Abstract
With the recent completion of a high-quality sequence of the human genome, the challenge is now to understand the functional elements that it encodes. Comparative genomic analysis offers a powerful approach for finding such elements by identifying sequences that have been highly conserved during evolution. Here, we propose an initial strategy for detecting such regions by generating low-redundancy sequence from a collection of 16 eutherian mammals, beyond the 7 for which genome sequence data are already available. We show that such sequence can be accurately aligned to the human genome and used to identify most of the highly conserved regions. Although not a long-term substitute for generating high-quality genomic sequences from many mammalian species, this strategy represents a practical initial approach for rapidly annotating the most evolutionarily conserved sequences in the human genome, providing a key resource for the systematic study of human genome function.
Keywords: comparative genomics, genome sequencing, genome analysis, phylogenetics, mammalian evolution
Comprehensive identification of functional elements in the human genome represents a central and ambitious goal in genomics (1). We currently have only rudimentary knowledge about such elements (apart from protein-coding sequences), and it is thus impossible to identify them directly from the human genome sequence. A powerful and unbiased approach for detecting candidates for such functionally important sequences is to compare orthologous regions from multiple related species to identify those regions that are evolving slowly and are thus likely to be under purifying selection. The crucible of evolution is a very sensitive assay for function: Selection will robustly reject mutations that decrease the fitness of a mammal to 99.9% of normal (2), whereas such a decrease is undetectable in typical laboratory tests.
The first opportunity to compare entire mammalian genomes came with the sequencing of the mouse (3) and subsequently the rat (4) genomes. Strikingly, sequence comparisons between the human genome and either rodent genome revealed that ≈5% of each of these genomes appears to be under purifying selection. Specifically, this analysis involved comparing (i) the distribution of calculated conservation scores for bases (assessed in small windows) across the entire genome with (ii) the distribution of the conservation scores for bases within transposable element fossils predating the divergence of humans and rodents (called ancestral repeats, which are thought to be nonfunctional and thus evolving at the background rate of neutral evolution). The former distribution showed a clear excess of bases with higher-than-average conservation scores, corresponding to about 5% of the genome. These results were surprising because it had been tacitly assumed that the predominant functional sequences in the mammalian genome were those directly encoding proteins, but these account for <2% of the genome. The full nature of the remaining ≈3% of the genome remains a mystery; presumably, they include gene-regulatory elements, RNA genes, chromosomal structural elements, and other as-yet-unknown functional elements.
Although the human-rodent sequence comparisons allow an overall estimate of the amount of the human genome that is functionally important (more precisely, under purifying selection), such analyses are inadequate for accurately identifying most of the functional elements. Some regions can be clearly identified as under strong constraint, such as “ultra-conserved sequences” with 100% identity over hundreds of bases across several species (5). However, most regions have intermediate conservation scores; some of these are functional elements and some represent the right tail of the distribution of neutrally evolving sequences. These alternatives can be distinguished by analyzing sequences from additional species (6), so that functional sequences stand out against the background of neutral evolution. In particular, the signal-to-noise ratio increases as one expands the comparison to an evolutionary tree with more species and longer total branch length.
How many mammals must be sampled for identifying functional elements in the mammalian genome? The answer depends on the precise goal(s) being pursued. Several studies have investigated this issue (7-10).
For example, Kellis et al. (10) described the ability to perform systematic identification of gene-regulatory elements in yeast, consisting of weakly conserved six-base sequences that occur multiple times in the genome. They extrapolated that similar results could be obtained for the human genome with sequence data from species constituting an evolutionary tree that provides a total branch length D = ≈4.
More generally, Eddy (11) considered the identification of individual sequence elements. He reported formulas for calculating the number N of mammalian species related by an evolutionary tree with equal branches of length d that would be needed to detect a given type of element, as a function of the element's length L, the conservation rate ω among the species, and the desired false-positive and false-negative rates. Considering highly conserved sequences (with each base evolving at a rate ω = 20% of the neutral rate), his results show that elements with L ≥ 50 bases can be detected with only human and mouse sequences (total branch length D = 0.45). Detection of elements with L = 8 bases could be accomplished with species providing a total branch length D =≈4 (for example, 40 species each with d = 0.1 from a common root). Detection of single bases under purifying selection (L = 1) could be achieved with a total branch length D = ≈32 (for example, 320 species at d = 0.1 from a common root).
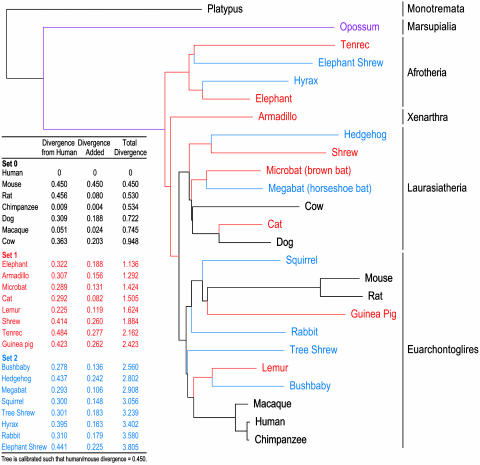
Based on such analyses, a reasonable starting point would be to obtain sequence from a set of mammals that provides a total branch length D = ≈4, with the aim of identifying functional elements eight bases or more in length. Fig. 1 shows one possible choice of species, displayed in an evolutionary tree that indicates the phylogenetic relationships and branch lengths. The species are divided into three sets: seven mammals for which high-redundancy genomic sequence has already been generated (total branch length D =≈0.95); eight additional mammals (set 1) that increase the branch length to D = ≈2.4; and eight further mammals (set 2) that increase the total to D =≈3.8. Ultimately, it would be desirable to have high-quality near-complete genomic sequence for all 16 of these additional mammals. However, this would require at least 8-fold sequence redundancy of each genome, or nearly 400 gigabases (Gb) of raw sequence. Given current capacities and costs, such an effort would require a large investment of resources and a considerable period of time.
Fig. 1.
Phylogenetic tree of eutherian mammals proposed for genome sequencing. Various sets of eutherian mammals are shown: set 0 consisting of seven species for which high-redundancy genomic sequence is already available (black), set 1 consisting of an additional eight species proposed for sequencing (red), and set 2 consisting of a further eight species proposed for sequencing (blue). The tree also shows a marsupial (purple), for which genomic sequence is available, and a monotreme (gray). The Inset table lists each species, the branch length (divergence) relative to human (in substitutions per site), the increase in total branch length (D) provided by adding each species to those above, and the total branch length provided by that species combined with those above. Details about the phylogenetic tree and the associated branch lengths are provided in the supporting information, which is published on the PNAS web site and at www.nisc.nih.gov/data.
We thus sought to explore an initial approach to obtain a substantial portion of the information at lower cost and in less time. Specifically, we investigated the utility of generating lower-redundancy sequence of each genome. Simple mathematical modeling (12) predicts that roughly 2-fold average redundancy should cover ≈86% (1 - e-2) of bases in each mammalian genome, thereby providing considerable (albeit incomplete) data. Increasing the amount of sequencing to about 8-fold average redundancy would increase the proportion of each genome covered to >99% (1 - e-8) as well as enhance the continuity and accuracy of the resulting assembled sequences, but the associated costs would be roughly 4-fold greater for a modest gain in coverage.
Before embarking on a low-redundancy sequencing strategy, it is essential to demonstrate the practical utility of such data for comparing mammalian genomes. We thus sought to investigate two key questions. First, can low-redundancy sequence be accurately and completely positioned relative to the orthologous sequence in the human genome, thereby allowing fine-scale alignment and meaningful comparative analyses? Second, are the resulting alignments sufficient for identifying the most highly conserved sequences in the human genome? To investigate such issues, we directly compare the performance of low-redundancy sequence versus high-quality finished sequence.
Alignment of Individual Sequence Reads
We first investigated the ability to align low-redundancy sequence to the human genome, extending similar studies performed previously (13-15). We began by considering the case of aligning individual sequence reads (roughly 650 bases) to the entire human genome sequence. This is a worst-case scenario, because (i) most sequence reads will be paired with a second sequence read generated from the opposite end of the subclone insert (“paired-end reads”) (16), thereby providing two nearby sequences that can be positioned concurrently; and (ii) most sequence reads will be assembled into larger sequence contigs and scaffolds that can be mapped to the human genome (see below). However, the alignment of individual reads provides a useful starting point for analysis.
Alignment of Simulated Sequences. We began by studying simulated data because this allows the most rigorous assessment of alignment accuracy. Simulated sequence reads were generated from hypothetical mammalian genomes at various evolutionary distances from the human genome (d, measured in average number of substitutions per base). Specifically, the simulated reads were produced by taking random 650-base segments of the human genome sequence and subjecting them to random “mutations” at appropriate densities (see supporting information). By generating the data sets in silico, each simulated read corresponds to a known location in the human genome, and the orthologous position of each base is known with certainty.
The simulated reads were positioned relative to the human genome sequence by using an approach that considers both the alignment score of the best match for each read and the increment over the second-best match (“S1-S2 method”; see supporting information). This strategy helps to avoid incorrectly positioning sequences to paralogous (as opposed to orthologous) regions; it comes at a cost of slightly lower sensitivity, but brings with it higher specificity, which is particularly desirable for genomes containing large amounts of related sequences (e.g., gene families and paralogous duplicons). Matches were detected with either the blastz computer program (17) or a hardware-optimized Smith-Waterman algorithm (TimeLogic, Carlsbad, CA).
We explored how the ability to align reads to the entire human genome declines as the evolutionary distance from human increases. The proportion of reads that could be correctly positioned relative to the human genome sequence was found to be extremely high (≥95%) for species at a distance d ≤ 0.8 from human. The proportion falls off dramatically for longer branch lengths (e.g., 73% for d = 1.0 and 4% for d = 1.2) because the number of false-positive matches in a 3-Gb genome grows too large.
Because the largest pairwise distance between eutherian mammals is <0.5 (Fig. 1), these results suggest that it should be possible to align sequence reads from any eutherian mammal to the entire human genome with good sensitivity and specificity.
Alignment of Actual Sequences. The main difference in analyzing actual (as opposed to simulated) sequence data is that many sequences in other mammalian species have no ortholog in the human genome. Such sequences reflect DNA that was inserted after divergence from the human lineage or deleted in the human lineage. For example, only ≈40% of the mouse (3) or rat (4) genomes has orthologous counterparts in the human genome, which sets an upper bound on the proportion of sequence from a given mammalian species that can possibly be aligned to the human genome sequence (≈40% in the case of rodents). The question is then: How closely can one come to this upper bound?
We analyzed 650-base sequences selected randomly from the current mouse genome sequence assembly (see supporting information). Using the same procedure as above, we found that 35% of these “reads” could be aligned to the human genome, with 99% of these aligning to the correct orthologous position within the genome. Scrutiny of the sequences that fail to align to the human genome sequence confirms that virtually all reflect segments that are wholly or partially absent in the human genome (see supporting information). We additionally studied two specific 1-Mb regions of the mouse genome (see supporting information) that differ in their proportion of orthologous sequence relative to the human genome (57% and 40% of bases, respectively). The respective proportion of reads that could be correctly aligned to the human genome was 57% and 34%, showing that the proportion of alignable reads is closely related to the proportion of orthologous sequence.
We similarly examined seven additional mammals (cat, dog, pig, cow, rat, mouse, and hedgehog) for which significant stretches of high-quality finished sequence was available (from ENCODE regions (18) ENm001 and ENm005, with only the first region studied for the latter two species). The results for each species are summarized in Table 1. The proportion of reads that aligned to a location within the human genome ranges from 76.7% for dog (d = 0.31; 98.2% aligning to the correct location) to 35.9% for hedgehog (d = 0.44; 97.3% aligning to the correct location); these findings are consistent with the fraction of bases that align with finished sequence data (data not shown). For all species in Table 1, at least 97% of the reads align to the correct location, indicating that the vast majority of orthologous sequence reads generated from any eutherian mammal can be accurately aligned to the human genome sequence.
Table 1. Alignment of sequence reads from various mammals to the human sequence of two ENCODE regions.
| ENCODE region ENm001
|
ENCODE region ENm005
|
||||||
|---|---|---|---|---|---|---|---|
| Species | Distance to human | No. of reads | Aligned to human, % | Correctly aligned to human, % | No. of reads | Aligned to human, % | Correctly aligned to human, % |
| Cat | 0.29 | 15,152 | 75.8 | 97.0 | 16,143 | 63.9 | 97.3 |
| Dog | 0.31 | 11,714 | 76.7 | 98.2 | 17,525 | 67.5 | 97.8 |
| Pig | 0.34 | 13,682 | 76.1 | 97.7 | 17,673 | 58.8 | 97.5 |
| Cow | 0.36 | 15,511 | 64.9 | 98.0 | 14,404 | 58.2 | 98.2 |
| Mouse | 0.45 | 15,847 | 46.9 | 98.4 | ND | ND | ND |
| Rat | 0.46 | 19,240 | 41.1 | 97.3 | 12,725 | 36.9 | 98.0 |
| Hedgehog | 0.44 | 15,779 | 35.9 | 97.3 | ND | ND | ND |
The sequences of two ENCODE regions, ENm001 and ENm005 [http://genome.ucsc.edu/ENCODE/regions.html (18)], were generated from the indicated seven species (see www.nisc.nih.gov). For each species, the phylogenetic distance relative to human is given in average number of substitutions per base (see Fig. 1). Columns show the total number of high-quality sequence reads processed for alignment to the entire human genome (using blastz and the S1-S2 scoring method; see supporting information), the percentage of reads that aligned to the human genome, and the percentage of aligning reads that correctly aligned to the orthologous region of the human genome. ND, not determined.
Assembly of Low-Redundancy Sequence
Aligning individual sequence reads is, as noted above, a worst-case scenario. Sequence reads can be assembled into contigs and scaffolds, which should be easier to align. To test this possibility, we constructed assemblies of the mouse genome based on random paired-end sequences providing 2- and 3-fold redundancy (see supporting information).
The resulting N50 scaffold lengths (defined as the length L at which 50% of the assembled sequence falls within contigs of size ≥ L) are large: 42 kb and 288 kb, respectively. Furthermore, the assembly quality appears to be good. Specifically, we compared the assemblies to 2 Mb of high-quality finished mouse genomic sequence: only two ≈2-kb contigs were misassembled and two ≈1-kb contigs were incorrectly oriented. Such errors would have only minimal impact on alignment and comparative analyses.
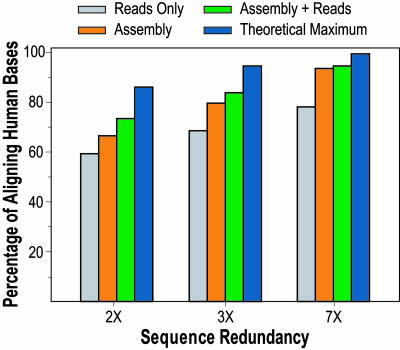
Fig. 2 illustrates the results of aligning assembled sequence generated at various levels of redundancy. The analysis focused on six finished BAC sequences, comprising ≈1 Mb of the mouse genome (see supporting information). This finished mouse sequence was aligned to the human genome, with ≈27% of the bases in the orthologous human region aligning. We then determined the proportion of these bases contained in mouse-human alignments when using mouse genome assemblies generated with lower-redundancy data. With 1-fold redundancy of unassembled sequence reads, ≈40% of these bases align (data not shown). The theoretical maximum based on Poisson sampling is 63% (1 - e-1), but this is not achieved because sequence reads consisting of repetitive sequences cannot be accurately positioned. With 2-fold redundancy, the proportion of aligning bases is ≈60% with unassembled reads, ≈67% with assembled contigs containing two or more reads, and ≈74% with assembled contigs plus singleton unassembled reads (Fig. 2). The latter is relatively close to the theoretical maximum under Poisson sampling of 86% (1 - e-2). This situation incrementally improves with 3-fold redundancy. These results indicate that the vast majority of bases that align with finished sequence can be aligned by using relatively low-redundancy sequence (e.g., 2- to 3-fold).
Fig. 2.
Characterization of alignments with various levels of sequence redundancy. Six regions of the mouse genome were studied, for which finished sequence was generated from bacterial artificial chromosome (BAC) clones (see supporting information for details). The sequence reads corresponding to these regions were extracted from whole-genome shotgun sequence data generated for the mouse genome (3). Subsets of these reads providing various levels of sequence redundancy (2×, 3×, and 7×) were then selected and assembled. The various data sets were then aligned to the human genome. The bar graph depicts the percentage of aligning bases, defined as the number of aligned human bases relative to the number obtained with finished mouse sequence. The different bars reflect alignments with all sequence reads before assembly (reads only); assembled sequence contigs containing two or more reads (assembly); assembled sequence contigs plus the remaining unassembled singleton reads (assembly + reads); and theoretical maximum attainable with indicated level of redundancy [calculated from the Lander-Waterman equation (12)].
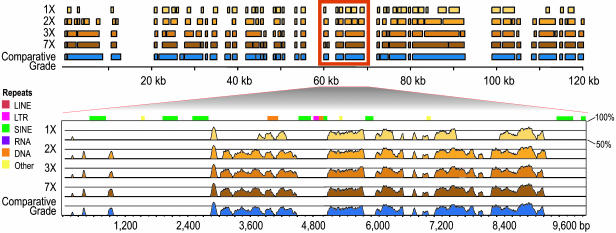
Fig. 3 further illustrates this point, depicting alignments between hedgehog and human sequences obtained with hedgehog sequence reads providing various levels of redundancy. The difference between low-redundancy sequence data (e.g., 2-fold) and high-quality sequence (e.g., 7-fold or finished) is relatively small.
Fig. 3.
Alignments obtained with various levels of sequence redundancy. High-quality sequence of a hedgehog BAC was used to illustrate alignments with lower-redundancy sequence data (see supporting information for details). “Comparative-grade” finished sequence (23) was generated for a BAC containing 120 kb of hedgehog genomic DNA. The data generated for that BAC were used to create subsets of sequence reads that provided various levels of sequence redundancy (1×, 2×, 3×, and 7×). The comparative-grade sequence and each of the unassembled subsets of sequence reads were then aligned to the orthologous region of the human genome, with the results shown at the top, as displayed with the Apollo viewer (24). An expanded view of a 10-kb interval is shown below, as displayed with the MultiVista viewer (25). Regions of the human genome without a hedgehog alignment largely reflect deletions in the hedgehog lineage or insertions in the human lineage since the most recent common ancestor of humans and hedgehogs. The divergence of hedgehog relative to human is estimated to be 0.44 (substitutions per site; see Fig. 1), roughly equivalent to that of mouse.
We also studied mouse genome assemblies generated with lower redundancies. The assembly with 1-fold redundancy has a dramatically lower N50 scaffold length than that with 2-fold redundancy (2 kb vs. 42 kb, respectively). Accordingly, the former has little utility in independently confirming the accurate placement of reads in the genome or in identifying problematic complex regions. In addition, most bases in the genome are covered by at most one read. The nucleotide accuracy of such a lower-redundancy assembly is thus substantially reduced, which may complicate analyses of functional elements (e.g., by introducing spurious insertions-deletions in coding regions).
In summary, genome assemblies with 2- or 3-fold redundancy provide substantial long-range information that can aid comparative analyses. Assemblies with 1-fold redundancy have much more limited utility.
Identification of Conserved Regions
We next explored the ability to detect highly conserved regions in the human genome with low-redundancy sequence data. To address this issue, it is necessary to define a “highly conserved region”; various definitions and algorithms have been used (7, 19-21). Here, we used the multispecies conserved sequences (MCSs) as described by Margulies et al. (7).
We examined the ENCODE region ENm001 (22), for which comparative-grade sequence is available from 11 mammals. [Comparative-grade finished sequence is assembled with at least 8-fold sequence redundancy and then refined to eliminate gross misassemblies and ensure correct order and orientation of sequence contigs (23).] Using these multispecies sequences, we identified a reference set of MCSs (7).
We then assessed the ability to detect these MCSs by using reconstructed data sets that had lower redundancies (0.5- to 6-fold) and sequences from fewer species. Each data set was assembled into contigs and scaffolds (but not further refined), aligned to the orthologous targeted region of the human genome sequence, and used to identify MCSs (see supporting information). With thresholds for detection chosen to achieve 97% specificity (7), we assessed the sensitivity of MCS detection for each data set.
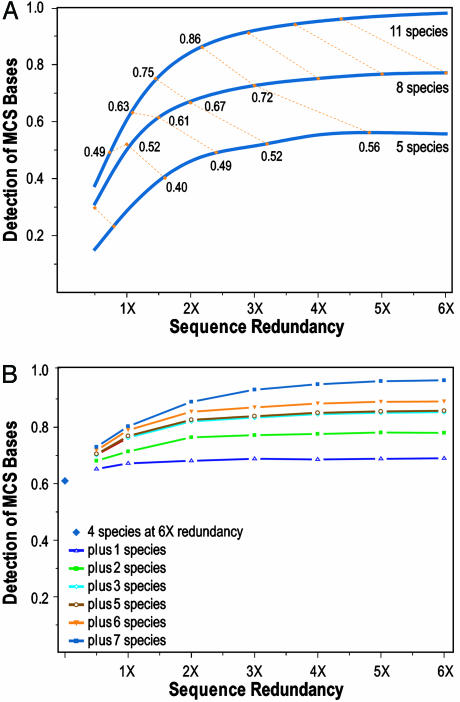
Fig. 4A compares the results for all 11 mammals and then subsets of 8 and 5 mammals, in each case analyzed at various levels of redundancy (and intermediate values interpolated). As expected, the sensitivity increases with the number of species sequenced and with higher redundancy (although the incremental gains are small beyond roughly 3-fold redundancy). The key issue concerns the tradeoff between species number and redundancy for a fixed total amount of sequencing. The iso-read curves in Fig. 4A connect points corresponding to equivalent total numbers of sequence reads. Considering the situations with different redundancies for 8 mammalian genomes, one can ask whether it would be better to distribute the same total sequence across more species or concentrate it over fewer species. With 1-fold redundancy of 8 mammals, the sensitivity of MCS detection (52%) is higher than the iso-read equivalent for 11 mammals (49%). At 1.5-fold redundancy of 8 mammals, the sensitivity is similar to that for 11 mammals. At 2-fold redundancy of 8 mammals, the sensitivity with 11 mammals is somewhat higher. At higher levels of redundancy, it is clearly better to distribute the reads across more mammals. In the cases examined, the sensitivity with 8 mammals is higher than that for the iso-read equivalents with 5 mammals. These results suggest that the efficiency of MCS detection decreases with redundancies greater than roughly 2-fold; it is thus better to obtain additional species' sequences than to obtain deeper redundancies.
Fig. 4.
Identification of MCSs with various levels of sequence redundancy. (A) The finished sequences of ENCODE region ENm001 (7, 18, 22) from 11 mammalian species were used to identify a reference set of MCSs. MCS detection was then repeated with subsets of the data providing several lower levels of sequence redundancy (using both assembled sequence contigs and unassembled reads) for different subsets of mammals. The threshold was set to ensure 97% specificity for detecting the reference set of MCS bases. The performance of detecting MCSs with 5 (lemur, dog, horse, hedgehog, and mouse), 8 (the previous 5 plus pig, armadillo, and rabbit), or 11 (the previous 8 plus cat, cow, and rat) species is depicted by three smoothed curves statistically fit to the actual data (see supporting information). The yellow dotted lines connect “iso-read” equivalents (see text for details) for 5, 8, and 11 mammals, calculated for discrete increments of redundancy for 8 mammals (0.5- through 6-fold redundancy), with the indicated numbers reflecting the sensitivity of MCS detection for that data point. (B) Analogous studies were performed with region ENm005, but starting with 6-fold redundant sequence from 4 species for which whole-genome sequence is already available (dog, rat, mouse, and chicken) and then adding sequence from 7 additional species [in the order cat, cow, pig, fugu plus Tetraodon (pufferfish), galago, and baboon].
Analogous results were obtained for ENCODE region ENm005, where we analyzed the effect of starting with already available sequence (at 6-fold redundancy) from four vertebrate species and then adding low-redundancy sequence from additional species one at a time (see Fig. 4B).
The precise results will vary with the specific definition of a highly conserved region, the species used, and the genomic regions studied. However, the above findings provide useful guidance in fashioning an overall strategy.
A Proposed Initial Sequencing Strategy
The above analyses demonstrate that (i) the vast majority of orthologous sequence in a mammalian genome can be aligned to the correct position in the human genome, even when using relatively short sequence stretches (i.e., individual sequence reads), and (ii) the resulting alignments can be used effectively for identifying the most highly conserved regions of the human genome (i.e., MCSs). These findings lend support to an initial strategy aimed at accelerating identification of functional elements in the human genome.
Given limited sequencing capacity, what is the best choice for the level of sequence redundancy generated for each species? For concreteness, we consider a sequencing capacity sufficient to generate 32-fold redundancy of a typical mammalian genome (≈100 Gb total). We consider several alternatives:
With a high redundancy (e.g., 7-fold), the resulting assemblies and alignments to the human genome should be excellent. However, one could generate data from fewer than 5 mammals, and the power to detect conserved regions would thus be greatly compromised.
A redundancy of 2- or 3-fold appears to be a more efficient option. One could sequence 16 mammalian genomes at 2-fold redundancy (or ≈11 at 3-fold). The resulting assemblies should provide good continuity (in terms of N50 scaffold length), good alignability, and good power to detect conserved regions.
The redundancy could be further reduced to 1-fold, allowing sequence to be obtained from 32 mammals. The above results indicate that there would be a small gain in the sensitivity of detecting conserved regions. However, the genome assemblies would be dramatically lower in quality with respect to continuity and accuracy (see above). This lowering could have negative consequences for many comparative analyses. The tradeoff is complex and deserving of more study, but it is our judgment that these deficiencies more than offset the modest gains in MCS detection.
Accordingly, we propose an initial strategy involving the shotgun sequencing of ≈16 additional eutherian mammalian genomes at roughly 2-fold redundancy. A possible choice of mammals is shown in Fig. 1. The precise details of the plan should be refined on the basis of additional data and analyses. For example, the redundancy might be tuned up or down after analyses of the sequences from an initial subset of mammals; such results would provide more definitive information than that available to date.
The choice of species also deserves continuing consideration. The species in Fig. 1 were chosen with several goals in mind: (i) maximizing total branch length, to increase the sensitivity of detecting conserved regions; (ii) representation of all major branches of the mammalian clade; (iii) selection of species with biological or medical utility, when possible; and (iv) availability of DNA samples (see supporting information). The tree in Fig. 1 provides a total branch length of D = 3.81 (a total of 0.95 from the 7 mammals with already-available sequence and an average of 0.18 from each of the additional 16 mammals). Given the expected coverage of the alignable human bases provided by these additional mammalian sequences (≈74% for 2-fold redundancy, as measured above), the proposed sequence data would provide an average branch length of D =≈3.1 [= 0.95 + (2.86 × 0.74)] at a typical alignable human base. This result falls short of the target of D =≈4 cited above, but it represents a reasonable start toward the goal.
We should emphasize that the proposed plan is not intended as a substitute for eventually acquiring high-quality sequence data from these mammals, but rather as a pragmatic first step to facilitate systematic studies of the biological function of conserved elements. To allow for generation of additional sequence at a later date, it would be sensible to store sufficient DNA samples from the individuals selected for sequencing.
Our proposal has the limitation that it is designed only to find those elements that are conserved across eutherian mammals. It would not, for example, identify elements that are specific to primates. To find such elements, one could design a similar plan focused only on primate sequences. However, many more species would be required because the typical branch length from the common primate ancestor is much shorter. Accordingly, such a project might be best undertaken when sequencing costs have fallen considerably below current levels.
In summary, there is a growing convergence between evolutionary and experimental studies of humans. With the explosive growth in the ability to sequence genomes, evolutionary comparison is likely to become one of the most powerful tools for biomedical research. The proposed strategy aims to firmly put us on a path toward harnessing that potential.
Supplementary Material
Acknowledgments
We thank the members of the NISC Comparative Sequencing Program (particularly R. Blakesley, G. Bouffard, J. McDowell, B. Maskeri, and N. Hanson), the National Human Genome Research Institute-appointed working group providing advice about the use of comparative sequencing to help annotate the human genome (R. Waterston, Chair; with additional input provided by S. O'Brien, W. Murphy, and E. Teeling), various colleagues for scientific input and helpful advice (D. Bofelli, J. Butler, M. Kamal, and B. Handsaker), and D. Haussler, F. Collins, and R. Waterston for critical feedback about the manuscript.
Author contributions: E.H.M., J.P.V., K.L.-T., E.D.G., E.S.L., J.C.M., and M.C. designed research; E.H.M., J.P.V., D.B.J., K.L.-T., J.L.C., J.C.M., and M.C. performed research; E.H.M., J.P.V., N.C.S.P., D.B.J., J.C.M., and M.C. contributed new reagents/analytic tools; E.H.M., J.P.V., W.M., D.B.J., K.L.-T., E.D.G., E.S.L., J.C.M., and M.C. analyzed data; and E.H.M., J.P.V., W.M., K.L.-T., E.D.G., E.S.L., J.C.M., and M.C. wrote the paper.
Abbreviations: NISC, National Institutes of Health Intramural Sequencing Center; BAC, bacterial artificial chromosome; MCS, multispecies conserved sequence.
References
- 1.Collins, F. S., Green, E. D., Guttmacher, A. E. & Guyer, M. S. (2003) Nature 422, 835-847. [DOI] [PubMed] [Google Scholar]
- 2.Ohta, T. (1976) Nature 263, 74-76. [DOI] [PubMed] [Google Scholar]
- 3.International Mouse Genome Sequencing Consortium (2002) Nature 420, 520-562. [DOI] [PubMed] [Google Scholar]
- 4.Rat Genome Sequencing Project Consortium (2004) Nature 428, 493-521.15057822 [Google Scholar]
- 5.Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W. J., Mattick, J. S. & Haussler, D. (2004) Science 304, 1321-1325. [DOI] [PubMed] [Google Scholar]
- 6.Cooper, G. M. & Sidow, A. (2003) Curr. Opin. Genet. Dev. 13, 604-610. [DOI] [PubMed] [Google Scholar]
- 7.Margulies, E. H., Blanchette, M., NISC Comparative Sequencing Program, Haussler, D. & Green, E. D. (2003) Genome Res. 13, 2507-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margulies, E. H., NISC Comparative Sequencing Program & Green, E. D. (2004) Cold Spring Harbor Symp. Quant. Biol. 68, 255-263. [DOI] [PubMed] [Google Scholar]
- 9.Eddy, S. (1998) in Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids, eds. Durbin, R., Eddy, S., Krogh, A. & Mitchison, G. (Cambridge Univ. Press, Cambridge, U.K.), pp. 192-231.
- 10.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241-254. [DOI] [PubMed] [Google Scholar]
- 11.Eddy, S. R. (2005) PLoS Biol. 3, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lander, E. S. & Waterman, M. S. (1988) Genomics 2, 231-239. [DOI] [PubMed] [Google Scholar]
- 13.Bouck, J., Miller, W., Gorrell, J., Muzny, D. & Gibbs, R. (1998) Genome. Res. 8, 1074-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, R., Bouck, J. B., Weinstock, G. M. & Gibbs, R. A. (2001) Genome Res. 11, 1807-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirkness, E. F., Bafna, V., Halpern, A. L., Levy, S., Remington, K., Rusch, D. B., Delcher, A. L., Pop, M., Wang, W., Fraser, C. M., et al. (2003) Science 301, 1898-1903. [DOI] [PubMed] [Google Scholar]
- 16.Green, E. D. (2001) Nat. Rev. Genet. 2, 573-583. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz, S., Kent, W. J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R. C., Haussler, D. & Miller, W. (2003) Genome Res. 13, 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ENCODE Project Consortium (2004) Science 306, 636-640. [DOI] [PubMed] [Google Scholar]
- 19.Tagle, D. A., Koop, B. F., Goodman, M., Slightom, J. L., Hess, D. L. & Jones, R. T. (1988) J. Mol. Biol. 203, 439-455. [DOI] [PubMed] [Google Scholar]
- 20.Cooper, G. M., Brudno, M., Stone, E. A., Dubchak, I., Batzoglou, S. & Sidow, A. (2004) Genome Res. 14, 539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siepel, A. & Haussler, D. (2004) Mol. Biol. Evol. 21, 468-488. [DOI] [PubMed] [Google Scholar]
- 22.Thomas, J. W., Touchman, J. W., Blakesley, R. W., Bouffard, G. G., Beckstrom-Sternberg, S. M., Margulies, E. H., Blanchette, M., Siepel, A. C., Thomas, P. J., McDowell, J. C., et al. (2003) Nature 424, 788-793. [DOI] [PubMed] [Google Scholar]
- 23.Blakesley, R. W., Hansen, N. F., Mullikin, J. C., Thomas, P. J., McDowell, J. C., Maskeri, B., Young, A. C., Benjamin, B., Brooks, S. Y., Coleman, B. I., et al. (2004) Genome Res. 14, 2235-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis, S. E., Searle, S. M. J., Harris, N., Gibson, M., Lyer, V., Richter, J., Wiel, C., Bayraktaroglir, L., Birney, E., Crosby, M. A., et al. (2002) Genome Biol. 3, RESEARCH0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayor, C., Brudno, M., Schwartz, J. R., Poliakov, A., Rubin, E. M., Frazer, K. A., Pachter, L. S. & Dubchak, I. (2000) Bioinformatics 16, 1046-1047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.