Abstract
Six cytotoxic and antimicrobial metabolites of a new bromo-phenazinone class, the marinocyanins A-F (1–6), were isolated together with the known bacterial metabolites 2-bromo-1-hydroxyphenazine (7), lavanducyanin (8, WS-9659A) and its chlorinated analog WS-9659B (9). These metabolites were purified by bioassay-guided fractionation of the extracts of our MAR4 marine actinomycete strains CNS-284 and CNY-960. The structures of the new compounds were determined by detailed spectroscopic methods and marinocyanin A (1) was confirmed by crystallographic methods. The marinocyanins represent the first bromo-phenazinones with an N-isoprenoid substituent in the skeleton. Marinocyanins A-F show strong to weak cytotoxicity against HCT-116 human colon carcinoma and possess modest antimicrobial activities against Staphylococcus aureus and amphotericin-resistant Candida albicans.
Keywords: Bromophenazinones, MAR4 actinomycete, Meroterpenoids
1. Introduction
Marine microorganisms are a prolific source of novel secondary metabolites. In particular, actinomycetes from the marine environment have produced a wide variety of structurally unique and biologically active molecules.1 While members of the obligate marine genus Salinispora have proven particularly useful in this regard,2 other groups of marine-derived actinomycetes have also yielded unusual classes of secondary metabolites. One noteworthy example is a streptomycete lineage designated MAR4.3 These bacteria encompass considerable taxonomic diversity and are enriched in the production of hybrid isoprenoid secondary metabolites relative to related streptomycetes.4 These observations are supported at the genomic level, with MAR4 strains averaging five ABBA prenyl-transferases per strain relative to less than one for other streptomycetes.5 MAR4 hybrid isoprenoids encompass a diversity of structures including prenylated polyketides, phenazines, and pyrroles, and are of considerable interest due to their potent biological activities.3–5
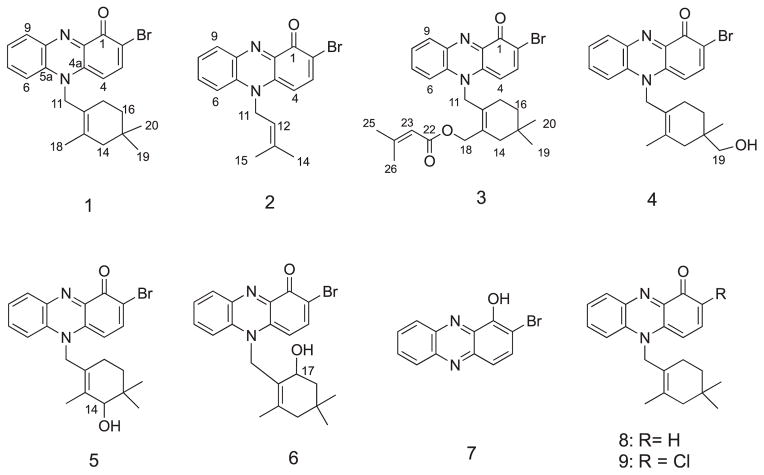
In the course of our chemical and biological screening of marine actinomycete strains, we have investigated the chemistry of two MAR4 actinomycetes, strains CNY-960 and CNS-284, which were isolated from marine sediments collected in the Solomon Islands and in Palau, respectively. Culture extracts of these related strains were active in cytotoxicity bioassays and subsequent bioassay-guided fractionation of the extracts, followed by the repeated HPLC purification of the active materials, led to the isolation of six new N-isoprenoid bromo-phenazinone antibiotics, marinocyanins A-F (1–6) along with the known metabolites 2-bromo-1-hydroxyphenazine (7), lavanducyanin (8, WS-9659A) and its chloro analog WS-9659B (9). In this paper, we report on the isolation of the marinocyanins A-F and provide details of their spectral properties, structure elucidation and biological properties (See Chart 1).
Chart 1.
Structures of phenazinone meroterpenoid metabolites isolated in this study.
2. Results and discussion
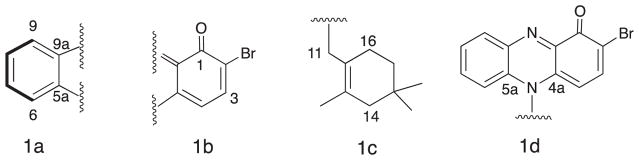
Marinocyanin A (1) was isolated as a dark blue crystal (mp 160 °C). The low resolution ESIMS spectrum of 1 showed two pseudomolecular ion peaks at m/z 411.1 [M+H]+, and 413.1 [M+H+2]+ with intensities of 1:1, which illustrated the isotopes for one bromine atom in the molecule. The molecular formula of 1 was assigned as C22H23BrN2O based on the HRESI-TOFMS measurement of the [M+H]+ pseudomolecular peak at m/z 411.1056 (calcd. 411.1063). The UV spectrum of 1 exhibited absorption maxima at 238 and 330 nm with an additional broad absorption between 720 nm, suggesting that 1 possessed a highly conjugated chromophore. IR spectral data for this metabolite showed absorption bands at 2950, 1489 and 1627 cm−1 indicating the presence of an aromatic nucleus and a conjugated carbonyl group, respectively. The structure of 1 was initially assigned based upon analysis of 1D and 2D-NMR data in CD3OD. The 13C NMR spectrum (Table 1) showed the presence of 22 carbon signals and analysis of the DEPT NMR data revealed that the compound contained 8 sp,3 13 sp2 and one carbonyl carbons (Table 1). The observed signals from 13 olefinic or aromatic carbons indicated the presence of 7 double bonds in 1, including one C=N functionality. These carbons, along with the one carbonyl, accounted for 8 of the 12° of unsaturation implied by the molecular formula, thus indicating the presence of four rings. The 1H NMR spectrum of marinocyanin A resolved all six aromatic protons, and gCOSY experiments revealed that they belonged to two isolated spin systems, one involving four contiguous protons (δ 8.01, 8.02, 7.72, 8.30) while the other involved two adjacent protons (δ 8.10, 6.32) (Fig. 1). In addition to these, the NMR spectrum also showed four methylene signals at δ 5.42, 1.89, 1.18, 1.40 and three methyl singlets at δ 1.99, 0.82 (2 × CH3). Interpretation of 2D COSY, HSQC and HMBC NMR data allowed all protons and carbons to be assigned. COSY data illustrated a methylene proton signal 16-H2 (δ 1.18) that was coupled to an adjacent methylene pair 17-H2 (δ 1.40). The latter showed long-range proton couplings with the methylene protons 11-H2 (δ 5.42); whereas the methylene protons 16-H2 (δ 1.18) showed a long-range coupling with 14-H2 (δ 1.89). In the HMBC spectrum the singlet methyl protons from C-19 and C-20 (δ 0.82, 6H) showed 2- and 3-bond correlations to C-14 (δ 46.8), C-15 (δ 29.7), and C-16 (δ 35.7). In addition, 2- and 3-bond couplings of the C-18 olefinic methyl group protons with C-12, C-13, and C-14 were observed. The connectivity between the C-11 and C-12 was also confirmed by long-range HMBC correlations from the C-11 protons to C-12, C-13, C-18, and C-17. These analyses suggested the presence of a terpenoid cyclolavandulyl constellation (Fig. 1, c).
Table 1.
NMR Spectroscopic Assignments for Marinocyanins A-C (1–3) in CD3OD at 500 MHz. Detailed COSY and HMBC data for 1–3 can be found in the Supplemental Information.
| C/H# | Marinocyanin A (1)
|
Marinocyanin B (2)
|
Marinocyanin C (3)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| δH (J Hz) | δC | δH (J Hz) | δC | δH (J Hz) | δC | ||||
| 1 | 171.8 | C | 171.8 | C | 171.4 | C | |||
| 2 | 108.9 | C | 108.1 | C | 108.2 | C | |||
| 3 | 8.10 d (8.5) | 147.1 | CH | 8.25 d (8.8) | 147.1 | CH | 8.21 d (8.5) | 147.1 | CH |
| 4 | 6.32 d (8.5) | 94.5 | CH | 6.46 d (8.8) | 94.4 | CH | 6.57 d (8.5) | 93.7 | CH |
| 4a | 135.2 | C | 134.4 | C | 135.2 | C | |||
| 5 | |||||||||
| 5a | 132.5 | C | 132.8 | C | 136.1 | C | |||
| 6 | 8.01 d (8.0) | 116.1 | CH | 8.00 d (8.2) | 116.5 | CH | 8.15 d (8.5) | 115.7 | CH |
| 7 | 8.02 t (8.0) | 137.6 | CH | 8.09 t (8.2) | 137.7 | CH | 8.07 t (8.5) | 136.9 | CH |
| 8 | 7.72 t (8.0) | 127.6 | CH | 7.79 t (8.2) | 127.6 | CH | 7.79 t (8.5) | 126.7 | CH |
| 9 | 8.30 d (8.0) | 133.8 | CH | 8.40 d (8.2) | 133.8 | CH | 8.43 d (8.5) | 133.1 | CH |
| 9a | 138.5 | C | 138.1 | C | 137.8 | C | |||
| 10 | |||||||||
| 10a | 145.3 | C | 145.2 | C | 145.0 | C | |||
| 11 | 5.42 br s | 51.9 | CH2 | 5.38 d (5.3) | 48.5 | CH2 | 5.75 br s | 50.6 | CH2 |
| 12 | 121.6 | C | 5.21 t (5.3) | 116.9 | CH | 127.3 | C | ||
| 13 | 131.9 | C | 139.5 | C | 131.0 | C | |||
| 14 | 1.89 br s | 46.8 | CH2 | 2.02 s | 18.5 | CH3 | 2.00 s | 41.7 | CH2 |
| 15 | 29.7 | C | 1.80 s | 25.6 | CH3 | 27.9 | C | ||
| 16 | 1.18 t (6.6) | 35.7 | CH2 | 1.22 t (6.6) | 34.3 | CH2 | |||
| 17 | 1.40 br s | 23.5 | CH2 | 1.54 br s | 22.9 | CH2 | |||
| 18 | 1.99 s | 19.4 | CH3 | 4.84 s | 62.6 | CH2 | |||
| 19 | 0.82 s | 28.1 | CH3 | 0.85 s | 27.2 | CH3 | |||
| 20 | 0.82 s | 28.1 | CH3 | 0.85 s | 27.2 | CH3 | |||
| 21 | |||||||||
| 22 | 166.7 | C | |||||||
| 23 | 5.82 s | 115.2 | CH | ||||||
| 24 | 158.6 | C | |||||||
| 25 | 1.99 s | 26.3 | CH3 | ||||||
| 26 | 2.25 s | 19.1 | CH3 | ||||||
Fig. 1.
Substructures 1a–d defined by 2D NMR analysis for marinocyanin A (1).
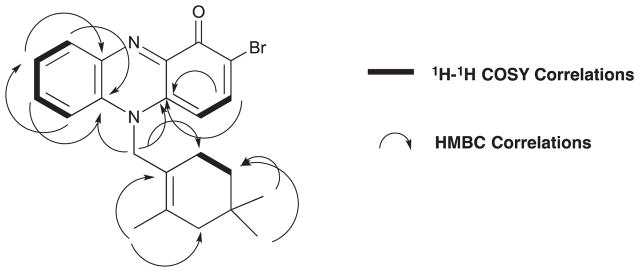
The two spin systems 1a and 1b, obtained from gCOSY NMR data analysis, were connected based on key HMBC correlations. Long range HMBC NMR correlations from H-6 (δ 8.01) to C-9a (δ 138.5), and C-8 (δ 127.6), from H-9 (δ 8.30) to C-5a (δ 132.5) and C-7 (δ 137.6), and from H-4 (δ 6.32) to C-2 (δ 108.5) and C-10a (δ 145.3), and from H-3 (δ 8.10) to C-1 (δ 171.8), C-2 (δ 108.9), and C-4a (δ 135.2) confirmed the presence of the phenazinone unit. Based on the molecular formula and the C-2 13C NMR shift, the bromine atom was placed at this position on the aromatic ring, thus allowing the assignment of this subunit as 2-bromophenazin-1-one (Fig.1, d, C-1 to C-10a). The C-11 methylene protons (δ 5.42) of the cyclolavandulyl component showed 3- and 4-bond HMBC correlations to C-4a (δ 135.2) and C-5a (δ 132.5) through the nitrogen atom. Thus, subunits 1c and 1d could be connected to complete the structure assignment of marinocyanin A (1) (See Fig. 2).
Fig. 2.
Key 2D NMR 1H-1H COSY and HMBC correlations used to establish the NMR-based structure of marinocyanin A (1).
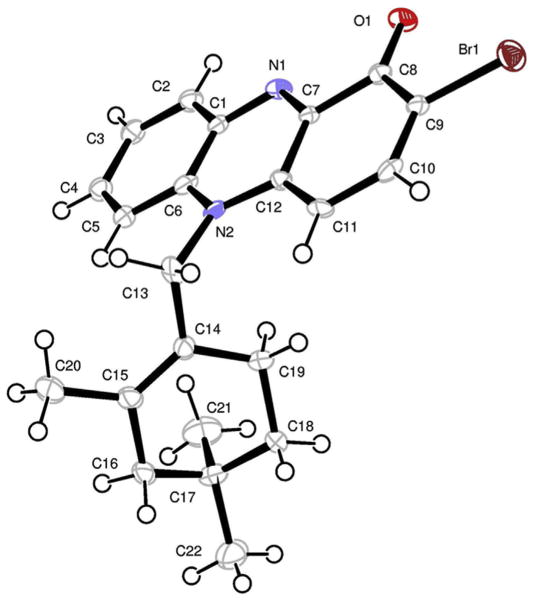
The structure assigned by interpretation of NMR data was further confirmed by X-ray crystallographic analysis (Fig. 3). Crystallization of 1 yielded orthorhombic crystals by slow evaporation of a solvent mixture of methanol, water and dichloromethane. The final X-ray structure fully confirmed the assignments made by NMR analysis.
Fig. 3.
Final X-ray computer-generated drawing for marinocyanin A (1).
Marinocyanin B (2) was isolated as a deep blue powder that analyzed for the molecular formula C17H15BrN2O by interpretation of HRESI-TOFMS data ([obsd M+H]+ at m/z 343.0440), and by comprehensive analysis of NMR data (Table 1). The UV absorption spectrum of 2 showed bands at 238 and 330 nm and a longer wavelength broad adsorption at 740 nm, which by analogy to 1, were suggestive of the presence of the bromo-phenazinone chromophore. Comparison of the 1H and 13C NMR spectroscopic data of 2 with marinocyanin A (1) revealed the identical bromo-phenazinone core, except the cyclolavandulyl ring was substituted with a C5 isoprenyl group. Comprehensive NMR analysis, utilizing data from COSY, HMQC, and HMBC experiments, allowed the complete assignment of the proton and carbon signals for 2, leading to the assignment of structure 2 for marinocyanin B.
Marinocyanin C (3) was also obtained as deep blue powder. The molecular formula of 3 was assigned as C27H29BrN2O3 by HRESI-TOFMS analysis ([obsd M+H]+ at m/z 509.1434), and by comprehensive analysis of NMR data. The UV absorption bands at 240, 330, and the characteristic broad band at 750 nm, confirmed that 3 also possessed the bromo-phenazinone functionality as in the marinocyanins A and B. However, a new IR absorption band at 1711 cm−1 suggested the presence of an ester carbonyl group in the molecule. The 1H and 13C NMR spectra of 3 showed features similar to those of 1 and 2, but in this case the NMR data were more complex, indicating overall that 3 possessed a modified cyclolavandulyl ring component (Table 1). In the 1H NMR spectrum of 3, the signals attributed to the full marinocyanin A structure were clear. However, new signals for two methyl groups (δ 1.99 s) and (δ 2.25 s) and an olefinic proton (δ 5.82 s) were observed. In addition to these, the methyl proton signal at C-18 in 1 was converted to a hydroxymethylene group in 3 (δ 4.84 s). These observations suggested that esterification of the C-18 hydroxy group had occurred. In the 13C NMR spectrum of marinocyanin C (3), five additional signals, two methyl carbons (δ 26.3 and 19.1), two olefinic carbons (δ 115.2, and 158.6), and an ester carbonyl carbon (δ 166.7) were detected. From detailed analysis of HMBC NMR data, the acyl group was assigned as a 3,3-dimethylacrylic acid residue. Thus, extensive interpretation of the 1D and 2D NMR spectral data, combined with comparison of data with those of marinocyanin A (1), established the structure of marinocyanin C.
Marinocyanin D (4) was also isolated as a deep blue powder that analyzed for the molecular formula C22H23BrN2O2 on the basis of HRESI-TOFMS analysis. A pseudomolecular ion peak at 427.1012 amu ([M+H]+, error 0.9 ppm) indicated that 4 contained the same number of degrees of unsaturation as 1. The UV absorption spectrum of 4 showed similar bands at 238, 330 and 745 nm, which were analogous to the absorptions of all bromo-phenazinones discovered. IR spectral data showed absorption bands at 3374, 2950, 1489 and 1627 cm−1 indicating the presence of a hydroxy group, an aromatic nucleus and a conjugated carbonyl group, respectively. Detailed analysis of 1H and 13C NMR data suggested that marinocyanin D (4) had the same carbon skeleton as marinocyanin A (1), but differed only in the substitution on the cyclolavandulyl ring (Table 2). Further analysis of NMR data showed that one of the methyl groups on C-15 was replaced by a hydroxymethylene group (δC 70.6, δH 3.23, 3.18). As in 1, comprehensive analysis of 2D NMR data allowed the full planar structure of 4 to be assigned. Unfortunately, the limited sample size obtained, and the tendency for decomposition, did not allow determination of the configuration of the C-15 carbon.
Table 2.
NMR spectroscopic data for marinocyanin D (4), E (5), and F (6) in CD3OD at 500 MHz.
| C/H# | 4
|
5
|
6
|
|||
|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 171.5, C | 171.5, C | 171.2, C | |||
| 2 | 108.2, C | 107.8, C | 107.5, C | |||
| 3 | 147.5, CH | 8.24, d (8.5) | 147.2, CH | 8.23, d (8.9) | 145.8, CH | 8.22, d (8.8) |
| 4 | 94.5, CH | 6.47, d (8.5) | 94.1, CH | 6.42, d (8.9) | 94.6, CH | 6.49, d (8.8) |
| 4a | 135.1, C | 134.9, C | 135.3, C | |||
| 5 | ||||||
| 5a | 133.2, C | 133.6, C | 133.3, C | |||
| 6 | 116.7, CH | 8.09, d (8.6) | 116.2, CH | 8.04, d (8.8) | 117.7, CH | 8.21, d (8.8) |
| 7 | 137.6, CH | 8.10, dd (8.6) | 137.4, CH | 8.07, dd (8.8, 1.5) | 136.9, CH | 8.04, dd (8.8) |
| 8 | 127.7, CH | 7.80, m | 127.4, CH | 7.78, d (8.8) | 126.6, CH | 7.76, dd (8.8) |
| 9 | 134.2, CH | 8.44, d (8.6) | 133.9, CH | 8.43, dd (8.8, 1.5) | 133.1, CH | 8.41, d (8.8) |
| 9a | 137.6, C | 137.6, C | 138.1, C | |||
| 10 | ||||||
| 10a | 144.9, C | 144.8, C | 144.9, C | |||
| 11 | 51.8, CH2 | 5.58, br s | 51.7, CH2 | 5.50, br s | 49.2, CH2 | 5.91, d (16.5) |
| 5.41, d (16.5) | ||||||
| 12 | 121.3, C | 124.4, C | 124.9, C | |||
| 13 | 134.4, C | 134.1, C | 135.2, C | |||
| 14 | 41.7, CH2 | 2.08, d (16.5) | 77.7, CH | 3.50, s | 46.2, CH2 | 2.08, d (16.5) |
| 1.88, d (16.5) | 1.88, d (16.5) | |||||
| 15 | 34.2, C | 32.9, C | 29.8, C | |||
| 16 | 30.9, CH2 | 1.34, m | 30.1, CH2 | 1.28, m | 45.5, CH2 | 1.56, m |
| 1.18, m | 1.27, m | |||||
| 17 | 22.9, CH2 | 1.46 br s | 23.3, CH2 | 1.45, br s | 65.1, CH | 3.38, m |
| 18 | 20.0, CH3 | 2.00, s | 17.0, CH3 | 2.09, s | 18.7, CH3 | 1.99, s |
| 19 | 70.6, CH2 | 3.23, d (12.5) | 24.3, CH3 | 0.86, s | 30.2, CH3 | 0.95, s |
| 3.18, d (12.5) | ||||||
| 20 | 22.1, CH3 | 0.80, s | 24.7, CH3 | 0.80, s | 25.3, CH3 | 0.75, s |
Marinocyanin E (5) was isolated as a deep blue solid that analyzed for the molecular formula C22H23BrN2O2 by HRESI-TOFMS and NMR methods, which indicated it was isomeric with 4. The 1H and 13C NMR spectra of marinocyanin E (5) displayed most of the key resonances seen in the spectra of 1 and 4. The most noticeable difference in the 1H NMR spectrum of 5 was the replacement of the methylene proton signal at C-14 by an oxymethine proton signal (Table 2). Furthermore, a new signal assignable to an oxymethine carbon appeared at δ 77.7. These data suggested 5 was the C-14 hydroxy analogue of 1, which was subsequently confirmed by complete 2D NMR analysis. As with compound 4 insufficient material was obtained to allow assignment of the configuration at C-14 and to record confident optical rotation data.
Marinocyanin F (6) was isolated also as a deep blue solid and its molecular formula assigned as C22H23BrN2O2 by analysis of HRESITOFMS and NMR data, which indicated that 6 was isomeric with 4 and 5. The 1H NMR spectrum of marinocyanin F (6) was slightly different to that of 4 and 5, with the only difference being the position of substitution of the hydroxy group in the cyclolavandulyl ring. The HMBC spectrum of 6 illustrated that protons 11a, 16a, and 16b showed correlations to C-17, which suggested that the hydroxy group was located at C-17 in 6 instead of C-14 or C-19 as in 4 and 5. Detailed analysis of UV, ESIMS, 1D and 2D NMR spectral data established the structure 6 for marinocyanin F (Table 2). Because of the small sample size available and the relative instability of the molecule we were unable to acquire confident optical rotation data or to pursue assigning the configuration at the C-17 chiral center.
Known compounds 7–9 were identified as 2-bromo-1-hydroxyphenazine,6,7 WS-9659A14 (lavanducyanin) and the C-2 chlorinated analog WS-9659B14 by comparison with their reported spectroscopic data. Spectroscopic data for 7 and 8 can be found in the Supplemental Information.
The marinocyanins showed varying degrees of in vitro antifungal and cancer cell cytotoxicities. Marinocyanin A (1) was the most potent antifungal agent showing an in vitro minimum inhibitory concentration (MIC) of 0.95 μM against amphotericin-resistant Candida albicans, whereas the other marinocyanins were far less active (Table 3). Marinocyanins A and B (1, 2) showed the most potent in vitro cytotoxicity against HCT-116 human colon carcinoma with IC50 values of 0.049 and 0.029 μM, respectively. A comparison of the structural differences between compounds 1–6 suggests that the potencies significantly diminish with modifications in the terpenoid ring system. In earlier testing, marinocyanins A and B, as well as 2-bromo-1-hydroxyphenazine (7) showed significant activities against NFκB, aromatase, and quinone reductase (QR).7,17
Table 3.
Bioactivities for compounds 1–8 against HCT-116 human colon carcinoma, the bacterium Staphylococcus aureus, and the pathogenic yeast Candida albicans.
| Compound | HCT-116, IC50 (μM) | Staphylococcus aureus MIC (μM) | Candida albicans MIC (μM) |
|---|---|---|---|
| 1 | 0.049 | 2.37 | 0.95 |
| 2 | 0.029 | 33.92 | 5.79 |
| 3 | 0.078 | 30.71 | 3.90 |
| 4 | 10.56 | 36.62 | 14.67 |
| 5 | 7.28 | 36.62 | 14.67 |
| 6 | 17.14 | 36.62 | 14.67 |
| 7 | 2.92 | 56.93 | 114.2 |
| 8 | 2.41 | 2.92 | 5.96 |
The MIC of vancomycin is 0.27 μM for S. aureus and the MIC of amphotericin B is 0.084 μM for C. albicans.
3. Conclusions
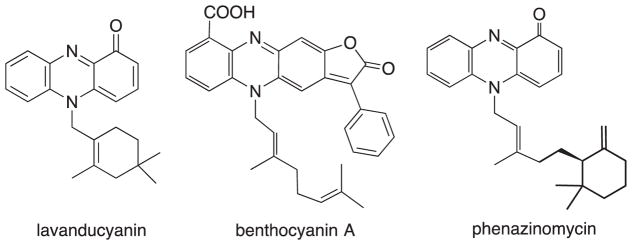
Naturally occurring phenazines from bacteria have been known since the middle of the nineteenth century. Phenazine derivatives have been isolated from terrestrial Pseudomonas spp.8 a Streptomyces sp.9 as well as a marine Streptomyces sp.,10 typically as bacterial pigments. More than 100 natural phenazines are known and most of them have shown antimicrobial, antitumor, antiparasitic, antimalarial, and enzyme inhibiting activities.11 The biosynthesis of typical phenazines seems to involve the shikimic acid pathway or the oxidative dimerization of anthranilic acid with the loss of the carboxyl carbon where appropriate.12 N-substituted phenazinones are known as a minor group among the phenazine pigments, the simplest analog being N-methyl pyocyanin.13 The marinocyanins are structurally closely related to known groups of metabolites, including lavanducyanin (WS-9655A), and 2-chlorolavanducyanin (WS-9659B),14 and members of the benthocyanin15 and phenazinomycin16 families. All of these similar compounds are N-isoprenoids with oxidation of one of the phenazine aromatic rings (See Fig. 4).
Fig. 4.
Known meroterpenoids related to the marinocyanins.
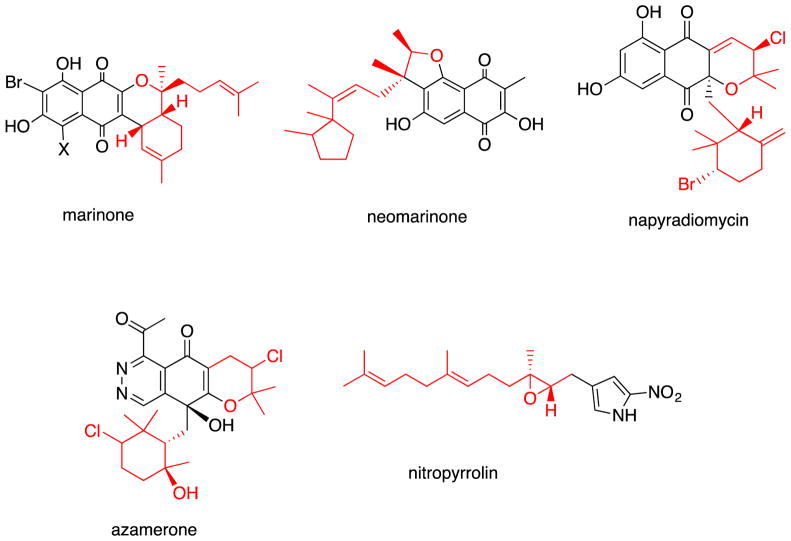
Finally, the marinocyanins were isolated from the fermentation broths of marine actinomycetes belonging to a new streptomycete taxonomic group, which we have designated as MAR4. We have encountered more than 12 discrete MAR4 strains, chemical analysis of which has shown the production of other meroterpenoids of the marinone, neomarinone, napyradiomycin, azamerone and nitropyrrolin classes (Fig. 5).
Fig. 5.
Select examples of MAR4 metabolites with the terpenoid components highlighted in red.
4. Experimental section
4.1. Isolation, identification, and cultivation of strains CNS-284 and CNY-960
Strain CNS-284 was isolated from a sediment sample collected at a depth of 34 m in April 2004 in Palau. The strain was identified as a member of the MAR4 clade within the Streptomycetaceae based on 16S rDNA sequence analysis (GenBank Accession # KC261628). Strain CNY-960 was isolated from a sediment sample collected in the Solomon Islands on 16 March 2013, and also identified as a member of the MAR4 group by 16S rDNA sequence analysis (GenBank Accession # KC261628.1). The two strains share 98.7% 16S sequence identity suggesting they could be different species within the MAR4 lineage. Producing strain CNS-284 was cultured in 25 2.8 L Fernbach flasks containing 1L A1BFe culture medium (10 g of starch, 4 g of yeast extract, 2 g of peptone, 5 mL of Fe2(SO4)3·4H2O at 8 g/L, 5 mL of KBr at 20 g/L, 1 L seawater) for 7 days at 25–27 °C with shaking at 230 rpm. The fermentation broth was then extracted by addition of 20 g/L of XAD-7 resin. The flasks were slowly shaken for 1 h after which the resin was collected by filtration and extracted with acetone. The acetone extract was dried and taken up in ethyl acetate to generate the organic extract. Similarly, strain CNY-960 was cultured in 30 replicate 2.8 L Fernbach flasks each containing 1 L of fermentation medium A1 (10 g of starch, 2 g of peptone, 4 g of yeast extract, 900 mL of seawater and 100 mL of distilled water) while shaking at 170 rpm at 27 °C for 7–8 days. At the end of the fermentation period, the aqueous layer of the fermentation broth was extracted three times with ethyl acetate. The ethyl acetate extracts were combined, and dried in vacuo to yield 3.0 g of organic extract.
4.2. Isolation of the marinocyanins
The culture extract from strain CNS-284 (20 g) from resin extraction was subjected to silica normal-phase flash chromatography generating 8 fractions using a gradient solvent system consisting of isooctane, ethyl acetate and methanol (100% isooctane, 20% ethyl acetate/isooctane, 40% ethyl acetate/isooctane, 60% ethyl acetate/isooctane, 80% ethyl acetate/isooctane, 100% ethyl acetate, 10% methanol/ethyl acetate, 20% methanol/ethyl acetate). The active fractions 6 and 7 were purified by RP-HPLC [Prep Nova-Pak HR C18, 6 μm, 60 Å, 300 mm × 40 mm, flow rate 10 mL/min, detection at 254 nm, using 40% CH3CN/H2O for 10 min then a linear gradient for 15 min to 65% CH3CN/H2O and then a linear gradient to 100% CH3CN over 35 min] to afford three active sub-fractions 6B, 6D, 7C. These sub-fractions were then separately purified by RP HPLC (Phenomenex Luna C8 (2), 10 mm × 250 mm, 5 μm) with a gradient solvent system (0–40 min; 30–65% aqueous CH3CN, 40–50 min; 65–100% aqueous CH3CN) at 2.5 mL/min flow rate and UV detection of 254 nm. Fraction 6A afforded marinocyanin B (2, 20 mg), D (4, 1.5 mg), and E (5, 1.3 mg). Fraction 6D gave marinocyanin A (1, 15 mg), C (3, 1.7 mg), F (6, 1.2 mg), 2-bromo-1-hydroxyphenzine (7, 3.2 mg), and WS-9659 B (9, 0.8 mg). Fraction 7C afforded marinocyanin A (1, 30 mg), B (2, 10 mg), andWS-9659 A (8, 22 mg). The crude extract (3.0 g) from strain CNY-960 was fractionated by vacuum liquid chromatography (VLC) on silica gel, eluting with a step gradient of hexane, ethyl acetate and methanol solvent mixtures [increasing the ethyl acetate (10%–100%) in hexane followed by 20% methanol in ethyl acetate. The ethyl acetate/hexane [7:3] fraction (330 mg) was further fractionated using VLC on silica gel eluting with increasing amounts of ethyl acetate (20%–100%) in hexane, subsequently increasing to methanol (20%–30%) in ethyl acetate. Fractions 2–5 were combined and subjected to further purification by Sephadex LH-20 open column chromatography eluting with CH3OH/CH2Cl2 (1:1) to afford a cytotoxic dark blue powder (50.3 mg). Purification of this fraction was achieved by RP-HPLC (Grace Alltima C18 10 μm, 250 × 4.6 mm, 10 mm × 250 mm, 2 mL/min, refractive index detection) with CH3CN/H2O (3:1) to yield (21.9 mg) of marinocyanin A (1). LC-MS comparisons of the metabolites observed from strain CNY-960 were almost identical to those from CNS-284.
Marinocyanin A (1)
(45 mg), blue colored crystal (mp 160 °C); UV (CH3OH) λmax (log ε) 241 (3.43), 332 (4.45), 720 (2.46) nm; IR (neat) νmax 2950, 2914, 1627, 1563, 1489, 1464, 1346, 1265, 1120, 970, 756 cm−1; 1H NMR (500 MHz, CD3OD) and 13C NMR (125 MHz, CD3OD), see Table 1; ESIMS m/z 411 [M + H]+, 433 [M+Na]+, 843 [2M+Na]+; HRESI-TOFMS m/z 411.1046 (calcd for , 411.1073).
Marinocyanin B (2)
(30 mg), blue colored powder; UV (CH3OH) λmax (log ε) 241 (3.24), 332 (4.12), 740 (2.16) nm; IR (neat) νmax 2950, 1607, 1562, 1463, 1405, 1319, 1260, 1125, 856, 752 cm−1; 1H NMR (500 MHz, CD3OD) and 13C NMR (125 MHz, CD3OD), see Table 2; HRESI-TOFMS m/z 343 [M+H]+, 365 [M+Na]+, 707 [2M+Na]+; HRESI-TOFMS m/z 343.0440 (calcd for , 343.0442).
Marinocyanin C (3)
(1.7 mg), blue colored solid; UV (CH3OH) λmax (log ε) 225 (3.46), 332 (4.48), 750 (2.09) nm; IR (neat) νmax 2923, 1711, 1604, 1563, 1461, 1319, 1266, 1230, 1143, 1075, 759 cm−1; 1H NMR (500 MHz, CD3OD) and 13C NMR (125 MHz, CD3OD), see Table 3; HRESI-TOFMS m/z 509 [M + H]+, 531 [M+Na]+, 1039 [2M+Na]+; HRESI-TOFMS m/z 509.1434 (calcd for , 509.1434).
Marinocyanin D (4)
(1.5 mg), blue colored solid; UV (CH3OH) λmax (log ε) 241 (3.12), 331 (4.06), 745 (2.39) nm; IR (neat) νmax 3374, 2917, 1605, 1559, 1461, 1318, 1263, 1140, 1035, 739 cm−1; 1H NMR (500 MHz, CD3OD) and 13C NMR (125 MHz, CD3OD), see Table 2; HRESI-TOFMS m/z 427 [M+H]+, 449 [M+Na]+, 875 [2M+Na]+; HRESI-TOFMS m/z 427.1012 (calcd for , 427.1016).
Marinocyanin E (5)
(1.3 mg), blue colored solid; UV (CH3OH) λmax (log ε) 241 (3.23), 331 (4.12), 745 (2.15) nm; IR (neat) νmax 3414, 2923, 1624, 1562, 1488, 1319, 1265, 1237, 1124, 1031, 737 cm−1; 1H NMR (500 MHz, CD3OD) and 13C NMR (125 MHz, CD3OD), see Table 2; HRESI-TOFMS m/z 427 [M+H]+, 449 [M+Na]+, 875 [2M+Na]+; HRESI-TOFMS m/z 427.1019 (calcd for , 427.1016).
Marinocyanin F (6)
(1.2 mg), blue colored solid; UV (CH3OH) λmax (log ε) 240 (3.16), 330 (4.25), 745 (1.95) nm; IR (neat) νmax 3385, 2952, 1621, 1560, 1463, 1319, 1265, 1168, 1144, 910, 760 cm−1; 1H NMR (500 MHz, CD3OD) and 13C NMR (125 MHz, CD3OD), see Table 2; HRESI-TOFMS m/z 427 [M+H]+, 449 [M+Na]+, 875 [2M+Na]+; HRESI-TOFMS m/z 427.1011 (calcd for , 427.1016).
4.3. Antimicrobial and cancer cell cytotoxicity assays
Antimicrobial bioassays were conducted with the bacterium Staphylococcus aureus and the amphotericin-resistant yeast Candida albicans using standard Microbroth Dilution methods.18 Standard cancer cell cytotoxicity bioassays were conducted using HCT-116 human colon carcinoma applying the common MTT-based methods well documented in the literature.19
4.4. Crystallographic data for marinocyanin A (1)
A crystal of 1 (0.15 × 0.04 × 0.02 mm) was orthorhombic, space group Pbca, with cell dimensions a = 13.325 (3), b = 9.994 (2), c = 28.522 (7) Å. Data were collected in a nitrogen gas stream at 100 (2) K using phi and omega scans. Crystal-to-detector distance was 60 mm and exposure time was 120 s per frame using a scan width of 1.0°. Data collection was 98.3% complete to 25.00° in θ. A total of 27140 reflections were collected covering the indices, −15 = h<=15, −11 = k<=11, −33 = l<=33. Of the total, 3296 reflections were found to be symmetry independent, with an Rint of 0.1217. Indexing and unit cell refinement indicated a primitive, orthorhombic lattice. The space group was found to be Pbca (No. 61). The data were integrated using the Bruker SAINT software program and scaled using the SADABS software program. Solution by direct methods (SIR-2004) produced a complete heavy-atom phasing model consistent with the proposed structure. All non-hydrogen atoms were refined anisotropically by full-matrix least-squares (SHELXL-97).20,21 All hydrogen atoms were placed using a riding model. Their positions were constrained relative to their parent atom using the appropriate HFIX command in SHELXL-97. Crystallographic data for marinocyanin A (1) have been deposited with the Cambridge Crystallographic Data Centre as supplementary Publication No. CCDC 1498854. Copies of the data can be obtained free of charge via www.ccdc.cam.ac.uk/retrieving.html (or from the CCDC, 12 Union Road, Cambridge CB2 1EZ, U.K.; fax: +44 1223 336033; email: deposit@ccdc.cam.ac.uk).
Supplementary Material
Acknowledgments
This research is the result of financial support from the National Institutes of Health, Fogerty Center, under the International Cooperative Biodiversity Groups program (U19TW007401). We thank the government of the Solomon Islands and the Tetepare Descendants Association for permission to collect samples in their coastal waters, and thank C. A. Kauffman and S. Kelly (SIO) for assistance with culturing and performing the antifungal and cytotoxicity bioassays.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.tet.2017.03.003.
References
- 1.(a) Fenical W, Jensen PR. Nat Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]; (b) Fiedler H-P, Bruntner C, Bull AT, Ward AC, Goodfellow M, Potterat O, Puder C, Mihm G. Antonie Leeuwenhoek. 2005;87:37–42. doi: 10.1007/s10482-004-6538-8. [DOI] [PubMed] [Google Scholar]; (c) Murphy BT, Jensen PR, Fenical W. In: Chap 3 in Handbook of Marine Natural Products. Fattorusso E, Gerwick WH, Taglialatela-Scafati O, editors. Springer Science, Business Media B.V; 2012. [Google Scholar]; (d) Blunt JW, Copp BR, Keyzers RA, Munro MH, Prinsep MR. Nat Prod Rep. 2015;28:116–211. doi: 10.1039/c4np00144c. (and earlier versions in this series) [DOI] [PubMed] [Google Scholar]
- 2.Jensen PR, Moore BS, Fenical W. Nat Prod Rep. 2015;32:738–751. doi: 10.1039/c4np00167b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallagher KA, Fenical W, Jensen P. Curr Opin Biotechnol. 2010;21:794–800. doi: 10.1016/j.copbio.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Gallagher KA, Jensen PR. BMC Genomics. 2015;16(960):1–13. doi: 10.1186/s12864-015-2110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallagher KA, Rauscher K, Ioca LP, Jensen PR. Appl Envir Microbiol. 2013;79:6894–6902. doi: 10.1128/AEM.01814-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrero NV, Bai F, Perez C, et al. Org Biomol Chem. 2014;12:881–886. doi: 10.1039/c3ob42416b. [DOI] [PubMed] [Google Scholar]
- 7.Conda-Sheridan M, Marler L, Park E-J, et al. J Med Chem. 2010;53:8688–8699. doi: 10.1021/jm1011066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomashow LS, Weller DM. J Bacteriol. 1988;170:3499. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mazzola M, Cook RJ, Thomashow LS, Weller DM, Pierson LS., III Appl Environ Microbiol. 1992;58:2616–2624. doi: 10.1128/aem.58.8.2616-2624.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Chatterjee S, Vijayakumar EKS, Franco CMM, Maurya R, Blumbach J, Ganguli B. J Antibiot. 1995;48:1353–1354. doi: 10.7164/antibiotics.48.1353. [DOI] [PubMed] [Google Scholar]; (b) Gebhardt K, Schimana J, Krastel P, et al. J Antibiot. 2002;55:794–800. doi: 10.7164/antibiotics.55.794. [DOI] [PubMed] [Google Scholar]
- 10.Pusecker K, Laatsch H, Helmke E, Weyland H. J Antibiot. 1997;50:479–483. doi: 10.7164/antibiotics.50.479. [DOI] [PubMed] [Google Scholar]
- 11.(a) Beifuss U, Tietze M. Top Curr Chem. 2005;244:77–113. [Google Scholar]; (b) Marinlit . A Database of the Marine Natural Products and Microbial Natural Products Literature. United Kingdom: Royal Society of Chemistry; 2016. [Google Scholar]
- 12.(a) Birkofer L, Birkofer A. Naturwissenschaften. 1949;36:92. [Google Scholar]; (b) Ge Y, Huang X, Wang S, Zhang X, Xu Y. FEMS microbio Lett. 2004;237:41–47. doi: 10.1016/j.femsle.2004.06.028. [DOI] [PubMed] [Google Scholar]; (c) Carter RE, Richards JH. J Am Chem Soc. 1961;83:495–496. [Google Scholar]
- 13.Friedeheim E. J Biol Chem. 1931;91:355–368. [Google Scholar]
- 14.(a) Nakayama O, Shigematsu N, Katayama A, et al. J Antibiot (Tokyo) 1989;42:1230–1234. doi: 10.7164/antibiotics.42.1230. [DOI] [PubMed] [Google Scholar]; (b) Imai S, Furihata K, Hayakawa Y, Noguchi T, Seto H. J Antibiot (Tokyo) 1989;42:1196–1198. doi: 10.7164/antibiotics.42.1196. [DOI] [PubMed] [Google Scholar]
- 15.(a) Shin-Ya K, Furihata K, Kato Y, Hayakawa Y, Clardy J, Seto H. Tetrahedron Lett. 1991;32:943–946. [Google Scholar]; (b) Shin-Ya K, Furihata K, Teshima Y, Hayakawa Y, Seto H. J Org Chem. 1993;58:4170–4172. [Google Scholar]
- 16.Funayama S, Eda S, Komiyama K, Omura S, Tokunaga T. Tetrahedron Lett. 1989;30:3151–3154. [Google Scholar]
- 17.Kondratyuk TP, Park E-J, Yu R, et al. Mar Drugs. 2012;10:451–464. doi: 10.3390/md10020451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zgoda JR, Porter J. Pharm Biol. 2001;39:221–225. [Google Scholar]
- 19.McCauley J, Zivanovic A, Skropeta D. Methods Mol Biol. 2013;1055:191–205. doi: 10.1007/978-1-62703-577-4_14. [DOI] [PubMed] [Google Scholar]
- 20.Sheldrick GM. Acta Crystallogr Sect A. 1990;46:467–473. [Google Scholar]
- 21.Sheldrick GM. SHELXL-97. Germany: University of Göttingen; 1997. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.