.
The high percentage of CNV changes between histologically similar tumors arising in different tissues or organs suggests that patterns of CNV variations may be developed into methods that can be used to distinguish these tumor types. While the implementation and validation of such practical tests is beyond the scope of this manuscript, we have demonstrated that such applications can be developed and applied to clinical practice.
Supplementary Material
Acknowledgments
Funding information
This work was generously supported by the NCI Specialized Program in Research Excellence (SPORE) in Lung Cancer, P50CA70907. National Cancer Institute, USA. and the National Natural Science Foundation of China [31331351].
We thank Dr. Stephen Lam (British Columbia Cancer Agency, Vancouver, Canada) for collecting lung cancers from British Columbia. We also thank Dr. John Minna (Hamon Center for Therapeutic Oncology, University of Texas Southwestern Medical Center, Dallas, Texas, USA) and Dr. Ignacio WIstuba (Departments of Thoracic/Head and Neck Medical Oncology and Cancer Biology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA) for their generous support and encouragement.
Footnotes
Additional Supporting Information may be found online in the supporting information tab for this article.
References
- 1.Clinical Lung Cancer Genome P, Network Genomic M. A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5:209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rekhtman N, Tafe LJ, Chaft JE, et al. Distinct profile of driver mutations and clinical features in immunomarker-defined subsets of pulmonary large-cell carcinoma. Mod Pathol. 2013;26:511–522. doi: 10.1038/modpathol.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gazdar AF. Should we continue to use the term non-small cell lung cancer? Ann Oncol. 2010;21(Suppl 7):vii225–vii229. doi: 10.1093/annonc/mdq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aisner DL, Marshall CB. Molecular pathology of non-small cell lung cancer: a practical guide. Am J Clin Pathol. 2012;138:332–346. doi: 10.1309/AJCPFR12WJKCEEZZ. [DOI] [PubMed] [Google Scholar]
- 5.Gazdar AF. The evolving role of the pathologist in the management of lung cancer. Lung Cancer Manag. 2012;1:273–281. doi: 10.2217/lmt.12.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Travis WD, Brambilla E, Noguchi M, et al. Diagnosis of lung cancer in small biopsies and cytology: implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification. Arch Pathol Lab Med. 2013;137:668–684. doi: 10.5858/arpa.2012-0263-RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travis WD, Rekhtman N. Pathological diagnosis and classification of lung cancer in small biopsies and cytology: strategic management of tissue for molecular testing. Semin Respir Crit Care Med. 2011;32:22–31. doi: 10.1055/s-0031-1272866. [DOI] [PubMed] [Google Scholar]
- 8.Thunnissen E, Beasley MB, Borczuk AC, et al. Reproducibility of histopathological subtypes and invasion in pulmonary adenocarcinoma. An international interobserver study. Mod Pathol. 2012;25:1574– 1583. doi: 10.1038/modpathol.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thunnissen E, Noguchi M, Aisner S, et al. Reproducibility of histopathological diagnosis in poorly differentiated NSCLC: an international multiobserver study. J Thorac Oncol. 2014;9:1354–1362. doi: 10.1097/JTO.0000000000000264. [DOI] [PubMed] [Google Scholar]
- 10.Girard L, Rodriguez-Canales J, Behrens C, et al. An expression signature as an aid to the histologic classification of non-small cell lung cancer. Clin Cancer Res. 2016;22:4880–4889. doi: 10.1158/1078-0432.CCR-15-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilad S, Lithwick-Yanai G, Barshack I, et al. Classification of the four main types of lung cancer using a microRNA-based diagnostic assay. J Mol Diagn. 2012;14:510–517. doi: 10.1016/j.jmoldx.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Bishop JA, Benjamin H, Cholakh H, Chajut A, Clark DP, Westra WH. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010;16:610– 619. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 13.Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010:5. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B-Q, You J, Huang T, Cai Y-D. Classification of Non-Small Cell Lung Cancer Based on Copy Number Alterations. PLoS One. 2014;9:e88300. doi: 10.1371/journal.pone.0088300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramani RG, Jacob SG. Improved classification of lung cancer tumors based on structural and physicochemical properties of proteins using data mining models. PLoS One. 2013;8:e58772. doi: 10.1371/journal.pone.0058772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y, He J. Molecular classification of non-small cell lung cancer: diagnosis, individualized treatment, and prognosis. Front Med. 2013;7:157–171. doi: 10.1007/s11684-013-0272-4. [DOI] [PubMed] [Google Scholar]
- 17.Craddock N, Hurles ME, Cardin N, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y-T, Lin X, Chirieac LR, et al. Impact on disease development, genomic location and biological function of copy number alterations in non-small cell lung cancer. PLoS One. 2011;6:e22961. doi: 10.1371/journal.pone.0022961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tonon G, Wong KK, Maulik G, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louhimo R, Hautaniemi S. CNAmet: an R package for integrating copy number, methylation and expression data. Bioinformatics. 2011;27:887–888. doi: 10.1093/bioinformatics/btr019. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y-W, Zheng Y, Wang J-Z, et al. Integrated analysis of DNA methylation and mRNA expression profiling reveals candidate genes associated with cisplatin resistance in non-small cell lung cancer. Epigenetics. 2014;9 doi: 10.4161/epi.28601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughey JJ, Butte AJ. Robust meta-analysis of gene expression using the elastic net. Nucleic Acids Res. 2015;43:e79. doi: 10.1093/nar/gkv229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song K. Recognition of prokaryotic promoters based on a novel variable-window Z-curve method. Nucleic Acids Res. 2012;40:963–971. doi: 10.1093/nar/gkr795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langarizadeh M, Moghbeli F. Applying naive bayesian networks to disease prediction: a systematic review. Acta Inform Med. 2016;24:364–369. doi: 10.5455/aim.2016.24.364-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garnis C, Lockwood WW, Vucic E, et al. High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer. 2006;118:1556–1564. doi: 10.1002/ijc.21491. [DOI] [PubMed] [Google Scholar]
- 26.Pei J, Balsara BR, Li W, et al. Genomic imbalances in human lung adenocarcinomas and squamous cell carcinomas. Genes Chromosomes Cancer. 2001;31:282–287. doi: 10.1002/gcc.1145. [DOI] [PubMed] [Google Scholar]
- 27.Rao N, Moran C, Suster S. Tumors of the lung and pleura. In: Damjanov I, Fan F, editors. Cancer Grading Manual. 2. New York: Springer; 2013. pp. 31–41. [Google Scholar]
- 28.Lin J, Sun T, Ji L, et al. Oncogenic activation of c-Abl in non-small cell lung cancer cells lacking FUS1 expression: inhibition of c-Abl by the tumor suppressor gene product Fus1. Oncogene. 2007;26:6989–6996. doi: 10.1038/sj.onc.1210500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Zhao J, Chen XF, et al. CpG island methylator phenotype involving tumor suppressor genes located on chromosome 3p in non-small cell lung cancer. Lung Cancer. 2008;62:15–22. doi: 10.1016/j.lungcan.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Pastuszak-Lewandoska D, Kordiak J, Antczak A, et al. Expression level and methylation status of three tumor suppressor genes, DLEC1, ITGA9 and MLH1, in non-small cell lung cancer. Med Oncol. 33:75. doi: 10.1007/s12032-016-0791-3. [DOI] [PubMed] [Google Scholar]
- 31.Tam KW, Zhang W, Soh J, et al. CDKN2A/p16 inactivation mechanisms and their relationship to smoke exposure and molecular features in non-small cell lung cancer. J Thorac Oncol. 2013;8:1378–1388. doi: 10.1097/JTO.0b013e3182a46c0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deben C, Deschoolmeester V, Lardon F, Rolfo C, Pauwels P. TP53 and MDM2 genetic alterations in non-small cell lung cancer: Evaluating their prognostic and predictive value. Crit Rev Oncol Hematol. 2016;99:63–73. doi: 10.1016/j.critrevonc.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 33.Javid J, Mir R, Julka PK, Ray PC, Saxena A. Association of p53 and mdm2 in the development and progression of non-small cell lung cancer. Tumour Biol. 2015;36:5425–5432. doi: 10.1007/s13277-015-3208-6. [DOI] [PubMed] [Google Scholar]
- 34.Boelens MC, van den Berg A, Fehrmann RS, et al. Current smoking-specific gene expression signature in normal bronchial epithelium is enhanced in squamous cell lung cancer. J Pathol. 2009;218:182–191. doi: 10.1002/path.2520. [DOI] [PubMed] [Google Scholar]
- 35.Cui E, Deng A, Wang X, et al. The role of adiponectin (ADIPOQ) gene polymorphisms in the susceptibility and prognosis of nonsmall cell lung cancer. Biochem Cell Biol. 2011;89:308–313. doi: 10.1139/o11-005. [DOI] [PubMed] [Google Scholar]
- 36.Kang JU, Koo SH, Kwon KC, Park JW, Kim JM. Identification of novel candidate target genes, including EPHB3, MASP1 and SST at 3q26.2-q29 in squamous cell carcinoma of the lung. BMC Cancer. 2009;9:237. doi: 10.1186/1471-2407-9-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv Q, Shen R, Wang J. RBPJ inhibition impairs the growth of lung cancer. Tumour Biol. 2015;36:3751–3756. doi: 10.1007/s13277-014-3015-5. [DOI] [PubMed] [Google Scholar]
- 38.Umekawa K, Kimura T, Kudoh S, et al. Reaction of plasma adiponectin level in non-small cell lung cancer patients treated with EGFR-TKIs. Osaka City Med J. 2013;59:53–60. [PubMed] [Google Scholar]
- 39.Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiorentino FP, Tokgun E, Sole-Sanchez S, et al. Growth suppression by MYC inhibition in small cell lung cancer cells with TP53 and RB1 inactivation. Oncotarget. 2016;7:31014–31028. doi: 10.18632/oncotarget.8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Network TCGAR. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowman BM, Sebolt KA, Hoff BA, et al. Phosphorylation of FADD by the kinase CK1alpha promotes KRASG12D-induced lung cancer. Sci Signal. 2015;8:ra9. doi: 10.1126/scisignal.2005607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin BC, Desnoyers LR. FGF19 and cancer. Adv Exp Med Biol. 2012;728:183–194. doi: 10.1007/978-1-4614-0887-1_12. [DOI] [PubMed] [Google Scholar]
- 44.Cui Y, Liu J, Liu Y, et al. Upregulation of FGF19 in lung adenocarcinoma and predicts poor prognosis. Int J Clin Exp Pathol. 2016;9:7338–7344. [Google Scholar]
- 45.Katoh M, Katoh M. Comparative genomics on mammalian Fgf3-Fgf4 locus. Int J Oncol. 2005;27:281–285. [PubMed] [Google Scholar]
- 46.Paterson AL, Pole JC, Blood KA, et al. Co-amplification of 8p12 and 11q13 in breast cancers is not the result of a single genomic event. Genes Chromosomes Cancer. 2007;46:427–439. doi: 10.1002/gcc.20424. [DOI] [PubMed] [Google Scholar]
- 47.Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Semrad TJ, Mack PC. Fibroblast growth factor signaling in nonsmall-cell lung cancer. Clin Lung Cancer. 2012;13:90–95. doi: 10.1016/j.cllc.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 49.Micke P, Mattsson JS, Djureinovic D, et al. The impact of the fourth edition of the WHO classification of lung tumours on histological classification of resected pulmonary NSCCs. J Thorac Oncol. 2016;11:862–872. doi: 10.1016/j.jtho.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 50.Bishop JA, Teruya-Feldstein J, Westra WH, Pelosi G, Travis WD, Rekhtman N. p40 (DeltaNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod Pathol. 2012;25:405– 415. doi: 10.1038/modpathol.2011.173. [DOI] [PubMed] [Google Scholar]
- 51.Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 52.Gazdar AF. EGFR mutations in lung cancer: different frequencies for different folks. J Thorac Oncol. 2014;9:139–140. doi: 10.1097/JTO.0000000000000063. [DOI] [PubMed] [Google Scholar]
- 53.Gazdar A, Robinson L, Oliver D, et al. Hereditary lung cancer syndrome targets never smokers with germline EGFR gene T790M mutations. J Thorac Oncol. 2014;9:456–463. doi: 10.1097/JTO.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006;118:257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 55.Sunaga N, Shames DS, Girard L, et al. Knockdown of oncogenic KRAS in non-small cell lung cancers suppresses tumor growth and sensitizes tumor cells to targeted therapy. Mol Cancer Ther. 2011;10:336–346. doi: 10.1158/1535-7163.MCT-10-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.