Abstract
The composition of porcine milk oligosaccharides (PMO) was analyzed during early lactation and their relation to piglet gut microbiome was investigated. Pigs are considered ideal intestinal models to simulate humans because of the striking similarity in intestinal physiopathology to humans. The evolution of PMO was investigated in the milk from 3 healthy sows at prefarrowing, farrowing, and d 7 and 14 postpartum by Nano-LC Chip Quadrupole-Time-of-Flight mass spectrometer (Agilent Technologies, Santa Clara, CA). Previously sequenced metagenome libraries were reanalyzed to examine changes with specific gut bacterial populations. Over 30 oligosaccharides (OS) were identified in the milk, with 3′-sialyllactose, lacto-N-tetraose, α1–3,β1–4-d-galactotriose, 2′-fucosyllactose, and 6′-sialyllactose being the most abundant species (accounting for ~70% of the total OS). Porcine milk had lower OS diversity (number of unique structures) than human milk, and appeared closer to bovine and caprine milk. In agreement with previous studies, only 3 fucosylated OS were identified. Surprisingly, their contribution to total OS abundance was greater than in bovine milk (9 vs. 1%). Indeed, fucosylated PMO increased during lactation, mirroring a similar trend observed for neutral and type I OS content during early lactation. Taken together, these results suggest that, in terms of abundance, PMO are closer to human milk than other domestic species, such as bovine and caprine milks. Metagenomic sequencing revealed that fucose-consuming bacterial taxa in the gut microbiota of piglets were qualitatively but not quantitatively different between nursing and weaning stages, suggesting that both the composition and structure of dietary glycans may play a critical role in shaping the distal gut microbiome. The similarity of both intestinal physiopathology and milk OS composition in human and porcine species suggests similar effects on gastrointestinal development of early nutrition, reinforcing the use of the pig intestinal model to simulate human intestinal models in the clinical setting.
Keywords: porcine milk, oligosaccharide, gut microbiota
INTRODUCTION
Breast milk is the optimal source of nutrition for the first few months of an infant’s life, as it provides all the necessary nutrients for optimal growth and development. Its composition has been shaped by natural selection, which acts on both the infant and the mother, maximizing infant survival, growth, and development while minimizing the metabolic costs of lactation for the mother (Zivkovic et al., 2011). As a result of this natural selective pressure, breast milk evolved to have a unique composition where lipids and lactose are the most abundant components, followed by oligosaccharides (OS) and proteins (German et al., 2008). Despite OS being the third most abundant components in milk (Kunz et al., 2000), it was long thought they had no biological significance. Recent studies pointed out many and diverse biological activities afforded by milk OS, including prebiotic activity, antiadhesion effects, anti-inflammatory properties, glycome-modifying activity, and roles in brain development, as well as influencing the growth-related characteristics of intestinal cells (Hickey, 2012; Lane et al., 2012; ten Bruggencate et al., 2014). Taken together, these bioactivities favor the establishment of a protective and healthful intestinal environment (Smilowitz et al., 2014). Milk OS are typically composed of 3 to 10 repeating monosaccharide units, including glucose (Glc), galactose (Gal), N-acetyl-glucosamine (GlcNAc), N-acetyl-galactosamine (GalNAc), fucose (Fuc), and sialic acids (NeuAc/ NeuGc). The core unit of bovine milk and other domestic milk OS can be either lactose [Gal(β1–4)Glc] or N-acetyl-lactosamine [Gal(β1–4)GlcNAc; Aldredge et al., 2013]. Based on their chemical composition, OS are classified as acidic (decorated by sialic acid, NeuAc/NeuGc) or neutral (containing GlcNAc, HexNAc, or fucose, lacking sialic acid).
Breast milk composition is not static but changes during lactation. The OS content can be as high as 23 g/L in colostrum and rapidly decrease during lactation to values a low as 5 to 10 g/L late in nursing (Newburg, 2013). Human milk OS (HMO) have been widely studied, with more than 200 structures partially characterized (Wu et al., 2010, 2011) displaying a predominance of neutral fucosylated (50–70%) compared with sialylated OS (10–25%; Niñonuevo et al., 2008). The concentration of OS in milk from other mammalian species is lower than that in human milk, with a concentration of 1 g/L in bovine colostrum and even lower concentrations in mature bovine milk (Fong et al., 2011); however, a great diversity of OS structures has been reported for domestic animals. A bioinformatic library of 55 structures was assembled for bovine milk OS (BMO; Aldredge et al., 2013), whereas 39 OS were described in porcine milk (Tao et al., 2010; Albrecht et al., 2014), 20 OS in caprine milk (Meyrand et al., 2013; Albrecht et al., 2014), and only 12 OS in camel milk (Alhaj et al., 2013; Albrecht et al., 2014). Conversely, the ratio of fucosylated to sialylated OS in milk from domestic animals is shifted toward sialylated OS, which predominate the overall composition (70%; Urashima et al., 2013).
Swine are important agricultural species and are considered to be an excellent model for nutritional studies because their digestive system, immune system physiology, and anatomical structure are similar to those of humans (Guilloteau et al., 2010). Only a few studies to date have evaluated porcine milk oligosaccharides (PMO) changes during early lactation (Tao et al., 2010; Albrecht et al., 2014). The present study characterized the composition and variation of PMO during the first 2 wk of lactation and examined how PMO composition and of the gut metagenome of suckling pigs interact to shape the nursing pig microbiome.
MATERIALS AND METHODS
Materials
Sodium borohydride was from Sigma-Aldrich (St. Louis, MO). Commercial OS standards of lacto-N-tetraose (LNT), lacto-N-neotetraose (LNnT), lacto-N-hexaose (LNH), and lacto-N-neohexaose (LNnH) were purchased from V-Labs (Covington, LA). All solvents used for sample preparation were HPLC-MS grade (Fisher Scientific, Fair Lawn, NJ). The DNA was extracted from feces using the Zymo Research Fecal DNA kit (Zymo Research, Irvine, CA) according to the manufacturer’s instructions, with modifications as described previously (Frese et al., 2015). Nonporous graphitized carbon solid-phase extraction (GCC-SPE; 2,000 µg binding capacity) was from Glygen Corp. (Columbia, MD). Nanopure water (18.2 MΩ/cm, 25°C) was used for the analytical work.
Collection of Porcine Samples
The University of California, Davis Institutional Animal Care and Use Committee approved all animal experiments under study before beginning the study, and this study has been described in detail previously (Frese et al., 2015). Three healthy Yorkshire or Hampshire adult pregnant multiparous sows from the University of California, Davis swine herd were selected for this study. Infant pigs were allowed to nurse freely until 21 d of age and did not consume sow feed. After 21 d, half the nursing pigs were moved to separate housing (away from the sow) and fed a standard starter feed (Hubbard Feeds, Mankato, MN). Animals were given ad libitum access to water and feed. Milk samples (2 mL) were collected by manual expression into a sterile collection container 1 d before farrowing (precolostrum), at farrowing (d 0), and at d 7 and 14 postfarrowing. Samples were immediately frozen at −80°C until analysis.
To avoid differences in sample preparation and analytical techniques that often hinder comparison of OS structures and abundances with published literature values, 2 separate samples of pooled human colostrum and pooled bovine colostrum were obtained at the University of California, Davis from other ongoing studies and analyzed simultaneously using the same analytical procedure.
OS Isolation and Purification
Milk OS were isolated and purified as previously described, with minor modification (Barile et al., 2010). Briefly, frozen milk samples were completely thawed, and a 0.5-mL aliquot of each sample was mixed with an equal volume of nanopure water and centrifuged at 14,000 × g in a microfuge for 30 min at 4°C to remove lipids. The top fat layer was removed, and 4 volumes of chloroform:methanol (2:1, vol/vol) were added, vigorously mixed, and the resulting emulsion was centrifuged at 4,000 × g for 30 min at 4°C. The upper methanol layer containing OS was transferred to a tube, 2 volumes of cold ethanol were added, and the solution was frozen for 1 h at −30°C, followed by centrifugation for 30 min at 4,000 × g and 4°C to precipitate the denatured protein. The supernatant (OS-rich fraction) was collected and freeze-dried using a speed vacuum centrifuge. Native milk OS were reduced to alditol forms using 2.0 M sodium borohydride and incubated at 65°C for 1 h. Standard solutions of the OS LNT, LNnT, LNH, and LNnH (100 µg/L) were reduced in identical conditions. Oligosaccharides were purified from the mixture by GCC-SPE. Prior to use, each GCC-SPE cartridge was activated with 3 column volumes of 80% acetonitrile (ACN) and 0.1% trifluoroacetic acid (TFA, vol/vol) and equilibrated with 3 column volumes of nanopure water. The carbohydrate-rich solution was loaded onto the cartridge, and salts and mono- or disaccharides were removed by washing with 10 column volumes of nanopure water. The OS were eluted with a solution of 40% ACN with 0.1% TFA (vol/vol) in water and dried in speed vacuum centrifuge at 35°C overnight.
Characterization by LC Quadrupole-Time-of-Flight MS
Prior to MS analysis, dried OS samples were reconstituted in 100 µL of nanopure water. Mass spectrometry analysis was performed with an Agilent 6520 accurate-mass Quadrupole-Time-of-Flight (QTOF) LC/MS with a microfluidic nanoelectrospray chip (Agilent Technologies, Santa Clara, CA) as previously described (Wu et al., 2011). The microfluidic chip contained one enrichment and one analytical column, both packed with graphitized carbon. Chromatographic elution was performed with a binary gradient of 3% ACN/0.1% formic acid in water (solvent A) and 90% ACN/0.1% formic acid in water (solvent B). The column was initially equilibrated with a flow rate of 0.3 µL/min for the nanopump and 4 µL/min for the capillary pump. The 65-min gradient was programmed as follows: 0 to 2.5 min, 0% B; 2.5 to 20 min, 0 to16% B; 20 to 30 min, 16 to 44% B; 30 to 35 min, 44 to 100% B; 35 to 45 min, 100% B; and 45 to 65 min, 0% B. Data were acquired in the positive ionization mode with a 450 to 2,500 m/z range. The electrospray capillary voltage was 1,700 to 1,900 V. The acquisition rate was 0.63 spectra/s for both MS and MS/MS modes. Automated precursor selection was employed based on ion abundance, performing up to 6 MS/MS spectra per individual MS when precursor was above ion abundance threshold. The precursor isolation window was selected to be narrow (1.3 m/z) to improve accuracy. Fragmentation energy was set at 1.8 V/100 Da with an offset of −2.4 V. Internal calibration was continuously performed by infusing 2 reference masses: 922.009 and 1221.991 m/z (ESI-TOF Tuning Mix G1969–85000, Agilent Technologies).
QTOF Data Analysis
The Molecular Feature Extraction function of Mass Hunter Qualitative Analysis Version B.06.00 (Agilent Technologies) was used to generate a list of deconvoluted masses selected to be in a range of 450 to 1,500 m/z with a ≥1,000 height count and a typical isotopic distribution of small biological molecules. Charge states allowed were restricted to single and double species. Oligosaccharide compositions were determined from the deconvoluted mass list with in-house software, and all OS compositions were confirmed by MS/MS analysis. Following MS/MS identity validation and assessment of reproducible retention times (RT), individual peaks for each OS were automatically integrated using the Targeted Feature Extractor from MassHunter Profinder Version B.06.00 (Agilent Technologies). The RT window allowed for compound matching was restricted to ±0.5 min and ±0.25% of the RT at each time point.
Metagenomic and 16S rRNA Sequencing
In a previous study (Frese et al., 2015), fecal samples were collected rectally from nursing animals (n = 3), suckling the sows from whom milk was collected, using a sterile cotton swab wet with sterile PBS (pH 7.0) at 14 and 35 d after birth. The DNA was extracted from fecal sampling swabs using a Zymo Research Fecal DNA Miniprep kit, and the resulting DNA was used to characterize the microbial communities by molecular methods. Metagenomic sequencing was conducted using barcoded 100-bp single read shotgun libraries and pooled for sequencing across triplicate runs on an Illumina MiSeq (Illumina, San Diego, CA) at the University of California, Davis Genome Center, with libraries prepared from samples from 3 animals before (at 14 d of life) and after weaning (at 35 d of life). The data sets were previously analyzed (Frese et al., 2015) and were reanalyzed here with respect to the novel MS findings described herein. Functionally binned reads were taxonomically identified using Kraken (Wood and Salzberg, 2014). Data sets from this and previous studies are publicly available in MG-RAST (http://metagenomics.anl.gov).
RESULTS AND DISCUSSION
Characterization of Porcine Colostrum OS
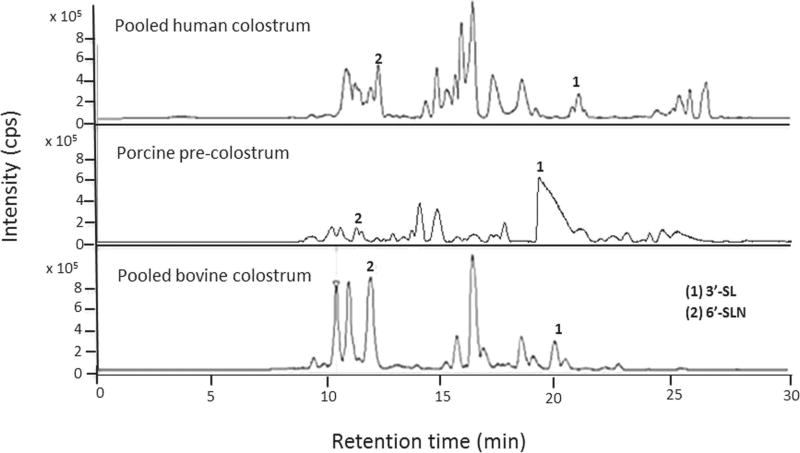
Mass spectrometry is now widely accepted as an ideal tool for the characterization of OS in mammalian milks and biological samples (Niñonuevo et al., 2008; Aldredge et al., 2013; DeLeoz et al., 2013). Leveraging the reproducible RT and accurate mass of the Nano-LC-Chip–QTOF MS (Agilent Technologies) system, we identified over 30 PMO and assessed similarities with HMO and BMO. Figure 1 presents the comparison of OS from pooled human colostrum, porcine precolostrum, and pooled bovine colostrum. Two major OS in common among the 3 types of milk were identified using in-house bioinformatics libraries as 3′-sialyllac- tose (634.2189 m/z: peak 1) and 6′-sialyllactosamine (675.2455 m/z. peak 2).
Figure 1.
Comparison of the base peak chromatograms of oligosaccharides extracted from pooled human colostrum, porcine precolostrum, and pooled bovine colostrum. Peak identification: peak 1 = 3′-sialyllactose (3′-SL; 636.23 m/z); peak 2 = 6′-sialyllactosamine (6′-SLN; 677.25 m/z); cps = counts per second.
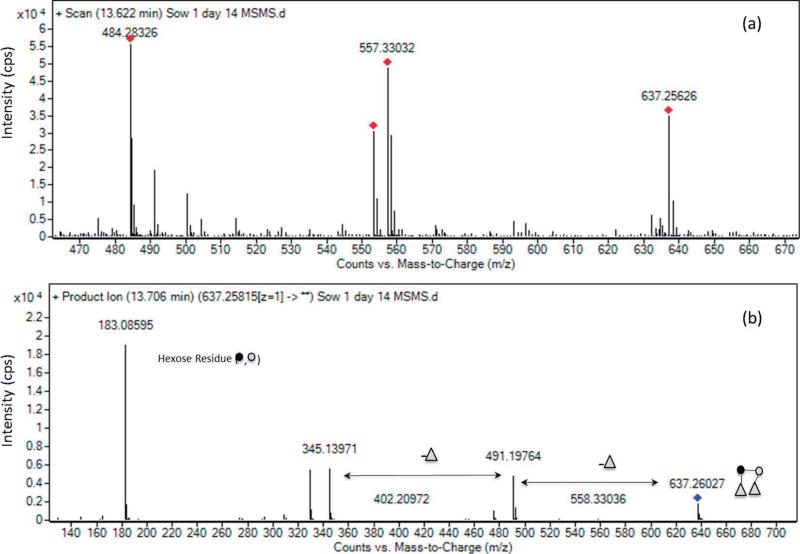
Table 1 presents the details of the 33 OS identified in porcine milk from 3 sows at a lactation period ranging from prefarrow to d 14 postfarrow. Interestingly, we identified lactodifucotetraose (636.24 m/z), an OS typical of human milk and never before identified in porcine milk (see Figure 2 for the MS/MS confirmation). The number of OS identified is within the range described in literature (Tao et al., 2010; Albrecht et al., 2014), differences may due to genetic variability in sows from different studies. Similarly to human milk, only a small number of OS (i.e., 5 compounds) represented nearly 70% of the remaining OS abundances. The most abundant PMO were 3′-sialyllactose (26%), LNT/ LNnT (20%), triose (9%), 2′-fucosyllactose (7%), and 6′-sialyllactosamine (5%; see Table 1, Supplemental Figure S1; http://dx.doi.org/10.3168/jds.2016-10966). Interestingly, this work identified a higher percentage of neutral OS (≈50%) for porcine milk compared with human milk (20%), and the absence of Neu5Gc-containing OS in porcine milk.
Table 1.
Details of milk oligosaccharides identified in porcine milk and comparison with human milk (HM) and bovine milk (BM)1
| Number | Exact mass (expected) |
Exact mass (calculated) |
Hex | HexNAc | Fuc | Neu5Ac | Neu5Gc | Retention time (min) |
Relative abundance2 |
HM presence3 |
BM presence3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 676.255 | 676.253 | 1 | 1 | 0 | 1 | 0 | 11.34 | 0.11 | ○ | ○ |
| 2 | 676.255 | 676.253 | 1 | 1 | 0 | 1 | 0 | 13.6 | 2.40 | ● | □ |
| 3 | 676.254 | 676.253 | 1 | 1 | 0 | 1 | 0 | 18.68 | 0.10 | ● | ● |
| 4 | 635.231 | 635.227 | 2 | 0 | 0 | 1 | 0 | 13.65 | 5.36 | ● | ● |
| 5 | 635.228 | 635.227 | 2 | 0 | 0 | 1 | 0 | 20.48 | 26.05 | ● | □ |
| 6 | 490.19 | 490.190 | 2 | 0 | 1 | 0 | 0 | 11.65 | 7.32 | □ | ● |
| 7 | 490.191 | 490.190 | 2 | 0 | 1 | 0 | 0 | 13.73 | 0.53 | ○ | ● |
| 8 | 636.239 | 636.248 | 2 | 0 | 2 | 0 | 0 | 13.71 | 2.59 | ● | ○ |
| 9 | 750.291 | 750.290 | 2 | 2 | 0 | 0 | 0 | 13.48 | 2.55 | ○ | ● |
| 10 | 506.185 | 506.185 | 3 | 0 | 0 | 0 | 0 | 10.39 | 8.84 | ○ | □ |
| 11 | 506.185 | 506.185 | 3 | 0 | 0 | 0 | 0 | 14.01 | 1.34 | ○ | ● |
| 12 | 797.281 | 797.280 | 3 | 0 | 0 | 1 | 0 | 21.41 | 0.79 | ○ | ● |
| 13 | 797.281 | 797.280 | 3 | 0 | 0 | 1 | 0 | 23.29 | 1.57 | ○ | ● |
| 14 | 709.265 | 709.264 | 3 | 1 | 0 | 0 | 0 | 11.94 | 0.85 | □ | ● |
| 15 | 709.265 | 709.264 | 3 | 1 | 0 | 0 | 0 | 14.27 | 20.03 | ● | ● |
| 16 | 709.264 | 709.264 | 3 | 1 | 0 | 0 | 0 | 18.04 | 0.66 | ○ | ● |
| 17 | 709.265 | 709.264 | 3 | 1 | 0 | 1 | 0 | 25.67 | 0.45 | ○ | ● |
| 18 | 1,000.356 | 1,000.359 | 3 | 1 | 0 | 1 | 0 | 16.49 | 0.21 | ● | ● |
| 19 | 1,000.360 | 1,000.359 | 3 | 1 | 0 | 1 | 0 | 22.38 | 3.62 | ● | ○ |
| 20 | 1,000.361 | 1,000.359 | 3 | 2 | 0 | 0 | 0 | 25.98 | 0.41 | ● | ○ |
| 21 | 912.343 | 912.343 | 3 | 2 | 0 | 0 | 0 | 13.05 | 0.38 | ○ | ● |
| 22 | 912.344 | 912.343 | 3 | 2 | 0 | 0 | 0 | 13.71 | 0.37 | ○ | ● |
| 23 | 912.344 | 912.343 | 3 | 2 | 0 | 0 | 0 | 14.73 | 0.29 | ○ | ○ |
| 24 | 912.343 | 912.343 | 3 | 2 | 0 | 0 | 0 | 16.21 | 0.28 | ○ | ○ |
| 25 | 912.343 | 912.343 | 3 | 2 | 0 | 0 | 0 | 16.65 | 0.20 | ○ | ○ |
| 26 | 668.238 | 668.237 | 4 | 0 | 0 | 0 | 0 | 14.00 | 1.85 | ○ | ● |
| 27 | 871.318 | 871.317 | 4 | 1 | 0 | 0 | 0 | 15.00 | 3.53 | ○ | ● |
| 28 | 871.318 | 871.317 | 4 | 1 | 0 | 0 | 0 | 17.46 | 1.94 | ○ | ● |
| 29 | 871.316 | 871.317 | 4 | 1 | 0 | 0 | 0 | 25.56 | 0.37 | ○ | ○ |
| 30 | 1,162.414 | 1,162.412 | 4 | 2 | 1 | 0 | 0 | 25.65 | 0.32 | ○ | ○ |
| 31 | 1,074.397 | 1,074.396 | 4 | 2 | 0 | 0 | 0 | 18.04 | 3.81 | ○ | ● |
| 32 | 1,365.493 | 1,365.492 | 4 | 2 | 0 | 1 | 0 | 24.53 | 0.65 | ● | ● |
| 33 | 1,365.492 | 1,365.492 | 4 | 2 | 0 | 1 | 0 | 25.91 | 0.06 | ● | ○ |
All numbers below each monosaccharide type (Hex, HexNAc, and so on) indicate how many monosaccharides are found in each oligosaccharide. For example, oligosaccharide N1 with mass 676.255 is composed of 1 Hex, 1 HexNAc, and 1 Neu5Ac. Hex = hexose; Fuc = fucose; HexNAc = N-acetylhexosamine; Neu5Ac = N-acetylneuraminic acid; Neu5Gc = N-glycolylne uramini c acid.
Relative abundances expressed as percentage of total oligosaccharides in all samples analyzed.
Presence of oligosaccharides in HM and BM: open squares, >15%; solid circles, 0.1–15%; open circles, not present.
Figure 2.
Confirmation of the oligosaccharide lactodifucotetraose (637.258 m/z) in porcine milk by MS/MS: (a) precursor spectrum; (b) product spectrum. Dark circle = glucose; light circle = galactose; triangle = fucose; cps = counts per second. Color version available online.
As expected, we noted significantly less structural diversity in PMO compared with HMO, yielding only 1 or 2 oligosaccharides with the same composition, similar to that reported for BMO (Zivkovic and Barile, 2011). Human milk is reported to contain more than 200 OS, whereas bovine milk or colostrum has up to 55 (Aldredge et al., 2013; Albrecht et al., 2014) and porcine milk 39 OS (Tao et al., 2010; Albrecht et al., 2014; Cheng et al., 2015). Porcine milk contains 2′-fu-cosyllactose and type I OS (LNT, lacto-N-fucopentaose I, lacto-N-fucopentaose II, lacto-N-difucohexose I) in higher abundance, whereas type II OS (whose core units are LNnT or LNnH, lacto-N-fucopentaose III) are present in lower abundances. No type I OS have been identified in bovine milk (Urashima et al., 2013). To identify the type of OS in the porcine milk analyzed, samples were spiked with reduced standard solutions of LNT, LNnT, LNH, and LNnH. Results (Supplemental Figure S2; http://dx.doi.org/10.3168/jds.2016-10966), which allowed us to identify the isomers LNnH, LNT and LNnT in the samples, suggest that porcine milk OS are structurally closer to OS in human milk than those in bovine milk. This work characterized 3 fucosylated OS, which is in agreement with Albrecht et al. (2014) but is lower than by Tao et al. (2010). Regardless of the lower structural diversity, the contribution of fucosylated OS to the total OS is high compared with bovine milk (9.1 vs. 1%). Overall, the composition and abundance of PMO suggest porcine milk is closer to human milk than bovine milk based on the higher abundance of fucosylated OS structures, the lower abundance of sialylated structures, and the presence of LNT at a high abundance (20%).
Changes in PMO During Early Lactation
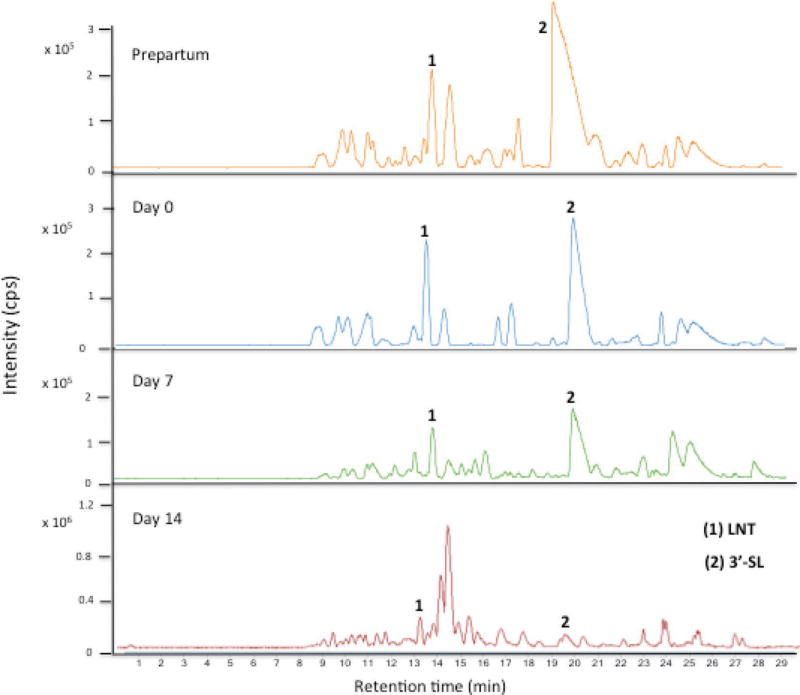
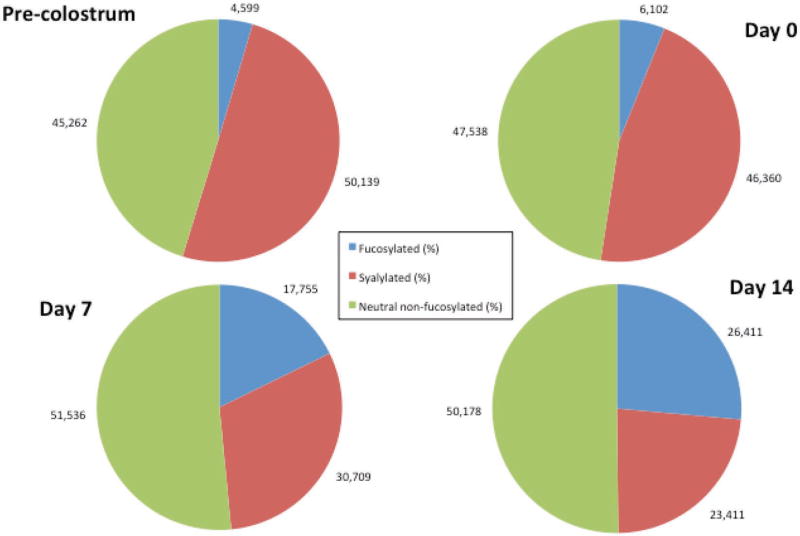
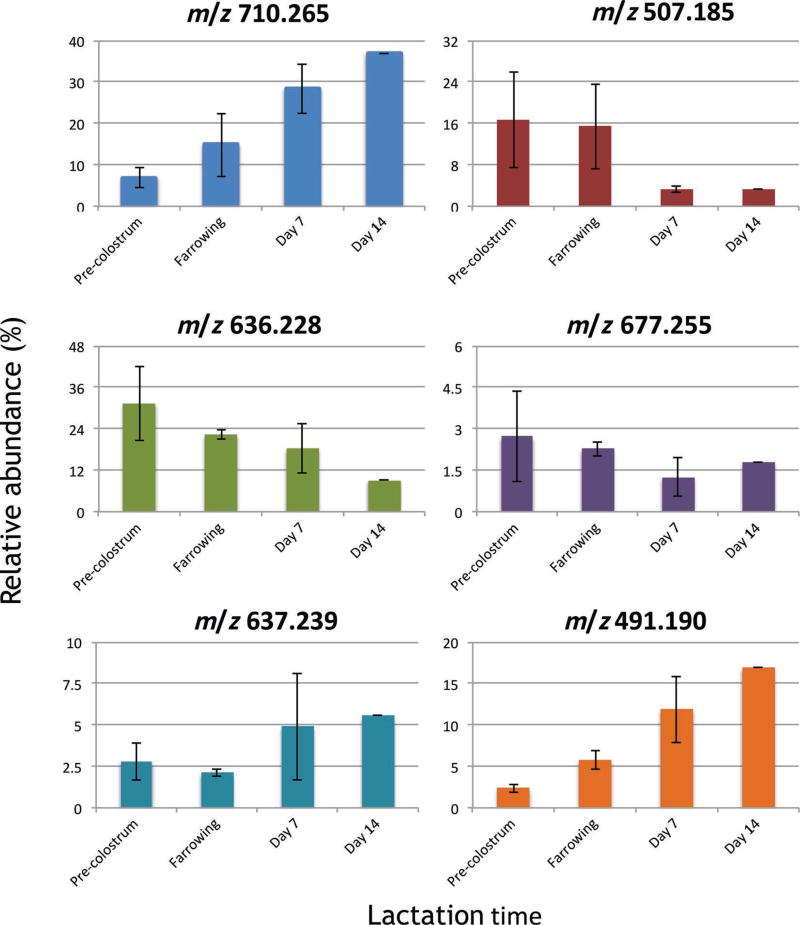
Porcine milk OS base peak chromatograms obtained from precolostrum, farrowing, and d 7 and 14 samples are shown in Figure 3. The PMO profile and abundance revealed a high variability during lactation (Tao et al., 2010), either increasing or decreasing with lactation. The peak with RT between 20.0 and 20.5 min corresponding to 3′-sialyllactose was the most abundant, as previously described for porcine milk (Tao et al., 2010), human milk (Thurl et al., 1996; Smilowitz et al., 2013), and bovine milk (Tao et al., 2009). Importantly, the OS at RT ≈13.5 was identified as LNT, which is abundant in human milk but only present at trace level in bovine milk. Relevant shifts in OS composition occurred as lactation progressed (see Figure 4). In precolostrum and farrowing stage samples, sialylated and nonfucosylated neutral OS were the most abundant and had similar abundances, whereas fucosylated OS were present in low abundance (4–6%). A significant decrease (P < 0.05) of sialylated OS correlated with a significant increase (P < 0.05) of fucosylated OS at d 14 is observed. Figure 5 shows the variation of the most abundant neutral nonfucosylated (top panel, 710.265 m/z; 507.185 m/z), sialylated (central panel, 636.228 m/z, 3′-sialyllactose; 677.225 m/z, 6′-sialyllactose), and fucosylated OS (bottom panel, 637.239 m/z; 491.190 m/z). The most abundant sialylated OS (central panel) decreased during early lactation (−30 to −60%), the abundance of the main fucosylated OS (bottom panel) increased during the same time period (98 to 615%), and neutral OS abundances (top panel) did not show a clear pattern, with some greatly increasing and others decreasing (+430 to −71%); LNT actually increased in porcine milk during lactation.
Figure 3.
Base peak chromatogram of porcine milk from the same sow at prepartum and d 0, 7, and 14 postpartum. Peak identification: peak 1 = lacto-N-tetraose (LNT; 710.27 m/z); peak 2 = 3′-sialyllactose (3′-SL; 636.23 m/z); cps = counts per second. Color version available online.
Figure 4.
Porcine milk oligosaccharide (OS) variation during various stages of lactation. Results are expressed as the average percentage (%) of total OS from 3 sows. Medium gray (blue) = fucosylated OS; dark gray (red) = sialylated OS; light gray (green) = neutral nonfucosylated OS. Color version available online.
Figure 5.
Variation of selected porcine oligosaccharides (OS) from 3 sows during early lactation. Results are expressed as mean ± SD of at least 2 sows, except in samples for d 14, where only 1 sample was available. Upper panel = neutral nonfucosylated OS; middle panel = sialylated OS; lower panel = fucosylated OS. Color version available online.
A decrease in sialylated OS over lactation was also reported in bovine milk (Tao et al., 2009; Barile et al., 2010), whereas in human milk the abundance is lower (10–25%) and stable across lactation (Niñonuevo et al., 2008). In the first days of lactation, PMO and BMO had a similar pattern, with highly sialylated OS being the most abundant; however, as lactation advanced, the similarity between PMO and HMO increased as a result of the fucosylation increment and sialylation decreased in PMO.
Milk OS are known to have at least 2 important potential functions: prevention of pathogen binding to the intestinal epithelial and stimulation of beneficial bacterial growth (Newburg et al., 2005; Yu et al., 2013). Both fucosylation and sialylation play significant roles in those functions (Bode and Jantscher-Krenn, 2012; Charbonneau et al., 2016), whereas sialylated OS are essential for neuronal development (ten Bruggencate et al., 2014). Given that PMO are structurally closer to HMO than to BMO, it is interesting to consider whether milk OS are tailor-made for the postnatal development of the specific type of animal gastrointestinal system and, thus, the more similar the gastrointestinal systems are, the closer their milk OS compositions will be (Albrecht et al., 2014). This hypothesis needs to be validated by performing studies using comparable instrumentation and similar methodology to further characterize other mammalian milks and compare their composition and abundances to human milk.
PMO and the Gut Microbiome
Considering the possible dramatic effect of OS on the neonatal gut microbiome, elucidation of the common structures and abundances of these compounds in milk over the lactation period is a critical step in understanding how these structures differentially affect the gut microbiome at a given time. We previously described the distal gut microbiome of animals born to the sows profiled in this study of OS. That study showed how composition of dietary glycans dramatically shape the nursing gut microbiome from both taxonomic and functional standpoints during nursing and after weaning (Frese et al., 2015). Congruent with findings that previously noted sialic acid and HexNAc as dominant components of sow milk, we found these were crucial in shaping the different communities comprising the gut microbiome before weaning and that the abundance of these OS was high in the milk fed to the nursing pigs during the various stages of lactation in our study. Nearly half of the glycans in sow milk analyzed in the present study were in a sialylated form, which agrees with the previous report by Tao et al. (2010) with regard to PMO. In profiling the gut metagenome of nursing pigs, we found that the consumption of free sialic acid was largely dominated by the Enterobacteriaceae, a family that makes up a large fraction of the total community during the nursing period (Frese et al., 2015). Accordingly, this sialic acid-consuming microbiota disappeared at weaning, when these structures decreased in the diet and were replaced by dietary starches.
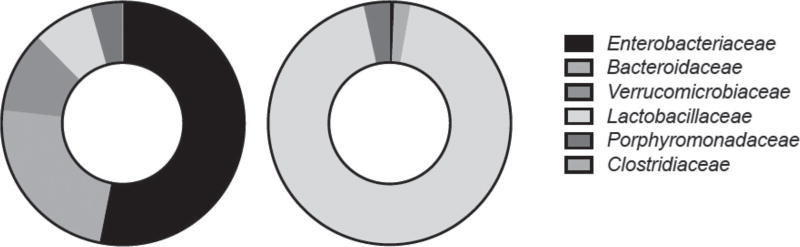
In the present study, we detected a surprising abundance of fucosylated glycans over lactation, despite detecting a smaller number of total structures than presented in previous reports (Tao et al., 2010). However, we did not previously examine how the fecal microbiome may be associated with fucose catabolism. In the present study, we reanalyzed these previous metagenomics-sequencing libraries to consider how fucose may affect the fecal metagenome. In contrast with our previous observations of microbial genes associated with Neu5Ac and HexNAc consumption (Frese et al., 2015), fucose catabolism pathways were not significantly different before and after weaning. However, the identity of fucose-consuming taxa changed dramatically at weaning, where Enterobacteriaceae comprised more than 53% of total assigned reads for metagenomic reads predicted to encode a fucose permease during nursing to 0.6% after weaning (see Figure 6). This suggests that both the composition and construction of dietary glycans may play a critical role in shaping the distal gut microbiome. Mirroring the interactions with sialic acid reported previously (Frese et al., 2015), Enterobacteriaceae were found to encode genes related to the consumption of free fucose (Figure 6) but lack the enzymatic capacity to liberate these monomers from PMO, which may also support the hypothesis that microbial cross-feeding is shaping these gut communities. Preferences among carbohydrate sources have been reported for Enterobacteriaceae (Chang et al., 2004), and this certainly offers a competitive advantage in the ecosystem for potential pathogens (Ng et al., 2013).
Figure 6.
Distribution of metagenomic reads from pig fecal samples annotated as fucose-permease enzymes in nursing (left circle) or weaned (right circle) animals. Weaned animals are piglets with diet changed at d 21, from sow milk to commercial standard starter food.
Although sialic acids are not found in the plant kingdom, fucose is a major component of many plant and animal glycans (Ma et al., 2006), which may explain the qualitative but not quantitative differences between nursing and weaning gut taxa consuming fucose. Thus, it is interesting to consider how these structures may have evolved to shape the gut microbiome of nursing pigs before domestication, given recently observed differences among wild, captive, and domestic Suidae (Ushida et al., 2015), or perhaps how breeding has shaped porcine milk OS production.
Conclusions
Nano-LC Chip QTOF MS-MS technology was used to identify 33 OS structures in porcine milk from precolostrum to d 14 of lactation. Porcine milk OS composition and evolution during early lactation were similar to those of human milk (despite lower diversity and number of structures), as indicated by the relative increase in fucosylated compounds and decrease in sialylated compounds as lactation progressed, accompanied by high amounts of neutral and type I OS. The consumption of milk OS was previously found to be reflected in the enzymatic potential of the gut metagenome, which is expanded here in light of our new findings related to the abundance of fucosylated structures. These findings suggest that porcine milk could be an ideal source for extraction of human milk OS mimics. The present study reinforces the pig model as one of the best human intestinal models. The similarity of both intestinal physiopathology and milk OS composition could lead to studies in the neonatal pig that would model how early nutrition intervenes in the gastrointestinal development in human infants.
Supplementary Material
Acknowledgments
This work was supported by the Bill and Melinda Gates Foundation (Seattle, WA), National Institutes of Health awards R01AT007079 and R01AT008759 (Bethesda, MD), and the Peter J. Shields Endowed Chair in Dairy Food Science (University of California–Davis). SAF was supported by National Institutes of Health Postdoctoral Fellowship F32AT008533. DAM and DB are founders of Evolve Biosystems (Davis, CA), a company focused on diet-based manipulation of the gut microbiota. SAF is now employed by Evolve Biosystems.
References
- Albrecht S, Lane JA, Mariño K, Al Busadah KA, Carrington SD, Hickey RM, Rudd PM. A comparative study of free oligosaccharides in the milk of domestic animals. Br. J. Nutr. 2014;111:1313–1328. doi: 10.1017/S0007114513003772. http://dx.doi.org/10.1017/S0007114513003772. [DOI] [PubMed] [Google Scholar]
- Aldredge DL, Geronimo MR, Hua S, Nwosu CC, Lebrilla CB, Barile D. Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures. Glycobiology. 2013;23:664–676. doi: 10.1093/glycob/cwt007. http://dx.doi.org/10.1093/glycob/cwt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhaj OA, Taufik E, Handa Y, Fukuda K, Saito T, Urashima T. Chemical characterisation of oligosaccharides in commercially pasteurised dromedary camel (Camelus dromedarius) milk. Int. Dairy J. 2013;28:70–75. http://dx.doi.org/10.1016/jidairyj.2012.08.008. [Google Scholar]
- Barile D, Marotta M, Chu C, Mehra R, Grimm R, Lebrilla CB, German JB. Neutral and acidic oligosaccharides in Holstein-Friesian colostrum during the first 3 days of lactation measured by high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. J. Dairy Sci. 2010;93:3940–3949. doi: 10.3168/jds.2010-3156. http://dx.doi.org/10.3168/jds.2010-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode L, Jantscher-Krenn E. Structure-function relationships of human milk oligosaccharides. Adv. Nutr. 2012;3:383S–391S. doi: 10.3945/an.111.001404. http://dx.doi.org/10.3945/an.111.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D-E, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, Conway T. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau MR, O’Donnell D, Blanton LV, Totten SM, Davis JCC, Barratt MJ, Cheng J, Guruge J, Talcott M, Bain JR, Muehlbauer MJ, Ilkayeva O, Wu C, Struckmeyer T, Barile D, Mangani C, Jorgensen J, Fan YM, Maleta K, Dewey KG, Ashorn P, Newgard CP, Lebrilla CB, Mills DA, Gordon JI. Sialylated milk glycans promote growth in gnotobiotic mice and pigs with a stunted Malawian infant gut microbiota. Cell. 2016;164:859–871. doi: 10.1016/j.cell.2016.01.024. http://dx.doi.org/10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LK, Wang LX, Xu QS, Huang LJ, Zhou DS, Li Z, Li SG, Du YG, Yin H. Chitooligosaccharide supplementation improves the reproductive performance and milk composition of sows. Livest. Sci. 2015;174:74–81. http://dx.doi.org/10.1016/j.livsci.2015.02.003. [Google Scholar]
- DeLeoz MLA, Wu S, Strum JS, Niñonuevo MR, Gaerlan SC, Mirmiran M, German JB, Mills DA, Lebrilla CB. A quantitative and comprehensive method to analyze human milk oligosaccharide structures in the urine and feces of infants. Anal. Bioanal. Chem. 2013;405:4089–4105. doi: 10.1007/s00216-013-6817-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong B, Ma K, McJarrow P. Quantification of bovine milk oligosaccharides using liquid chromatography-selected reaction monitoring-mass spectrometry. J. Agric. Food Chem. 2011;59:9788–9795. doi: 10.1021/jf202035m. http://dx.doi.org/10.1021/jf202035m. [DOI] [PubMed] [Google Scholar]
- Frese SA, Parker K, Calvert CC, Mills DA. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome. 2015;3:28. doi: 10.1186/s40168-015-0091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German JB, Freeman SL, Lebrilla CB, Mills DA. Human milk oligosaccharides: Evolution, structures and bioselectivity as substrates for intestinal bacteria. In: Bier DM, German JB, Lönnerdal B, editors. Nestlé Nutrition Workshop Series: Pediatric Program. Karger; Basel, Switzerland: 2008. pp. 205–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilloteau P, Zabielski R, Hammon HM, Metges CC. Nutritional programming of gastrointestinal tract development. Is the pig a good model for man? Nutr. Res. Rev. 2010;23:4–22. doi: 10.1017/S0954422410000077. http://dx.doi.org/10.1017/S0954422410000077. [DOI] [PubMed] [Google Scholar]
- Hickey RM. The role of oligosaccharides from human milk and other sources in prevention of pathogen adhesion. Int. Dairy J. 2012;22:141–146. http://dx.doi.org/10.1016/j.idairyj.2011.09.012. [Google Scholar]
- Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annu. Rev. Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. http://dx.doi.org/10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- Lane JA, Mariño K, Naughton J, Kavanaugh D, Clyne M, Carrington SD, Hickey RM. Anti-infective bovine colostrum oligosaccharides: Campylobacter jejuni as a case study. Int. J. Food Microbiol. 2012;157:182–188. doi: 10.1016/j.ijfoodmicro.2012.04.027. http://dx.doi.org/10.1016/j.ijfoodmicro.2012.04.027. [DOI] [PubMed] [Google Scholar]
- Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158R–184R. doi: 10.1093/glycob/cwl040. http://dx.doi.org/10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- Meyrand M, Dallas DC, Caillat H, Bouvier F, Martin P, Barile D. Comparison of milk oligosaccharides between goats with and without the genetic ability to synthesize αs1-casein. Small Rumin. Res. 2013;113:411–420. doi: 10.1016/j.smallrumres.2013.03.014. http://dx.doi.org/10.1016/j.smallrumres.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburg DS. Glycobiology of human milk. Biochemistry (Mosc.) 2013;78:771–785. doi: 10.1134/S0006297913070092. http://dx.doi.org/10.1134/S0006297913070092. [DOI] [PubMed] [Google Scholar]
- Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. http://dx.doi.org/10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. http://dx.doi.org/10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niñonuevo MR, Perkins PD, Francis J, Lamotte LM, LoCascio RG, Freeman SL, Mills DA, German JB, Grimm R, Lebrilla CB. Daily variations in oligosaccharides of human milk determined by microfluidic chips and mass spectrometry. J. Agric. Food Chem. 2008;56:618–626. doi: 10.1021/jf071972u. http://dx.doi.org/10.1021/jf071972u. [DOI] [PubMed] [Google Scholar]
- Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. Breast milk oligosaccharides: Structure-function relationships in the neonate. Annu. Rev. Nutr. 2014;34:143–169. doi: 10.1146/annurev-nutr-071813-105721. http://dx.doi.org/10.1146/annurev-nutr-071813-105721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilowitz JT, O’Sullivan A, Barile D, German JB, Lönnerdal B, Slupsky CM. The human milk metabolome reveals diverse oligosaccharide profiles. J. Nutr. 2013;143:1709–1718. doi: 10.3945/jn.113.178772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao N, DePeters EJ, German JB, Grimm R, Lebrilla CB. Variations in bovine milk oligosaccharides during early and middle lactation stages analyzed by high-performance liquid chromatography-chip/mass spectrometry. J. Dairy Sci. 2009;92:2991–3001. doi: 10.3168/jds.2008-1642. http://dx.doi.org/10.3168/jds.2008-1642. [DOI] [PubMed] [Google Scholar]
- Tao N, Ochonicky KL, German JB, Donovan SM, Lebrilla CB. Structural determination and daily variations of porcine milk oligosaccharides. J. Agric. Food Chem. 2010;58:4653–4659. doi: 10.1021/jf100398u. http://dx.doi.org/10.1021/jf100398u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Bruggencate SJ, Bovee-Oudenhoven IM, Feitsma AL, van Hoffen E, Schoterman MH. Functional role and mechanisms of sialyllactose and other sialylated milk oligosaccharides. Nutr. Rev. 2014;72:377–389. doi: 10.1111/nure.12106. http://dx.doi.org/10.1111/nure.12106. [DOI] [PubMed] [Google Scholar]
- Thurl S, Müller-Werner B, Sawatzki G. Quantification of individual oligosaccharide compounds from human milk using high-pH anion-exchange chromatography. Anal. Biochem. 1996;235:202–206. doi: 10.1006/abio.1996.0113. [DOI] [PubMed] [Google Scholar]
- Urashima T, Taufik E, Fukuda K, Asakuma S. Recent advances in studies on milk oligosaccharides of cows and other domestic farm animals. Biosci. Biotechnol. Biochem. 2013;77:455–466. doi: 10.1271/bbb.120810. [DOI] [PubMed] [Google Scholar]
- Ushida K, Tsuchida S, Ogura Y, Toyoda A, Maruyama F. Domestication and cereal feeding developed domestic pig- type intestinal microbiota in animals of suidae. Anim. Sci. J. 2015;87:835–841. doi: 10.1111/asj.12492. http://dx.doi.org/10.1111/asj.12492. [DOI] [PubMed] [Google Scholar]
- Wood DE, Salzberg SL. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Grimm R, German JB, Lebrilla CB. Annotation and structural analysis of sialylated human milk oligosaccharides. J. Proteome Res. 2011;10:856–868. doi: 10.1021/pr101006u. http://dx.doi.org/10.1021/pr101006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J. Proteome Res. 2010;9:4138–4151. doi: 10.1021/pr100362f. http://dx.doi.org/10.1021/pr100362f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z-T, Chen C, Newburg DS. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology. 2013;23:1281–1292. doi: 10.1093/glycob/cwt065. http://dx.doi.org/10.1093/glycob/cwt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic AM, Barile D. Bovine milk as a source of functional oligosaccharides for improving human health. Adv. Nutr. 2011;2:284–289. doi: 10.3945/an.111.000455. http://dx.doi.org/10.3945/an.111.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. USA. 2011;108:4653–4658. doi: 10.1073/pnas.1000083107. http://dx.doi.org/10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.