Abstract
This study focused on the screening of small-molecule inhibitors that target signal transducers and activators of transcription 3 (Stat3) in human breast carcinoma. The constitutive activation of Stat3 is frequently detected in human breast cancer cell lines as well as clinical breast cancer specimens and may play an important role in the oncogenesis of breast carcinoma. Activated Stat3 may participate in oncogenesis by stimulating cell proliferation, promoting tumor angiogenesis, and resisting apoptosis. Because a variety of human cancers are associated with constitutively active Stat3, Stat3 represents an attractive target for cancer therapy. In this study, of the nearly 429,000 compounds screened by virtual database screening, chemical samples of top 100 compounds identified as candidate small-molecule inhibitors of Stat3 were evaluated by using Stat3-dependent luciferase reporter as well as other cell-based assays. Through serial functional evaluation based on our established cell-based assays, one compound, termed STA-21, was identified as the best match for our selection criteria. Further investigation demonstrated that STA-21 inhibits Stat3 DNA binding activity, Stat3 dimerization, and Stat3-dependent luciferase activity. Moreover, STA-21 reduces the survival of breast carcinoma cells with constitutive Stat3 signaling but has minimal effect on the cells in which constitutive Stat3 signaling is absent. Together, these results demonstrate that STA-21 inhibits breast cancer cells that express constitutively active Stat3.
Keywords: breast carcinoma, dimerization, STA-21
Signal transducers and activators of transcription 3 (Stat3) is activated in response to cytokines and growth factors (1). JAKs, Src, and EGF receptor are Stat3 upstream regulators (2-4). The main domains of Stat3 protein include the tetramerization and leucine zipper at the N terminus, the DNA-binding domain, and the SH2 transactivation domain at the C-terminal end. The SH2 region is responsible for the binding of Stat3 to the tyrosine-phosphorylated receptors and for the dimerization that is necessary for DNA binding and gene expression (5). Stat3 is activated by phosphorylation at tyrosine residue 705 (Tyr-705), which leads to dimer formation, nuclear translocation, recognition of Stat3-specific DNA binding elements, and activation of target gene transcription (1, 5).
The constitutive activation of Stat3 is frequently detected in breast carcinoma cell lines but not in normal breast epithelial cells (6, 7). It has been reported that ≈60% of breast tumors contain persistently activated Stat3 (8). Stat3 has been classified as an oncogene because activated Stat3 can mediate oncogenic transformation in cultured cells and tumor formation in nude mice (9). Stat3 may participate in oncogenesis by stimulating cell proliferation, promoting angiogenesis, and conferring resistance to apoptosis induced by conventional therapies (10-13). The possible downstream targets on which Stat3 promotes its oncogenesis may be through up-regulation of antiapoptotic factors (Bcl-2, survivin, Mcl-1, and Bcl-XL), cell-cycle regulators (cyclin D1 and c-myc), and inducer of tumor angiogenesis (VEGF) (9, 13-17). Activated Stat3 signaling directly contributes to malignant progression of cancer. Stat3 oncogenic function acts through the prosurvival proteins such as survivin, Mcl-1, Bcl-2, and Bcl-XL and results in inhibition of apoptosis (14, 18-20). Blockade of Stat3 signaling inhibits cancer cell growth, indicating that Stat3 plays a role in the survival or growth of tumor cells (12, 18, 19, 21-24).
Because Stat3 is frequently activated in breast cancer (8), it represents an attractive target for the development of new anti-cancer therapy in the treatment of breast cancer. In one approach, peptide-based Stat3 inhibitors were designed to target the Stat3 SH2 domain and were effective in blocking the Stat3 function in vitro (25). In another approach, compounds have been used to inhibit Stat3 upstream regulators Janus kinases (JAKs), especially JAK2 (26). In our opinion, direct inhibition of Stat3 by using drug-like, nonpeptide small molecules has several advantages, including blocking all the activity mediated by Stat3 activation and compounds with better cell permeability and better in vivo stability and bioavailability. Based on high-resolution x-ray 3D crystal structure of Stat3β homodimer (27), the SH2 domain is critical for Stat3 dimerization, which is a decisive event for the activation of Stat3 (9). Therefore, we hypothesize that a small molecule that binds to the Stat3 SH2 domain may directly block Stat3 dimerization and its activity. In this work, we report the discovery of a Stat3 small-molecule inhibitor through virtual database screening.
Materials and Methods
Structure-Based Virtual Screening. To identify potential candidate compounds that can disrupt the Stat3 dimerization, the crystal structure of Stat3β solved at 2.25-Å resolution (27) was retrieved from the Protein Data Bank (PDB ID code 1BG1) (28) and was used in this study. The chemical databases used in our virtual screening included the National Cancer Institute (NCI) database, the Merck Index, and the Sigma-Aldrich and Ryan Scientific (Isle of Palms, SC) catalogs. Collectively, these four databases offered a collection of ≈429,000 small-molecule organic compounds. The public NCI database provides 3D structural models available from NCI. The other three chemical catalogs only provide 2D chemical structures; their 3D structural models were generated by using the corina program (Version 2.6, Molecular Networks, Erlangen, Germany) with the standard settings.
The molecular docking program dock (Version 4.0) (29) was used to perform the virtual screening. The binding cavity on the Stat3 SH2 domain was the region targeted for docking. The sybyl software (Version 6.9, Tripos Associates, St. Louis) was used to assign the standard AMBER (refers to a set of molecular mechanical force fields for the simulation of biomolecules) atomic partial charges on the Stat3β protein and the Gasteiger-Hückel atomic partial charges on each ligand molecule to be docked. The parameters used by dock, which controlled how it performed docking in our work, are summarized in the supporting information, which is published on the PNAS web site. Each molecule in our databases with a molecular weight between 200 and 700 was docked into the targeted binding site in Stat3. The top 10% scored compounds from each database, as selected by dock, were extracted and combined together to provide a total of 35,000 candidate compounds. Based on the binding models of these compounds predicted by dock, the x-score program (Version 1.1) (30) was applied to obtain an estimate of the binding affinities of these compounds of Stat3. The preselected 35,000 compounds then were reranked according to their binding affinities as estimated by x-score. Of the best-scored 200 compounds selected by x-score from the 35,000 compounds, physical samples of 100 compounds were obtained from NCI or purchased from Sigma-Aldrich or Ryan Scientific.
Cell Lines and Culture. Human breast cancer cell lines MDA-MB-231, MDA-MB-435s, MDA-MB-453, MDA-MB-468, and MCF7, human ovarian carcinoma cell line Caov-3, and human skin fibroblasts were cultured in DMEM containing 10% FBS and appropriate antibiotics in a 5% CO2 incubator at 37°C. For testing different compounds, the cells were exposed to the compounds for 48 h at a final concentration of 20 or 30 μM, respectively. Then, the cells were harvested and subjected to flow-cytometry analysis as described in refs. 21 and 31.
Plasmid Transfection into MDA-MB-435s Cells. The plasmids, pCMV-Stat3-Flag (kind gift from James Darnell, Rockefeller University, New York) and pCMV-Stat3-hemagglutinin (HA) (constructed by Vector Core Facility, University of Michigan, Ann Arbor, MI) for expression of Stat3-Flag and Stat3-HA tagged proteins, were cotransfected into MDA-MB-435s cells by using Lipofectamine 2000 reagent (Invitrogen). After 48-h transfection, the cells were treated with 20 μM STA-21 for 24 h and then harvested for immunocytochemistry and Western blot analysis.
Establishment of Stat3-Dependent Luciferase Reporter in Breast Cancer Cells. Plasmid pLucTKS3 contains seven copies of Stat3-binding site in thymidine kinase minimal promoter (32) and its activation specifically depends on Stat3 status in the cell environment. pLucTKS3 and pLucSV40 luciferase reporter plasmids were transfected into Caov-3 and MDA-MB-435s cell lines, respectively, by using Lipofectamine 2000 reagent. The stable clones, which showed high luciferase activity, were selected for screening Stat3 inhibitors. The selected clones were exposed to Stat3 inhibitors at a final concentration of 20 μM for 48 h, and luciferase activity was measured by using a Promega luciferase kit, following the manufacturer's instructions.
EMSA Assay. To test the effect of candidate compounds on Stat3 DNA binding activity, a gel EMSA was performed as described in ref. 25.
Immunocytochemistry. The cells transfected by pCMV-Stat3-Flag and pCMV-Stat3-HA plasmids were exposed to 20 μM STA-21 for 24 h, then fixed with 100% methanol for 30 min at -20°C. After 3× washes with PBS, anti-HA (rabbit, Santa Cruz Biotechnology) and/or anti-Flag (mouse, Sigma) antibodies (Abs) were added to the cells and incubated for 1 h at 37°C. The cells were washed 3× with PBS, then secondary Abs for anti-rabbit IgG-FITC and/or anti-mouse IgG-rhodamine were added and incubated for 1 h. After 3× washes by PBS, the cells were observed by using a fluorescence microscope.
Results
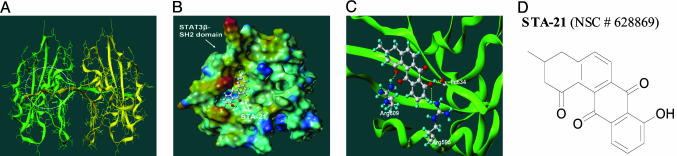
Discovery of STA-21 Through Structure-Based Virtual Database Screening. The 3D structure of STAT3β homodimer shows that the dimerization of Stat3 occurs between two SH2 domains (Fig. 1A) (27, 28). These two SH2 domains are hinged together by a loop segment (from Ala-702 to Phe-716) from each monomer. The phosphoryl Tyr-705 critical for the biological function of Stat3 locates right on this loop segment and binds, together with several adjacent amino acid residues (Leu-706, Thr-708, and Phe-710), to a cavity on the SH2 domain of the other monomer. The targeted region defined in our virtual screening covers where the Tyr-705-Phe-710 peptide segment binds. We hypothesize that a small molecule that binds to this region will compete with the Tyr-705-containing peptide, consequently blocking the dimerization of Stat3.
Fig. 1.
Schematic diagrams of modeling of structure-based virtual database screening. (A) SH2 domain dimerization interface of the STAT3β protein. The structure is based on Protein Data Bank entry 1BG1. Two SH2 domains are colored differently. The circled region indicates the target PTR-binding site used in our virtual screening study. (B) Predicted binding model of STA-21 to the STAT3β SH2 domain. STA-21 is rendered by a ball-and-stick model. The molecular surface of STAT3β SH2 domain is colored with the electrostatic potentials: red for most positively charged regions and blue for most negatively charged regions. (C) Specific hydrogen bonds formed between the STAT3β SH2 domain and STA-21. The binding model was predicted by dock. Only the residues that form hydrogen bonds with STA-21 are shown in explicit atomic models. (D) STA-21 structure. A-C were generated by using sybyl.
With the aid of structure-based virtual screening, we narrowed our interests from a total of 429,000 compounds to 200 top candidate compounds and were able to obtain the chemical samples of 100 compounds. We first tested these 100 compounds by using in vitro cell luciferase assay (see supporting information). Of the 100 compounds tested, the most promising compound is STA-21 (National Service Center no. 628869) obtained from NCI. STA-21, a natural product, is a deoxytetrangomycin, an angucycline antibiotic with a molecular weight of 306 (Fig. 1D). The binding mode of STA-21 to Stat3 was predicted by the dock program and was refined by structural organization using the AMBER force field implemented in sybyl software. The refined model, shown in Fig. 1 B and C, predicts that STA-21 binds at the same site where the Tyr-705-containing peptide binds and forms a number of hydrogen bonds with nearby residues, including Arg-595, Arg-609, and Ile-634 (Fig. 1C).
STA-21 Inhibited Stat3-Dependent Luciferase Reporter Activity in Carcinoma Cells with Constitutive Stat3 Signaling. The selected inhibitors were evaluated by using a Stat3 luciferase reporter system. Both MDA-MB-435s breast carcinoma and Caov-3 ovarian carcinoma cells express constitutively activated Stat3 (31, 33). We established cloned cells from these two cell lines by stable transfection of a Stat3-dependent luciferase reporter, pLucTKS3 (32). As a result of persistently activated Stat3, both cell clones showed high luciferase activity. Of the 100 small molecules tested, STA-21 showed remarkably inhibitory effect on Stat3-induced luciferase activity in Caov-3 cloned cells (Fig. 2A). The inhibition of STA-21 on Stat3 activation was further confirmed by using MDA-MB-435s cloned cells stably transfected with pLucTKS3 (Fig. 2B). For MDA-MB-435s cloned cells, after the exposure to 20 μM STA-21 for 48 h, luciferase activity was decreased by >5-fold (Fig. 2B). This result indicates that STA-21 may prevent Stat3 binding to pLucTKS3-containing Stat3-binding sites and inhibit luciferase activity in the cloned cells. In contrast, STA-21 did not affect luciferase activity in the clones transfected with simian virus 40 luciferase reporter that was absent of the Stat3-binding site (Fig. 2B), which indicated that STA-21 could not reduce luciferase activity by itself in the absence of Stat3 DNA-binding sites. As controls, we also listed several compounds that do not inhibit Stat3 luciferase activity as shown in Fig. 2A.
Fig. 2.
STA-21 inhibits Stat3-dependent luciferase activity in cancer cells. (A) The clone from Caov-3 carcinoma cells stably transfected with pLucTKS3 Stat3-dependent luciferase reporter was used for initial Sat3 small-molecule inhibitor screening. The cloned cells were treated with 20 μM STA-21 as well as other small-molecule compounds for 48 h, and then the cells were harvested for luciferase activity analysis. (B) The clones from MDA-MB-435s cells stably transfected with pLucTKS3 Stat3-dependent luciferase reporter or simian virus 40 (SV40) luciferase reporter were treated with 20 μM STA-21 for 48 h. Luciferase activity was measure by using a Promega luciferase kit according to the manufacturer's instructions. The results were based on the means and standard deviations from three separate experiments.
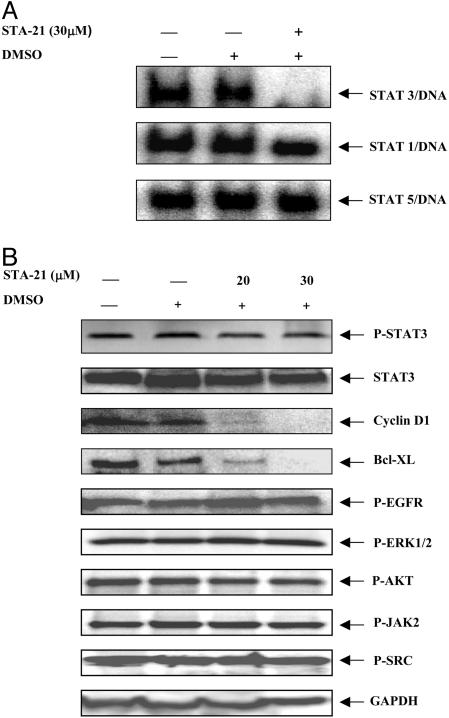
STA-21 Inhibited Stat3 but Not Its Upstream Regulators. We further examined whether or not STA-21 could reduce Stat3 DNA binding activity. In MDA-MB-435s breast carcinoma cells with constitutive Stat3 signaling, high Stat3 DNA binding activity was observed (Fig. 3A). In contrast, MCF10A and TERT breast cells without constitutive Stat3 signaling did not show Stat3 DNA binding activity (data not shown). STA-21 inhibited Stat3 DNA binding activity (Fig. 3A) and its downstream antiapoptotic factors in breast carcinoma cells with constitutive Stat3 signaling (Fig. 3B). In our study, MDA-MB-468 breast carcinoma cells were treated by STA-21 at a final concentration of 20 or 30 μM, respectively. STA-21 inhibited Stat3-regulated downstream factors, Bcl-XL and cyclin D1. Interestingly, the phosphorylation of Stat3 upstream regulators JAK2 (P-JAK2), Src (P-Src), and EGF receptor (P-EGFR) were not affected by STA-21 (Fig. 3B). Combined with the results of STA-21 inhibition to Stat3, but not Stat1 and Stat5, DNA binding activity in MDA-MB-435s cells (Fig. 3A), the inhibition of Stat3 by STA-21 may be through direct inhibition at the Stat3 protein level and is the least possibility of the inhibition of the Stat3 upstream regulators. In addition, STA-21 did not affect the phosphorylation of AKT (P-AKT) and ERK (P-ERK) (Fig. 3B).
Fig. 3.
STA-21 inhibits Stat3 DNA binding activity and Stat3-regulated antiapoptotic factors. (A) MDA-MB-435s cell nucleus extract was incubated with 30 μM STA-21 for 30 min at room temperature and then incubated with r-[32P]ATP-labeled consensus binding sequence for 20 min at room temperature. The reaction mixtures were resolved on 8% polyacrylamide gel. (B) The lysates from MDA-MB-468 cells treated with indicated concentrations of STA-21 for 48 h were resolved on 10% SDS/PAGE, then immunoblotted with Abs as indicated.
STA-21 Significantly Inhibited the Growth and Survival of Breast Carcinoma Cells with Constitutive Stat3. Because STA-21 inhibits Stat3-dependent luciferase and DNA binding activities, we next examined whether STA-21 inhibited the growth and survival of breast cancer cells with constitutive Stat3 signaling. After the cells were exposed to 20 or 30 μM STA-21 for 48 h, STA-21 showed remarkable inhibition of the survival of the breast carcinoma cells MDA-MB-231, MDA-MB-435s, and MDA-MB-468 that express persistently activated Stat3 (Fig. 4A). However, STA-21 had minimal inhibitory effect on MCF7 and MDA-MB-453 breast carcinoma cells and human skin fibroblasts that have no constitutive Stat3 signaling (Fig. 4B). Combined with the data from a cell viability assay (MTT, data not shown), STA-21 demonstrated strong potential to inhibit the growth and survival of breast cancer cells that express for constitutively active Stat3.
Fig. 4.
STA-21 inhibits the survival of breast carcinoma cells with constitutive Stat3 signaling but not in cells without constitutive Stat3 signaling. (A) The phosphorylation of Stat3 at Tyr-705 in different cell lines. (B) The cell lines were treated with STA-21 at the indicated concentrations for 48 h, and then cells were harvested and analyzed for the SubG1 profile that indicated apoptotic cells on a FACScan Flow Cytometer (Becton Dickinson). The results were based on the means and standard deviations from three separate experiments.
Stat3 Nucleus Translocation and Protein Dimerization Were Abrogated by STA-21 in MDA-MB-435s Breast Carcinoma Cells. The plasmids pCMV-Stat3-Flag and pCMV-Stat3-HA for expression of Stat3-Flag- and Stat3-HA-tagged proteins were cotransfected into MDA-MB-435s breast carcinoma cells. The results showed that the nucleus translocation of Stat3-Flag and Stat3-HA proteins was blocked by STA-21 (Fig. 5). The cells were treated with 20 μM STA-21 for 24 h, then immunostained with anti-rabbit HA IgG-FITC (green) and/or anti-mouse Flag IgG-rhodamine (red). From FITC and rhodamine separate staining, STA-21 seemed to inhibit Stat3-Flag or -HA nucleus translocation as shown in Fig. 5 E and F. In the cells without STA-21 treatment, strong orange staining was observed in the nucleus by using combined FITC and rhodamine staining, indicating that Stat3-Flag- and Stat3-HA-tagged proteins colocalized into the nucleus and two colors merged to become orange (Fig. 5D). However, when treated with 20 μM STA-21, much weaker orange staining was observed in the whole cell (Fig. 5G), suggesting that STA-21 blocked nucleus translocation of Stat3-Flag and -HA proteins. As a control, DMSO used as a solvent had no effect on the control cells (data not shown).
Fig. 5.
STA-21 inhibits Stat3 translocation and dimerization in breast carcinoma cells. The MDA-MB-435s cells cotransfected with pCMV-Stat3-Flag and pCMV-Stat3-HA plasmid were exposed to 20 μM STA-21 compound for 24 h, then fixed with 100% methanol. After the fixed cells were stained with anti-HA (rabbit, Santa Cruz Biotechnology) and/or anti-Flag (mouse, Sigma) Abs, secondary anti-rabbit IgG-FITC and/or anti-mouse IgG-rhodamine Abs were added. The cells were observed by using a fluorescence microscope. (A) Untransfected and untreated MDA-MB-435s cells. (B and E) Untreated (B) and STA-21-treated (E) cells cotransfected by pCMV-Stat3-Flag and pCMV-Stat3-HA plasmids were immunostained with anti-Flag IgG-rhodamine. (C and F) Untreated (C) and STA-21 treated (F) transfected cells were immunostained with anti-HA IgG-FITC. (D and G) The transfected cells were coimmunostained with both anti-HA IgG-FITC and anti-Flag IgG-rhodamine. (D) Untreated cells showed bright orange color. (G) STA-21 treated cells showed weak orange staining and separate green and red color. (Magnification: ×400.) (H) The MDA-MB-435s cells were cotransfected with pCMV-Stat3-Flag and pCMV-Stat3-HA plasmids and were exposed to 20 μM STA-21 compound for 24 h, and then cell lysates were immunoprecipitated with anti-HA or anti-Flag Abs, respectively, as described previously. Cells were resolved on 10% SDS/PAGE and then immunoblotted with anti-HA, anti-Flag or anti-Sat3 Abs as described in Materials and Methods.
We further investigated the effect of STA-21 on Stat3 in vivo dimerization in breast cancer cell line MDA-MB-435s. After the exposure to 20 μM STA-21 for 24 h, the cells were harvested, and the lysates from the cells expressing Stat3-Flag- and Stat3-HA-tagged proteins were immunoprecipitated with an anti-Flag or anti-HA Ab, respectively. The immunoprecipitated reaction mixtures were resolved on a 10% SDS/PAGE and immunoblotted with anti-HA, anti-Flag, or anti-Stat3 Abs, respectively. The results showed that STA-21 abrogated Stat3 dimerization between Stat3-Flag and Stat3-HA proteins in MDA-MB-435s cancer cells (Fig. 5H).
Discussion
Stat3 is a nuclear protein with oncogenic function, and tyrosine phosphorylation at Tyr-705 causes it to dimerize and then translocate to the nucleus and bind to specific promoter sequences on its target genes. Because the dimerization of Stat3 is a decisive event for its activation, blocking the dimerization and nucleus translocation of Stat3 using cell-permeable, nonpeptide small molecules is a very attractive approach for the inhibition of the Stat3 signaling pathway in breast cancer. In this study, we used a computational screening approach to identify potential small-molecule inhibitors of Stat3 from a database of 429,000 small molecules. Of the 100 candidate compounds tested, STA-21 showed remarkable inhibition of Stat3 dimerization, DNA binding, and nucleus translocation as well as the Stat3-regulated genes such as Bcl-XL and cyclin D1. STA-21 only slightly inhibited Stat3 phosphorylation at Tyr-705 in MDA-MB-468 breast carcinoma cells but did not inhibit the phosphorylation of the Stat3 upstream regulators in the same cell line. We also observed slight reduction of Stat3 phosphorylation after exposure to 30 μM STA-21 in MDA-MB-435s breast carcinoma cells (data not shown). The different effect of STA-21 on Stat3 phosphorylation between MDA-MB-468 and MDA-MB-435 breast carcinoma cells might be due to different expression of tyrosine kinases, such as Src, JAK2, gp130, and EGF receptor, which regulate Stat3 phosphorylation between MDA-MB-468 and MDA-MB-435s breast carcinoma cell lines depends on the gp130/JAK2/Stat3 signaling pathway. By inhibiting Stat3 dimerization, STA-21 also might partially prevent certain Stat3 upstream tyrosine kinases from phosphorylating Stat3. However, additional experiments are needed to test this hypothesis.
Moreover, STA-21 reduced the survival of breast carcinoma cells with constitutively active Stat3 but had minimal toxicity to the cells without constitutive Stat3 signaling. Based on these results, we conclude tentatively that inhibition of Stat3 by STA-21 in the cells with constitutive Stat3 signaling results in cell death, due to the irreversible disruption of Stat3 survival pathway that the cells mainly depend on. STA-21 represents a lead compound that inhibits the constitutively Stat3 signaling in human breast cancer cells.
Supplementary Material
Acknowledgments
We thank the Drug Synthesis and Chemistry Branch, the Developmental Therapeutics Program, NCI, and the National Institutes of Health for providing the chemical sample of STA-21. This work was supported in part by Department of Defense Breast Cancer Research Program Concept Grant DAMD17-03-1-0508 (to S.W.) and University of Michigan Comprehensive Cancer Center NCI Core Grant P30 CA46592.
Author contributions: H.S., S.W., and J.L. designed research; H.S. and R.W. performed research; H.S., R.W., and S.W. contributed new reagents/analytic tools; H.S. and J.L. analyzed data; and H.S., R.W., S.W., and J.L. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HA, hemagglutinin; NCI, National Cancer Institute; PTR, phosphoryl tyrosine; Stat3, signal transducers and activators of transcription 3.
References
- 1.Darnell, J. J., Kerr, I. & Stark, G. (1994) Science 264, 1415-1421. [DOI] [PubMed] [Google Scholar]
- 2.Bromberg, J. F., Horvath, C. M., Besser, D., Lathem, W. W. & Darnell, J. E. J. (1998) Mol. Cell. Biol. 18, 2553-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sartor, C., Dziubinski, M., Yu, C., Jove, R. & Ethier, S. (1997) Cancer Res. 57, 978-987. [PubMed] [Google Scholar]
- 4.Garcia, R., Bowman, T., Niu, G., Yu, H., Minton, S., Muro-Cacho, C., Cox, C., Falcone, R., Fairclough, R., Parsons, S., et al. (2001) Oncogene 20, 2499-2513. [DOI] [PubMed] [Google Scholar]
- 5.Zhong, Z., Wen, Z. & Darnell, J. (1994) Science 264, 95-98. [DOI] [PubMed] [Google Scholar]
- 6.Garcia, R., Yu, C., Hudnall, A., Catlett, R., Nelson, K., Smithgall, T., Fujita, D., Ethier, S. & Jove, R. (1997) Cell Growth Differ. 8, 1267-1275. [PubMed] [Google Scholar]
- 7.Bowman, T., Garcia, R., Turkson, J. & Jove, R. (2000) Oncogene 19, 2474-2488. [DOI] [PubMed] [Google Scholar]
- 8.Dechow, T., Pedranzini, L., Leitch, A., Leslie, K., Gerald, W., Linkov, I. & Bromberg, J. (2004) Proc. Natl. Acad. Sci. USA 101, 10602-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bromberg, J., Wrzeszcznska, M., Devgan, G., Zhao, Y., Pestell, R., Albanese, C. & Darnell, J. J. (1999) Cell 98, 295-303. [DOI] [PubMed] [Google Scholar]
- 10.Catlett-Falcone, R., Dalton, W. & Jove, R. (1999) Curr. Opin. Oncol. 11, 1-8. [DOI] [PubMed] [Google Scholar]
- 11.Catlett-Falcone, R., Landowski, T., Oshiro, M., Turkson, J., Levitzki, A., Savino, R., Ciliberto, G., Moscinski, L., Fernandez-Luna, J., Nunez, G., et al. (1999) Immunity 10, 105-115. [DOI] [PubMed] [Google Scholar]
- 12.Alas, S. & Bonavida, B. (2003) Clin. Cancer Res. 9, 316-326. [PubMed] [Google Scholar]
- 13.Wei, L. H., Kuo, M. L., Chen, C. A., Chou, C. H., Lai, K. B., Lee, C. N. & Hsieh, C. Y. (2003) Oncogene 22, 1517-1527. [DOI] [PubMed] [Google Scholar]
- 14.Real, P., Sierra, A., De Juan, A., Segovia, J., Lopez-Vega, J. & Fernandez-Luna, J. (2002) Oncogene 21, 7611-7618. [DOI] [PubMed] [Google Scholar]
- 15.Puthier, D., Bataille, R. & Amiot, M. (1999) Eur. J. Immunol. 29, 3945-3950. [DOI] [PubMed] [Google Scholar]
- 16.Niu, G., Wright, K.L., Huang, M., Song, L., Haura, E., Turkson, J., Zhang, S., Wang, T., Sinibaldi, D., Coppola, D., et al. (2002) Oncogene 21, 2000-2008. [DOI] [PubMed] [Google Scholar]
- 17.Kiuchi, N., Nakajima, K., Ichiba, M., Fukada, T., Narimatsu, M., Mizuno, K., Hibi, M. & Hirano, T. (1999) J. Exp. Med. 189, 63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoki, Y., Feldman, G. & Tosato, G. (2003) Blood 101, 1535-1542. [DOI] [PubMed] [Google Scholar]
- 19.Epling-Burnette, P., Liu, J., Catlett-Falcone, R., Turkson, J., Oshiro, M., Kothapalli, R., Li, Y., Wang, J., Yang-Yen, H., Karras, J., et al. (2001) J. Clin. Invest. 107, 351-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen, M., Kaestel, C., Eriksen, K., Woetmann, A., Stokkedal, T., Kaltoft, K., Geisler, C., Ropke, C. & Odum, N. (1999) Leukemia 13, 735-738. [DOI] [PubMed] [Google Scholar]
- 21.Burke, W., Jin, X., Liu, R., Huang, M., Reynolds, R. K. & Lin, J. (2001) Oncogene 20, 7925-7934. [DOI] [PubMed] [Google Scholar]
- 22.Mora, L., Buettner, R., Seigne, J., Diaz, J., Ahmad, N., Garcia, R., Bowman, T., Falcone, R., Fairclough, R., Cantor, A., et al. (2002) Cancer Res. 62, 6659-6666. [PubMed] [Google Scholar]
- 23.Ni, Z., Lou, W., Leman, E. & Gao, A. (2000) Cancer Res. 60, 1225-1228. [PubMed] [Google Scholar]
- 24.Rahaman, S., Harbor, P., Chernova, O., Barnett, G., Vogelbaum, M. & Haque, S. (2002) Oncogene 21, 8404-8413. [DOI] [PubMed] [Google Scholar]
- 25.Turkson, J., Ryan, D., Kim, J., Zhang, Y., Chen, Z., Haura, E., Laudano, A., Sebti, S., Hamilton, A. & Jove, R. (2001) J. Biol. Chem. 276, 45443-45455. [DOI] [PubMed] [Google Scholar]
- 26.Blaskovich, M., Sun, J., Cantor, A., Turkson, J., Jove, R. & Sebti, S. (2003) Cancer Res. 63, 1270-1279. [PubMed] [Google Scholar]
- 27.Becker, S., Groner, B. & Muller, C. (1998) Nature 394, 145-151. [DOI] [PubMed] [Google Scholar]
- 28.Berman, H., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T., Weissig, H., Shindyalov, I. & Bourne, P. (2000) Nucleic Acids Res. 28, 235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ewing, T., Makino, S., Skillman, A. & Kuntz, I. (2001) J. Comput. Aided Mol. Des. 15, 411-428. [DOI] [PubMed] [Google Scholar]
- 30.Wang, R., Lai, L. & Wang, S. (2002) J. Comput. Aided Mol. Des. 16, 11-26. [DOI] [PubMed] [Google Scholar]
- 31.Song, H., Sondak, V., Barber, D., Reid, T. & Lin, J. (2004) Int. J. Oncol. 24, 1017-1026. [PubMed] [Google Scholar]
- 32.Turkson, J., Bowman, T., Garcia, R., Caldenhoven, E., De Groot, R. P. & Jove, R. (1998) Mol. Cell. Biol. 18, 2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song, H., Ethier, S., Dziubinski, M. & Lin, J. (2004) Biochem. Biophys. Res. Commun. 314, 143-150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.