Abstract
It is estimated that approximately one billion people are at risk of infection with obligate intracellular bacteria, but little is known about the underlying mechanisms that govern their life cycles. The difficulty in studying Chlamydia spp., Coxiella spp., Rickettsia spp., Anaplasma spp., Ehrlichia spp. and Orientia spp. is, in part, due to their genetic intractability Recently, genetic tools have been developed; however, optimizing the genomic manipulation of obligate intracellular bacteria remains challenging. In this Review, we describe the progress in, as well as the constraints that hinder, the systematic development of a genetic toolbox for obligate intracellular bacteria. We highlight how the use of genetically manipulated pathogens has facilitated a better understanding of microbial pathogenesis and immunity and how the engineering of obligate intracellular bacteria could enable the discovery of novel signalling circuits in host–pathogen interactions.
Bacteria have historically been divided into two distinct groups: extracellular bacteria, which exist as free-living organisms in their environmental niches, and intracellular bacteria, which infect and replicate inside host cells. Facultative intracellular bacteria, including Salmonella spp., Francisella spp., Legionella pneumophila, Listeria monocytogenes, Yersinia spp. and many others, can replicate in either niche. Conversely, obligate intracellular bacteria, including Chlamydia spp., members of the order Rickettsiales (Anaplasma spp., Ehrlichia spp., Rickettsia spp. and Orientia spp.) and Coxiella burnetii, generally require a host cell for replication (BOX 1).
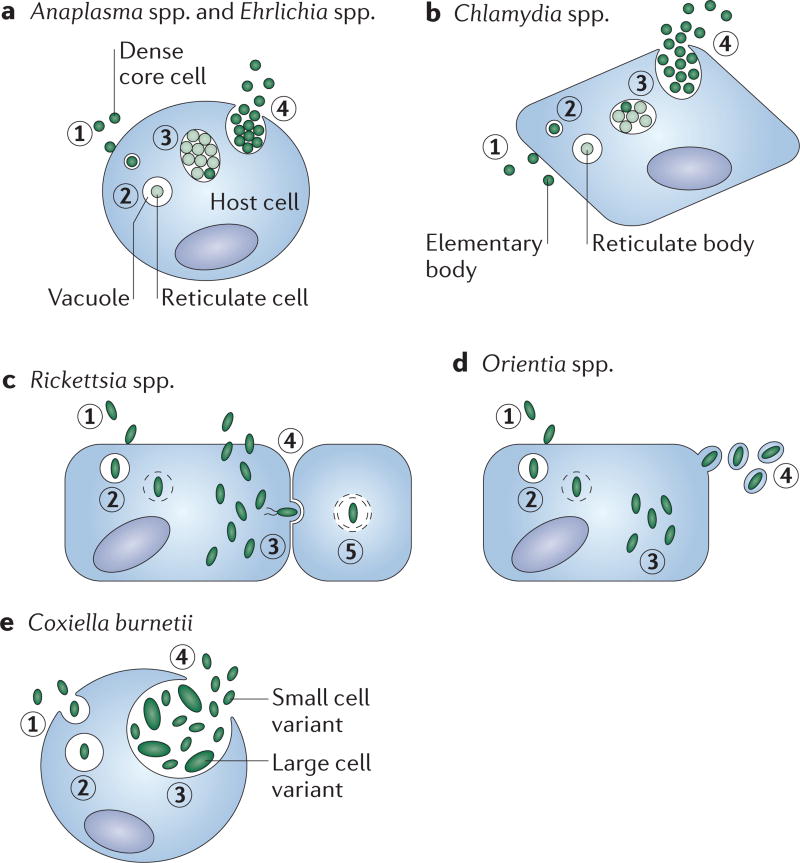
Box 1 | Lifecycle of obligate intracellular bacteria.
The life cycles of Anaplasma spp. and Ehrlichia spp. (see the figure, part a) and Chlamydia spp., (see the figure, part b) are biphasic and characterized by cyclical infectious and replicative forms. Eukaryotic host cells internalize the infectious particles (step 1), termed dense core cells (for Anaplasma spp. and Ehrlichia spp.) and elementary bodies (for Chlamydia spp.). Once inside the host cell, the elementary body differentiates into the replicative morphotype (step 2), known as the reticulate cell (for Anaplasma spp. and Ehrlichia spp.) and reticulate body (for Chlamydia spp.). Reticulate bodies replicate inside a vacuole (step 3), in which they eventually differentiate back to the elementary body morphotype and exit the host cell through extrusion, lysis or possibly other unknown mechanisms (step 4)58,69. The life cycles of members of the Rickettsiaceae feature cytosolic replication (see the figure, parts c,d)66. Members of the Rickettsiaceae induce their uptake by host cells (step 1) and, once internalized, they must escape the phagosome (step 2) before replication. Next, cytosolic bacteria replicate and redistribute themselves intracellularly (step 3). To complete their life cycles, Rickettsia spp. lyse the host cell (step 4) or infect neighbours through intercellular spread (step 5), whereas Orientia spp. bud from the host cell (step 4, figure part d)66, 95Coxiella burnetii (see the figure, part e) is phagocytosed by the host (step 1). The phagosome matures into the Coxiella-containing vacuole (step 2), in which the bacteria differentiate into the large cell variant and replicate (step 3). Last, the bacteria differentiate back into the infectious small cell variant and lyse the cell (step 4)96,97.
Pathogenic obligate intracellular bacteria pose a substantial public health threat. In this Review, we describe, in detail, the best-studied and clinically relevant pathogens (TABLE 1), which include Coxiella spp., pathogenic Chlamydia spp. and arthropod-transmitted members of the Rickettsiales. For decades, facultative intracellular bacteria, such as Salmonella spp., Francisella spp. and L. pneumophila, have been used as models to study host– pathogen interactions1, whereas obligate intracellular bacteria have been underappreciated, despite their requirement to silently establish a replicative niche and remodel the host environment. By infecting host cells, cell biologists and immunologists can uncover novel signalling cascades that have remained uncharacterized, in part, owing to the historical focus on facultative intracellular bacteria2. Thus, obligate intracellular bacteria provide an unparalleled opportunity to discover novel principles of both pathogen and host biology.
Table 1.
Disease pathogenesis of selected obligate intracellular bacteria
| Coxiella burnetii |
Chlamydia Trachomatis |
Anaplasma phagocytophilum |
Ehrlichia chaffeensis | Rickettsia spp. |
Orientia tsutsugamushi |
|
|---|---|---|---|---|---|---|
| Disease | Q fever97 | Genital infection and trachoma58 | Anaplasmosis69 | Ehrlichiosis69 | Spotted fever and typhus66 | Scrub typhus98 |
| Clinical presentation | Mild-to-severe pneumonia and hepatitis; it may progress to chronic infection97 | Asymptomatic-to-severe fever, malaise, leukopenia and increased liver enzymes69 | Mild-to-severe fever, malaise, leukopenia, increased liver enzymes69 and occasional CNS symptoms101 | Fever, respiratory and CNS symptoms, and organ failure66 | Fever, disseminated intravascular coagulation and organ failure98 | |
| Distribution | Global97 | Global58 | The Americas, Europe and Asia102 | The Americas and Asia69 | Global103 | Asia, Oceania and Chile98,104 |
| Epidemiology | Ubiquitous in animals; potential for outbreaks among agricultural workers97 | 130 million new genital infections annually105 and 40 million people with active trachoma (230 million at risk)106 | 2,600 reported cases annually in the United States107 with increasing incidence108 but limited global estimates | 3 cases per million people annually in the United States; increasing incidence108 but limited global estimates | Historically devastating outbreaks, global estimates limited and new species constantly emerging109,110 | 1 million infections per year; 1 billion people at risk109 |
| Transmission | Inhalation38 | Contact with infected fluids58 | Tick vector69 | Tick vector69 | Arthropod vector66 | Mite vector98 |
| Major mammalian host cell | Alveolar macrophage38 | Epithelial cells58 | Granulocytes and endothelial cells69 | Monocytes and macrophages69 | Endothelial cells66 | multiple98 |
| Organs affected |
|
Genital tract and eyes58 | Inflammatory lesions in organs and liver damage72 | Multiple101 | Multiple66 | Multiple98 |
| Notes | Highly virulent97; lung endothelium and epithelium are minor targets of infection97 | Serovars A–C cause trachoma and serovars D–K cause genital infection58 | Lacks LPS and peptidoglycan69; macrophages are minor targets of infection72; antigenic variation69 | Lacks LPS and peptidoglycan; antigenic variation69 | SFG, typhus group, transitional and ancestral groups66 | ~40% of genome contains repeated sequences85,111 |
| Genetic tractability | Genetically tractable; axenic medium7 | Serovar L2 is genetically tractable24 | Transposon library13 | Transposon library15 | Mutagenesis41,42,49,53 and shuttle vectors29,30 | No reports |
CNS, central nervous system; LPS, lipopolysaccharide; SFG, spotted fever group.
Compared with facultative intracellular bacteria, relatively little is known about the underlying principles of bacterial physiology, host–pathogen interactions and the mechanisms that govern infection by obligate intracellular bacteria. Among the reasons for this discrepancy is the difficulty in studying bacterial signalling networks in vivo. For arthropod-transmitted members of the Rickettsiales, a lack of a basic understanding of vector physiology, immune response and microbial interactions also remains a major hurdle. More than 20 years have passed since the first attempt to transform an obligate intracellular bacterium3 (FIG. 1). The refinement of genetic manipulation methods for all obligate intracellular bacteria has proven exceedingly difficult, mostly because these microorganisms only proliferate inside of host cells. Their limited genetic toolbox often precludes the sophisticated structure–function analyses that are routinely carried out in facultative intracellular bacteria and hinders progress in public health and basic science. In this Review article, we provide an overview of the advances in, and challenges of, genetically engineering obligate intracellular bacteria. We also highlight examples in which the use of genetically manipulated pathogens has improved our understanding of microbial pathogenesis and immunity. Finally, we provide an outlook on the most pressing scientific issues that could be addressed in regard to host–pathogen interactions using engineered obligate intracellular bacteria.
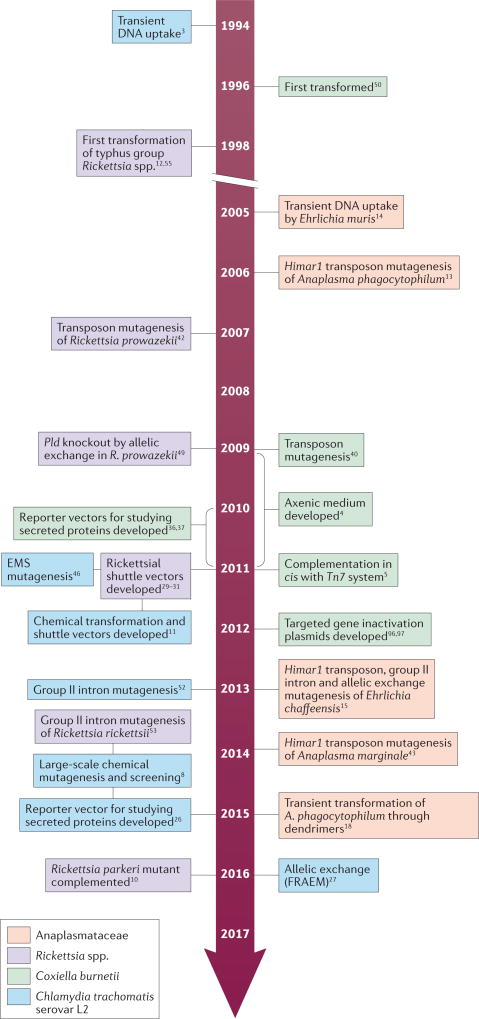
Figure 1. Advances in the genetic manipulation of obligate intracellular bacteria.
EMS, ethyl methanesulfonate; FRAEM, fluorescence-reporteda allelic exchange mutagenesis; Pld, phospholipase D.
Genetic tools: methods and limitations
Until recently, the intracellular lifestyle of obligate intracellular bacteria has precluded the development of practical genetic tools. Prior to 2009, when an axenic medium that supports extracellular growth was developed, genetic tools for C. burnetii were limited4 (FIG. 1). Sophisticated genetic tools were rapidly developed once extracellular growth was possible, owing to the faster growth rate and ease of selecting clonal transformants5. Chlamydia spp. and members of the Rickettsiales must still be cultured and manipulated intracellularly, which poses substantial technical hurdles. In addition, obligate intracellular bacteria often grow slower than facultative intracellular bacteria. For example, Rickettsia prowazekii has a replication time of 8–12 h, which is 2–3 times longer than L. pneumophila6. Obligate intracellular bacteria must also be purified from host cells before most transformation methods can be applied, because chemical reagents typically cannot cross both host and pathogen membranes. The purification steps are laborious, inefficient and tend to damage bacteria, which might render them non-infectious. As mutants must be selected and propagated in host cells, mutating genes that are essential for cell invasion and survival remains problematic. Last, complementation of a mutated gene with the wild-type gene under the control of its own promoter remains a major bottleneck that has only been overcome for three obligate intracellular bacteria: C. burnetii7, Chlamydia trachomatis serovar L2 (REFS 8,9) and Rickettsia parkeri10. Despite these difficulties, there has been substantial progress in the development of genetic tools for obligate intracellular bacteria (BOX 2).
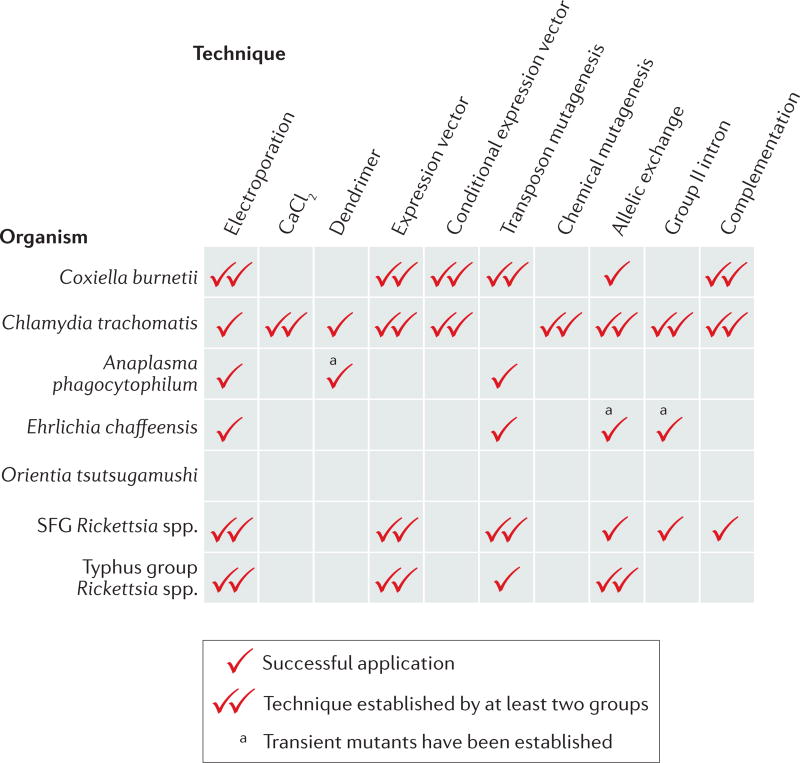
Box 2 | Genetic tools for obligate intracellular bacteria.
Although all obligate intracellular bacteria adopt intracellular lifestyles, they are not equally amenable to genetic manipulation. Coxiella burnetii is technically no longer an obligate intracellular bacterium owing to the development of an axenic medium that enabled its host-free cultivation in the laboratory5. Most genetic tools have been developed for Chlamydia trachomatis serovar L2, which is now considered to be genetically tractable24. However, genome manipulation of clinically relevant C. trachomatis serovars and other Chlamydia spp. remains challenging. In the order Rickettsiales, Rickettsia spp. (spotted fever group (SFG) and typhus group) have the most sophisticated genetic tools available, and a recent article was the first to report genetic complementation10. Fewer genetic tools have been developed for members of the Anaplasmataceae family (Anaplasma spp. and Ehrlichia spp.), and no successful genetic manipulation has been reported for Orientia tsutsugamushi (in the family Rickettsiaceae). The differential ability to genetically manipulate obligate intracellular bacteria may be due, in part, to the biological differences between species (for example, the ability to form plaques or the presence of endogenous plasmids). A lack of adoption of some genetic tools may be caused by the small size of the scientific community that researches obligate intracellular bacteria, which limits the number of groups that contribute to the development and refinement of genomic techniques. In the figure, one tick indicates successful application of a genetic tool (columns) for each major species (rows). Two ticks indicate that the technique has been published by at least two independent research groups.
Transformation methods
The first step in almost all methods of genetic manipulation is transformation. Three transformation methods have been reported: chemical transformation, electroporation and polyamidoamine dendrimers (PAMAM dendrimers) mixed with a plasmid vector (FIG. 2a). Chemical transformation in a solution of calcium chloride has been used routinely to transform C. trachomatis serovar L2 since the development of a groundbreaking standard protocol in 2011 (REF. 11) (FIG. 1). Other obligate intracellular bacteria are transformed through the electroporation of cell-free bacteria3,5,12–15. Following transformation, bacteria are mixed with host cells that they must infect to survive. As chemical transformation or electroporation requires cell-free infectious bacteria, efficiency and the rate of mutant recovery would probably be improved if obligate intracellular bacteria could be manipulated within the host cell. Thus, attempts have been made to transform Chlamydia spp. and Anaplasma phagocytophilum with PAMAM dendrimers (FIG. 2a).
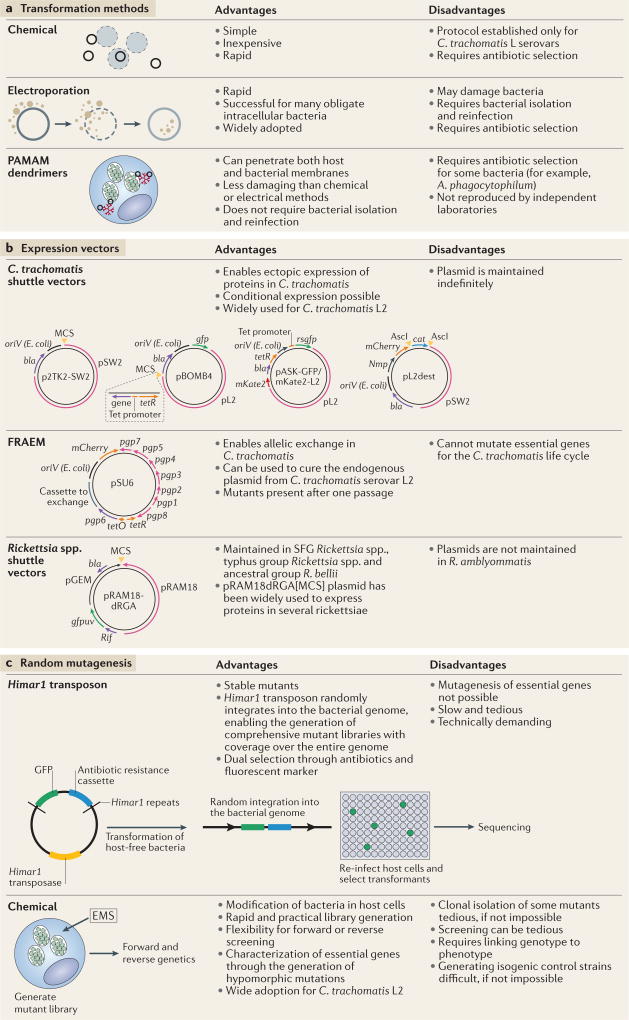
Figure 2. Transformation methods, expression vectors and random mutagenesis.
a Chemical transformation with calcium chloride is used to transform Chlamydia trachomatis serovar L2. Electroporation is primarily used to transform members of the order Rickettsiales13,15,33. Plasmids complexed with polyamidoamine (PAMAM) dendrimers provide an alternative method to transform obligate intracellular bacteria. b | Shuttle vectors have been developed for C. trachomatis and Rickettsia spp. In both cases, portions of endogenous plasmids (pSW2 and pL2 for C. trachomatis, and pRAM18 for Rickettsia spp.) are fused to Escherichia coli plasmid backbones to enable replication. The shuttle vectors include E. coli origins of replication (oriV), fluorescent markers (GFP and GFP optimized for excitation by ultraviolet light (GFPuv) or red fluorescent protein (mCherry)), antibiotic selection genes (β-lactamase (bla), chloramphenicol (cat), spectinomycin (aadA) and rifampicin resistance (rif)) and multiple cloning sites (MCS)21,22,29. Two chlamydial shuttle vectors, pBOMB4 and pASK-GFP/mKate2-L2, can be modified to include a tetracycline-inducible promoter for conditional gene expression22,23. The shuttle vector pL2dest enables the expression of proteins that are fused with a β-lactamase reporter to study protein secretion26. The fluorescence-reported allelic exchange mutagenesis (FRAEM) vector pSU6 enables allelic exchange in C. trachomatis by behaving as a suicide vector in the absence of tetracycline27. c | The Himar1 transposase randomly inserts the transposon into the bacterial genome. Successful transformants are selected with antibiotics and the expression of a fluorescent protein13. The mutants are selected and sequenced to determine insertion sites. In chemical mutagenesis, mutations are caused by DNA-alkylating agents (for example, ethyl methanesulfonate (EMS)). Mutants are selected, pooled and subjected to forward or reverse genetics screens8A. phagocytophilum, Anaplasma phagocytophilum; R. amblyommatis, Rickettsia amblyommatis; R. bellii, Rickettsia bellii; SFG, spotted fever group; tetO, tetracycline operator; tetR, tetracycline repressor.
In proof-of-principle experiments, complexes comprising plasmid DNA and PAMAM dendrimers were added directly to infected host cell monolayers to reintroduce the cloned C. trachomatis plasmid into two plasmid-free strains, C. trachomatis serovar L2 (25667R)16 and Chlamydia pneumoniae AR-39 (REF. 17). These chlamydial transformants were stable over several passages. In addition, fluorescein isothiocyanate-conjugated (FITC) dendrimers were evaluated for their ability to transform RF/6A primate endothelial cells infected with A. phagocytophilum; indeed, vacuoles that contained fluorescent A. phagocytophilum indicated that the dendrimers penetrated the host cell and the vacuole. In subsequent experiments, intracellular A. phagocytophilum was transformed to express GFP that was optimized for excitation by ultraviolet light (GFPuv) through dendrimers that were complexed with the transforming plasmid that integrated into the bacterial genome. However, these transformants were only retained for a few passages in medium that contained antibiotics, possibly owing to the disruption of essential genes18. Although dendrimer-enabled transformation still warrants further technical development, it could enable the introduction of markers that can be used to readily identify individual mutants in a pool, thus enabling the high-throughput screening of more recalcitrant species (for example, A. phagocytophilum). Currently, dendrimers are not routinely used to transform obligate intracellular bacteria, despite the attractive possibility to transform these bacteria inside host cells.
Shuttle vectors
Shuttle vectors are important genetic tools because they can be easily manipulated in Escherichia coli and maintained for the ectopic expression of genes under investigation in C. trachomatis, Rickettsia spp. and C. burnetii. C. trachomatis shuttle vectors were derived from the endogenous 7.5 kb plasmid of C. trachomatis serovar LGV L2 (REF. 19). This plasmid (pL2) encodes eight genes that have diverse poorly understood roles in the infection cycle of C. trachomatis11,20. A pL2 variant, pSW2, contains a deletion in the first coding sequence20. These plasmids were used to generate the shuttle vectors p2TK2-SW2 (pSW2 parent)21 and pBOMB4 (pL2 parent)22 by fusing them to an E. coli origin of replication and β-lactamase (bla) for selection (FIG. 2b). p2TK2-SW2 and pBOMB4 are used for the routine ectopic expression of proteins in C. trachomatis and have been modified to include fluorescent markers, multiple cloning sites and/or tetracycline-inducible promoters21–25. Other modified versions of the pSW2 plasmid include the vector pL2dest, which was generated to enable the direct testing of potential secreted proteins with the TEM β-lactamase reporter assay26. Until recently, it was challenging to directly manipulate the chlamydial genome through allelic exchange because the transformed bacterium retained both the targeted and wild-type gene on a stably maintained plasmid27. A technical breakthrough occurred when a suicide vector was developed. The pSU6 plasmid enabled fluorescence-reported allelic exchange mutagenesis (FRAEM) in C. trachomatis by putting pgp6, which is required for plasmid maintenance, under the control of a tetracycline-induced promoter27 (FIG. 2b). Once tetracycline is removed from the medium, pgp6 is no longer expressed and the plasmid is not maintained. The pSU6 suicide vector enabled routine allelic exchange, which was previously limited to small DNA segments and a few selection markers in Chlamydia psittaci28. The utility of the pSU6 suicide vector was illustrated by the replacement of tryptophan synthase alpha subunit (trpA), which is involved in tryptophan biosynthesis, and three type III secretion system (T3SS) effectors with a gfp and bla cassette27. For each of the mutants, exclusively GFP-fluorescing inclusions were observed in the next passage following the removal of tetracycline, which indicates the loss of the plasmid backbone (which encodes red-fluorescent mCherry) and the integration of the allelic exchange cassette into the chlamydial genome27. As the pSU6 suicide vector was only recently developed, it has not yet been widely adopted by researchers in the Chlamydia field; however, the potential to generate targeted gene knockouts in C. trachomatis serovar L2 through allelic exchange is an exciting development24.
Shuttle vectors for Rickettsia spp. were derived from three endogenous plasmids of the spotted fever group (SFG) Rickettsia amblyommatis, namely pRAM18, pRAM23 and pRAM32, although only pRAM18 has been used extensively29. These shuttle vectors contain a rifampicin resistance cassette, a fluorescent marker, portions of standard E. coli vectors that confer antibiotic resistance and regions of the endogenous R. amblyommatis plasmids that contain dnaA and parA29, which are required for the replication and maintenance of DNA (FIG. 2b). The pRAM18dRGA and pRAM32dRGA plasmids were transformed into the SFG Rickettsia conorii, R. parkeri, Rickettsia montanensis, Rickettsia monacensis and R. amblyommatis, the typhus group R. prowazekii30 and Rickettsia typhi31, and the ancestral group Rickettsia bellii. With the exception of R. amblyommatis, all of the bacteria acquired a fluorescent signal, which indicated successful transformation and expression from the shuttle vector29. Failure to establish pRAM18dRGA in the parent R. amblyommatis confirms that plasmids that share the same partitioning system are incompatible in rickettsiae32.
A multiple cloning site was inserted into pRAM-18dRGA, which facilitated the ligation of a gene that encodes the red fluorescent protein mCherry under the control of an Anaplasma marginale promoter (yielding pRAM18dRGA[AmTrCh]). Both R. montanensis and R. conorii were transformed with this shuttle vector and acquired red fluorescence, which demonstrates the functionality of this construct29,33. Interestingly, R. conorii transformed with pRAM18dRGA[AmTrCh] was maintained in cell culture and within an experimentally infected animal without the administration of antibiotics34, which indicates that the plasmids were stable for the duration of these experiments in the absence of antibiotic selection. Furthermore, rickA from R. monacensis was overexpressed in R. bellii from the multiple cloning site of the shuttle vector, which resulted in significant changes in adhesion to cells and intracellular motility35. Finally, pRAM18dRGA[Sca4] complemented surface cell antigen 4 (sca4) in an R. parkeri mutant10.
Shuttle vectors for C. burnetii were created following the establishment of the axenic medium. Two reporter plasmids that enabled for β-lactamase–gene fusions, pCBTEM36 and pJB-CAT-BlaM37, were independently generated. C. burnetii uses the Dot/Icm type IV secretion system to remodel its intracellular niche (Supplementary information S1 (figure)), and detailed knowledge of substrates was gained through TEM β-lactamase reporter assays38. The pJB-CAT plasmid backbone was used to generate other shuttle vectors that enabled the ectopic expression of tagged proteins in C. burnetii and complementation of mutated genes7. Although the copy numbers of shuttle vectors are difficult to control and may cause polar effects, they provide a convenient method to examine gene function directly in obligate intracellular bacteria.
Mutagenesis
Random mutagenesis methods include transposon insertion and chemical mutagenesis (FIG. 2c). Himar1 is a mariner transposon that is derived from a transposable element isolated from the stable fly Haematobia irritans39 that has been successfully used to mutagenize various organisms, including several obligate intracellular bacteria, such as C. burnetii40, Rickettsia spp.41,42, Anaplasma spp.13,43 and Ehrlichia chaffeensis15. The Himar1 transposase system encodes the transposase and the transposon on one or two suicide plasmids that are electroporated into host cell-free bacteria13,42,44. The Himar1 transposase randomly integrates the transposon, which contains an antibiotic resistance cassette and fluorescent markers flanked by repeats, into AT dinucleotide sites in the bacterial genome through a cut-and-paste mechanism13. Genetic markers in the transposon facilitate selection, visual detection and the identification of insertion loci in the bacterial genome. Transposase systems have also been used to complement C. burnetii mutations in cis7.
Chemical mutagenesis provides an alternative to the Himar1 transposon system, but without selection or detection markers. To date, only Chlamydia abortus (formerly C. psittaci var. ovis) and C. trachomatis have been successfully mutagenized using DNA-alkylating agents to generate libraries8,45–47. Chemical mutagenesis does not require bacterial transformation, and obligate intracellular bacteria do not need to be purified from the host cell before application of the mutagen. Another advantage of chemical mutagenesis is that hypomorphic mutations, which decrease gene function instead of completely inactivating it, can be generated, enabling the characterization of genes that are involved in essential functions25. Linking mutations to observed phenotypes can be laborious because it is technically demanding to identify, select and recover mutants in the absence of molecular tags24. Nonetheless, as Chlamydia spp. are naturally competent, carrying out linkage analysis can be carried out using lateral gene transfer24,25. Co-infection with a rifampicin-resistant mutant and a spectinomycin-resistant wild-type strain produces recombinant mutants that are resistant to both antibiotics. The recombinant clones are then genotyped and assessed for phenotype48. Repeated rounds of co-infection, phenotypic screening and genotyping are carried out until a single mutation can be linked to a phenotype. Other strategies involve using temperature-sensitive mutants to carry out genetic mapping, which does not require the generation of antibiotic- resistant strains of C. trachomatis47. Libraries of mutant C. trachomatis strains obtained through ethyl methanesulfonate mutagenesis (EMS mutagenesis) have been screened using various approaches, including temperature, forward genetics and reverse genetics46–48. Although linking phenotypes to mutations is tedious, chemical mutagenesis has proven to be an important technique for elucidating the function of genes in C. trachomatis serovar L2. For example, EMS mutagenesis has led to the characterization of chlamydial metabolic features and the identification of an effector that is responsible for mediating the assembly of filamentous actin (F-actin) around the inclusion8.
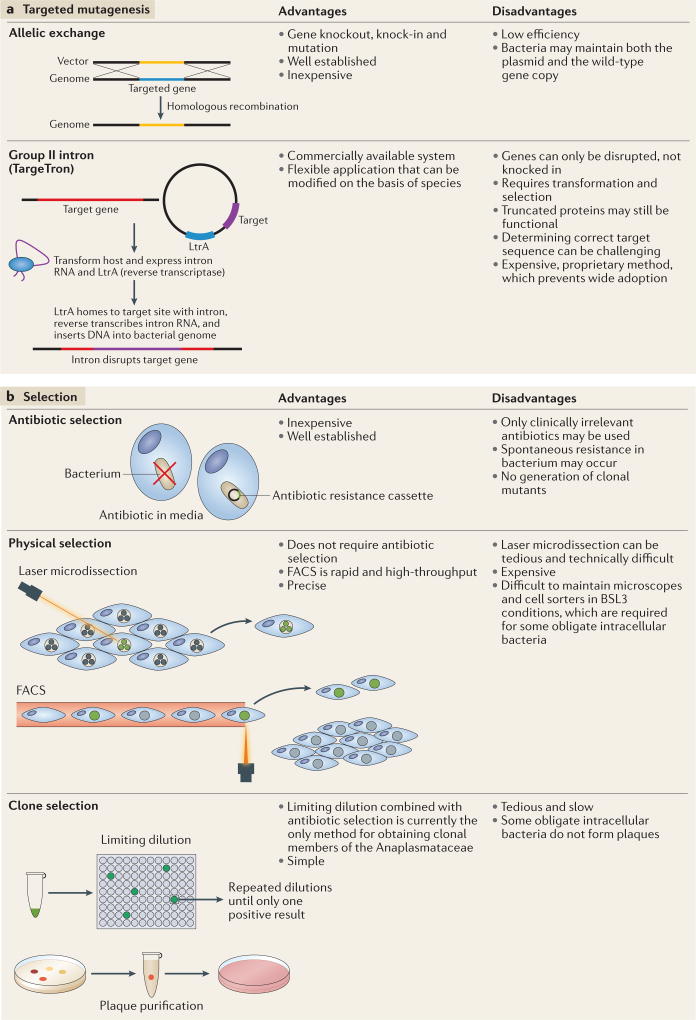
Targeted mutagenesis includes allelic exchange and mobile group II introns. Allelic exchange is routinely used to generate mutants in facultative intracellular bacteria and extracellular bacteria; only recently has this technique been successfully applied to obligate intracellular bacteria (FIG. 3a). Early attempts to mutagenize Chlamydia spp. resulted in allelic exchange of the ribosomal RNA (rRNA) operon with a synthetic 16S rRNA gene that contained point mutations, which rendered the mutant bacterium resistant to spectinomycin and kasugamycin28. This technique has only been successful in C. psittaci and has been limited to replacing small gene segments28. The recent development of the pSU6 suicide vector facilitated allelic exchange in C. trachomatis serovar L2 (REF. 27) (FIG. 2b).
Figure 3. Targeted mutagenesis and selection.
a Targeted mutagenesis enables the alteration of specific bacterial genes. Allelic exchange can be used to introduce point mutations, and to insert and delete specific genes. Group II intron technology (TargeTron) enables introns to be specifically inserted into bacterial genes to disrupt gene function through LtrA, a multifunctional protein derived from Lactococcus lactis, which reverse transcribes and splices the intron and cleaves the recipient DNA for intron insertion51. TargeTrons have been successfully applied to generate transient mutants of Ehrlichia chaffeensis15 and stable mutants of Chlamydia trachomatis and Rickettsia rickettsii52,53. b | Two methods are currently used to distinguish mutants from wild-type bacteria: antibiotic selection and physical selection. Mutants can be physically separated from wild-type bacteria by fluorescence-activated cell sorting (FACS) and laser microdissection. One final step is obtaining clonal mutants, which can be isolated by limiting dilution, or, in some cases, plaque purification5. BSL3, biological safety level 3.
Allelic exchange has also been carried out in R. prowazekii to inactivate the phospholipase D gene (pld), which is a putative virulence factor49. The pld ORF and surrounding bases were cloned into the pBluescript SKII+ plasmid and an internal segment was replaced with a rifampicin resistance cassette49. Transformation was achieved using a linear DNA fragment that was amplified from the construct, as it was less likely to result in the insertion of the entire plasmid into the rickettsial genome49. The resulting Δpld R. prowazekii mutant strain was used to determine whether the disruption of pld leads to attenuated virulence in R. prowazekii49. In E. chaffeensis, two types of recombinant linear DNA segment that targeted a hypothetical protein were generated. One segment was prepared to introduce an antibiotic resistance cassette to disrupt the gene, which resulted in an increase in the size of the bacterial genome. To avoid polar effects, a second recombinant segment was created to remove a segment of the disrupted gene, such that the resulting construct was approximately the size of the wild-type gene. Mutants were detected independently of the recombination methods, but they persisted for only a few days in culture15. Last, allelic exchange was used to transform C. burnetii to ampicillin resistance50, but this technique is not widely used in the Coxiella field, perhaps, in part, owing to the success of other genetic tools.
Mobile group II introns facilitate the selective integration of DNA that is reverse-transcribed from RNA into specific sites in the bacterial genome51. The selectivity of insertion is conferred through the sequence of the intron RNA, and commercial systems, such as TargeTron, have been successfully used to genetically modify E. chaffeensis, C. trachomatis serovar L2 and Rickettsia spp. through insertional mutagenesis (FIG. 3a). In E. chaffeensis, TargeTron constructs were assembled for six different genes, including those that encode outer membrane proteins15. Unfortunately, the resulting mutants were only detectable for less than a week and stable mutant lines could not be selected. By contrast, TargeTron constructs were used to mutate the C. trachomatis serovar L2 effector inclusion membrane protein A (incA)52, which is thought to promote the fusion of inclusions inside cells that are infected with C. trachomatis52. Recently, Chlamydia promoter of cell survival (CpoS) was disrupted by TargeTron, confirming the phenotype of a chemically mutagenized strain9. In another example, a premature stop codon and a rifampicin resistance cassette were introduced to inactivate outer membrane protein A (ompA) in virulent Rickettsia rickettsii str. Sheila Smith53. There are only a few cases of the successful application of TargeTron constructs to generate stable targeted mutants of obligate intracellular bacteria. This could be attributed to the proprietary TargeTron technology or the availability of other tools (for example, allelic exchange).
Recovery and selection
Three methods are currently used to distinguish mutants from the wild-type bacteria: antibiotic selection, fluorescence-activated cell sorting (FACS) and laser microdissection (FIG. 3b). Antibiotic selection is a standard technique that enables the survival of only those bacteria that acquired a resistance cassette that encodes factors that promote survival when exposed to xenobiotics. Only clinically irrelevant antibiotics are permitted for use by regulatory agencies, owing to the potential for the escape of resistant strains into the environment. These regulations further limit the small range of antibiotics that are effective against obligate intracellular bacteria. Moreover, different species, strains, biovars and serovars may be differentially susceptible to antibiotics24, and resistance can develop after continued exposure in cell culture. Cells infected with labelled C. trachomatis and Rickettsia spp. that express a fluorescent protein have been sorted by FACS33,54–56. Similarly, single cells that are infected with labelled C. trachomatis have been successfully isolated through laser microdissection57. However, the technical demands and cost of specialized equipment, coupled with biosafety limitations (for example, maintenance of cell sorters in biosafety level 3 laboratories), prevent these techniques from being widely used. Although the selection of transformants can be tedious and potentially expensive, a major hurdle to overcome is the isolation of clonal mutants. Micromanipulation of cells that are infected with C. burnetii has been used to extract single Coxiella-containing vacuoles, which can then infect sterile host cells or be cultivated in axenic medium5,40. As there are no axenic media available for most obligate intracellular bacteria, simple clone picking and expansion are not possible. Some strains of Chlamydia spp. and Rickettsia spp. can be plaque purified to obtain clonal transformants, but not all obligate intracellular bacteria form visible plaques. Therefore, this method is not widely applicable5. Alternatively, clonal transformants can be recovered through time-consuming limiting dilution5 (FIG. 3b).
New functional insights
Chlamydia trachomatis effectors
Chlamydia spp. reside in a membrane-bound inclusion that becomes remodelled by integral membrane proteins (Supplementary information S1 (figure)). These inclusion proteins (Incs) are secreted by the T3SS and regulate membrane trafficking, cell signalling and cytoskeletal rearrangements to promote the formation and maintenance of inclusions58. C. trachomatis has more than 50 putative Inc proteins, some of which are expressed at unique stages of the developmental cycle59.
Despite the importance of Incs for infection with Chlamydia spp., detailed knowledge at the mechanistic level was limited until recently, owing to the genetic intractability of these microorganisms. Recently, a mutant library of approximately 1,000 EMS-mutagenized isolates of C. trachomatis was generated8. A microscopy-based forward-genetics screen was used to identify two mutants that had abnormal F-actin assembly, as deficits in this function were likely due to mutations in proteins that control inclusion maturation. The sequencing of one mutant (strain M407) and genetic linkage analysis led to the identification of a nonsense mutation in CTL0184 (C307T/Q103*). The assembly of F-actin was restored in strain M407 complemented with the p2TK2-SW2 shuttle vector that encoded wild-type CTL0184 under the control of its endogenous promoter.
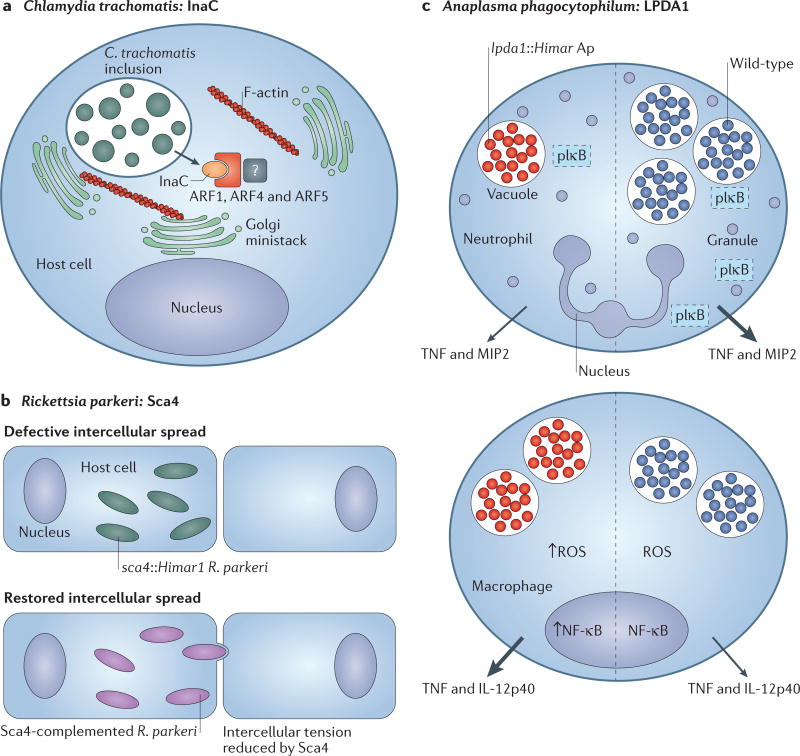
CTL0184 was subsequently investigated and renamed inclusion membrane protein for actin assembly (InaC; FIG. 4a). Co-immunoprecipitation assays identified two classes of host molecule, 14-3-3 scaffolding proteins and ADP-ribosylation factors (ADP-ribosylation factor 1 (ARF1), ARF4 and ARF5), as InaC interacting partners. InaC and F-actin assembly were shown to be required for the redistribution of the Golgi body, which is thought to be a source of lipids for inclusion remodelling58. ARFs are Golgi-associated GTPases that regulate membrane trafficking60. However, the InaC-mediated enlistment of ARFs was not important for lipid acquisition in the cell culture system8, and the mechanism by which InaC regulates the assembly of F-actin and Golgi distribution together with ARFs and 14-3-3 proteins remains to be identified. Other intracellular pathogens, such as L. pneumophila and typhus group Rickettsia spp., recruit host ARFs through their RalF effector proteins61–63. L. pneumophila uses ARF1 to control the delivery of endoplasmic reticulum (ER)-derived vesicles to the maturing Legionella-containing vacuole63. Interestingly, the typhus group Rickettsia species R. prowazekii61 and R. typhi hijack ARFs to manipulate the rearrangement of the actin cytoskeleton at the plasma membrane, which has been shown to lead to pathogen uptake in R. typhi62. Thus, it is likely that C. trachomatis, similarly to other intracellular pathogens, uses ARFs to manipulate host vesicle trafficking and actin cytoskeleton dynamics during the establishment of the inclusion.
Figure 4. Discoveries in microbial pathogenesis facilitated by genetic tools.
a Chemical mutagenesis of Chlamydia trachomatis led to the identification of inclusion membrane protein for actin assembly (InaC), which recruits f ilamntous actin (F-actin) and induces Golgi redistribution around the inclusion in manner that is dependent on ADP ribosylation factor 1 (ARF1), ARF4 and ARF5 (REF. 8). b | Rickettsia parkeri disseminates intercellularly through a mechanism that is not used by facultative intracellular bacteria. Screening of a Himar1 transposon mutagenesis library of R. parkeri identified a mutant deficient in intercellular spread named surface cell antigen 4 (sca4)Himar1 (REF. 10). The sca4, Himar1 mutant was complemented by pRAM18dRGA[Sca4], which restored intercellular spread. Sca4 was found to inhibit vinculin binding to α-catenin (not shown), thereby reducing intercellular force transduction and enabling the intercellular spread of R. parkeri. c | Himar1 transposon mutagenesis was used to generate an Anaplasma phagocytophilum mutant library. One mutant exhibited a single transposon insertion in the dihydrolipoamide dehydrogenase 1 (lpda1) gene. Compared with wild-type infection, the lpda1, Himar1 A. phagocytophilum mutant infected neutrophils less well, and activated nuclear factor-κB (NF-κB) poorly (as measured by decreased phosphorylated inhibitory subunit of NF-κB (pIκB)), and elicited the production of less tumour necrosis factor (TNF) and macrophage inflammatory protein 2 (MIP2; also known as CXCL2) in neutrophils70. Transient infection of macrophages by the lpda1, Himar1 A. phagocytophilum strain correlated with enhanced nuclear NF-κB activity, which led to the increased production of reactive oxygen species (ROS) and the release of proinflammatory cytokines70. Mice that were infected with the lpda1, Himar1 mutant or wild-type A. phagocytophilum HZ exhibited differences in immunopathology and cell-specific outcomes, which suggests that LPDA1 may have a role in inhibiting excessive host immune activation70. Arrow size correlates with relative quantity of cytokines secreted. IL-12p40, interleukin-12 subunit p40.
The Inc protein CTL0481, which was renamed Chlamydia promoter of survival (CpoS), was also recently identified in a forward genetic screen for C. trachomatis mutants that enhanced the death of infected cells9. Loss-of-function mutations in cpoS led to the induction of necrosis and apoptosis midway through the infection cycle, which correlated with an exaggerated type I interferon (IFN) response. In agreement with this, complementation of the chemically mutagenized strain with a p2TK2-SW2-derived plasmid that encodes cpoS did not elicit host cell death. Furthermore, inactivation of cpoS using a TargeTron construct in a wild-type strain of C. trachomatis serovar L2 induced cell death. Unexpectedly, the release of type I IFNs was uncoupled from cell death, although the cyclic dinucleotide sensor stimulator of interferon genes (STING) was required. The interacting partners of CpoS included multiple RAB proteins that control ER-to-Golgi protein trafficking, which suggests that it might modulate membrane trafficking at the inclusion9. Last, as STING and calcium flux are suggested to be linked64,65, and the depletion of ER calcium stores partially rescued the cell death phenotype of CpoS-deficient mutants, CpoS-mediated STING and cell death inhibition may be attributed to calcium homeostasis9.
Rickettsial dissemination, A. phagocytophilum moonlighting and E. chaffeensisgrowth
SFG rickettsiae are cytosolic obligate intracellular bacteria that cause vascular and endothelial tissue damage66 (TABLE 1; Supplementary information S1 (figure)). They disseminate intracellularly in the nutrient-rich host cytosol and move from cell to cell without entering the extracellular environment10. Largely on the basis of electron microscopy and studies in L. monocytogenes, it was assumed that SFG rickettsiae coopted host actin with a single bacterial effector to propel themselves through plasma membranes into a recipient cell, in which they quickly escaped into the cytoplasm10. However, R. parkeri mutants that are defective in actin-based motility revealed that R. parkeri uses two different actin-polymerizing proteins, RickA and Sca2, which are not used by other motile intracellular bacteria41. This, together with basic phenotypic differences with the L. monocytogenes model (such as a shorter time in protrusions, a lack of actin tails and a quicker transition from protrusion to vesicle), suggests an alternative mechanism for intercellular spread10.
A Himar1 mutagenesis library of R. parkeri was screened for mutants that formed small plaques, as these were likely to be deficient in some aspect of intercellular spread10,41 (FIG. 4b). One mutant had a transposon insertion in sca4, which led to the production of a truncated Sca4 protein10. Sca4 was predicted to be secreted67 and shown to bind to vinculin68, but its role in infection remained unclear. A series of traction force microscopy experiments showed that Sca4 decreased intercellular tension to enable R. parkeri to spread into the donor cell independently of actin-based motility10 (FIG. 4b). Importantly, normal intercellular spread of sca4::Himar1 was restored after complementation with the pRAM18dRGA[Sca4] expression vector10.
A. phagocytophilum is a tick-borne pathogen that causes granulocytic anaplasmosis in humans (HGA) and other mammals69 (TABLE 1; Supplementary information S1 (figure)). Himar1 transposon mutagenesis enabled the generation of a mutant library of A. phagocytophilum to study disease pathogenesis13. Library screening led to the identification of an A. phagocytophilum mutant that contains a single transposon insertion in the dihydrolipoamide dehydrogenase 1 (lpda1) gene70 (FIG. 4c), which encodes a metabolic enzyme that has been implicated to have alternate functions in virulence71. In a murine model of infection, the A. phagocytophilum mutant lpda1::Himar1 induced several clinical abnormalities, including more pronounced peripheral blood neutropenia and higher erythrocyte counts70. Increased levels of IFNγ, splenomegaly and splenic extramedullary haematopoiesis were observed in mice that were infected with the lpda1::Himar1 mutant strain but not with the wild-type bacterium. Transient infection of macrophages by the lpda1::Himar1 A. phagocytophilum strain correlated with enhanced nuclear factor-κB (NF-κB) activity, which led to an increase in the production of reactive oxygen species (ROS) and the release of pro-inflammatory cytokines70. Thus, the normal expression of LpdA1 in wild-type bacteria may prevent enhanced immune activation in macrophages, which are increasingly being recognized as the immune cells that are responsible for the cytokine storm that is characteristic of A. phagocytophilum infection72. These findings suggest that protein moonlighting, a phenomenon whereby a protein carries out more than one function71, may be used as a strategy of immune evasion by A. phagocytophilum.
Transposon-based mutagenesis of E. chaffeensis, an emerging tick-borne pathogen, has become valuable for vaccine development (TABLE 1; Supplementary information S1 (figure)). This approach was used to create mutations in four genes that are predicted to encode membrane proteins (Ech_0230, Ech_0379, Ech_0601 and Ech_0660)15. With the exception of Ech_0601, all mutations caused attenuation of the growth of the pathogen in both the reservoir (deer) and incidental (dog) hosts. Conversely, no effect on bacterial growth was observed in the tick vector Amblyomma americanum73. Two clonally-purified attenuated mutants that had insertions in the Ech_0379 and Ech_0660 genes provided protection against wild-type infection in both deer and dogs74. Thus, refining genetic tools for practical application in obligate intracellular bacteria could accelerate vaccine development and protect vulnerable populations.
Correcting virulence factor paradigms
Classified as a select agent, R. prowazekii is considered a potential bioweapon and no vaccine against R. prowazekii exists in the United States; hence, research into the virulence of R. prowazekii that leads to an effective vaccine is vital to protect public health49. Comparative genomics approaches that examined the differences between virulent and avirulent strains of R. prowazekii identified several putative virulence genes75. One example was pld, which, when expressed in a surrogate system, enabled Salmonella enterica subsp. enterica serovar Typhimurium to escape the phagosome76. Several years later, allelic exchange was used to replace wild-type pld of R. prowazekii with a mutated copy49. No differences in growth kinetics, rickettsial burden or ability to escape the phagosome were observed between the virulent wild-type R. prowazekii str. Madrid Evir and the pld mutant in vitro49. Nevertheless, in a guinea pig infection model, the Δpld mutant was attenuated compared with the virulent R. prowazekii strain. Guinea pigs that were infected with the Δpld mutant lost less weight and were afebrile compared with animals infected with virulent R. prowazekii49.
OmpA of R. rickettsii was considered a major virulence factor before the advent of genetic tools66,77. The original study relied on purified OmpA, which was shown to block the attachment of R. rickettsii when pre-incubated with host cells. Selective degradation and denaturation of OmpA also prevented R. rickettsii from adhering to host cells77. Uncertainties in regard to the function of OmpA remained, and group II intron technology was recently used to insert a 5′ premature stop codon into ompA of the highly virulent R. rickettsii str. Sheila Smith53. Unexpectedly, no differences in virulence between the wild-type and mutant strains were detected in either cell culture or the guinea pig infection model53, refuting the notion that OmpA is required for, or enhances, the invasion and disease pathogenesis of R. rickettsii.
Advances in genetic tools have aided in clarifying the function of Chlamydia protease-like activity factor (CPAF). Following its identification, CPAF was shown to cleave host transcription factors that control the expression of major histocompatibility complex (MHC) class I and class II78, which ultimately leads to evasion of the immune system and undetected infection with Chlamydia spp.78. Subsequent studies reported that many other host proteins were cleaved by CPAF79. However, as genetic tools were unavailable, recombinant and ectopically expressed CPAF were used for validation of these substrates79. Thirteen years after the initial discovery of CPAF, a C. trachomatis mutant with defects in CPAF expression was generated80. Using this mutant, several of the phenotypes ascribed to CPAF were shown to occur independently of CPAF. Specifically, CPAF did not affect Golgi fragmentation, NF-κB activation, inhibition of apoptosis and protection from reinfection80. CPAF was described to cleave vimentin, which is an intermediate filament that surrounds the inclusion, and nuclear lamin-associated protein 1 (LAP1; also known as TOR1AIP1) following the disruption of the membrane80. CPAF was also required for the optimal development of elementary bodies80 and has unanticipated roles in infection, including in the regulation of T3SS effectors and bacterial survival in the lower genital tract81,82. Despite these new findings, other questions in regard to the role of CPAF in infection remain; for example, whether CPAF is secreted into the host cytosol or is sequestered in its active form in the inclusion83. Overall, genetic tools have enabled the chlamydial and rickettsial fields to correct artefactual results, which benefits researchers and the public alike, as new therapeutics and vaccines against obligate pathogens are desperately needed.
Concluding remarks
Much has been achieved in the genetic engineering of obligate intracellular bacteria since the first transformation of C. trachomatis3 (FIG. 1). The genetic toolbox for these organisms has grown to include transformation protocols; rickettsial, chlamydial and Coxiella shuttle vectors; random mutagenesis systems; and the ability to generate targeted knockouts through allelic exchange, group II intron technology and FRAEM (FIG 2,3). Despite these advances, limitations still impede the complete application of genetic tools. Chiefly among these is the lack of axenic media, which enables host-free growth of obligate intracellular bacteria. Challenges due to the intracellular lifestyle of obligate intracellular bacteria, tedious selection methods and the inability to efficiently obtain clonal populations limit the practicality of genetic manipulation. Routine complementation of mutagenized genes by expressing the wild-type gene under the control of its own promoter is still technically demanding. Nonetheless, the future of genetic manipulation of obligate intracellular bacteria is promising, as highlighted using the example of C. burnetii.
C. burnetii was strictly classified as an obligate intracellular bacterium until the formulation of an axenic medium that enabled its host-free cultivation in the laboratory4. This landmark development was facilitated through advances in genome sequencing and systematic refinements to the growth medium. No defect in infectivity has been shown for C. burnetii extracellular growth compared with host-cell cultivated bacteria4. C. burnetii grows much faster in axenic medium than in host cells and forms colonies, which enables selection and the subsequent expansion of clonal populations. Instead of several months, transformation experiments now take less than three weeks to complete, which has accelerated the development of genetic tools available to modify the genome of C. burnetii4,5 (FIG. 1). These genetic tools and the study of the metabolic requirements of C. burnetii have provided important insights into the composition and availability of nutrients in the parasitophorous vacuole.
One promising avenue of research is the rickettsial type IV secretion system (T4SS). Members of the Rickettsiales encode a vir-type P-type IV secretion system (rvh) and use this molecular scaffold, in part, to remodel the host environment84–86. The functional characterization of rvh, including the identification of substrates, has been hampered by the inability to easily generate Rvh-deficient species in the Rickettsiales. Duplications and repeats in rvh seem to have persisted despite evolutionary pressure towards genome reduction; thus, these features may be evidence of an alternative function or, conversely, a regulatory circuit86. We anticipate that mechanistic characterization of rvh, aided by mutagenesis using the TargeTron system or random approaches, will shed light onto rickettsial pathogenesis, virulence and host–pathogen interactions.
Another key question in the biology of obligate intracellular bacteria is how Chlamydia spp. and members of the Anaplasmataceae establish and protect their intracellular niches. Inc proteins that are secreted by Chlamydia spp. mediate these processes58. The functional relevance of each Inc protein, how they are regulated and in what manner they contribute to the establishment of this unique organelle, remain poorly understood. Although few Anaplasmataceae effectors have been characterized, some encode ankyrin repeat motifs that are common in eukaryotes and may have implications for virulence87. Ankyrin repeats mediate protein–protein interactions, provide scaffolding for signalling pathways and localize molecules to subcellular compartments in eukaryotes88. Ankyrin-repeat containing proteins (Anks) in A. phagocytophilum and E. chaffeensis were shown to mediate host epigenetic changes to promote bacterial survival69,89. Elucidating the function of conserved Anks in obligate intracellular bacteria can provide insight into host signalling pathways. Furthermore, characterizing mediators of inclusion development through genomic manipulation of obligate intracellular bacteria will reveal basic cell biology principles and ‘druggable’ targets.
Iron is an essential and limiting nutrient for both hosts and pathogens90. Remarkably, little is known about iron homeostasis in obligate intracellular bacteria, including sources, mechanisms of uptake and regulation during colonization of the mammalian host and the arthropod vector. A. marginale, a bovine pathogen that is transmitted by hard ticks, parasitizes erythrocytes. No iron regulatory elements, enzyme systems to degrade haem, surface receptors or siderophores have been identified in the genome of A. marginale91. Only a single orthologue to a bacterial transferrin (ferric binding protein A (FbpA)), a permease (FbpB), and a transporter that is thought to supply energy through ATP hydrolysis (FbpC) have been found92. Unlike similar iron transport systems, these genes are not encoded in a single operon, but rather are dispersed throughout the genome of A. marginale93, 94. How this minimal machinery is used for iron uptake and homeostasis is unknown, but routine targeted genetic manipulation could improve our understanding of bacterial iron acquisition.
The development of novel genetic tools, especially for members of the Rickettsiales, will be crucial to discover novel paradigms in microbial pathogenesis and immunity. As mutants of Chlamydia spp., Rickettsia spp., Orientia spp., Anaplasma spp., Ehrlichia spp. and Coxiella spp. are characterized, the research community will learn more about the fascinating lifestyles of these organisms. Although formerly thought of as just an exotic group of bacteria, the unfolding story of genetic engineering in obligate intracellular bacteria highlights the importance of intellectual curiosity, ingenuity and persistence to overcome scientific obstacles.
Supplementary Material
Acknowledgments
The authors apologize to those colleagues whose work could not be cited owing to the broad scope of the Review and space limitations. Work in the authors’ laboratories was supported by the US National Institutes of Health (Institutional Training Grant T32AI007540 to E.E.M.; R01 AI072683, R56 AI123346 and R21 AI122014 to J.A.C.; R01 AI100759 to R.H.V.; U19 AI084044 to P.M.B.; R0 1 AI070908 to R.R.G.; R01 AI020384 and R21 AI103272 to D.O.W.; R01 AI042792 to U.G.M. and K.A.B.; R01 AI072606 and R21 AI111086 to J.J.M.; R01 AI106859 and R21 AI115449 to J.W.M.; and R01 AI093653 and R01AI116523 to J.H.F.P.), the US Department of Agriculture (USDA-ARS 2090-32000-038-00D to S.M.N.), the Center of Excellence for Vector-Borne Diseases at Kansas State University (to R.R.G.) and the University of Maryland School of Medicine (to J.H.F.P.). The content of this Review is solely the responsibility of the authors and does not necessarily represent the official views of the US National Institute of Allergy and Infectious Diseases or the US National Institutes of Health.
Glossary
- Transformation
A technique that induces bacteria to take up exogenous DNA molecules, usually through chemical or electrical methods
- Polyamidoamine dendrimers
(PAMAM dendrimers). Highly branched polymers that can be used to deliver small molecules or DNA to cells
- Allelic exchange
A common method that is used to knock in, knock out or otherwise mutagenize a DNA segment that relies on homologous recombination between the wild-type gene and an exogenous DNA construct
- Fluorescence-reported allelic exchange mutagenesis
(FRAEM). A method for allelic exchange in Chlamydia trachomatis serovar L2 that can be monitored byobserving fluorescent chlamydial inclusions
- Dot/Icm type IV secretion system
A set of bacterial proteins that inject effector molecules into the eukaryotic host cytosol to remodel the intracellular niche
- Mariner transposon
An abundant class II transposable element first discovered in Drosophila spp. that integrates into a wide range of genomes
- Ethyl methanesulfonate mutagenesis
(EMS mutagenesis). A technique in which a DNA-alkylating agent (EMS) is applied to a population of cells to create a library of strains that contain random mutations
- Mobile group II introns
Mobile bacterial ribozymes that self-splice, reverse transcribe the spliced RNA into DNA, and then integrate the DNA into the bacterial chromosome
- Necrosis
A type of inflammatory cell death that occurs spontaneously after damage to a cell
- Apoptosis
A mode of non-inflammatory programmed cell death
- Stimulator of interferon genes
(STING). An endoplasmic reticulum-associated cytosolic intracellular pattern recognition molecule that senses cyclic dinucleotides and induces the production of type I interferons
- Vinculin
A mammalian cytoskeletal protein that anchors the cell membrane to the actin cytoskeleton
- Granulocytic anaplasmosis
A mild-to-severe tick-borne infectious disease caused by Anaplasma phagocytophilum, which infects neutrophils and myeloid cells, that is characterized by fever, thrombocytopenia, leukopenia and liver damage
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Vance RE. Immunology taught by bacteria. J. Clin. Immunol. 2010;30:507–511. doi: 10.1007/s10875-010-9389-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw DK, McClure EE, Wang X, Pedra JHF. Deviant behavior: tick-borne pathogens and inflammasome signaling. Vet. Sci. 2016;3:27. doi: 10.3390/vetsci3040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tam JE, Davis CH, Wyrick PB. Expression of recombinant DNA introduced into Chlamydia trachomatis by electroporation. Can. J. Microbiol. 1994;40:583–591. doi: 10.1139/m94-093. [DOI] [PubMed] [Google Scholar]
- 4.Omsland A, Heinzen RA. Life on the outside: the rescue of Coxiella burnetii from its host cell. Annu. Rev. Microbiol. 2011;65:111–128. doi: 10.1146/annurev-micro-090110-102927. This review describes the development of an axenic medium for C. burnetii, which enabled the rapid development of genetic tools for this microorganism. [DOI] [PubMed] [Google Scholar]
- 5.Beare PA, Sandoz KM, Omsland A, Rockey DD, Heinzen RA. Advances in genetic manipulation of obligate intracellular bacterial pathogens. Front. Microbiol. 2011;2:97. doi: 10.3389/fmicb.2011.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connor TJ, Adepoju Y, Boyd D, Isberg RR. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc. Natl Acad. Sci. USA. 2011;108:14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beare PA. Genetic manipulation of Coxiella burnetii. Adv. Exp. Med. Biol. 2012;984:249–271. doi: 10.1007/978-94-007-4315-1_13. [DOI] [PubMed] [Google Scholar]
- 8.Kokes M, et al. Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia. Cell Host Microbe. 2015;17:716–725. doi: 10.1016/j.chom.2015.03.014. This article reports the generation of a large-scale chemical mutagenesis library for C. trachomatis serovar L2 and provides a systematic framework for screening mutants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sixt BS, et al. The Chlamydia trachomatis inclusion membrane protein CpoS counteracts STING-mediated cellular surveillance and suicide programs. Cell Host Microbe. 2017;21:113–121. doi: 10.1016/j.chom.2016.12.002. This study describes a chlamydial effector that was discovered through chemical mutagenesis, verified through multiple genetic tools and shed light on the basic processes of eukaryotic host cell death. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamason RL, et al. Rickettsia Sca4 reduces vinculin-mediated intercellular tension to promote spread. Cell. 2016;167:670–683.e10. doi: 10.1016/j.cell.2016.09.023. This article reports the mechanism of intercellular spread for R. parkeri and the first published case of complementation of a rickettsial mutant with a shuttle vector. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, et al. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 2011;7:e1002258. doi: 10.1371/journal.ppat.1002258. This protocol establishes a robust transformation method for C. trachomatis serovar L2 that is currently used extensively by the field. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rachek LI, Tucker AM, Winkler HH, Wood DO. Transformation of Rickettsia prowazekii to rifampin resistance. J. Bacteriol. 1998;180:2118–2124. doi: 10.1128/jb.180.8.2118-2124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsheim RF, et al. Transformation of Anaplasma phagocytophilum. BMC Biotechnol. 2006;6:42. doi: 10.1186/1472-6750-6-42. This study describes the generation of a Himar1 transposon mutagenesis library for A. phagocytophilum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long SW, Whitworth TJ, Walker DH, Yu X-J. Overcoming barriers to the transformation of the genus Ehrlichia. Ann. NY Acad. Sci. 2005;1063:403–410. doi: 10.1196/annals.1355.072. [DOI] [PubMed] [Google Scholar]
- 15.Cheng C, et al. Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog. 2013;9:e1003171. doi: 10.1371/journal.ppat.1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra MK, Gérard HC, Whittum-Hudson JA, Hudson AP, Kannan RM. Dendrimer-enabled modulation of gene expression in Chlamydia trachomatis. Mol. Pharm. 2012;9:413–421. doi: 10.1021/mp200512f. [DOI] [PubMed] [Google Scholar]
- 17.Gérard HC, et al. Dendrimer-enabled DNA delivery and transformation of Chlamydia pneumoniae. Nanomedicine. 2013;9:996–1008. doi: 10.1016/j.nano.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Oki AT, et al. Dendrimer-enabled transformation of Anaplasma phagocytophilum. Microbes Infect. 2015;17:817–822. doi: 10.1016/j.micinf.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas NS, Lusher M, Storey CC, Clarke IN. Plasmid diversity in Chlamydia. Microbiology. 1997;143:1847–1854. doi: 10.1099/00221287-143-6-1847. [DOI] [PubMed] [Google Scholar]
- 20.Seth-Smith HMB, et al. Coevolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics. 2009;10:239. doi: 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agaisse H, Derré IA. C. trachomatis cloning vector the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C trachomatis promoter. PLoS ONE. 2013;8:e57090. doi: 10.1371/journal.pone.0057090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauler LD, Hackstadt T. Expression and targeting of secreted proteins from Chlamydia trachomatis. J. Bacteriol. 2014;196:1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickstrum J, Sammons LR, Restivo KN, Hefty PS. Conditional gene expression in Chlamydia trachomatis using the Tet system. PLoS ONE. 2013;8:e76743. doi: 10.1371/journal.pone.0076743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bastidas RJ, Valdivia RH. Emancipating Chlamydia: advances in the genetic manipulation of a recalcitrant intracellular pathogen. Microbiol. Mol. Biol. Rev. 2016;80:411–427. doi: 10.1128/MMBR.00071-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sixt BS, Valdivia RH. Molecular genetic analysis of Chlamydia species. Annu. Rev. Microbiol. 2016;70:179–198. doi: 10.1146/annurev-micro-102215-095539. [DOI] [PubMed] [Google Scholar]
- 26.Mueller KE, Fields KA. Application of β-lactamase reporter fusions as an indicator of effector protein secretion during infections with the obligate intracellular pathogen Chlamydia trachomatis. PLoS ONE. 2015;10:e0135295. doi: 10.1371/journal.pone.0135295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller KE, Wolf K, Fields KA. Gene deletion by fluorescence-reported allelic exchange mutagenesis in Chlamydia trachomatis. mBio. 2016;7:e01817–15. doi: 10.1128/mBio.01817-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binet R, Maurelli AT. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc. Natl Acad. Sci. USA. 2009;106:292–297. doi: 10.1073/pnas.0806768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burkhardt NY, et al. Development of shuttle vectors for transformation of diverse Rickettsia species. PLoS ONE. 2011;6:e29511. doi: 10.1371/journal.pone.0029511. This article reports the generation of shuttle vectors that can be used to ectopically express proteins in diverse Rickettsia spp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood DO, et al. Establishment of a replicating plasmid in Rickettsia prowazekii. PLoS ONE. 2012;7:e34715. doi: 10.1371/journal.pone.0034715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hauptmann M, et al. GFPuv-expressing recombinant Rickettsia typhi: a useful tool for the study of pathogenesis and CD8+ T cell immunology in Rickettsia typhi infection. Infect. Immun. 2017 doi: 10.1128/IAI.00156-17. http://dx.doi.org/10.1128/IAI.00156-17. [DOI] [PMC free article] [PubMed]
- 32.Million-Weaver S, Camps M. Mechanisms of plasmid segregation: have multicopy plasmids been overlooked? Plasmid. 2014;75:27–36. doi: 10.1016/j.plasmid.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riley SP, Macaluso KR, Martinez JJ. Electrotransformation and clonal isolation of Rickettsia species. Curr. Protoc. Microbiol. 2015;39:3A.6.1–3A.6.20. doi: 10.1002/9780471729259.mc03a06s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riley SP, et al. Nonselective persistence of a Rickettsia conorii extrachromosomal plasmid during mammalian infection. Infect. Immun. 2016;84:790–797. doi: 10.1128/IAI.01205-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliver JD, Burkhardt NY, Felsheim RF, Kurtti TJ, Munderloh UG. Motility characteristics are altered for Rickettsia bellii transformed to overexpress a heterologous rickA gene. Appl. Environ. Microbiol. 2014;80:1170–1176. doi: 10.1128/AEM.03352-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen C, et al. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc. Natl Acad. Sci. USA. 2010;107:21755–21760. doi: 10.1073/pnas.1010485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voth DE, et al. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J. Bacteriol. 2011;193:1493–1503. doi: 10.1128/JB.01359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moffatt JH, Newton P, Newton HJ. Coxiella burnetii: turning hostility into a home. Cell. Microbiol. 2015;17:621–631. doi: 10.1111/cmi.12432. [DOI] [PubMed] [Google Scholar]
- 39.Lampe DJ, Churchill ME, Robertson HM. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 40.Beare PA, et al. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J. Bacteriol. 2009;191:1369–1381. doi: 10.1128/JB.01580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed SC, Lamason RL, Risca VI, Abernathy E, Welch MD. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr. Biol. 2014;24:98–103. doi: 10.1016/j.cub.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z-M, Tucker AM, Driskell LO, Wood D. O. Mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl. Environ. Microbiol. 2007;73:6644–6649. doi: 10.1128/AEM.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crosby FL, et al. Knockout of an outer membrane protein operon of Anaplasma marginale by transposon mutagenesis. BMC Genomics. 2014;15:278. doi: 10.1186/1471-2164-15-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felsheim RF, et al. Transformation of Anaplasma marginale. Vet. Parasitol. 2010;167:167–174. doi: 10.1016/j.vetpar.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodolakis A. In vitro, in vivo properties of chemically induced temperature-sensitive mutants of Chlamydia psittaci var. ovis: screening in a murine model. Infect. Immun. 1983;42:525–530. doi: 10.1128/iai.42.2.525-530.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kari L, et al. Generation of targeted Chlamydia trachomatis null mutants. Proc. Natl Acad. Sci. USA. 2011;108:7189–7193. doi: 10.1073/pnas.1102229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brothwell JA, et al. Interrogating genes that mediate Chlamydia trachomatis survival in cell culture using conditional mutants and recombination. J. Bacteriol. 2016;198:2131–2139. doi: 10.1128/JB.00161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen BD, Valdivia RH. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc. Natl Acad. Sci. USA. 2012;109:1263. doi: 10.1073/pnas.1117884109. This study details the chemical mutagenesis procedure for Chlamydia trachomatis serovar L2 and describes the linkage analysis conducted to link mutation and phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Driskell LO, et al. Directed mutagenesis of the Rickettsia prowazekii pld gene encoding phospholipase D. Infect. Immun. 2009;77:3244–3248. doi: 10.1128/IAI.00395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suhan ML, Chen SY, Thompson HA. Transformation of Coxiella burnetii to ampicillin resistance. J. Bacteriol. 1996;178:2701–2708. doi: 10.1128/jb.178.9.2701-2708.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb. Perspect. Biol. 2011;3:a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson CM, Fisher DJ. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS ONE. 2013;8:e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noriea NF, Clark TR, Hackstadt T. Targeted knockout of the Rickettsia rickettsii OmpA surface antigen does not diminish virulence in a mammalian model system. mBio. 2015;6:e00323–15. doi: 10.1128/mBio.00323-15. This article uses targeted mutagenesis to show that a classical virulence factor in R. rickettsii is not important for virulence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Driskell LO, Tucker AM, Woodard A, Wood RR, Wood DO. Fluorescence activated cell sorting of Rickettsia prowazekii-infected host cells based on bacterial burden and early detection of fluorescent rickettsial transformants. PLoS ONE. 2016;11:e0152365. doi: 10.1371/journal.pone.0152365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Troyer JM, Radulovic S, Azad AF. Green fluorescent protein as a marker in Rickettsia typhi transformation. Infect. Immun. 1999;67:3308–3311. doi: 10.1128/iai.67.7.3308-3311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alzhanov DT, Suchland RJ, Bakke AC, Stamm WE, Rockey DD. Clonal isolation of Chlamydia-infected cells using flow cytometry. J. Microbiol. Methods. 2007;68:201–208. doi: 10.1016/j.mimet.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 57.Podgorny OV, et al. Isolation of single Chlamydia-infected cells using laser microdissection. J. Microbiol. Methods. 2015;109:123–128. doi: 10.1016/j.mimet.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 58.Elwell C, Mirrashidi K, Engel J. Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 2016;14:385–400. doi: 10.1038/nrmicro.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lutter EI, Martens C, Hackstadt T. Evolution and conservation of predicted inclusion membrane proteins in chlamydiae. Comp. Funct. Genomics. 2012;2012:362104. doi: 10.1155/2012/362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alix E, et al. The capping domain in RalF regulates effector functions. PLoS Pathog. 2012;8:e1003012. doi: 10.1371/journal.ppat.1003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rennoll-Bankert KE, et al. Which way in? The RalF Arf-GEF orchestrates Rickettsia host cell invasion. PLoS Pathog. 2015;11:e1005115. doi: 10.1371/journal.ppat.1005115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kagan JC, Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 2002;4:945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- 64.Prantner D, Perkins DJ, Vogel SN. AMP-activated kinase (AMPK) promotes innate immunity and antiviral defense through modulation of stimulator of interferon genes (STING) signaling. J. Biol. Chem. 2017;292:292–304. doi: 10.1074/jbc.M116.763268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hare DN, et al. Membrane perturbation-associated Ca2+ signaling and incoming genome sensing are required for the host response to low-level enveloped virus particle entry. J. Virol. 2015;90:3018–3027. doi: 10.1128/JVI.02642-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sahni SK, Narra HP, Sahni A, Walker DH. Recent molecular insights into rickettsial pathogenesis and immunity. Future Microbiol. 2013;8:1265–1288. doi: 10.2217/fmb.13.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gillespie JJ, et al. Secretome of obligate intracellular. Rickettsia. FEMS Microbiol. Rev. 2015;39:47–80. doi: 10.1111/1574-6976.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park H, Lee JH, Gouin E, Cossart P, Izard T. The rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin. J. Biol. Chem. 2011;286:35096–35103. doi: 10.1074/jbc.M111.263855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rikihisa Y. Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat. Rev. Microbiol. 2010;8:328–339. doi: 10.1038/nrmicro2318. [DOI] [PubMed] [Google Scholar]
- 70.Chen G, et al. Anaplasma phagocytophilum dihydrolipoamide dehydrogenase 1 affects host-derived immunopathology during microbial colonization. Infect. Immun. 2012;80:3194–3205. doi: 10.1128/IAI.00532-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Henderson B, Martin A. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 2011;79:3476–3491. doi: 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dumler JS. The biological basis of severe outcomes in Anaplasma phagocytophilum infection. FEMS Immunol. Med. Microbiol. 2012;64:13–20. doi: 10.1111/j.1574-695X.2011.00909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheng C, Nair AD, Jaworski DC, Ganta RR. Mutations in Ehrlichia chaffeensis causing polar effects in gene expression and differential host specificities. PLoS ONE. 2015;10:e0132657. doi: 10.1371/journal.pone.0132657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nair AD, et al. Attenuated mutants of Ehrlichia chaffeensis induce protection against wild-type infection challenge in the reservoir host and in an incidental host. Infect. Immun. 2015;83:2827–2835. doi: 10.1128/IAI.00487-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang JZ, Hao JF, Walker DH, Yu XJ. A mutation inactivating the methyltransferase gene in avirulent Madrid E strain of Rickettsia prowazekii reverted to wild type in the virulent revertant strain Evir. Vaccine. 2006;24:2317–2323. doi: 10.1016/j.vaccine.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 76.Whitworth T, Popov VL, Yu XJ, Walker DH, Bouyer DH. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape. Infect. Immun. 2005;73:6668–6673. doi: 10.1128/IAI.73.10.6668-6673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H, Walker DH. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb. Pathog. 1998;24:289–298. doi: 10.1006/mpat.1997.0197. [DOI] [PubMed] [Google Scholar]
- 78.Zhong G, Fan P, Ji H, Dong F, Huang Y. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 2001;193:935–942. doi: 10.1084/jem.193.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen AL, Johnson KA, Lee JK, Sutterlin C, Tan M. CPAF: a chlamydial protease in search of an authentic substrate. PLoS Pathog. 2012;8:e1002842. doi: 10.1371/journal.ppat.1002842. This study shows that several substrates of a chlamydial protease are artefacts of sample processing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Snavely EA, et al. Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog. Dis. 2014;71:336–351. doi: 10.1111/2049-632X.12179. This article shows that several substrates of a chlamydial protease are artefactual through the use of a CPAF-processing-defective C. trachomatis serovar L2 strain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patton MJ, et al. Chlamydial protease-like activity factor and type III secreted effectors cooperate in inhibition of p65 nuclear translocation. mBio. 2016;7:e01427–16. doi: 10.1128/mBio.01427-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang Z, et al. The Chlamydia-secreted protease CPAF promotes chlamydial survival in the mouse lower genital tract. Infect. Immun. 2016;84:2697–2702. doi: 10.1128/IAI.00280-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bavoil PM, Byrne GI. Analysis of CPAF mutants: new functions, new questions (the ins and outs of a chlamydial protease) Pathog. Dis. 2014;71:287–291. doi: 10.1111/2049-632X.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dunning Hotopp JC, et al. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2006;2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cho N-H, et al. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc. Natl Acad. Sci. USA. 2007;104:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gillespie JJ, et al. The Rickettsia type IV secretion system: unrealized complexity mired by gene family expansion. Pathog. Dis. 2016;74:ftw058. doi: 10.1093/femspd/ftw058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rikihisa Y, Lin M. Anaplasma phagocytophilum and Ehrlichia chaffeensis type IV secretion and Ank proteins. Curr. Opin. Microbiol. 2010;13:59–66. doi: 10.1016/j.mib.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jernigan KK, Bordenstein SR. Ankyrin domains across the tree of life. PeerJ. 2014;2:e264. doi: 10.7717/peerj.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lina TT, et al. Hacker within! Ehrlichia chaffeensis effector driven phagocyte reprogramming strategy. Front. Cell. Infect. Microbiol. 2016;6:58. doi: 10.3389/fcimb.2016.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 91.Brayton KA, et al. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc. Natl Acad. Sci. USA. 2005;102:844–849. doi: 10.1073/pnas.0406656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dhungana S, et al. The influence of the synergistic anion on iron chelation by ferric binding protein, a bacterial transferrin. Proc. Natl Acad. Sci. USA. 2003;100:3659–3664. doi: 10.1073/pnas.0536897100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khun HH, Deved V, Wong H, Lee BC. fbpABC gene cluster in Neisseria meningitidis is transcribed as an operon. Infect. Immun. 2000;68:7166–7171. doi: 10.1128/iai.68.12.7166-7171.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Doyle CK, Zhang X, Popov VL, McBride JW. An immunoreactive 38-kilodalton protein of Ehrlichia canis shares structural homology and iron-binding capacity with the ferric ion-binding protein family. Infect. Immun. 2005;73:62–69. doi: 10.1128/IAI.73.1.62-69.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ge Y, Rikihisa Y. Subversion of host cell signaling by Orientia tsutsugamushi. Microbes Infect. 2011;13:638–648. doi: 10.1016/j.micinf.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 96.Beare PA, Larson CL, Gilk SD, Heinzen RA. Two systems for targeted gene deletion in Coxiella burnetii. Appl. Environ. Microbiol. 2012;78:4580–4589. doi: 10.1128/AEM.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van Schaik EJ, Chen C, Mertens K, Weber MM, Samuel JE. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat. Rev. Microbiol. 2013;11:561–573. doi: 10.1038/nrmicro3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Valbuena G, Walker DH. Approaches to vaccines against Orientia tsutsugamushi. Front. Cell. Infect. Microbiol. 2013;2:170. doi: 10.3389/fcimb.2012.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Menon S, et al. Human and pathogen factors associated with Chlamydia trachomatis-related infertility in women. Clin. Microbiol. Rev. 2015;28:969–985. doi: 10.1128/CMR.00035-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hu VH, Holland MJ, Burton MJ. Trachoma: protective and pathogenic ocular immune responses to Chlamydia trachomatis. PLoS Negl. Trop. Dis. 2013;7:e2020. doi: 10.1371/journal.pntd.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dumler JS, Madigan JE, Pusterla N, Bakken JS. Ehrlichioses in humans: epidemiology, clinical presentation, diagnosis, and treatment. Clin. Infect. Dis. 2007;45:S45–S51. doi: 10.1086/518146. [DOI] [PubMed] [Google Scholar]
- 102.Stuen S, Granquist EG, Silaghi C. Anaplasma phagocytophilum — a widespread multi-host pathogen with highly adaptive strategies. Front. Cell. Infect. Microbiol. 2013;3:31. doi: 10.3389/fcimb.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Merhej V, Angelakis E, Socolovschi C, Raoult D. Genotyping, evolution and epidemiological findings of Rickettsia species. Infect. Genet. Evol. 2014;25:122–137. doi: 10.1016/j.meegid.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 104.Weitzel T, et al. Endemic scrub typhus in South America. N. Engl. J. Med. 2016;375:954–961. doi: 10.1056/NEJMoa1603657. [DOI] [PubMed] [Google Scholar]
- 105.Newman L, et al. Global estimates of the prevalence and incidence of four curable sexually transmitted Infections in 2012 based on systematic review and global reporting. PLoS ONE. 2015;10:e0143304. doi: 10.1371/journal.pone.0143304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Taylor HR, Burton MJ, Haddad D, West S, Wright H. Trachoma. Lancet. 2014;384:2142–2152. doi: 10.1016/S0140-6736(13)62182-0. [DOI] [PubMed] [Google Scholar]
- 107.Sanchez E, Vannier E, Wormser GP, Hu LT. Diagnosis, treatment, and prevention of Lyme disease, human granulocytic anaplasmosis, and babesiosis: a review. JAMA. 2016;315:1767–1777. doi: 10.1001/jama.2016.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dahlgren FS, Mandel EJ, Krebs JW, Massung RF, McQuiston JH. Increasing incidence of Ehrlichia chaffeensis and Anaplasma phagocytophilum in the United States, 2000-2007. Am. J. Trop. Med. Hyg. 2011;85:124–131. doi: 10.4269/ajtmh.2011.10-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kelly DJ, Richards AL, Temenak J, Strickman D, Dasch GA. The past and present threat of rickettsial diseases to military medicine and international public health. Clin. Infect. Dis. 2002;34:S145–S169. doi: 10.1086/339908. [DOI] [PubMed] [Google Scholar]
- 110.Richards AL. Worldwide detection and identification of new and old rickettsiae and rickettsial diseases. FEMS Immunol. Med. Microbiol. 2012;64:107–110. doi: 10.1111/j.1574-695X.2011.00875.x. [DOI] [PubMed] [Google Scholar]
- 111.Nakayama K, et al. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 2008;15:185–199. doi: 10.1093/dnares/dsn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.