Abstract
Oocyte differentiation is a highly dynamic and intricate developmental process whose mechanistic understanding advances female reproduction, fertility, and ovarian cancer biology. Despite the many attributes of the zebrafish model, it has yet to be fully exploited for the investigation of early oocyte differentiation and ovarian development. This is partly because the properties of the adult zebrafish ovary make it technically challenging to access early stage oocytes. As a result, characterization of these stages has been lacking and tools for their analysis have been insufficient. To overcome these technical hurdles, we took advantage of the juvenile zebrafish ovary, where early stage oocytes can readily be found in high numbers and progress in a predictable manner. We characterized the earliest stages of oocyte differentiation and ovarian development and defined accurate staging criteria. We further developed protocols for quantitative microscopy, live time-lapse imaging, ovarian culture, and isolation of stage-specific oocytes for biochemical analysis. These methods have recently provided us with an unprecedented view of early oogenesis, allowing us to study formation of the Balbiani body, a universal oocyte granule that is associated with oocyte survival in mice and required for oocyte and egg polarity in fish and frogs. Despite its tremendous developmental significance, the Bb has been little investigated and how it forms was unknown in any species for over two centuries. We were able to trace Balbiani body formation and oocyte symmetry breaking to the onset of meiosis. Through this investigation we revealed novel cytoskeletal structures in oocytes and the contribution of specialized cellular organization to differentiation. Overall, the juvenile zebrafish ovary arises as an exciting model for studies of cell and developmental biology. We review these and other recent advances in vertebrate oogenesis in an accompanying manuscript in this issue of Developmental Biology. Here, we describe the protocols for ovarian investigation that we developed in the zebrafish, including all experimental steps that will easily allow others to reproduce such analysis. This juvenile ovary toolbox also contributes to establishing the zebrafish as a model for post-larval developmental stages.
Keywords: Oocyte polarity, oogenesis, ovary development, zebrafish, Balbiani body, animal-vegetal axis, chromosomal bouquet, meiosis, symmetry-breaking, centrosome, quantitative image analysis, live time-lapse imaging, ovarian culture, immune-fluorescence, in situ hybridization, oocyte isolation
Introduction
The investigation of oogenesis is important to germ cell biology and reproduction, but also more generally as a model for organogenesis. While early oogenesis has been described extensively in Drosophila, its mode of oogenesis and ovarian development is not representative of vertebrates, although some aspects are conserved. In the mouse, the general progression of oogenesis has been characterized and a number of mutant genes studied that disrupt oogenesis [1–3]. The zebrafish has yet to be fully exploited for studies of oogenesis, in part due to a lack of molecular and morphological characterization and insufficient tools of analysis. As a consequence, later oogenesis stages have been better described than early ones, and information on the earliest steps in oocyte development, as well as their ovarian context have been lacking.
A three-dimensional (3D) high resolution cellular view is key to the morphological analysis of developmental processes and to analyze comprehensively cellular features of differentiation. The adult zebrafish ovary provides challenges to such analysis in early stage oocytes, which are very small. The adult zebrafish ovary is thick and lobular in morphology and contains a non-synchronous mix of oocytes of multiple stages. Many are late stage oocytes, which are very large (200–730 µm in diameter) and opaque. The earliest stage oocytes are very small (8–30 µm) and transparent. This presents several technical hurdles in analyzing early stage oocytes. First, the small and transparent early oocytes are difficult to identify among the larger opaque ones. Secondly, the thickness of the adult ovary makes it challenging to obtain uniform penetration of antibodies and vital dyes, which is required for high resolution quantitative imaging. Thirdly, the thick ovary also reduces the quality of image acquisition by confocal microscopy. Thus, early oogenesis in the zebrafish has been less accessible to whole mount advanced microscopy and predominantly addressed by analysis on tissue sections (i.e., [4–6]), which lack 3D information. Due to these limitations, the descriptive and mechanistic understanding of oocyte development during early oogenesis has been lacking.
To circumvent these issues, we and others have used the juvenile ovary as a model for early oogenesis. In contrast to the adult ovary, the juvenile ovary is flat and elongated. It contains only young developing oocytes that are transparent. The oocytes in a given juvenile ovary are generally in similar stages that progress weekly. This provides excellent conditions for staining and confocal imaging techniques, where oocytes of the desired stages are readily found in high numbers. We took advantage of these properties of the juvenile ovary and established methods for quantitative microscopy and live time-lapse imaging, providing an excellent view of oogenesis processes. We have defined accurate staging criteria for early oocytes. We have also developed a short-term ovarian culture protocol, expanding the experimental repertoire for the use of pharmacological drugs that complement the genetic tools available in the zebrafish. Finally, we developed a method for isolating stage-specific oocytes for biochemical analysis. This toolbox can in turn be applied to genetic loss-of-function studies of oogenesis. Thus, the juvenile ovary brings great advantages to the zebrafish as a model for studies of early vertebrate oogenesis.
We characterized the developmental progression in juvenile ovaries between 4–10 weeks post-fertilization (wpf). Differences in environmental conditions between fish facilities, such as feeding regimen, lead to differences in juvenile fish growth rates. In addition, growth can vary between individuals raised in the same tank, under consistent conditions. A more accurate standardized measurement of post-embryonic development was determined as the standard length (SL; the length from snout to the base of the tail in mm, excluding the tail fin) [7]. We monitored SL of fish raised in our lab between 4–10 wpf. At 4 wpf 72% of fish (n=32) had SL=8–11 mm, and 28% SL=6–8 mm. At 5–6 wpf 92% of fish (n=143) had SL=10–14 mm and 8% were over 15 mm. At 7–8 wpf 93% (n=96) had SL=15–21 mm and 7% were under 15 mm. We found that ovaries of fish SL=8–10 mm contain mostly germline stem cells, oogonia and oocytes at the onset of meiosis. Ovaries of fish SL=10–20 mm additionally contained young stage I oocytes at early diplotene that are ≤60 µm in diameter. These findings are consistent with previous reports [8]. Therefore, ovaries of a specific SL range can be selected to analyze oocyte stages of interest.
To determine if the early oogenesis stages in the juvenile ovary reflect oogenesis in the adult, we analyzed early oocyte stages in the adult ovary. We dissected adult ovaries manually removing all large opaque oocytes (stage III and IV, vitellogenic stages) and analyzed ovarian tissue pieces that contained younger oocytes (stage I and II). We confirmed that the key morphological features of early oogenesis, such as oocyte stages and sizes, nuclear shape, chromosomal condensation, nucleoli number and position, Balbiani body formation and general cellular organization (all described below) are consistent between juvenile and adult oogenesis.
In addition to the advantageous physical properties, juvenile ovaries offer more powerful statistical analysis. In zebrafish, the immature gonad of all fish initially develops as an ovary. This gonad has bi-potentiality and later either further develops as an ovary or transitions to a testis. The process of sex determination defines the fate of the gonad to either continue to develop as an ovary or transition to a testis [8–10]. Sex determination in laboratory strains depends on environmental cues and is not completely understood, but the transition mechanism to testis is partly known [9–11]. In this process, apoptosis causes oocyte loss. While the signal is unknown, oocytes likely signal to the somatic follicle cells to express cyp19a1a, the Aromatase enzyme that converts testosterone to estrogen, and to down regulate the expression of antimullerian hormone (amh) [9,10]. Oocyte loss causes down regulation of cyp19a1a expression and induces expression of amh [9,10]. Amh then induces spermatogenesis and the gonad develops as a testis [9,10]. The initial signal for oocyte apoptosis is unknown. The use of ovaries prior to sex determination takes advantage of the fact that all individuals initially develop as females, making ovaries 2-fold more accessible and allowing for larger sample sizes in experiments.
It is important to avoid juvenile ovaries that are transitioning to testes, as they contain apoptotic oocytes that do not reflect normal oogenesis progression. We found that such transitioning ovaries are rare (~10%, n>33) in fish of SL<15 mm and gradually more frequent in fish of SL=15–21 mm, consistent with previous reports [8]. Therefore, we use ovaries of fish with SL<15 mm. Ovaries transitioning to testes are morphologically distinct [9,10], and are easy to identify during dissection, as they are more similar to testis in morphology, being thinner, less transparent and with fewer clearly visible oocytes. In addition, gonads transitioning to testes appear in confocal microscopy analysis to have very few sparsely isolated oocytes, and contain more loose tissue. Degenerating apoptotic nuclei are also evident and easily detected by DAPI staining as amorphous speckles that are smaller and brighter than normal nuclei. Such gonads should be discarded from experiments.
It is possible that sex determination processes begin in gonads of fish of SL<15 mm, before showing signs of apoptosis or transition. However, in examining hundreds of ovaries, we never observed different types or categories of ovaries (aside from the transitioning ones described above), nor have others reported them [8–10]. Additionally, our observations for early oocyte stages are consistent between juvenile and adult ovaries, indicating that they reflect normal oogenesis, as also reported by others [8]. Therefore, it is most likely that if these processes take place, they do not affect early oogenesis prior to transitioning to testes.
Altogether, the zebrafish juvenile ovary arises as an exciting genetic model for the study of cell and developmental biology of the ovary, germ cell development, reproduction, and organogenesis. In addition, while the zebrafish is vastly studied as a model for embryonic and larval development, zebrafish post-larval development is addressed to a much lesser extent. Our work joins that of others in contributing to establish a comprehensive juvenile zebrafish model. In the accompanying manuscript in this issue of Developmental Biology (ref), we review recent advances in the field, and discuss the zebrafish contributions that were allowed by the protocols described here. Here, we include accurate oocyte staging criteria, and all experimental steps from collecting ovaries to various imaging techniques and isolation of oocytes for biochemical analysis.
Methods
Oocyte staging criteria
Oogenesis in zebrafish has been divided into four principle stages based on ultrastructural analysis [12]. Stage I (oocyte diameter of 7–140 µm) spans the earliest stages of oocyte development from the onset of meiosis through an elongated and arrested meiotic I prophase, in which the oocyte dramatically increases in size. During stage II (oocyte diameter of 140–340 µm), the vitelline envelope begins to form and cortical granules are generated that exocytose following egg activation in the cortical reaction that modifies the vitelline envelope (i.e. the chorion). Oocytes of stage III (diameter 340–690 µm) are referred to as the vitellogenic stage, when vitellogenin (yolk) protein accumulates. Oocytes then resume and complete meiosis I during stage IV (diameter 690–730 µm), referred to as oocyte maturation, and once ovulated become a fully mature egg (diameter of 730–750 µm). All stages from I to IV are typically found in the adult ovary.
Stage I of oogenesis spans many key events of meiosis and oocyte differentiation, such as chromosomal pairing and recombination, oocyte polarization and Balbiani body formation, and changes in cellular organization from cyst to follicles of individual oocytes. Stage I was previously divided into stages Ia and Ib, based on the folliculogenesis state of the oocyte; individual oocytes with follicle cells around them are referred to as stage Ib, while all preceding stages are referred to as stage Ia [12]. Several key events of meiosis occur during stage Ia, which were previously characterized further using morphological criteria [13]. A more definitive marker for these meiotic sub-stages is telomere distribution dynamics. At the onset of meiosis (leptotene stage) telomeres are loaded on the nuclear envelope (NE) and their rotation on the NE facilitates chromosomal homology searches for pairing. Telomeres then cluster to one pole of the NE (zygotene stage, bouquet formation), eventually stabilizing pairing. Following the bouquet, homologous recombination occurs (pachytene stage) and telomeres disperse radially on the NE and unload back into the nucleus (diplotene). We expanded on the previous morphological criteria, utilizing telomere distribution dynamics as a molecular marker for these stages in whole mount ovaries [14]. We established accurate staging criteria to distinguish between key events during stage I of oogenesis using telomere dynamics, nuclear morphological criteria, and oocyte size, specifically identifying meiotic progression steps.
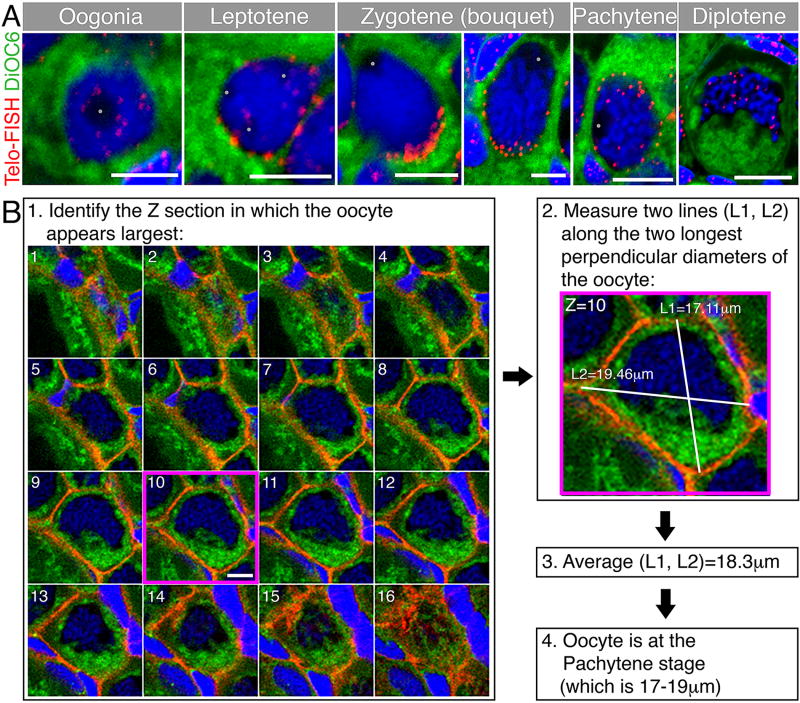
We summarize the staging criteria in Table 1, and provide images of representative oocytes demonstrating some of these criteria for each stage (Fig. 1A). Analysis of oocytes by whole mount allows for more accurate oocyte size measurements. Since oocytes are not usually perfect spheres in the ovary, it is difficult to measure and calculate their absolute size. As an indication for oocyte size we defined an approximate oocyte diameter. This diameter is generated by measuring two perpendicular lines (or close to perpendicular) across the widest diameters of the oocyte using the Z section with the largest dimensions. The average of these two lines is the approximate diameter of the oocyte (Fig. 1B), which we found allows for consistent staging of oocytes, across different ovaries. We characterized the organization of oocytes in germline cysts and nests, and their individual folliculogenesis, each of which coincides with inherent meiotic stages. We therefore propose that stage Ia and Ib nomenclature be refined further to specify the meiotic stage [13], i.e., st.Ia-Leptotene, st.Ia-Zygotene, and so on.
Table 1. Criteria for accurate staging of early meiotic oocytes.
Characteristics are details of specific oocyte stages, including size, morphological features, molecular markers and cellular organization, defining them as criteria for correct oocyte staging. An updated nomenclature for these stages is provided. The features described are depicted schematically in Fig. 6A.
| Stage | Nomenclature | Mitosis or meiotic function |
Size (diameter in µm) |
DNA condensation |
Telomere distribution |
Nucleolus | Nuclear shape |
Centrosome | Cellular organization |
|---|---|---|---|---|---|---|---|---|---|
| Oogonia | Oog | mitotic | 9–11 | Not condensed | Random intranuclear | 1–3 in the center of the nucleus | Round | Perinuclear | Cyst |
| Leptotene | St.IA-Leptotene | Double strand breaks formation and beginning of synapse | 8–9 | Does not appear condensed | Radial on nuclear envelope | 1–3 in the center or periphery | Round | Perinuclear | Nest (likely a cyst) |
| Early Zygotene | St.IA-Early Zygotene | Synapse | 10–12 | Does not appear condensed | Tightly clustered in one pole of the nuclear envelope | 1 in the periphery of the nucleus | Round | Perinuclear, apposing telomere cluster | Nest (likely a cyst) |
| Late Zygotene | St.IA-Late Zygotene | Synapse | 13–16 | Condensed into chromosomes | Clustered in one pole of the nuclear envelope, but more loose | 1 in the periphery of the nucleus | Round | Perinuclear, apposing telomere cluster | Nest (likely a cyst) |
| Pachytene | St.IB-Pachytene | Double strand breaks repair and Homologous recombination | 17–19 | Condensed into chromosomes | Radial on nuclear envelope | 1 in the periphery of the nucleus | Nuclear cleft | In the nuclear cleft cytoplasm | Individual (starting to accumulate somatic follicle cells) |
| Diplotene | St.IB-Diplotene | Chiasmata persist, meiosis arrested in dyctate | 20+ | Condensed – “lampbrush chromosomes” starting at diplotene | Random intranuclear | Multiple in the periphery of the nucleus (“perinucleolar stage” starting at diplotene) | Nuclear cleft (most prominent at 20–25µm and gradually receding, complete sphere at ~45–50µm) | In the nuclear cleft cytoplasm, dissociate at >25µm | Individual (continuing folliculogenesis) |
Figure 1. Representative oocytes of early stages and a method to measure oocyte size for staging.
(A) Representative early oocytes demonstrating staging criteria from Table 1, including telomere distribution as a marker of meiosis progression. Telomeres (Telo-FISH, red) are distributed in the nucleus of the oogonia, then loaded radially on the NE of the leptotene stage. At the early zygotene stage (left zgotene panel), telomeres are tightly clustered on one side of the NE. This clustering is less tight in the later zygotene stage (right zygotene panel). Telomeres redistribute radially at the pachytene stage and start to unload from the NE. At the diplotene stage telomeres are intranuclear. The general cytoplasm is detected by the DiOC6 lipid dye (green), and chromosomes are stained with DAPI (blue). Note staging criteria, such as oocyte size, DNA condensation, nucleoli (grey dots) number, and nuclear morphology across stages. Scale bars are 5 µm for oogonia-zygotene, and 10 µm for pachytene and diplotene. Modified from [14]. (B) A method to measure oocyte size in three dimensions in whole mount ovaries. For convenience, the oocyte shown is stained for cytoplasmic membranes (β-catenin, red), DiOC6 (green) and DAPI (blue). (1) Along the Z axis, find the optical section in which the oocyte appears the largest (section #10, magenta frame). Scale bar (only shown on section #10) is 10 µm. (2) In this optical section, measure the length of two perpendicular, or close to perpendicular, lines along the longest diameters of the oocyte (L1, L2). (3) The average of L1 and L2 defines the approximate diameter that indicates the size of the measured oocyte. (4) Determine what oocyte stage in Table 1 corresponds to the size of the measured oocyte, and determine stage according to size and additional staging criteria in Table 1.
Visualization of DNA, mRNA and protein
Dissection, fixation and microsurgery of ovaries
The ovary is held in place by connective tissue that is stretched along the anterior-posterior axis. Upon removal of non-fixed juvenile ovaries, they curl up, losing their flat morphology that is ideal for imaging. To maintain the overall ovary morphology, ovaries are fixed within the fish; the body wall muscle of one lateral side is removed from the fish, followed by the head and tail. The ovaries are not removed, but fixed within the exposed trunk piece (Fig. 2–3). Fine dissection of the ovary is performed post-fixation (Fig. 2–6 to 2–10).
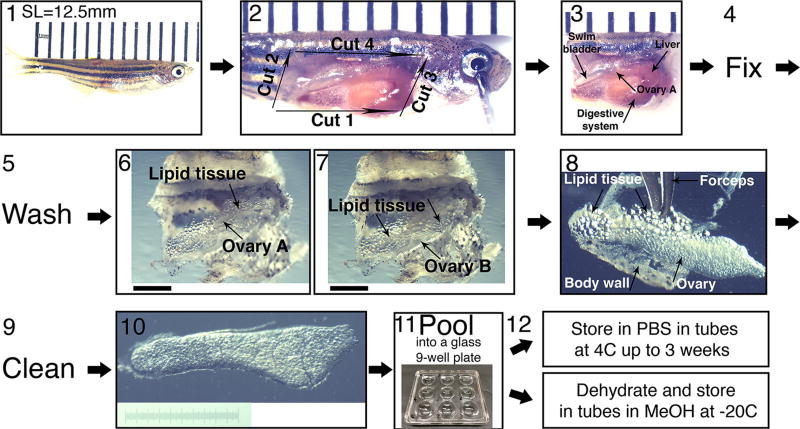
Figure 2. Collection of ovaries from juvenile fish and their fine dissection.
(1) SL measurement. A representative 5 wpf fish of SL=12.5 mm is shown. Fish is laid on a ruler; bars are 1 mm apart. The image is stitched from two smaller frames. (2) The four initial dissection cuts from step 4 in the protocol, exposing the fish internal organs. Bars as in (1). (3) The head and tail are removed and the entire trunk piece is fixed. If ovaries are removed from the fish prefixation they curl up. Fixing the entire trunk piece preserves the ovaries morphology. The ovary closer to the image plane (ovary A) and other organs are indicated with arrows. Bars as in (1). (4) The trunk piece is fixed overnight and (5), washed 2–3× in 1×PBS, and the ovaries are fine 26 dissected (6–10). (6–7) The swim bladder, liver and digestive system, are removed from the trunk piece, exposing the ovaries and surrounding lipid tissue. (6) is focused on ovary A and (7) on ovary B. Scale bars are 1 mm. (8–9) The ovaries are removed from the fish, and are cleaned from all lipid tissue and body wall remnants (arrows). (10) An example of a clean ovary. Scale bar is 1 mm (graded 10 µm). (11) Ovaries can be pooled in a glass well-plate and (12) stored in the appropriate conditions for the downstream application.
Protocol
Euthanize fish in ice-cold water.
Place the fish on a dissecting dish, and under a dissecting microscope measure its SL. SL=10–15 mm is ideal (Fig. 2-1). Smaller fish in this range have younger stage I oocytes, while larger fish in this range have progressively later stage I oocytes.
Pin the fish to the plate with one pin through the eye and another through the base of the tail.
- Under a dissecting microscope make the following cuts (Fig. 2-2):
- From the cloaca to the gills along the ventral midline.
- From the cloaca dorsally.
- From the anterior tip of cut #i dorsally.
- These cuts create a flap of body wall and its musculature. Raise this flap and cut it at its dorsal edge, along the anterior-posterior axis, to remove completely.
Cut off the head and tail of the fish and discard.
Transfer the trunk piece (Fig. 2–3) that includes the exposed visceral organs into cold 4% paraformaldehyde (PFA). One can place 15–20 trunks in one 15 ml conical tube of PFA.
Fix overnight (o/n; ~16 hours) at 4°C with nutation.
Pour the trunk pieces and PFA into a large plastic petrie dish on ice, remove the PFA with a transfer pipette and add 1×PBS.
Wash fixed tissue 2 times (2×) in 1×PBS.
Transfer the trunk to a glass petrie dish with fresh 1×PBS to finely dissect out the ovaries using forceps.
Under a dissecting microscope remove the liver, digestive system and swim bladder (Fig. 2–3 before, and Fig. 2–6 after removal).
Find the two ovaries and cut their connection points to the rest of the body at the anterior and posterior tips. Gently pull out the ovaries along the anterior-posterior axis, without directly touching the ovary, but holding the lipid tissue around it, lifting it away from the body. During the preparation of the trunk piece for fixation, one ovary (ovary A, in focus in Fig. 2–6) might adhere to the removed body wall and be lost. The other ovary (ovary B, in focus in Fig. 2–7) can still be collected.
Gently hold the ovary in place at the bottom of the dish.
Separate the two ovaries, cutting at their posterior tip, where they merge into the oviduct.
Gently clean the ovaries from the connective tissue and lipid cells that surround it. Hold the ovary from one tip, and pull the connective tissue and lipid cells away to the other side. Avoid directly touching the ovary when pulling, and hold through the lipid tissue (Fig. 2–8). The cleaner the ovary is (Fig. 2–10), the better the antibody penetration and imaging will be.
Pool all dissected ovaries into a glass 9-well plate with fresh ×1 PBS on ice. One can pool 10–16 ovaries per well depending on their size. Pooling all dissected ovaries per genotype helps later to divide them between experimental groups or storage conditions (PBS or MeOH) in more efficiently designing experiments based on the number of ovaries available. In addition, even in fish of similar SL there is some variability of ovarian size. Pooling ovaries can help to distribute ovaries more uniformly between experimental groups for more reproducible results.
Comments
Step 4: In removing the flap of body wall as described here, the ovary close to the body wall (ovary A in Fig. 2–6) remains attached to it ~10–20% of the time and thus may be removed with the body wall. If dissecting ovaries from a precious genotype where both ovaries are needed from all fish, skip cuts ii to iv and proceed to Step 5. Then dissect out both ovaries after fixation.
Keep PFA on ice while dissecting the fish. When all fish are ready, transfer to a nutator in a cold room.
Once the ovaries are dissected out of the fish body and until all lipid cells are removed, they will float and might be lost. Gently hold the ovaries and keep them at the bottom of the dish with one forcep while dissecting with the other. Remove some lipid cells before separating the two ovaries to prevent the one that is not being worked on from floating.
To transfer dissected ovaries, use a wide glass pasteur pipette. Burn the edge of the pipette to smoothen to prevent it from rupturing the ovaries. Forceps can also be used gently.
Materials
Dissecting dish for fine dissection of fixed ovaries (EMS #70540).
Forceps #5.
Micro-scissors (FST #15000-00).
-
PFA – only use 16% PFA ampules (Thermo Scientific #28309). Make fresh 4% PFA in 1× PBS every time.
***Except for staining of microtubules when microtubule stabilizing buffer (MSB) [15] is used, then use 37% Formaldehyde to make 4% Formaldehyde in 1× MSB. The MSB is acidic and not compatible with PFA.
-
10× PBS – must be pH 7.4.
*** all solutions are made with autoclaved sterile milliQ water.
Storage of dissected fixed ovaries
Storage conditions differ according to the subsequent application, and in an antibody specific manner. For mRNA in situ hybridization (ISH) or DNA ISH, always store ovaries in MeOH at −20° C. For fluorescent immunohistochemistry (IHC), ovaries are stored in either 1× PBS at 4°C in the dark, or in MeOH at −20°C. For IHC, each antibody should be tested in these different conditions. While MeOH improves penetration of antibodies, it might precipitate epitopes perturbing their detection and in our hands is not compatible with all antibodies. The following is a list of the conditions for several commercially available primary antibodies that worked in our hands (company; working concentration; post-fix ovarian storage): mAb414 (Fig. 3; Abcam; 1:1,000; either PBS or MeOH), LamB1 (Fig. 3B; Abcam; 1:400; optimal in PBS, but works in MeOH), γ Tubulin (Sigma-Aldrich; 1:400; either PBS or MeOH), Acetylated tubulin (Sigma- Aldrich; 1:200; PBS), β -Catenin (Sigma-Aldrich; 1:1,000; MeOH).
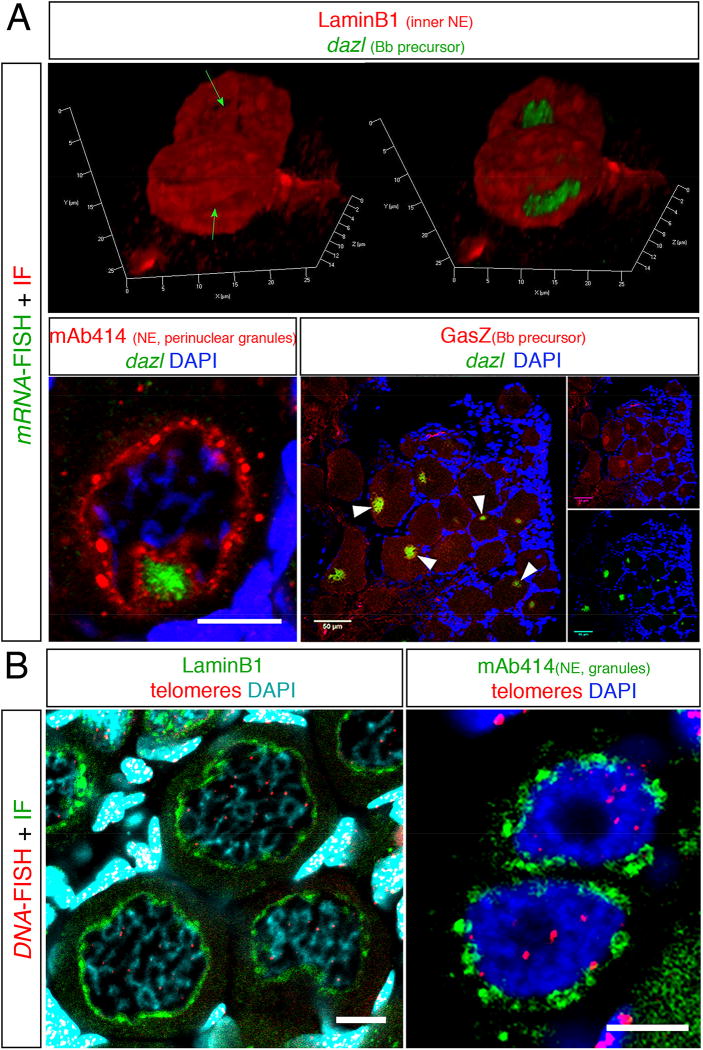
Figure 3. Examples of staining combinations of IHC and FISH protocols.
(A) The combination of dazl mRNA FISH with IHC of various proteins. (top) A snapshot form a 3D movie showing dazl in the nuclear cleft in two pachytene-early diplotene oocytes, as detected with the Lamin B1 protein. In the left image the dazl chanel is omitted exposing the nuclear cleft (green arrows). (bottom left) dazl localization in the nuclear cleft as detected with mAb414 in 30–35 µm oocytes. Scale bar=10 µm. (bottom right) Colocalization of dazl and the GasZ protein in the Bb (white arrowheads) of oocytes in whole ovary. Scale bar=50 µm. (B) The combination of DNA FISH with IHC of various proteins. (left) DNA FISH detecting telomeres by a probe for telomere repeats sequences (Telo-FISH) combined with IF for LaminB1 in a group of diplotene 30–40 µm oocytes. Note telomere detection at presumptive chromosomal edges, which at this stage should be still connected by chiasmata. Partial projection is shown. Scale bar=10 µm. (right) The distribution of mAb414 positive perinuclear granules in two oogonia where telomeres are found intranuclearly. Scale bar=5 µm. Panels A(left) and A(right) are modified from Elkouby et al, 2016 [14].
Protocol
PBS storage:
Transfer ovaries into a 0.5 ml safe-lock tube with fresh 1×PBS. Store in the dark at 4°C. Ovaries should be used within three weeks.
MeOH storage:
In the glass 9-well plate, replace approximately 2/3 of the PBS with fresh cold (keep on ice) 100% MeOH. The ovaries will appear white and will sink to the bottom of the well.
Wash twice for 1 minute with fresh cold 100% MeOH.
Transfer ovaries into a 0.5 ml safe-lock tube with fresh cold 100% MeOH.
Store at −20°C. Imaging quality remains very good after several months of storage.
Fluorescent immunohistochemistry staining (IHC)
Protocol
Before you begin:
If ovaries are stored in MeOH:
Rehydrate ovaries at room temperature (RT) for 3–5min each in:
75%MeOH: 25%PBS
50%MeOH: 50%PBS
25%MeOH: 75%PBT
100% PBT
100% PBT
If ovaries are stored in PBS:
Wash 2× 3–5minutes in PBT (to make sure removing all residual MeOH)
Wash 5× 20 minutes (min) at RT in PBT. Extensive washes are required to maximize antibody permeability and to achieve optimally uniform staining, which is key for quantitative imaging.
Block in Blocking Solution for 1.5–2 hr at RT.
Replace Block with primary antibody in blocking solution and incubate o/n at 4°C.
Wash 4× 20 min at RT in PBT.
Add secondary antibody (1:500) in fresh blocking solution and incubate at RT for 1 hr 45 min. From this point on protect from light, i.e. keep in dark.
Wash 4× 20 min at RT in PBT.
-
Nuclear counterstaining with DAPI:
DAPI solution (1:1000 of 300 µM stock in PBT) 50 min at RT.
Wash 2× 5 min PBT.
Wash 2× 5 min 1× PBS.
Comments
All staining steps are done with nutation.
Since juvenile ovaries are very small, thin and transparent, it is challenging to replace solutions without losing the ovaries. Always use 0.5 ml safe-lock tubes (actual volume is ~750 µl). To aspirate solution from the tube, use a syringe and a 27-gauge needle. Place the needle with its bevel side against the tube and aspirate gently and smoothly making sure not to aspirate the ovaries. After every step with nutation, allow the ovaries to sink to the bottom of the well/tube for a minute before aspirating. To add the new solution, gently add 680 µl. This volume allows for efficient nutation without the ovaries floating to the surface or the tube edge, risking their loss. Take extra care when solutions with no detergent are used (ovaries are more transparent and tend to sink more slowly).
Staining with the lipid dye DiOC6 (1:5000, Molecular Probes), which mostly detects mitochondria in early oocytes, can be done in the same solution with the DAPI staining.
All secondary antibodies were used at a working concentration of 1:500 dilution.
Materials
PBT (0.3% TritonX-100 (Fisher #BP151-100) in 1×PBS)
Block solution (0.3% TritonX-100, 10% Fetal Bovine Serum (FBS), 1×PBS). Make fresh every time.
DAPI dilactate (soluble in water; Molecular Probes, #D3571).
Safe-Lock tubes 0.5 ml (Eppendorf, #022363611).
PrecisionGlide Needles 27-gauge × 1.25 inch, 0.4 mm × 30 mm (BD #305136).
1 ml syringe.
mRNA fluorescent in situ hybridization (FISH)
Fluorescent ISH protocols provide high resolution intracellular signal that can be combined with IHC. We found the hybridization chain reaction (HCR) method (2nd generation DNA HCR; Molecular Instruments) [16] to work very successfully, although other methods such as the RNAScope [17] should work as well. The key for every method is a careful adjustment of the hybridization temperature used, in order to generate an ideal signal to noise ratio. Test at least three different temperatures in 5°C increments around the manufacturer’s recommendation. For the HCR method protocol, follow the manufacturer’s instructions, described in Choi et al., 2014 [16]. Optimal hybridization temperature for dazl mRNA was 50°C (Figs. 3A).
Comments
Use 100 µl of hybridization solution and hybridization buffer washes per tube.
In all formamide-based hybridization buffers, ovaries are even more transparent and tend to float. Do not nutate, and take extra caution when replacing solutions.
DNA fluorescent in situ hybridization (FISH) for telomeres, “Telo-FISH”
Protocol
- Rehydrate ovaries 3–5min each in:
- 75%MeOH: 25%PBS
- 50%MeOH: 50%PBS
- 25%MeOH: 75%PBT
- 100% PBT
- 100% PBT
Wash 4× 20 min in PBT.
Wash 3× 5 min in PBS Tween. ***If FISH is combined with the IHC protocol, skip step 2 and start at this step, and make sure to protect from light throughout.
5 min 50% PBS Tween: 50% hybridization buffer. No nutation.
5 min 25% PBS Tween: 75% hybridization buffer. No nutation.
Pre-hybridization: replace solution with 150 µl 100% RT hybridization buffer, and incubate in a water bath preheated to 80°C for 3.5 min. No nutation.
During that time, prepare probe solution. Dissolve probe to a final concentration of 88 nM in preheated (80°C) hybridization buffer. Probe stock solution is stored at −80°C, thaw a fresh aliquot immediately before use and directly add to the preheated hybridization buffer.
Hybridization: replace solution with 150 µl hybridization buffer w/probe, and incubate at 80° C for 10 min. No nutation.
Remove tubes from water bath and incubate for 2 hours at RT. No nutation.
Add 150 µl of 50% hybridization buffer: 50% 2×SSC. This will result in a 75% hybridization buffer. Incubate 5 min. No nutation.
Wash 5 min with 50% hybridization buffer: 50% 2×SSC. No nutation.
Wash 5 min with 25% hybridization buffer: 75% 2× SSC. No nutation.
Wash 2× 10 min 2×SSC.
Wash 3–5 min 50% 2×SSC: 50% PBS.
Wash 3–5 min 25% 2×SSC: 75% PBS.
Wash 2× 3–5 min PBT.
Counter stain with DAPI (+/− DiOC6) as in steps #9–11 in the IHC protocol.
Mount as described below in section “Mounting ovaries for confocal microscopy”.
Comments
This protocol is modified from the PNA-Bio company’s instructions.
All steps are done with nutation, unless specifically noted otherwise.
The hybridization buffer contains 70% formamide and is viscous. The ovaries become transparent and float in the tube. When adding hybridization buffer containing solutions, do not add the entire volume at once. Gently add a drop or two at a time on the surface of the solution in the tube. This may help the ovaries sink. Be extra careful not to aspirate ovaries when changing wash solutions. Leave a small volume of the replaced solution at the bottom of the tube. The extensive washes will wash effectively.
The high pre-hybridization and hybridization temperature is required to denature the genomic dsDNA to allow probe access.
The gradual transfer from SSC to PBS helps to preserve the ovary and oocyte morphology.
This protocol should work for DNA-FISH of potentially every genomic target. In this case, a short probe targeting repetitive telomeric sequences was used. For non-repetitive targets, more probes spanning longer sequences of the target might be required for a good signal. This would probably require adjustment of probe concentrations and hybridization conditions.
Materials
PBS Tween (0.1% Tween 20 in 1×PBS).
Hybridization buffer (70% Formamide, 1 mM Tris pH 7.2, 8.5% MgCl2 buffer, 1× Blocking solution, 0.1% Tween 20).
MgCl2 buffer (25 mM magnesium chloride, 9 mM citric acid, 82 mM sodium hydrogen phosphate, pH 7)
Blocking solution (Blocking reagent (Roche) in 100 mM Maleic acid, pH 7.5).
Telo-probe (5’-CCCTAACCCTAACCCTAA-3’, Cy3- conjugated; PNA-Bio).
2×SSC
Combinations of mRNA-FISH or DNA-FISH with IHC
Two conditions in the mRNA-FISH and DNA-FISH protocols, incubation at high temperature and the high formamide concentration in the hybridization buffer, can be harmful to antibody stainings. Formamide may destroy the epitope of interest, and high temperatures may denature the epitope-antibody complex. A successful way to combine the protocols is by performing the IHC first to prevent epitope loss by formamide (examples in Fig. 3; [18,19]). To avoid denaturing of the epitope-antibody complex, a quick fixation is performed after the IHC and before the FISH protocol to cross-link the epitope and antibody in the complex [18,19]. To maintain conditions appropriate for preserving mRNA, some solutions are modified in the IHC [18,19]. Even in combining the protocols in the following way, not all antibodies and epitopes are compatible with the mRNA/DNA-FISH protocol conditions. Always test your antibody in performing IHC alone to validate the signal before attempting to combine with the FISH protocol. MeOH storage post-fixation is also required for successful mRNA/DNA-FISH, so test your antibody to determine if it can successfully detect the epitope in these conditions.
Protocol
Begin after step #8 in the IHC protocol.
Wash 30 min in PBT.
Fix for 15–20 min in fresh 4% PFA at RT.
Wash 3× 5 min in 1×PBS.
Proceed immediately to mRNA/DNA-FISH protocol, or store o/n at 4°C in the dark.
Start mRNA/DNA-FISH protocol.
- For DAPI/DiOC6 staining after mRNA/DNA-FISH, gradually replace SSC based solutions with PBS:
- 5 min 50% SSC: 50% PBS
- 5 min 25% SSC: 75% PBT
- 2× 5 min 100% PBT.
Stain with DAPI/DiOC6 as in steps #9–11 in the IHC protocol.
Comments
If IHC precedes mRNA-FISH, make sure all tubes and tips are sterile.
If IHC precedes mRNA-FISH, add RNAasin (1:100, Sigma-Aldrich) to all blocking solutions, to protect against potential RNAases in the blocking serum.
SSC osmolarity is much higher than that of PBS. Changing directly from SSC to PBS might destroy cellular morphology. Replace SSC with PBS gradually as indicated.
Not all antibodies and epitopes are compatible with these conditions (hybridization at 45°C or 80°C and 50–70% formamide). Always test your antibody performing IHC alone to validate the signal before attempting to combine with FISH. MeOH storage post-fixation is also required for successful Telo-FISH, so test your antibody in these conditions.
Mounting ovaries for confocal microscopy
It is advantageous to mount ovaries between two cover slips, so that the ovary can be imaged from both sides if desired. Use a 24 × 40 mm cover slip as the “slide”, and an 18 × 18 mm cover slip as the “cover slip”. To prevent squishing the ovaries while keeping them tightly against the cover slip plane for ideal image acquisition, 100 µm Secure-seal spacers are used between the two cover slips (Fig. 4A). It is extremely important to use #1.5 cover slips. Incorrect glass thickness will distort the light path along the Z-axis causing spherical aberrations, resulting in artificially flattened or inflated cellular morphology. Confocal microscopes typically correct for the light passing through the glass of the cover slip, and are adjusted to a 0.15 mm glass.
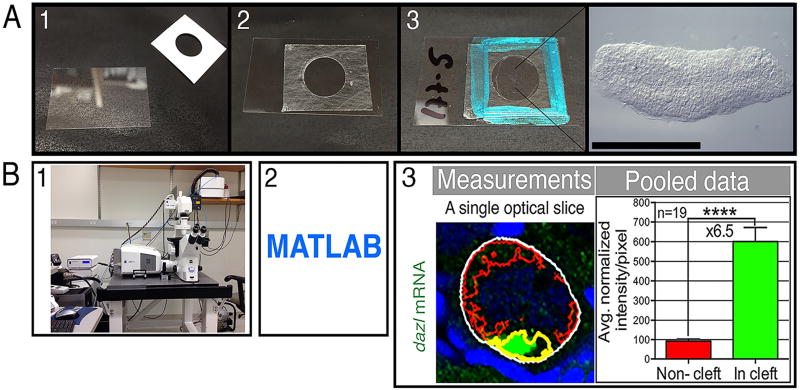
Figure 4. Mounting ovaries and quantitative imaging.
(A) Mounting set up. A microseal spacer is placed on a #1.5 24×40 mm cover slip (panels 1–2). After the ovaries are placed in vectashield with their flat side snuggly on the slip, cover with a #1.5 18×18 cover slip and seal with nail polish (panel 3). This setting allows one to image from both sides of the mounted ovaries. An example of a mounted ovary is shown (right image in panel 3), scale bar is 1 mm. (B) Quantitative imaging. After image acquisition (panel 1), custom programs in MATLAB (panel 2) are used for quantification. Panel 3 shows an example of such quantification of signal enrichment within the nuclear cleft. This example MATLAB code automatically identifies based on markers features of the oocyte, including the nucleus, the non-cleft cytoplasm (red outline) and the cleft cytoplasm (magenta outline) within the total oocyte volume (white outline) in all optical sections of the stack that spans the oocyte. It measures the intensities/area for each domain in each section. These data are then pooled for statistical analysis and results are plotted (pooled data). Images are modified from, and detailed protocol is described in Elkouby et al., 2016 [14]. The cleft cytoplasm outline here is artificially painted yellow for simplicity, the outline that is automatically generated by the program is in green and can be found in the original publication.
Protocol
At the end of IHC or FISH protocols, transfer ovaries to a glass well plate (Fig. 2–11).
Discard most of the PBS and add 70–100ul (total) Vectashield. Add drops from the edges of the PBS first to allow gradual mixing, and then on top of the solution to submerge ovaries.
Add a 7 ul drop of Vectashield to the slide.
Transfer ovaries to the slide with forceps.
Cover the ovaries with another 7 ul drop of Vectashield.
Use forceps to align ovaries so they lay with their lateral face flat on the slide.
Cover with a cover slip.
Seal with nail polish (Fig. 4-3).
Store in a slide folder at 4°C, protected from light.
Store o/n or longer before imaging. This will allow for complete equilibration of the ovaries in the vectashield that is necessary for imaging. Imaging sooner after mounting results in fuzzy images.
Comments
Depending on the size of the ovaries, at least three and up to seven ovaries can be mounted on a single slide.
Materials
Vectashield with DAPI (VectorLabs). To achieve good labeling of DNA, we found it necessary to use DAPI both in counterstaining and in the mounting medium. This is valuable since DNA morphology helps to properly stage young oocytes.
Coverslips 24×40 mm-1.5 Fisher #12-544-C.
Coverslips: 18×18 mm-1.5 Fisher #12-541-A.
Secure-seal spacers, one well, 13 mm diameter, 0.12 m deep (Molecular Probes #S24735)
Quantitative imaging
The protocols above provide high quality staining for protein, mRNA and DNA targets. The buffer detergent concentrations, incubation conditions and the extensive washes result in optimal and uniform penetration of detection reagents and excellent signal to noise ratio. Importantly, keeping the conditions consistent between samples and across experiments is key for quantitative imaging.
Once samples are processed uniformly, it is critical to ensure that image acquisition is uniform and consistent. The following are acquisition conditions we found optimal for juvenile ovaries.
Coverslips must be #1.5 to avoid spherical aberrations along the Z axis.
A 40× objective combines a large image field that samples many oocytes with high-resolution imaging of cellular features.
- Consistent pixel size. The XY resolution and zoom determines the pixel size of final images. For quantitative imaging, pixel size across images needs to be consistent. Set:
- XY=1104×1104 pixels
- Zoom=×0.8.
- 12-bit
- Consistent intensity and excitation. Set:
- pixel dwell time=0.59 seconds
- Sampling averaging=X2
- Laser power=7–11% depending on the antibody, but consistent per antibody.
- Gain=380–650 depending on the antibody, but consistent per antibody.
- Use the range indicator function to confirm below-saturation conditions.
-
Consistent Z resolution. Set:
Pinhole for every wavelength should result in the same Z thickness of 1.1 µm. The pinhole function uses the Airy unit, which differs between wavelength. The Airy unit should be set for each channel to provide an equal Z thickness for all channels.
Z increments for stacks were 0.53 µm and each Z section is 1.1µm thick; it should be approximately half the thickness.
Comments
We use the Zeiss LSM 710 confocal microscope. The precise settings above may differ between microscopes. However, they should be set to be consistent.
The Zeiss image acquisition software has a “reuse” function that allows for using the exact same acquisition conditions from an uploaded image file in scanning a new image. Once the settings for the acquisition of a certain staining signal have been determined, it is highly recommended to use the “reuse” function in all samples and experiments examining this signal.
Quantifying algorithms
The sample processing and image acquisition described above should result in high quality images, which are of consistent pixel number and size, as well as consistent light sampling that generates quantitatively comparable data between experimental conditions. This now can be used for quantitative analysis by various software programs and custom made MATLAB codes, designed for specific experimental questions. An example for such analysis is the “cleft analysis” used to measure intensities of proteins (IHC), mRNA (mRNA-FISH) and mitochondria (DiOC6 lipid dye) signals in the cytoplasm within the nuclear cleft versus the remaining cytoplasm in oocytes (Fig. 4B)[14].
Time-lapse Imaging of live oocytes/ovaries
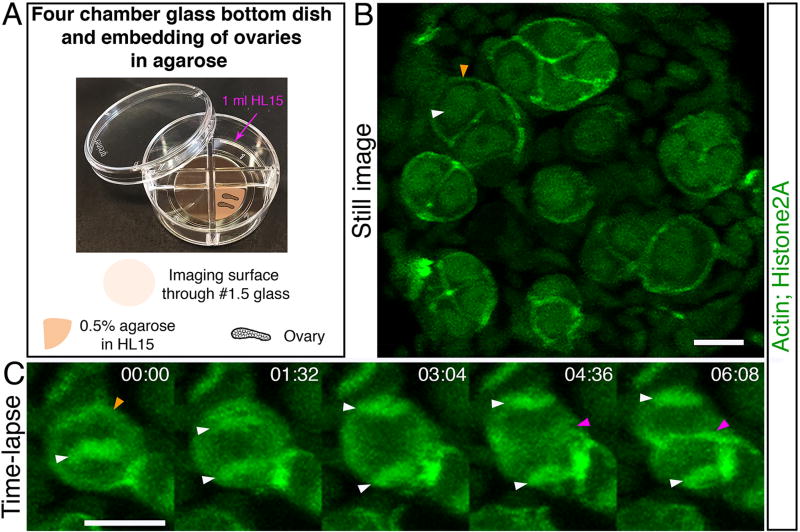
Live time-lapse imaging of oocytes is key for a high temporal resolution analysis and more complete understanding of dynamic biological processes. Many highly dynamic events are the basis of early oocyte differentiation and polarization, such as mitotic divisions of oogonia, chromosome and telomere dynamics of leptotene to pachytene meiotic oocytes, and the formations of the nuclear cleft during pachytene to early diplotene. However, in vertebrates, live time-lapse imaging of isolated mouse spermatocytes [20] and Xenopus isolated oocytes [21] have been reported, but such an analysis of young oocytes within the ovary has only recently been reported in Medaka [22], and has yet to be performed in frogs, zebrafish or mice. We have set up a protocol for isolation, culturing and live time-lapse imaging of the zebrafish juvenile ovary, using both fluorescently labeled transgenes and fluorescent vital dyes. Figure 5 shows an example of live time-lapse imaging of dividing mitotic oogonia progressing from metaphase to cytokinesis.
Figure 5. Live time-lapse imaging.
Ovaries of fish expressing Lifeact-GFP to label Actin and H2A-GFP to label chromatin via Histone2A [Tg (βact:Lifeact-GFP); Tg(h2afva:h2afva-GFP)] were collected and imaged live. (A). A four-chamber glass bottom dish used for mounting 27 cultured live ovaries for time-lapse imaging. (B) A still image from a time-lapse recording showing several 2 to 8-cell oogonial cysts. Image is a partial projection not showing all cells in each cyst. White arrowhead indicates chromatin detected by H2A-GFP labeling. Orange arrowhead indicates the cortex of the cell as detected by the Lifeact-GFP. Scale bar is 10 µm. (C) A dividing oogonial cell progressing from metaphase, to anaphase, and cytokinesis. Images are partial projections. Time is indicated in minutes (min). Scale bar is 10 µm. White arrowheads indicate the segregating chromosomes (H2A-GFP) from the metaphase plate and into individual daughter cells. The orange arrowhead indicates the cortex of the cell as detected by Lifeact-GFP at time 00:00 min. The magenta arrowhead indicates the appearance of a cleavage furrow in cytokinesis at time 04:36 min, and presumptive membrane between daughter cells at time 06:08 min Tg(βact:Lifeact-GFP) and Tg(h2afva:h2afva-GFP) were reported [33] [34], respectively.
Isolation and culture of the ovary
Protocol
-
1
Anesthetize fish in 0.02% Tricaine.
-
2
Place the fish on a dissecting dish, and measure its SL. SL=10–20mm is ideal. The smaller fish will have younger oocytes, with more progressively developing oocytes in larger fish.
-
3
Euthanize the fish by decapitation. Pin the fish to the plate through the base of the tail and hold its anterior tip down with a forceps.
Make the cuts as described above and in Fig 2-2:
-
5
Cut the head and tail of the fish and discard.
-
6
Transfer the trunk piece that includes the exposed visceral organs into a glass dish with Hank’s warmed to 28°C.
-
7
Remove the digestive system.
-
8
Find the two ovaries and cut their connection points to the rest of the body at the anterior and posterior tips. Gently pull out the ovaries along the anterior-posterior axis, without directly touching the ovary, but holding the lipid tissue around it, lifting it away from the body.
-
9
Gently hold the ovary in place at the bottom of the plate.
-
10
Separate the two ovaries, cutting at their posterior tip, where they merge into the oviduct.
-
11
Gently clean the ovaries from the connective tissue and lipid cells that surround it. Hold the ovary from one tip, and pull the connective tissue and lipid cells away to the other side. Avoid directly touching the ovary when pulling. The cleaner the ovary is, the better vital dyes penetrate and imaging will be.
-
12
Pool all dissected ovaries in a glass well plate with fresh HL-15 warmed to 28°C. Keeping the live ovaries at 28°C is critical. During the dissection of each ovary keep the plate at 28°C and only take out when the next ovary is ready. Immediately place back at 28°C with the newly dissected ovary.
-
13
If imaging a transgene, verify that the ovaries are positive for the signal using a fluorescent dissecting scope. This will not provide intracellular resolution, but will sort out negative or weak signal ovaries and spare their extra unnecessary processing.
-
14
Let ovaries rest for 30 min at 28°C before mounting, which is described below.
-
15
Ovaries can be cultured for several hours to o/n.
Comments
Do all steps from anesthetizing the fish to placing the clean ovary at 28°C as fast as possible.
The live non-fixed ovary is very soft and gentle. Take extra caution not to damage the tissue. It is not always possible to successfully obtain both ovaries from every fish.
Once the ovaries are dissected out and until all lipid cells are removed, they will float and might be lost. Gently hold the ovaries and keep them at the bottom of the dish with one forceps while dissecting with the other. Remove some lipid cells before separating the two ovaries to prevent the floatation of the one that is not being worked on.
Once the live ovaries are dissected out of the body, they curl up. Only gently straighten them for cleaning. They will be laid flat later when mounting.
Make fresh Hank’s and HL-15 for every experiment.
Materials
Tricaine (Sigma-Aldrich). Make 0.4% sterile stock.
Dissecting tools are as described for dissection of fixed ovaries.
Hank’s solution (0.137 M NaCl, 5.4 mM KCl, 0.25 mM Na2H PO4, 0.44 mM KH2 PO4, 1.3 mM CaCl2, 1.0 mM Mg SO4, 4.2 mM NaH CO3). Follow the Zebrafish Book for making and storage [23].
HL-15 solution (60% Hanks, 40% L-15, 1:100 GlutaMax).
2× L-15, without L-glutamine and Phenol Red (Lonza, 12-669E). Store at 4° C. L-glutamine is not stable and is added fresh from a stock (GlutaMax). Phenol Red is auto-florescent and will increase background in imaging.
-
GlutaMax 100× (Gibco #35050-061). Store at RT.
*** all solutions are made with autoclaved sterile milliQ water.
Mounting cultured live ovaries and staining with vital dyes
Protocol
Boil 1% low-melt agarose/Hank’s in a microwave and let cool down for 10 min on the bench.
Take 1 ml and place at 42°C until you are ready to mount, to prevent it from solidifying.
- Make 500 µl of:
- 190 µl Hank’s
- 300 µl 2× L-15
- 10 µl GlutaMax
- Keep at 28°C.
When ready to mount, add 500 µl of 1% low-melt agarose/Hank’s from step 2, to the 500 µl solution from step 3, to make final 0.5% low-melt agarose/HL-15. Mix well. At the average temperature of 35° C, the solution will remain liquid for a while.
Rinse a four-compartment glass bottom dish with Hank’s (Fig. 5A).
Gently spread evenly in each compartment of the glass bottom dish that will be used (Fig. 5A) 100 µl of the final mounting solution from step 4.
Let cool for 1 minute. It will make the solution a little viscous, which will later help to flatten the ovaries and keep them at the bottom tightly against the imaging glass.
Place one ovary into the mounting solution in the dish. Gently push it down to the bottom and use the outside surface of the forceps to flatten the curled up ovary. Ideally, an ovary should lay straight with its flat side tightly against the glass (Fig. 5A).
Repeat step 8 with more ovaries, placing 2–3 ovaries per compartment.
When the agarose solidifies, add 1 ml HL-15 per compartment. Gently pipette the solution against the wall of the compartment.
Place the dish at 28°C to rest for 30 min.
Make vital dye solution in warm (28°C) HL-15, protecting it from light.
Gently aspirate the HL-15 from the dish and replace with 1 ml vital dyes/HL-15.
Incubate for 2–3 hours at 28°C before imaging, protecting it from light.
If imaging a fluorescent protein transgene only, steps 12–14 are not required.
Comments
This protocol can be used for drug treatments of live ovaries, followed by live imaging or fixation and IHC.
The four-compartment dish allows for use of smaller volumes. Importantly, control and experimental groups can be contained in different compartments of the same dish, allowing for maximally equivalent conditions.
Use glass bottom dish of a #1.5 glass, for correct imaging.
Staining with vital dyes can alternatively be performed before mounting. Replace the HL-15 with HL-15 media that includes vital dyes in the glass well plate. Incubate 1 hour at 28° C. Mount as described in steps 1–9. When the agarose solidifies add HL-15 that includes the vital dye and place at 28° C, protected from light.
Materials
Hank’s solution.
HL-15 solution.
2× L-15.
GlutaMax.
Low-melt agarose (Lonza; gelling temperature: 26–30°C). Make a 1% stock in Hank’s, keep at RT.
Cellview - cell culture dish with glass bottom, 35mm, four compartments (Greinier Bio-One GmbH, #627871).
Hoechst H33342 (Sigma-Aldrich, #14533). Final solution 6.66 µM.
Mitotracker Red CMXRos (Molecular Probes, #M7512). Final solution 500 nM.
DiOC6 (Molecular Probes, #D273). Final solution 0.001 mg/ml.
Live imaging
The key for imaging of live ovaries is keeping the temperature at ~28°C and making sure the sample has reached equilibrium before starting to image. To keep temperature constant, it is necessary to use an imaging environment chamber and an objective heater. Even when using an environmental chamber, the objective remains at RT, which is usually cold in microscope rooms, and touches the samples. A cold objective acts as a robust heat sink and cools down the sample rapidly. An objective heater that can be set to specific temperatures solves this issue.
Protocol
Turn on the microsocope and set up a temperature-regulated imaging chamber. We use a chamber that is assembled on the microscope stage and has a bottom and lid. We set the temperature to 28°C for the bottom and 30°C for the lid.
Set up the temperature of the objective heater to 29°C.
Once the chamber and heater have reached the set temperatures, load your dish.
Use a 60× objective and find the sample and set the focus.
Incubate the dish for ~40 min. This will allow the sample to equilibrate to the chamber environment. A non-equilibrated sample results in a focal drift during imaging.
Start imaging.
- For efficient imaging:
- Penetration of vital dyes is not always maximal through the entire depth of the tissue, so look for an area of interest nearer to the surface of the ovary.
- Focus on the area of interest, use the smallest Z depth required to span the area of interest, and use shorter exposure times.
- Use the lowest power possible to avoid photo-bleaching and tissue damage.
- Movies can be recorded for several hours.
- For imaging of rapid processes typical time-lapse intervals are 15–30 seconds for 5–30 minutes in length. Multiple such movies made sequentially from different regions of a single ovary can be recorded for several hours.
Comments
We use the Olympus IX81 spinning disc confocal microscope. Settings may differ for other microscopes.
We have also been successful in performing live time-lapse imaging using the Zeiss LSM710 confocal microscope, and its environmental chamber set to bottom 29°C and top 30°C. Temperature fluctuations vary between different rooms and different environmental chambers vary in their ability to hold temperature, and in this case an objective heater was not necessary. We used a 40× water immersion objective (immersion water pre-warmed to 28°C), and imaging settings as described above for imaging of fixed samples, keeping the laser power as low as possible. Time-lapse intervals ranged between 30–100 sec.
Materials
Environmental imaging chamber.
Objective heater with control box.
Isolation of stage-specific oocytes for molecular or biochemical analysis
Since all stages of ovarian development contain multiple stages of oocytes, performing biochemical analysis on whole ovaries is not always informative. Stage-specific functions in oogenesis will be reflected in stage-specific gene expression or post-translational protein modifications, for example, which could be masked by their absence in a large cohort of oocyte stages. Furthermore, any one ovary can vary greatly in the proportion of oocytes at particular stages, so whole ovary analysis can lead to varying results between ovaries or even in groups of ovaries. We developed a protocol to isolate stage-specific oocytes from ovaries. In this protocol, oocytes are first isolated from the ovary by enzymatic digestion of connective tissue and extracellular matrix (ECM) components, and then separated by size using combinations of sieve sizes.
Protocols for isolation by digestion [24] and sieve separation [25] are successful in Xenopus, which use collagenases I only for digestion. However, the ECM of mouse and bovine follicles contains a variety of collagen types as well as hyaluronic acid [26,27], as likely do zebrafish ovaries. In addition, we tested digestion alone by collagenase I, collagenase II, or Hyaluronidase and found that each single treatment was inefficient, requiring long incubation times that eventually lyse the oocytes. We therefore optimized the use of a cocktail of all three enzymes, resulting in efficiently isolated intact oocytes.
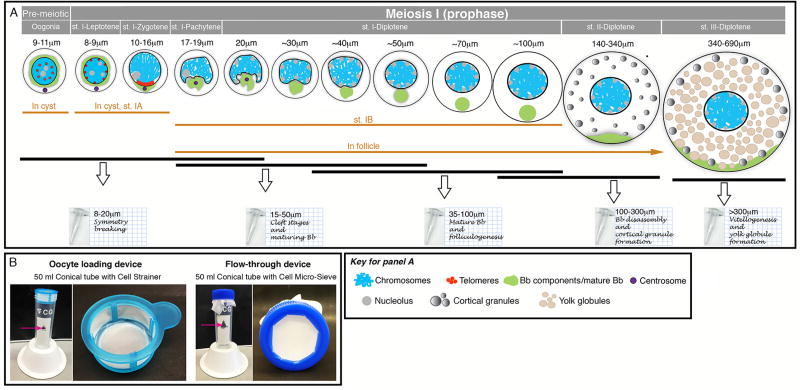
This protocol is successful in isolating all stages from oogonia to stage III oocytes, and is based on separating oocyte stages by size. Our physical separation resolution does not allow for the specific isolation of each distinct prophase sub-stage of stage I oocytes, but these can be pooled in groups of stages that are isolated together. In this way, we isolated oocytes as follows (Fig. 6):
Group 1. Size: 8–20 µm diameter; stages: Oogonia, st.I-Leptotene, st.I-Zygotene, st.IPachytene; key biological events: onset of meiosis, symmetry-breaking, chromosomal bouquet and synapsis, and recombination.
Group 2. Size: 15–50 µm diameter; stages: st.I-Pachytene, early st.I-Diplotene, mid-st.IDiplotene; key biological events: folliculogenesis of individual oocytes, nuclear cleft stages, maturing Bb, recombination and clearance of most DNA double strand breaks.
Group 3. Size: 35–100 µm diameter; stages: mid-st.I-Diplotene, late st.I-Diplotene; key biological events: folliculogenesis of individual oocytes and mature Bb formed.
Group 4. Size: 100–300 µm diameter; stages: late st.I and st.II, Bb disassembly, vegetal pole RNA and protein localization via Bb and late pathway [28], animal pole RNA localization, and cortical granule formation.
Group 5. Size: >300 µm diameter; stages: st.III, vitellogenesis and yolk globule formation. Oocytes over 300 µm include stages III and IV. Oocytes of stage IV undergo maturation during which the vitellogenin yolk proteins are cleaved and the oocyte becomes transparent [12,29]. Stage IV oocytes are found less frequently in dissected adult ovaries. Here we collected all oocytes >300 µm, and manually selected opaque oocytes to enrich for st. III oocytes. If stage IV oocytes are desired, more ovaries can be pooled, or females can be raised in tanks containing males (empirically found to have more st. IV) and the transparent oocytes selected. Alternatively, st. III oocytes can be matured in vitro using described protocols [30–32] to obtain st. IV oocytes.
Once oocytes of desired stages are collected, RNA, protein, or chromatin can be extracted for subsequent biochemical or molecular biology applications.
Figure 6. Isolation of stage specific oocytes.
(A) A depiction of the grouped stages of isolated oocytes. Schematics of oocytes stages from premeiotoc oogonia through st.III are shown (not to scale). Above each stage, the size and stages are indicated. The morphological criteria for each stage as described in Table 1 are depicted schematically here (see key) and with oocyte diameter size above each stage. The cellular organization for the stages, whether in cyst (Oogonia and St.IA), or in follicle (St. IB) is indicated below the cartoons. Black bars indicate the range of stages that are pooled in each group of isolated oocytes. The tubes at the bottom indicate the size range and biological significance of each group. (B) Loading and flow through devices for the isolation protocols. Two left panels: loading device assembled from a 50 ml conical tube and a commercial cell strainer. Two right panels: flow through device assembled from a 50 ml conical tube, a rimmed 50 ml conical tube’s cap, and a Cell Micro-Sieve. The same conical tube is used throughout the protocol, and the two devices alternate at different steps of the protocols. Magenta arrows indicate the wholes cut in the tubes that are used for adding solutions without disassembling the Cell Micro-Sieve.
Protocol
You will need a single 50 ml conical tube for the collection and washing of the desired oocyte stages, and two alternate sieve devices: an oocyte loading device and a flow-through device.
- Prepare in advance for steps 5–10:
- 50 ml conical tube: cut a hole in the side of the tube at approximately the 30 ml mark (Fig 6B). Cut out the center of a cap of a 50 ml conical tube, leaving a rim and set aside.
- Oocyte loading device: place a Cell Strainer in the 50 ml tube (Fig. 6B). If a Cell Micro-Sieve is used instead, place a square (cut with excess size) over the opening of the 50 ml tube, push in with your finger to make a small basket with minimal folds and close with the rimmed cap.
- Flow through device: Cut Cell Micro-Sieves of desired mesh size to a square to fit over a 50 ml conical tube opening. Place the Cell Micro-Sieve square tightly on the 50 ml tube and close with the rimmed cap (Fig. 6B). Make sure the Cell Micro-Sieve fits snugly and is tight and flat to avoid squashing the collected oocytes.
Pool two adult ovaries into a 2 ml tube with HL15 warmed to 28°C.
Replace HL-15 with 2 ml of digestive HL-15 (28°C).
Incubate up to 10 min at RT. Every 1–2 min invert the tube several times. Connective tissue is digested and individual oocytes are released. Monitor carefully and stop the digestion as soon as oocytes are separated and no (or minimal) clumps are apparent. Incubation with digestive enzymes for too long damages the oocytes.
Pour the 2 ml volume of the digestive HL-15 with the separated oocytes onto a loading device (step 1d) with a cell strainer of the upper size limit of the desired group. To make sure to collect all separated oocytes, fill the tube with fresh HL-15 and pour onto the loading device. All oocytes smaller than that size are collected in the tube. Either cell strainers or Cell Micro-Sieves can be used (cell strainers are ready for use, but come in limited mesh sizes. Sieves come in a wider mesh size range).
Remove the loading device and add fresh HL-15 to a total of ~20–25 ml in the 50 ml conical tube to dilute the digestive HL-15.
Install the flow-through device (from step 1c) with a Cell Micro-Sieve of the lower size limit of the desired group. Place the sieve material over the tube and screw on the rimmed cap to make the flow through device. Smaller oocytes will flow through.
Carefuly tip the tube so smaller size oocytes flow through the flow-through device to discard them and the digestive HL-15. Make sure the hole in the side of the 50 ml tube faces up while tipping.
Add fresh ~20–25ml HL-15 through the hole.
Repeat step #8 twice to wash 3× total. Do not exceed 3× washes, the more isolated oocytes touch the sieve the greater the chance for their lysis.
Add fresh HL-15. Incubate at 28°C for an hour to let the oocytes settle at the bottom of the tube.
- To transfer the oocytes into a 1.5 ml tube:
- For oocytes <100 µm only: centrifuge for 1–2 min at 800 rpm and then discard supernatant HL-15.
- For oocytes >100 µm only: step 11 is sufficient, do not centrifuge, directly discard supernatant HL-15.
- For either size, leave a little bit of the HL-15 in the tube. Gently resuspend the oocyte pellet in the HL-15 and transfer into a 1.5 ml tube.
Briefly spin down the 1.5 ml tube in a bench-top micro-centrifuge for 10–15 seconds and discard HL-15 supernatant, leaving just enough to thinly cover the oocyte pellet.
Snap-freeze in liquid nitrogen and store at −80°C.
Comments
Follicle cells are expected in samples of isolated oocytes. However, our preliminary RNA sequencing analysis shows high expression of germline specific markers and meiotic genes, as well as robustly differential expression between oocyte stages, but significantly lower expression of follicle cell specific genes. This suggests that follicle cells are underrepresented in the oocyte samples, likely because of the significantly higher mass of oocytes relative to follicle cells, and hence the follicle cells do not seem to hamper analysis.
If oocytes are not released sufficiently after ~6 min in the digestive HL-15, use a fine tip (P200 µl) to gently swirl the solution to assist physical separation, and continue inverting the tube.
To avoid damaging the oocytes, steps #4–9 need to be done quickly.
When using sieves avoid tight folds. Oocytes trapped in folds will lyse.
Always isolate one desired group at a time. Attempting to isolate more groups from the same digested ovaries often results in lysing the oocytes, due to multiple passes through sieves and longer processing time.
Make all solutions with autoclaved MilliQ water and make sure all tubes, bottles and cell sieves are sterile.
Oocyte stages within groups 4 and 5 can be further separated to smaller size range groups using appropriate sieve size combinations. In principle, using the appropriate sieve combination, one can isolate different oocyte size pools as desired.
Materials
HL-15. Can use L-15 with no L-glutamine, but with Phenol Red (Sigma-Aldrich, #L5520).
Digestive HL15 (3 mg/ml Collagenase I, 3 mg/ml Collagenase II, 1.6 mg/ml Hyaluronidase, in HL15).
Collagenase I (Sigma-Aldrich, #C0130).
Collagenase II (Sigma-Aldrich, #C6885).
Hyaluronidase (Sigma-Aldrich, #H4272).
Cell strainers in various pore sizes (BD Falcon).
Cell-Micro-Sieves in various pore sizes 12’ × 12’ sheets (BioDesign Inc. of NY).
Concluding remarks
We provide a comprehensive toolbox for the study of the juvenile zebrafish ovary. While the zebrafish has not yet been fully exploited for the study of oogenesis, it offers several attributes as an excellent model, including live time-lapse high resolution imaging in whole-mount ovaries, and easy accessibility to ovaries for great statistical power of analyses in a vertebrate species. The methods described here have allowed for an exceptional view of oogenesis in both wild type conditions and during functional analysis using mutants and pharmacological drugs in culture. We believe these methods will make similar oogenesis studies accessible to other researchers and contribute to expanding the field. In the future, it is possible that our characterization of juvenile oogenesis can be used for imaging ovaries of desired stages within live transparent (pigment-less) fish strains, such as casper by using two-photon microscopy. Finally, combined with recent genome editing technologies, this toolbox will prove yet more powerful.
Highlights.
Accessibility of early stage oocytes has been limited in zebrafish.
Accurate staging criteria for early oocyte differentiation.
Protocols for the investigation of the juvenile zebrafish ovary.
Live time-lapse imaging of oogonial progression.
Comprehensive methodology offers an unprecedented view of vertebrate oogenesis.
Acknowledgments
MCM and YME were supported by NIH R01GM117981.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bolcun-Filas E, Rinaldi VD, White ME, Schimenti JC. Reversal of female infertility by Chk2 ablation reveals the oocyte DNA damage checkpoint pathway. Science. 2014;343(6170):533–6. doi: 10.1126/science.1247671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgoyne PS, Mahadevaiah SK, Turner JM. The consequences of asynapsis for mammalian meiosis. Nat Rev Genet. 2009;10(3):207–16. doi: 10.1038/nrg2505. [DOI] [PubMed] [Google Scholar]
- 3.Jagarlamudi K, Rajkovic A. Oogenesis: transcriptional regulators and mouse models. Mol Cell Endocrinol. 2012;356(1–2):31–9. doi: 10.1016/j.mce.2011.07.049. [DOI] [PubMed] [Google Scholar]
- 4.Kosaka K, Kawakami K, Sakamoto H, Inoue K. Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis. Mech Dev. 2007;124(4):279–89. doi: 10.1016/j.mod.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Howley C, Ho RK. mRNA localization patterns in zebrafish oocytes. Mech Dev. 2000;92(2):305–9. doi: 10.1016/s0925-4773(00)00247-1. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Mari A, Yan YL, Bremiller RA, Wilson C, Canestro C, Postlethwait JH. Characterization and expression pattern of zebrafish Anti-Mullerian hormone (Amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr Patterns. 2005;5(5):655–67. doi: 10.1016/j.modgep.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn. 2009;238(12):2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maack G, Segner H. Morphological development of the gonads in zebrafish. Journal of Fish Biology. 2003;62(4):895–906. [Google Scholar]
- 9.Dranow DB, Tucker RP, Draper BW. Germ cells are required to maintain a stable sexual phenotype in adult zebrafish. Dev Biol. 2013;376(1):43–50. doi: 10.1016/j.ydbio.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Mari A, Canestro C, Bremiller RA, Nguyen-Johnson A, Asakawa K, Kawakami K, et al. Sex reversal in zebrafish fancl mutants is caused by Tp53-mediated germ cell apoptosis. PLoS Genet. 2010;6(7):e1001034. doi: 10.1371/journal.pgen.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson JL, Rodriguez Mari A, Braasch I, Amores A, Hohenlohe P, Batzel P, et al. Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS One. 2012;7(7):e40701. doi: 10.1371/journal.pone.0040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selman K, Wallace RA, Sarka A, Qi XP. Stages of Oocyte Development in the Zebrafish, Brachydanio-Rerio. Journal of Morphology. 1993;218(2):203–24. doi: 10.1002/jmor.1052180209. [DOI] [PubMed] [Google Scholar]
- 13.Leu DH, Draper BW. The ziwi promoter drives germline-specific gene expression in zebrafish. Dev Dyn. 2010;239(10):2714–21. doi: 10.1002/dvdy.22404. [DOI] [PubMed] [Google Scholar]
- 14.Elkouby YM, Jamieson-Lucy A, Mullins MC. Oocyte Polarization Is Coupled to the Chromosomal Bouquet, a Conserved Polarized Nuclear Configuration in Meiosis. PLoS Biol. 2016;14(1):e1002335. doi: 10.1371/journal.pbio.1002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solnica-Krezel L, Driever W. Microtubule arrays of the zebrafish yolk cell: organization and function during epiboly. Development. 1994;120(9):2443–55. doi: 10.1242/dev.120.9.2443. [DOI] [PubMed] [Google Scholar]
- 16.Choi HM, Beck VA, Pierce NA. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano. 2014;8(5):4284–94. doi: 10.1021/nn405717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross-Thebing T, Paksa A, Raz E. Simultaneous high-resolution detection of multiple transcripts combined with localization of proteins in whole-mount embryos. BMC Biol. 2014;12:55. doi: 10.1186/s12915-014-0055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaumeil J, Augui S, Chow JC, Heard E. Combined immunofluorescence, RNA fluorescent in situ hybridization, and DNA fluorescent in situ hybridization to study chromatin changes, transcriptional activity, nuclear organization, and X-chromosome inactivation. Methods Mol Biol. 2008;463:297–308. doi: 10.1007/978-1-59745-406-3_18. [DOI] [PubMed] [Google Scholar]
- 19.Namekawa SH, Lee JT. Detection of nascent RNA, single-copy DNA and protein localization by immunoFISH in mouse germ cells and preimplantation embryos. Nat Protoc. 2011;6(3):270–84. doi: 10.1038/nprot.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibuya H, Morimoto A, Watanabe Y. The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLoS Genet. 2014;10(12):e1004821. doi: 10.1371/journal.pgen.1004821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang P, Torres J, Lewis RA, Mowry KL, Houliston E, King ML. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol Biol Cell. 2004;15(10):4669–81. doi: 10.1091/mbc.E04-03-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura S, Kobayashi K, Nishimura T, Higashijima S, Tanaka M. Identification of germline stem cells in the ovary of the teleost medaka. Science. 2010;328(5985):1561–3. doi: 10.1126/science.1185473. [DOI] [PubMed] [Google Scholar]
- 23.Westerfield M. The zebrafish book : a guide for the laboratory use of zebrafish (Brachydanio rerio) Eugene, OR: M. Westerfield; 1993. [Google Scholar]
- 24.Gagnon JA, Mowry KL. Visualization of mRNA localization in Xenopus oocytes. Methods Mol Biol. 2011;714:71–82. doi: 10.1007/978-1-61779-005-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claussen M, Pieler T. Identification of vegetal RNA-localization elements in Xenopus oocytes. Methods. 2010;51(1):146–51. doi: 10.1016/j.ymeth.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Berkholtz CB, Lai BE, Woodruff TK, Shea LD. Distribution of extracellular matrix proteins type I collagen, type IV collagen, fibronectin, and laminin in mouse folliculogenesis. Histochem Cell Biol. 2006;126(5):583–92. doi: 10.1007/s00418-006-0194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodgers RJ, Irving-Rodgers HF, Russell DL. Extracellular matrix of the developing ovarian follicle. Reproduction. 2003;126(4):415–24. doi: 10.1530/rep.0.1260415. [DOI] [PubMed] [Google Scholar]
- 28.Escobar-Aguirre M, Elkouby YM, Mullins MC. Localization in Oogenesis of Maternal Regulators of Embryonic Development. In: Pelegri Francisco J, MD, Sutherland Ann., editors. Vertebrate Development - Maternal to Zygotic Control. USA: Springer, New York; 2016. In press. [Google Scholar]
- 29.Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, Mullins MC. Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev Cell. 2004;6(6):771–80. doi: 10.1016/j.devcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Nair S, Lindeman RE, Pelegri F. In vitro oocyte culture-based manipulation of zebrafish maternal genes. Dev Dyn. 2013;242(1):44–52. doi: 10.1002/dvdy.23894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seki S, Kouya T, Tsuchiya R, Valdez DM, Jr, Jin B, Hara T, et al. Development of a reliable in vitro maturation system for zebrafish oocytes. Reproduction. 2008;135(3):285–92. doi: 10.1530/REP-07-0416. [DOI] [PubMed] [Google Scholar]
- 32.Tokumoto T, Yamaguchi T, Ii S, Tokumoto M. In vivo induction of oocyte maturation and ovulation in zebrafish. PLoS One. 2011;6(9):e25206. doi: 10.1371/journal.pone.0025206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behrndt M, Salbreux G, Campinho P, Hauschild R, Oswald F, Roensch J, et al. Forces driving epithelial spreading in zebrafish gastrulation. Science. 2012;338(6104):257–60. doi: 10.1126/science.1224143. [DOI] [PubMed] [Google Scholar]
- 34.Pauls S, Geldmacher-Voss B, Campos-Ortega JA. A zebrafish histone variant H2A.F/Z and a transgenic H2A.F/Z:GFP fusion protein for in vivo studies of embryonic development. Dev Genes Evol. 2001;211(12):603–10. doi: 10.1007/s00427-001-0196-x. [DOI] [PubMed] [Google Scholar]