Abstract
TNF-α is a pivotal cytokine whose overproduction can be lethal. Previously, we identified a transcription factor, LPS-induced TNF-α factor (LITAF), that regulates TNF-α transcription. We now report the discovery and characterization of a regulatory cofactor that we call signal transducer and activator of transcription (STAT) 6(B) because of its considerable homology to STAT6 [here referred to as STAT6(A)]. The STAT6(B) gene expression was found to be activated by LPS. Furthermore, we show that cotransfection of STAT6(B) and LITAF induces an interaction between the two proteins, consequently forming a complex that subsequently translocates into the nucleus and up-regulates the transcription of cytokines. The effect of the complex on a panel of cytokines was tested. In addition, the specific role of LITAF in this complex was established with experiments, including RNA interference technology. Overall, these findings describe roles for LITAF, STAT6(B), and the LITAF-STAT6(B) complex in the regulation of inflammatory cytokines in response to LPS stimulation in mammalian cells.
Keywords: LPS-induced TNF-α factor, TNF-α transcriptional regulation
TNF-α, an important cytokine mediator of immune regulation and inflammation, can cause either beneficial or detrimental properties, through its proinflammatory and proapoptotic effects in various cell types (1). Many studies of TNF-α have focused on its regulation by transcription factors, such as NF-κB (2), Ets (3), NF-AT (4), AP-1 (5), cAMP response element-binding protein (6), and C/EBPβ (7).
We previously identified a factor named LPS-induced TNF-α factor (LITAF), which was shown to mediate TNF-α transcription (8). Inhibition of LITAF mRNA expression in THP-1 cells resulted in a reduction of TNF-α transcripts. We found that high levels of LITAF mRNA are expressed predominantly in the placenta, peripheral blood leukocytes, lymph nodes, and spleen. We also identified the DNA site within the TNF-α promoter that is essential for LITAF binding (9). These findings suggested that LITAF contributes to the activation of TNF-α gene expression in response to LPS. However, ELISA analysis showed that the TNF-α protein levels induced by a transient transfection of LITAF were not significantly higher compared with levels observed after LPS treatment. Several reasons can be advanced for this phenomenon. One explanation might be that because the TNF-α promoter contains not only the LITAF-binding site but also sites for NF-κB, activating protein 1, and others (2–7), corresponding transcription factors can also be induced by LPS to bind to the TNF promoter and regulate its gene expression. Alternatively, in some cases, cytokine gene expression is regulated by cofactors that singly reduce promoter activity but together enhance it to a significant extent (10, 11). Therefore, we hypothesized that LITAF might require the participation of a cofactor to substantially induce TNF-α gene expression. For this study, we used a yeast two-hybrid system to identify other mediators that might interact with LITAF in the regulation of TNF-α transcription. After high-stringency screenings, we identified a gene that was homologous to members of the signal transducer and activator of transcription (STAT) family. Because of its significant sequence homology to STAT6, we named the new clone STAT6(B) (GenBank accession no. AY615283) and STAT6 became STAT6(A). To date, seven members of the STAT family have been characterized (STAT1, 2, 3, 4, 5a, 5b, and 6) (12). These factors form homo- or heterodimers in the cytoplasm and then translocate into the nucleus where they activate the expression of target genes (13). These STAT family members constitute an important pathway in cytokine signaling (11–13).
Here, we report that LPS-induced STAT6(B) directly interacts with LITAF and forms a LITAF-STAT6(B) complex in cytoplasm. This complex then translocates into the nucleus, where it significantly up-regulates transcription of several inflammatory cytokines, including GRO, IL-1α, regulated on activation normal T cell expressed and secreted (RANTES), TNF-α, IFN-γ, monocyte chemoattractant protein 2 (MCP-2), and IL-10. Finally, short hairpin RNA (shRNA) introduced to silence the effect of LITAF in the LITAF-STAT6(B) complex resulted in a decrease of TNF-α gene expression and protein production. These findings suggest that LITAF plays a leading role in cytokine regulation after the LITAF-STAT6(B) complex translocates into the nucleus.
Materials and Methods
Cell Lines. All bacterial cloning constructs used Escherichia coli strain DH5α (Invitrogen). Yeast strain AH109 (BD Biosciences) was used to screen the library and to verify protein–protein interactions. THP-1 human monocytic cells were grown in RPMI medium 1640 with 10% FCS. U2OS cells were grown in DMEM with 10% FCS. All cultures were maintained in a humidified atmosphere of 5% CO2 at 37°C.
Macrophages. Macrophages were obtained from C57BL/6 mice (The Jackson Laboratory) and purified by conventional methods (14).
Plasmid Constructs. The human LITAF full-length DNA fragment (9) was subcloned into the vector pGBKT7 (Clontech), and named pGBKLITAF. The recombinant plasmid was used as a DNA-BD/bait and transformed into AH109 yeast cells, which were screened for growth on SD/-Trp medium.
A double-stranded oligomeric DNA fragment as a hemagglutinin tag was generated by annealing, using the following primer pairs: 5′-CCCAAGCTTACATGGCCTACCCCTACGACGTGCCCGACTACGCCTCCCTCGGATCCCG-3′ and 5′-cgggatccgagggaggcgtagtcgggcacgtcgtaggggtaggccatgtaagcttggg-3′. The DNA fragment was inserted into the HindIII/BamHI site of pcDNA3 (Invitrogen). This recombinant vector, named pcDNA3HA, was used as the backbone for all constructs. Full-length human LITAF DNA was generated by PCR with the following primer pairs: 5′-CGGGATCCGTGCAGACGGTCTACGTG-3′ and 5′-cggaattccagttggga cagtaatgg-3′, which was subcloned into pcDNA3HA. Human STAT6 DNA (GenBank accession no. NM_003153,) was purchased from Open Biosystems (Huntsville, AL). Full-length DNA was generated by using PCR with the following primer pairs: 5′-CGGGATCCATGTCTCTGTGGGGTCTG-3′ and 5′-gctctagatcaccaactggggttggccct-3′, which was subcloned into pcDNA3HA; this construct is referred to as STAT6(A) in this study. Full-length human STAT6(B) DNA was isolated from a human spleen match-maker cDNA library (Clontech) and then subcloned into pcDNA3HA. The STAT6(B) DNA fragments representing amino acids 1–150, amino acids 151–404, or amino acids 1–404 were generated by PCR with the following primer pairs: 5′-CGGGATCCATGGCCCGACGGAACCCTTCT-3′ and 5′-gctctagatcaagctgttgtgagaaggaaaag-3′, 5′-CGGGATCCGAACAGATGGGTAAGGAT GGC-3′ and 5′-gctctagatcaccaactggggttggccct-3′, or 5′-CGGGATCCATGGCCCGACGGAACCCTTCT-3′ and 5′-gctctagatcaccaactggggttggccct-3′, respectively, then subcloned into pcDNA3HA. The DNA, named pSHAG-LITAF (GenBank accession no. NM_004862), which transcribes LITAF-specific shRNA in mammalian cells, was purchased from Open Biosystems and was used to silence LITAF gene expression.
Yeast Two-Hybrid System. Human spleen pACT2-cDNAs (Clontech) were used as activation domain (AD) fusion constructs with a selection marker (-Leu) as were DNA from pGBKLITAF as DNA-BD/bait with a different selection maker (-Trp). Both pACT2-cDNAs (1 μg) and pGBKLITAF (1 μg) were cotransformed into AH109-competent yeast cells and cultured in the high-stringency medium with the selection markers SD/-Ade/-His/-Leu/-Trp/X-α-gal, then screened as described (Clontech).
Northern Blot Analysis of STAT6(B) Gene Expression. The 2,543-bp fragment of the DNA from the coding sequence of STAT6(A) was amplified by PCR with the following primer pairs: 5′-CGGGATCCATGTCTCTGTGGGGTCTG-3′ and 5′-gctctagatcaccaactggggttggccct-3′. The 450-bp fragment of the DNA from STAT6(B) (amino acids 1–150, Fig. 8, which is published as supporting information on the PNAS web site) was amplified by PCR with the following primer pairs: 5′-CGGGATCCATGGCCCGACGGAACCCTTCT-3′ and 5′-gctctagatcaagctgttgtgagaaggaaaag-3′. Both DNAs above were labeled with [32P]dCTP as described (15) and used for Northern hybridizations.
Isolation of LITAF RNA After Inducing shRNA in THP-1 Cells. THP-1 cells (5 × 106) matured by treatment with 200 nM phorbol 12-myristate 13-acetate (Sigma) were transfected using FuGENE 6 (Roche Molecular Biochemicals) with 0, 0.25, or 0.5 μg/ml pSHAG-LITAF DNA before stimulation with 0.1 μg/ml LPS (Sigma) for 3 h, then washed with PBS. The treated cells were grown in RPMI medium 1640 with 10% FCS and maintained in an atmosphere of 5% CO2 at 37°C overnight. Untreated THP-1 cells served as controls and were also cultured in the same conditions. The total RNA from treated cells was isolated by using the RNA STAT-60 kit according to the manufacturer's protocol (Tel-Test, Friendswood, TX).
Immunoprecipitation and Western Blot Analysis. Mouse macrophages were stimulated for 24 h at 37°C with 0.1 μg/ml LPS. Both the treated cells and untreated control cells were grown in RPMI medium 1640 with 10% FCS. After incubation, cells were rinsed once in ice-cold PBS and routinely lysed in lysis reagent (Promega) following the manufacturer's instructions. The cell lysates were incubated with 2 μg of the appropriate antibody for 2 h at 4°C, followed by incubation with 20 μl of protein A/G plus-agarose-Sepharose beads (SC-2003, Santa Cruz Biotechnology) for an additional 1 h. The beads were washed three times in PBS buffer and then suspended in SDS sample buffer heated at 95°C for 5 min. The eluted proteins were applied to SDS-polyacrylamide gels, and proteins were detected by Western blotting (9).
EMSA. EMSA was performed as described (9).
Transient Transfection and Luciferase Assay. The assay was performed as described (9, 16).
ELISA. THP-1 cells in a 96-well plate at 2 × 104 cells per well were induced to mature by addition of 200 nM phorbol 12-myristate 13-acetate (Sigma) and incubated at 37°C in an atmosphere of 5% CO2 for 20 h, then washed with PBS twice and stimulated with 0.1 μg/ml LPS (Sigma) or transfected by FuGENE 6 with DNAs of LITAF, STAT6(A), STAT6(B), or pSHAG-LITAF. TNF-α in the supernatant of each treated and untreated control cells were measured by ELISA (Abraxis, Warminster, PA).
Human Protein Cytokine Array. THP-1 cells in a 96-well plate at 2 × 104 cells per well were induced to mature by addition of 200 nM phorbol 12-myristate 13-acetate (Sigma) and incubated at 37°C in an atmosphere of 5% CO2 for 20 h, then washed with PBS and transfected by FuGENE 6 with DNA of LITAF, STAT6(B), or empty vector, or cotransfected with both LITAF and STAT6(B). The treated cells were grown in RPMI medium 1640 with 10% FCS. After incubation for 24 h at 37°C in an atmosphere of 5% CO2, the conditioned medium was harvested and centrifuged at 1,500 × g to remove cell debris. Meanwhile, the membranes included in a human protein cytokine array kit (RayBiotech, Norcross, GA) were blocked with a blocking buffer, and then 1 ml of medium from each culture of treated THP-1 cells was individually added and incubated at room temperature for 2 h. The membranes were then analyzed according to the manufacturer's instructions.
Results
Identification of a Gene, STAT6(B), Which Interacts with LITAF. We used a yeast two-hybrid system to identify mediators that might interact with LITAF in the regulation of TNF-α. First, LITAF full-length DNA was inserted into pGBKT7, with the resulting construct named pGBKLITAF. This recombinant DNA containing a selection marker (-Trp) was used as the bait, and a human spleen pACT2-cDNA library with a different selection marker (-Leu) was used for the AD fusion constructs. Both bait and AD DNAs were mixed and transformed into yeast AH109 cells. To eliminate false-positives, high-stringency conditions were used to select transformants on SD/-Trp/-Leu plus -Ade/-His medium. The transformants surviving in high-stringency medium were screened and only the AD DNA from the transformants was prepared. The DNA was reconfirmed by retransformation and rehybridization with pGBKLITAF in AH109 cells. A clone was isolated after high-stringency screenings. Sequence analysis of this clone showed that it contained an ORF (amino acids 1–404) and a poly(A) tail (Fig. 8). The DNA sequence and its ORF in the region from amino acid 151 to 404 were homologous to a known gene, STAT6 (GenBank accession no. NM_003153), referred to as STAT6(A) in this study, except for two amino acid differences: P-S at amino acid position 292 and M-T at amino acid position 324. However, the sequence in the region from amino acids 1 to 150 was completely different from STAT6(A), as shown in Fig. 9, which is published as supporting information on the PNAS web site. The 3′ UTR of this clone, from the stop codon (1,408 bp) to poly(A) tail (2,141 bp) was almost identical with the UTR of STAT6(A), with only three differences: base pairs were shifted, c-g at 1,906 bp and g-a at 1,935 bp, and a single deletion of a base pair (2,085–2,086 bp, Fig. 8).
To confirm the difference between STAT6(A) and STAT6(B), Northern blots were prepared with RNA samples from multiple human tissues and probed with both transcripts. As seen in Fig. 1, distinct transcripts of ≈3.5 kb for STAT6(A), and of ≈1.5 kb for STAT6(B), were identified. Although both were abundantly expressed in spleen tissue, STAT6(B) transcripts were more highly expressed in samples prepared from colon, testis, and peripheral blood leukocytes in comparison with STAT6(A). STAT6(B) chromosomal localization was then determined by in situ hybridization (data not shown). The fluorescent banding spots were observed on chromosome 12q13, which is close to the location of STAT6(A) (17).
Fig. 1.
Detection of the transcripts of STAT6(A) and STAT6(B) by Northern blot. Filters (Clontech) containing preblotted mRNA (2 μg of each) from different adult human tissues were separately hybridized with an α-32P-dCTP-labeled DNA probe of STAT6(A) (full-length) or STAT6(B) (amino acids 1–150) and then autoradiographed with Biomax MR film (Kodak).
Investigation of the Interaction Between LITAF and STAT6(B) by Immunoprecipitation. Although yeast two-hybrid experiments showed an association between LITAF and STAT6(B), more definitive evidence about their interaction is required. Thus, we used mouse macrophages for immunoprecipitation experiments designed to determine whether LITAF interacts with STAT6(B). STAT6 antibody was used to detect the product of STAT6(A) (120 kDa) or STAT6(B) (≈60 kDa) because the antibody specifically recognizes the carboxyl terminus of STAT6(A) of mouse or human, which is also shared by STAT6(B). Additionally, the antibody (Stat6, Santa Cruz Biotechnology) originally made to bind to STAT6(A) would be expected to bind murine STAT6(B) because the carboxyl terminus of STAT6(B) has >88% homology to mouse STAT6(A) (GenBank accession no. L47650). The cell lysates extracted from LPS-treated mouse macrophages were immunoprecipitated with the appropriate antibody followed by Western blotting. As shown in Fig. 2, lysates pulled down with either LITAF or STAT6(B) could be completely detected by LITAF or STAT6(B) antibody (Fig. 2, lanes 4 and 6). These findings suggest that full-length LITAF directly interacts with full-length STAT6(B). Interestingly, the result (Fig. 2) also provides a clue about the regulation of both LITAF and STAT6(B): they seem to be induced specifically by LPS. In addition, mouse STAT6(A) could not be detected in either non-LPS-treated or LPS-treated lysates (Fig. 2, lanes 1 and 2); therefore, the effects on mouse STAT6(A) appear not to be relevant in this pull-down assay and will not be considered further.
Fig. 2.
Immunoprecipitation of murine macrophages. The cell lysates from untreated (lanes 1, 3, and 5) or LPS-treated (100 ng/ml LPS) (lanes 2, 4, and 6) macrophages were immunoprecipitated with anti-LITAF (lanes 3 and 4) or anti-STAT6 (lanes 5 and 6), and then protein A/G beads (SC-2003, Santa Cruz Biotechnology) were added. The precipitates were subjected to Western blotting with the antibodies indicated by arrows.
LPS-Dependent STAT6(B) Gene Expression. Because STAT6(B) was identified as involved in interaction with LITAF, and both proteins were clearly observed after LPS stimulation in mouse macrophages (Fig. 2), we hypothesized that human STAT6(B) gene expression is also involved in LPS-dependent regulation of cytokines and LITAF. To test this hypothesis, matured THP-1 cells were stimulated with various doses of LPS, then cell lysate was harvested and subjected to Western blot analysis with the indicated antibodies: Stat6 (S-20), LITAF (611615), or actin (C-11) as control. As shown in Fig. 3, the protein level of STAT6(B) or LITAF was progressively increased in response to increasing doses of LPS. In contrast, STAT6(A) production was not significantly changed in response to LPS concentration. This result strongly suggests that both LITAF and STAT6(B), but not STAT6(A), are induced in an LPS-dependent manner.
Fig. 3.
Detection of LPS-induced LITAF, STAT6(A), or STAT6(B) production in THP-1 cells. Cells were untreated (lane 1) or pretreated with LPS at 5 (lane 2), 25 (lane 3), 50 (lane 4), 100 (lane 5), or 200 (lane 6) ng/ml for 3 h, then washed with PBS and maintained overnight. Cell protein lysates were extracted and subjected to Western blot analyses.
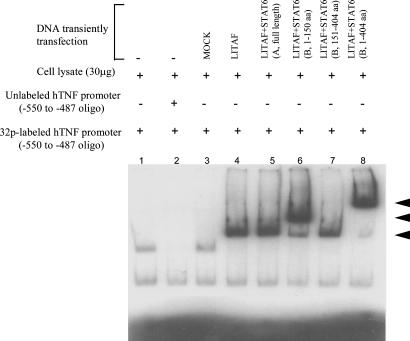
EMSA. To determine whether STAT6(B) contains a binding domain for LITAF, EMSA was performed. DNAs of full-length LITAF (Fig. 4, lanes 4–8), empty vector (lane 3), STAT6(A) (lane 5), and STAT6(B) fragment (lanes 6–8) were transfected or cotransfected into U2OS cells. The resulting cell extracts were then purified and assessed by EMSA as described below. The shifted protein bands shown in Fig. 4 indicated that STAT6(B) contains a LITAF-binding domain in the region from amino acids 1 to 150. This result suggests that overexpression of both LITAF and STAT6(B) could lead to formation of a LITAF-STAT6(B) heterodimer complex that can bind to the TNF promoter.
Fig. 4.
EMSA of the protein–DNA interaction. Protein (30 μg) extracted from each culture of transfected U2OS cells was added to the appropriate reaction buffer with the oligonucleotide DNA probe, 32P-ATP-labeled hTNF-α promoter DNA as described (9). Protein from untransfected cells was applied in lanes 1 and 2, and a 50-fold excess of unlabeled competitor was added (lane 2). The proteins from each condition of cells treated with 1 μg of DNA of empty vector alone (lane 3), full-length LITAF alone (lane 4), both full-length LITAF and STAT6(A) (lane 5), full-length LITAF plus a fragment of STAT6(B) (amino acids 1–150, lane 6), full-length LITAF plus a fragment of STAT6(B) (amino acids 151–404, lane 7), and full-length LITAF plus full-length STAT6(B) (amino acids 1–404, lane 8) were assessed by EMSA. The shifted DNA bands are indicated by arrows.
Translocation of LITAF/STAT6(B) Complex. Homodimers of STAT6(A) are known to translocate from cytoplasm to nucleus where they bind to target gene sequences and activate the expression of those genes (18). Because STAT6(B) was found to have a high degree of homology at the carboxyl terminus with STAT6(A), we hypothesized that a LITAF-STAT6(B) complex might also translocate into the nucleus after formation in cytoplasm. Thus, we measured fluctuations in protein levels of STAT6(B) and LITAF over time in cells known to produce TNF-α in a regulated manner. DNAs of both STAT6(B) (full-length) and LITAF (full-length) were cotransfected into THP-1 monocytic cells. Cell extracts harvested at various times after transfection were purified and subjected to Western blot analysis. As seen in Fig. 5, the protein levels of both LITAF (Fig. 5A) and STAT6(B) (Fig. 5B) gradually rose in both whole-cell extracts and nucleus at a similar rate. In contrast, 24 h after transfection, the amount of cytoplasmic STAT6(B) decreased sharply, and the level of cytoplasmic LITAF dropped ≈2-fold. We observed that both LITAF and STAT6(B) levels increased steadily in the nucleus after that time. This increase of LITAF and STAT6(B) levels in the nucleus coincided with a decline in their concentrations in the cytoplasm. We interpret these changes over time as suggesting that a LITAF-STAT6(B) protein complex may be forming and translocating into the cell nucleus 4–8 h after transfection. Interestingly, STAT6(A) did not seem to be affected by overexpression of LITAF because its concentration was constant in both whole-cell extracts and cytoplasm. In addition, STAT6(A) protein could not be found in the nucleus at any of the times selected. Finally, the absence of β-tubulin control from nuclear samples indicated that the nuclear extracts were not contaminated with proteins from the cytoplasm during the protein purification process (19).
Fig. 5.
Detection of translocation of LITAF-STAT6(B) complex in THP-1 cells. Protein extracts were collected from whole cells, cytoplasm, or nuclei after overexpression of both LITAF and STAT6(B). The alterations in the protein levels of exogenous LITAF or STAT6(B) and endogenous STAT6(A) over time were measured by Western blot with the following antibodies: LITAF (611615, BD Biosciences), Stat6 (S-20, Santa Cruz Biotechnology), or β-tubulin (C-20, Santa Cruz Biotechnology) as control.
Regulation of Cytokines by LITAF/STAT6(B) Complex. Because both LITAF and STAT6(B) were found in the nucleus (Fig. 5), we further investigated whether they act separately or in concert in mediating activation of TNF-α. While studying the complex, we used a cytokine array to look at effects on other cytokines (Fig. 6 A and B; see also Fig. 10, which is published as supporting information on the PNAS web site) By using an empty vector as a control (Fig. 10b1), DNA of LITAF (full-length, Fig. 10b2), STAT6(B) (full-length, Fig. 10b3) or both full-length of LITAF+STAT6(B) (Fig. 10b4) respectively, was transfected or cotransfected into THP-1 cells. Conditioned medium from each treated cell type was then prepared and incubated with membranes containing an array of 44 human protein cytokine antibodies (Fig. 10a). Autoradiographs were scanned, and the density of each cytokine at the corresponding position was determined. Each blot shown in Fig. 10b illustrate a single result that is representative of three replicates. The relative intensities of each cytokine were normalized to control spots on the same membrane. Major increases or decreases among cytokines are represented graphically in Fig. 6a. LITAF alone (Figs. 6 A and B and 10b2) up-regulated TNF-α (positions a-7 and a-8) and IL-1β (positions l-1 and l-2). STAT6(B) alone (Figs. 6 A and B and 10b3) up-regulated several different target cytokines such as IL-1α (Fig. 10a; positions k-1 and k-2), GRO (positions h-1 and h-2), IL-10 (positions h-3 and h-4), and RANTES (positions h-5 and h-6). Interestingly, the cytokines that were induced only modestly by either LITAF or STAT6(B) alone were more significantly up-regulated in the cells cotransfected with both LITAF and STAT6(B) (Figs. 6 A and B and 10b4): 2.1-fold for GRO, 3-fold for IL-1α, 3.2-fold for RANTES, 4-fold for TNF-α, 6.2-fold for IFN-γ, 8.5-fold for MCP-2, and 10-fold for IL-10.
Fig. 6.
Cytokine antibody array and Western blot of DNA-transfected THP-1 cells. (A) Human 44 cytokines were blotted onto a membrane and arrayed three times following the manufacturer's protocol. The intensities of the relative expression levels of cytokines were quantified by densitometry (VersaDoc imaging system, Bio-Rad). The β-gal gene was included in all transfections. The density value of each test sample was normalized to β-gal from the same lysates as described (15) and graphed. (B) Detection of LITAF/STAT6(B) complex-regulated gene expression in THP-1 cells. Proteins from THP-1 whole cells without DNA transfection (lane 1) or after DNA transfection with empty vector (lane 2), overexpression of LITAF alone (lane 3), STAT6(A) alone (lane 4), STAT6(B) alone (lane 5), and both LITAF and STAT6(B) (lane 6) were extracted and subjected to Western blot analyses. A total of 80 μg of protein extract was loaded per lane. Blots were probed with the following antibodies: Stat6 (S-20, Santa Cruz Biotechnology), LITAF (611615, BD Biosciences), IL-1α (R-20, Santa Cruz Biotechnology), IL-10 (M-18, Santa Cruz Biotechnology), MCP-2 (C-17, Santa Cruz Biotechnology), RANTES (C-19, Santa Cruz Biotechnology), or actin (C-11, Santa Cruz Biotechnology) as control.
To confirm these findings at the protein level, we selected the four most strongly affected cytokines, IL-1α, IL-10, MCP-2, and RANTES, and under the same experimental conditions, measured their protein levels by Western blots. As shown in Fig. 6b, these protein levels were not altered in response to overexpression of LITAF alone (lane 3) or of STAT6(A) alone (lane 4). However, modest induction of all except MCP-2 was observed after transient transfection of STAT6(B) alone (lane 5), whereas the combination of LITAF and STAT6(B) resulted in big increases in all of the cytokines tested (lane 6). These results are consistent with the array data shown in Fig. 6, although we note that the other RNA fluctuations observed in the array have not yet been confirmed by Western blot analysis.
Silencing of LITAF Expression by LITAF-Specific shRNA. Because the LITAF-STAT6(B) putative complex showed a greater capacity to regulate target cytokines in comparison with either LITAF or STAT6(B) alone, we considered the possibility that LITAF plays an important regulatory role in the complex. As a first step, we investigated whether LITAF could be silenced by delivery of LITAF-specific shRNA. Northern blot analysis was performed on total RNA extracted from LPS-stimulated THP-1 cells after transfection with 0.25 μg (lane 3) or 0.5 μg (lane 4) of pSHAG-LITAF. Meanwhile, proteins from each of the treated cell cultures were purified and subjected to Western blot analysis. As shown in Fig. 7 A and B, both the RNA level (Fig. 7A, lane 4) and the protein level (Fig. 7B, lane 4) of LITAF induced by LPS were greatly decreased in cells transfected with 0.5 μg/ml pSHAG-LITAF, demonstrating that the shRNA driven by pSHAG-LITAF effectively silenced LITAF gene expression.
Fig. 7.
Detection of shRNA-silenced LITAF gene expression. (A) Ten micrograms of total RNA extracts from LPS-stimulated THP-1 cells after transfection of 0 (lanes 1 and 2), 0.25 (lane 3), or 0.5 (lane 4) μg of pSHAG-LITAF-DNA to induce shRNA were isolated and subjected to Northern blot analysis with an α-32P-dCTP-labeled DNA probe of LITAF (full-length) or GAPDH (Clontech). (B) The protein extracts from the treated cells were probed with the following antibodies: LITAF (611615, BD Biosciences) or actin (C-11, Santa Cruz Biotechnology) as a constitutive control. (C) Transcriptional activity of a series of deletion constructs of TNF-α promoter DNA. U2OS cells were transiently transfected with LITAF alone, STAT6(B) alone, or empty vector (mock) as control or were cotransfected with LITAF plus pSHAG-LITAF, LITAF plus STAT6(B), or LITAF plus STAT6(B) plus pSHAG-LITAF. Subsequent luciferase production was measured as described (9) in triplicate assays. The β-gal gene was included in all transfections. The promoter activity of each test sample was normalized to β-gal in the same lysates as described (15) and graphed. (D) ELISA in THP-1 cells. Cells were transiently transfected with LITAF alone, STAT6(A) alone, STAT6(B) alone, or empty vector as control and cotransfected with LITAF plus STAT6(A), LITAF plus STAT6(B), LITAF plus pSHAG-LITAF, or LITAF plus STAT6(B) plus pSHAG-LITAF. After 16 h of incubation, supernatants from treated cells were collected, and TNF-α secreted from each was measured by ELISA, in triplicate assays. The β-gal gene was included in all transfections. The density value of each test sample was normalized to β-gal in the same lysates and graphed.
To characterize the functional importance of LITAF in the LITAF/STAT6B complex, we examined the effect of the complex, with and without silenced LITAF, on TNF-α promoter activity. U2OS cells containing the TNF-α promoter linked to luciferase were transiently transfected with LITAF alone, STAT6(B) alone, or both DNAs as described. TNF-α promoter activity was analyzed via luciferase activity. As shown in Fig. 7C, the LITAF-STAT6(B) cotransfection caused a 1.5-fold increase in TNF-α promoter activity, in comparison with transfection with LITAF alone. STAT6(B) alone did not substantially change luciferase activity. However, the ability of either LITAF alone or LITAF-STAT6(B) complex to increase TNF-α promoter activity was significantly inhibited by LITAF-shRNA. We also measured the TNF-α concentrations in the supernatants of these transfected THP-1 cells. As shown in Fig. 7D, 1.8-fold more TNF-α was produced after 16 h by cells containing the LITAF-STAT6(B) complex than by cells transfected with LITAF alone. The level of secreted TNF-α protein induced by either LITAF alone or LITAF-STAT6(B) complex was also significantly decreased by the delivery of shRNA in cells. These findings confirm, by luciferase assay and ELISA, that shRNA introduced to silence the effect of LITAF in the LITAF-STAT6(B) complex resulted in a decrease of TNF-α gene expression and protein production. They further suggest that LITAF might play a leading role in cytokine regulation after the LITAF-STAT6(B) complex translocates to the nucleus in the cells.
Discussion
In this study, yeast two-hybrid analysis was used to identify a clone that interacts with LITAF (Figs. 8 and 9) and subsequently functions as a cofactor in transcriptional regulation of TNF-α (Figs. 6 and 7). The cloned DNA contains an ORF (amino acids 1–404) and was found highly homologous to STAT6 in the region from amino acids 151–404; however, differed completely in the region from amino acids 1 to 150. We have named this clone STAT6(B) and have named STAT6 as STAT6(A). A similar occurrence was reported for STAT5, which encompasses two fraternal twins STAT5(A) and STAT5(B), yet with different biological activity (12). At this point, our data support that STAT6(B) may be generated from STAT6(A) by alternate splicing; however, the possibility that STAT6(B) is the product of a gene independent of STAT6(A) remains.
STAT family members share two major domains. The first domain is the oligomerization domain in the N terminus. This domain, containing ≈700 amino acids of tyrosine residues, is specifically phosphorylated by Janus kinases (Jaks) to allow STAT proteins to undergo dimerization and then to translocate from cytoplasm to nucleus, where they act as transcription factors. The second is the Src homology domain 2 (SH2) domain that allows the STATs to associate with an appropriate cytokine receptor in response to stimulation (12, 20). STAT6(A), a member of this family, has been well characterized. Upon the binding of IL-4 to its cognate receptor, latent, monomeric STAT6(A) protein is recruited to the α chain of the IL-4 receptor. Subsequently, STAT6(A) is phosphorylated on tyrosine 641 in its SH2 domain by Jak1 and/or Jak3, becoming an activated protein that homodimerizes through its oligomerization domain, migrates to the nucleus, binds specific cis-acting elements, and activates transcription of IL-4 responsive genes (21).
In contrast, we found that STAT6(B) is not affected by Jaks in response to IL-4 stimulation (data not shown), but rather, is expressed upon LPS stimulation. This occurrence is likely due to the absence of the oligomerization domain and SH2 domain in the N terminus.
It is well known that cytokines are involved in proinflammatory and antiinflammatory responses to pathogens and in cell differentiation and proliferation (22–26). Dysregulated cytokine gene transcription has been observed in many disorders of the immune system, and has, in some cases, been shown to be a critical event (27, 28). Therefore, it is very important to determine the mechanisms regulating cytokine gene expression, to better understand and control cytokine-mediated diseases. In this study, analysis by means of a cytokine antibody array (Fig. 6) demonstrated that the LITAF-STAT6(B) complex significantly up-regulated GRO, IL-1α, RANTES, TNF-α, IFN-γ, MCP-2, and IL-10. Interestingly, the analysis of these cytokine promoters show that most of them contain a CTCCC sequence upstream of their TATA signal, a specific DNA-binding site that is specific to LITAF on TNF-α promoter (9). It implies that LITAF-STAT6(B) complex can affect the promoter activity on these cytokines, although we do not know yet whether the complex regulates these cytokines through promoter binding activity or by other means.
TNF-α, an important mediator of immune regulation and inflammation, has been implicated as both a beneficial and an injurious factor through its proinflammatory and apoptotic effects in various cell types. Many studies of TNF-α have focused on the investigation of the transcriptional regulation by factors such as NF-κB (2), Ets (3), NF-AT (4), activating protein 1 (5), cAMP response element-binding protein (6), and C/EBPβ (7). However, it is still unclear whether these transcription factors translocate from the cytoplasm to the nucleus once induced. In this study, we show that STAT6(B) binds to LITAF, because STAT6(B) was identified on the basis of this binding. Analysis of protein levels over time after transfection of LITAF and STAT6(B) into THP-1 cells indicated that the LITAF-STAT6(B) complex is formed and begins translocating into the nucleus during a 4- to 8-h interval. LITAF and STAT6(B) protein levels continued to rise steadily in the nucleus after that. Interestingly, STAT6(A) was not affected by overexpression of LITAF, as indicated by a stable protein level in both the whole cell and cytoplasm. No STAT6(A) protein could be detected in the nucleus. This finding suggests that STAT6(A) neither associates with LITAF nor is involved in the LITAF pathway. Thus, we propose that the site of LITAF and STAT6(B) interaction most probably takes place in the region of amino acids 1–150 on STAT6(B). However, our current studies show that the overexpression of LITAF alone can up-regulate TNF-α production somewhat (8, 9), although less dramatically than LPS treatment. Indeed LPS treatment induces not only LITAF but also a number of other cytokine-regulating transcription factors, including NF-κB, Ets, NF-AT, and activating protein 1. These factors also have the ability to bind to the TNF-α promoter and regulate its expression. Potential contributions from endogenous LITAF and STAT6(B) also cannot be ruled out.
Recently, RNA interference technology has been improved by delivery of shRNAs from expression vectors in cells (29, 30). In this study, we have used a commercial expression vector that can express LITAF-specific shRNA in cells to silence LITAF gene expression. Based on the present data, we conclude that LITAF may play an important role in the regulation of TNF-α and other cytokines upon formation of a complex with STAT6(B) and highlight the potential role of STAT6B in the regulation of inflammatory processes.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health/National Institute of Dental and Craniofacial Research Grant DE14079 (to S.A.).
Author contributions: S.A. designed research; X.T., D.L.M., and S.A. performed research; S.A. contributed new reagents/analytic tools; X.T., D.LM., S.E.L., and S.A. analyzed data; and X.T., D.L.M., S.E.L., and S.A. wrote the paper.
Abbreviations: STAT, signal transducer and activator of transcription; LITAF, LPS-induced TNF-α factor; RANTES, regulated on activation normal T cell expressed and secreted; MCP, monocyte chemoattractant protein; shRNA, short hairpin RNA; AD, activation domain; SH2, Src homology domain 2; Jak, Janus kinase.
References
- 1.Beutler, B. & Cerami, A. (1989) Annu. Rev. Immunol. 7, 625–655. [DOI] [PubMed] [Google Scholar]
- 2.Collart, M. A., Baeuerle, P.& Vassalli, P. (1990) Mol. Cell. Biol. 10, 1498–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kramer, B., Wiegmann, K. & Kronke, M. (1995) J. Biol. Chem. 270, 6577–6583. [DOI] [PubMed] [Google Scholar]
- 4.Lee, M. H, Park, J., Chung, S. W., Kang, B. Y., Kim, S. H. & Kim, T. S. (2004) Int. Arch. Allergy Immunol. 134, 213–222. [DOI] [PubMed] [Google Scholar]
- 5.Vitiello, M., D'Isanto, M., Galdiero, M., Raieta, K., Tortora, A., Rotondo, P., Peluso, L. & Galdiero, M. (2004) Cytokine 27, 15–24. [DOI] [PubMed] [Google Scholar]
- 6.Johansson, C. C., Bryn, T., Yndestad, A., Eiken, H. G., Bjerkeli, V., Froland, S. S., Aukrust, P. & Tasken, K. (2004) AIDS 18, 171–179. [DOI] [PubMed] [Google Scholar]
- 7.Pope, R., Mungre, S., Liu, H.& Thimmapaya, B. (2000) Cytokine 12, 1171–1181. [DOI] [PubMed] [Google Scholar]
- 8.Myokai, F., Takashiba, S., Lebo, R. & Amar, S. (1999) Proc. Natl. Acad. Sci. USA 96, 4518–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang, X., Fenton, M. J. & Amar, S. (2003) Proc. Natl. Acad. Sci. USA 100, 4096–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joo, A., Aburatani, H., Morii, E., Iba, H. & Yoshimura, A. (2004) Oncogene 23, 726–734. [DOI] [PubMed] [Google Scholar]
- 11.Tomida, M. & Saito, T. (2004) Oncogene 23, 679–686. [DOI] [PubMed] [Google Scholar]
- 12.Scott, M. J., Godshall, C. J. & Cheadle, W. G. (2002) Clin. Diagn. Lab. Immunol. 9, 1153–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rane, S. G. & Reddy, E. P. (2002) Oncogene 21, 3334–3358. [DOI] [PubMed] [Google Scholar]
- 14.Shimomura, H., Matsuura, M., Saito, S., Hirai, Y., Isshiki, Y. & Kawahara, K. (2001) Infect Immun. 69, 3663–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pohjanpelto, P. & Holtta, E. (1996) EMBO J. 15, 1193–1200. [PMC free article] [PubMed] [Google Scholar]
- 16.Berns, K., Hijmans, E. M., Mullenders, J., Brummelkamp, T. R., Velds, A., Heimerikx, M., Kerkhoven, R. M., Madiredjo, M., Nijkamp, W., Weigelt, B., et al. (2004) Nature 428, 431–437. [DOI] [PubMed] [Google Scholar]
- 17.Leek, J. P., Hamlin, P. J., Bell, S. M. & Lench, N. J. (1997) Cytogenet. Cell Genet. 79, 208–209. [DOI] [PubMed] [Google Scholar]
- 18.Wick, K. R. & Berton, M. T. (2000) Mol Immunol. 37, 641–652. [DOI] [PubMed] [Google Scholar]
- 19.Lindeman, G. J., Gaubatz, S., Livingston, D. M. & Ginsberg, D. (1997) Proc. Natl. Acad. Sci. USA 94, 5095–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paul, W. E. (1997) Ciba Found. Symp. 204, 208–216. [DOI] [PubMed] [Google Scholar]
- 21.O'Shea, J. J., Pesu, M., Borie, D. C. & Changelian, P. S. (2004) Nat. Rev. Drug Discov. 3, 555–564. [DOI] [PubMed] [Google Scholar]
- 22.Szabo, S. J., Gold, J. S., Murphy, T. L. & Murphy, K. M. (1993) Mol. Cell. Biol. 13, 4793–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charpentier, J. & Mira, J. P. (2001) Arch Pediat. 4, 689–696. [Google Scholar]
- 24.Fernandez, M. I., Pedron, T., Tournebize, R., Olivo-Marin, J. C., Sansonetti, P. J. & Phalipon, A. (2003) Immunity 18, 739–749. [DOI] [PubMed] [Google Scholar]
- 25.Bickel, M., Axtelius, B., Solioz, C. & Attstrom, R. (2001) J. Clin. Periodontol. 28, 840–847. [DOI] [PubMed] [Google Scholar]
- 26.Fink, L., Seeger, W., Ermert, L., Hanze, J., Stahl, U., Grimminger, F., Kummer, W. & Bohle, R. M. (1998) Nat. Med. 4, 1329–1333. [DOI] [PubMed] [Google Scholar]
- 27.Tlaskalova-Hogenova, H., Stepankova, R., Hudcovic, T., Tuckova, L., Cukrowska, B., Lodinova-Zadnikova, R., Kozakova, H., Rossmann, P., Bartova, J., Sokol, D., et al. (2004) Immunol. Lett. 93, 97–108. [DOI] [PubMed] [Google Scholar]
- 28.Romagnani, S. (2002) Mol. Immunol. 38, 881–885. [DOI] [PubMed] [Google Scholar]
- 29.Xia, H., Mao, Q., Eliason, S. L., Harper, S. Q., Martins, I. H., Orr, H. T., Paulson, H. L., Yang, L., Kotin, R. M. & Davidson, B. L. (2004) Nat. Med. 10, 816–820. [DOI] [PubMed] [Google Scholar]
- 30.Paddison, P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. (2002) Genes Dev. 16, 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimri, G. P., Lee, X., Basile, G. & Campisi, J. (1995) Proc. Natl. Acad. Sci. USA 92, 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikita, T., Daniel, C., Wu, P. & Schindler, U. (1998) J. Biol. Chem. 273, 17634–17642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.