Abstract
NifB utilizes two equivalents of S-adenosyl methionine (SAM) to insert a carbide atom and fuse two substrate [Fe–S] clusters forming the NifB cofactor (NifB-co), which is then passed to NifEN for further modification to form the iron–molybdenum cofactor (FeMo-co) of nitrogenase. Here, we demonstrate that NifB from the methanogen Methanocaldococcus infernus is a radical SAM enzyme able to reductively cleave SAM to 5′-deoxyadenosine radical and is competent in FeMo-co maturation. Using electron paramagnetic resonance spectroscopy we have characterized three [4Fe–4S] clusters, one SAM binding cluster, and two auxiliary clusters probably acting as substrates for NifB-co formation. Nitrogen coordination to one or more of the auxiliary clusters in NifB was observed, and its mechanistic implications for NifB-co dissociation from the maturase are discussed.
NifB belongs to the superfamily of radical S-adenosyl methionine (SAM) enzymes.1 These enzymes are present in all forms of life and provide the most common way in biology to perform directed radical based chemistry by utilizing a [4Fe–4S] cluster to coordinate SAM and generate a 5′-deoxyadenosyl radical (5′dA•). The 5′dA• then typically abstracts an H atom from the substrate to initiate catalysis.2 In NifB, simple [Fe–S] clusters are converted into a nitrogenase-specific [Fe–S] cluster of high nuclearity, termed NifB-co.3 NifB-co is the source of most (or all) Fe and S present in FeMo-co along with its central C atom (Scheme 1).4–7 The insertion of carbide to fuse [Fe–S] clusters is a unique reaction among radical SAM enzymes.1
Scheme 1. Maturation of Nitrogenase FeMo Cofactora.
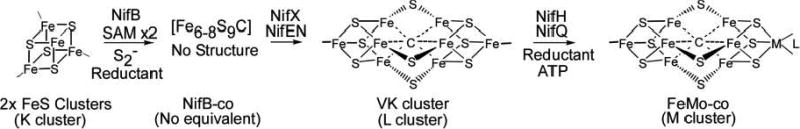
aRequirements for metallocluster transformation are indicated for each step of FeMo-co biosynthesis. M is Mo, and L represents homocitrate. The two nomenclatures used to refer to these metalloclusters are shown.
NifB proteins, first purified from Azotobacter vinelandii and Klebsiella oxytoca, exhibited spectroscopic and catalytic properties typical of radical SAM proteins and were shown to be competent in FeMo-co maturation.3,8 Further investigations were limited due to instability of the purified NifB when not fused with the downstream maturase NifEN.9–11 NifB from methanogens was shown to be more stable target for spectroscopic studies.12,13 Purification of NifB from the methanogens Methanocaldococcus infernus, Methanosarcina acetivorans, and Methanobacterium thermoautotrophicum confirmed their ability to use SAM as substrate to form dAdoH and SAH, the products of the radical SAM and demethylation reactions, respectively.12,14 These homologues were competent in FeMo-co maturation and were proposed to each have three [4Fe–4S] clusters based on the Fe content of fully reconstituted enzymes.
Here, we expand our understanding of NifB by characterizing the three [Fe–S] clusters of M. infernus NifB (MiNifB) through the use of site-directed variants and electron paramagnetic resonance (EPR) spectroscopy. Further, we detect N ligand(s) to at least one of these clusters and discuss the implications this may have on the release of NifB-co to NifEN.
The metin_0554 gene from M. infernus was heterologously expressed as a His10-tagged protein in E. coli as described.13 Anaerobically purified MiNifB ran as a single 35–kDa band in SDS-PAGE (Figure S1A), in good agreement with its predicted molecular weight of 37 543 Da and TOF/TOF mass spectrometry analysis (37 483 Da; Figure S2). The UV–vis spectrum of as-isolated MiNifB displayed features of [Fe–S] proteins (Figure S1B). Among these features, the shoulder at 320 nm and the peak at 400–420 nm are the most defining, as is the broad tail extending beyond 700 nm. This absorption feature is assigned to S → Fe charge transfer transitions characteristic of [4Fe–4S]2+ clusters. Consistently, as isolated MiNifB contained, on average, 3.3 Fe and 4 acid-labile S atoms per monomer (Table S1). In vitro reconstitution of [Fe–S] clusters was carried out either on as-isolated MiNifB or on samples previously stripped of Fe and S (apo-MiNifB). Incorporation of Fe and S was higher on apo-MiNifB samples (Table S1), and these preparations exhibited UV–vis spectra with the most prominent 320 and 420 nm features (Figure S1B).
As-isolated MiNifB was active in the NifB-dependent in vitro FeMo-co synthesis and apo-NifDK activation assay. Apo-NifDK activation was strictly dependent on addition of MiNifB or NifB-co (isolated from K. oxytoca cells and used here as control of NifB activity; Figure S1C). Addition of either apo-MiNifB or aerobically purified MiNifB did not activate apo-NifDK (data not shown). The capacity of MiNifB to transfer a methyl group from SAM, generate 5′dAdo•, and initiate FeMo-co synthesis were determined by measuring concomitant production of S-deoxyadenosyl-l-homocysteine (SAH), 5′-deoxyadenosine (dAdoH), and reconstituted NifDK in a single assay. SAH and AdoH production correlated with MiNifB-dependent activation of nitrogenase over time (Figure S1D). Interestingly, demethylation of SAM proceeded at higher rates than dAdoH formation (3:2 SAH to dAdoH ratio), and this difference was maintained over time. Incubation with 13CD3-SAM resulted in production of deuterated 5′-deoxyadenosine (dAdoD), demonstrating abstraction of a H atom from the methyl group of SAM (Figure S3).
The Fe and S content of reconstituted wild-type (WT) MiNifB (10 ± 0.2 Fe and 11.2 ± 0.4 S atoms per monomer, Table S1) was consistent with the presence of, at minimum, two [Fe–S] clusters in addition to the SAM-binding [4Fe–4S] cluster. The SAM-binding cluster is anchored to the enzyme by three cysteine thiolate side chains, identified by the canonical CX3CX2C motif. Substitution of these Cys residues (C36A/C40A/C43A) in variant SM (Table 1) resulted in reduction of Fe and S content, changes in the UV–vis spectrum, and inability to activate apo-NifDK (Figure 1A–D and Table S1). NifDK activation ability was also lost in the AM variant carrying amino acid substitutions in the C-terminal CXXC U-type motif, which is similar to those shown to bind [Fe–S] clusters for storage and transfer15,16 and in the DM variant with amino acid substitutions in both domains (Table 1 and Figure 1D). [Fe–S] cluster reconstituted DM preparations exhibited UV–vis features, as well as Fe and S content consistent with a [4Fe–4S] cluster (Figure 1C and Table S1), suggesting the presence of one additional [Fe–S] cluster-binding motif in MiNifB. Anaerobic size-exclusion chromatography of reconstituted WT and the SM, AM, and DM MiNifB variants indicated these proteins were monomers (Table S2), suggesting that each MiNifB monomer is able to coordinate all observed clusters.
Table 1. MiNifB Variant Nomenclature.
| name | variant | substitutions |
|---|---|---|
| WT | wild type | none |
| SM | SAM motif variant | C36A/C40A/C43A |
| AM | auxiliary motif variant | C260A/C263A |
| DM | double variant | C36A/C40A/C43A C260A/C263A |
Figure 1.
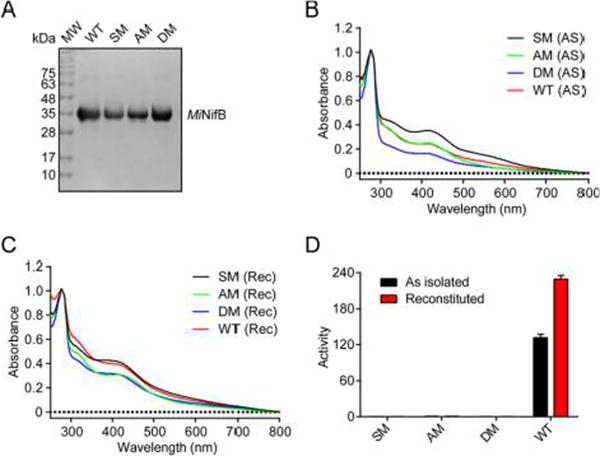
Biochemical properties of MiNifB variants lacking [Fe–S] cluster-binding motifs. (A) SDS-PAGE of pure MiNifB preparations. (B,C) UV–visible spectra of WT MiNifB (red), and the SM (black), AM (green), and DM (blue) variants in as-isolated (AS) (B) and reconstituted (Rec) (C) forms. Spectra were normalized to their absorbance at 279 nm. (D) MiNifB-dependent apo-NifDK reconstitution assays with MiNifB WT and variants. Activities are nmol of ethylene·min−1·mg−1 NifDK (mean ± SD; n ≥ 2).
To investigate the composition of the [Fe–S] clusters present in WT MiNifB, continuous wave ×-band (9.4 GHz) EPR analysis was performed on reconstituted enzyme preparations. WT and the SM and AM variants reduced by sodium dithionite (DTH), in the presence or absence of SAM, exhibited spectra with components representative of multiple S = 1/2 [Fe–S] clusters (Figure 2A–D and Figure S4). The EPR spectrum of the DM variant was consistent with a single [4Fe–4S] cluster (Figure 2E), in agreement with its Fe and S content (Table S1). The experimental spectra of WT and all the variants were simulated using the DM variant as starting point (see Table S3 for simulation parameters, Table S4 for spin quantitations, and Figure S5 for individual component spectra).
Figure 2.
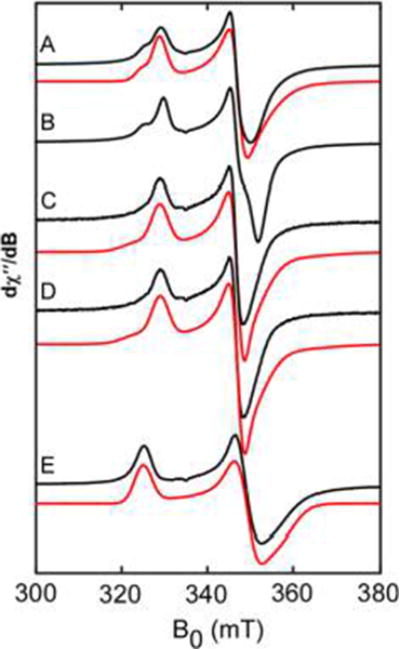
X-band CW EPR spectroscopy of DTH-reduced MiNifB WT and site-directed variants. (A) WT MiNifB (274 μM), (B) WT MiNifB in the presence of 3 mM SAM, (C) SM variant (159 μM), (D) AM variant (159 μM), and (E) DM variant (252 μM). Data are shown in black with composite spectral simulations in red.
Temperature-dependent EPR, which has proven useful to establish the nuclearity of [Fe–S] clusters,17,18 was utilized to identify individual species present. Data shown in Figure S6 suggest that the three clusters present in MiNifB are [4Fe–4S] clusters. Additionally, the g1 feature of each cluster becomes clear when examining the temperature dependence of the different spectra, providing spectroscopic handles for these clusters. Deconvolution through spectral subtractions was performed to support individual cluster assignments. Unique features for each of the species present, and the positions of g1 and g3 for each of the clusters were verified (Figure S7). The cluster present in the DM variant, termed AC2, showed features typical of a [4Fe–4S] cluster, such as a fairly axial g-tensor (g = [2.039 1.923 1.886]) and signal disappearance above 40 K. Three clusters were identified in the SM variant. The [4Fe–4S] cluster described above for the DM variant, an additional [4Fe–4S] cluster, termed AC1, with g-values of [2.062 1.917 1.875], and a third cluster that was only observed above 50 K and had a much more rhombic g-tensor of [2.058 1.985 1.909] consistent with a [2Fe–2S]. This [2Fe–2S] cluster was only apparent in the SM variant and was tentatively assigned to a degraded cluster, termed ACx. The EPR spectrum of the AM variant could be simulated with a combination of AC2 and a new [Fe–S] cluster with g-values of [2.038 1.931 1.916], which has been assigned to the SAM-binding cluster (RS). The g-values and temperature dependence of the EPR spectrum for RS are in agreement with reported SAM binding clusters.1
Examination of WT MiNifB by three pulse electron spin echo envelope modulation (3P-ESEEM) revealed nitrogen modulations to the echo from one or more of the [Fe–S] clusters (Figure 3, black trace). Incubation with isotopically enriched 15N-SAM, carried out to investigate changes in spectral features attributed to SAM coordination, resulted in no change in modulations (Figure 3, blue trace) as would be expected if this modulation derived from coupling to SAM nitrogen. This lack of 14N/ 15N SAM isotope sensitivity of the ESEEM spectra suggests proteinaceous nitrogen coordination to one or more of the [Fe–S] clusters. Due to its simplified EPR spectrum and cluster content, the DM variant was selected to investigate nitrogen coordination from the protein to the AC2 cluster. The nitrogen modulations observed in the DM variant (Figure 3, red trace) were very similar to those of the WT, suggesting that AC2 is coordinated by at least one nitrogen ligand. While the nitrogen modulations arising from AC2 are present in the spectra of WT MiNifB with peaks at 2 and 0.7 MHz (Figure 3), there was also evidence for additional nitrogen modulations from a second cluster (peaks at 2.3 and 1 MHz), which was tentatively assigned to AC1.
Figure 3.
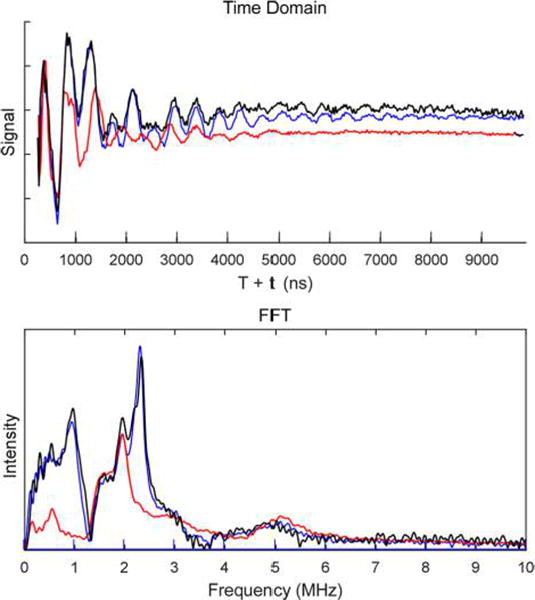
X-band (9.4 GHz) 3P-ESEEM time domain (top) and Fourier transform of the time domain (bottom) of MiNifB. Black trace, WT MiNifB reduced with 10 mM DTH in the presence of 3 mM 14N-SAM. Blue trace, WT MiNifB reduced with 10 mM DTH in the presence of 3 mM 15N-SAM. Red trace, DM variant reduced with 10 mM DTH in the absence of SAM.
NifB from M. infernus is functional in nitrogenase cofactor maturation and represents a simplified homologue (lacking the NifX domain) of NifB proteins from model diazotrophs A. vinelandii and K. oxytoca. These results are in agreement with those recently reported for M. acetivorans and M. thermoautotrophicum.14 EPR and biochemical analyses indicate the presence of three [4Fe–4S] clusters in MiNifB. One of these clusters is identified as the [4Fe–4S] SAM binding cluster (RS) by the canonical CX3CX2C motif and dAdoH formation assays. While the auxiliary clusters AC1 and AC2 may be present as conduits for electron transfer, our preferred model is that they are in-fact precursors to NifB-co. Based on the temperature behavior of EPR spectra, reporting information on the relaxation dynamics of the cluster, we propose that both auxiliary clusters are present as [4Fe–4S] clusters. While the temperature dependence of an [Fe–S] cluster can help discriminate between a [4Fe–4S] and [2Fe–2S] cluster (a [4Fe–4S] cluster having faster relaxation rates than a [2Fe–2S] cluster), this analysis can become clouded when multiple clusters are present and near each other causing increased relaxation rates from dipolar coupling between them. Therefore, the presence of one [2Fe–2S] cluster cannot be completely ruled out. The assignment of NifB-co as a fusion of two [4Fe–4S] clusters or one [4Fe–4S] and one [2Fe–2S] units is consistent with the previous proposal of NifB-co comprising at least the [6Fe–9S–C] core of FeMo-co.6
Based on the number of strictly conserved cysteines in MiNifB, there are not enough to fully coordinate the SAM-binding cluster plus two additional [4Fe–4S] clusters (Table S5). This would indicate that the two auxiliary clusters are either not saturated in Fe coordination or that some noncysteinyl residues may coordinate the auxiliary clusters as suggested by 3P-ESEEM. MiNifB has two conserved His residues as well as several conserved Arg residues. Coordination by His or Arg residues is well-known for [2Fe–2S] clusters and has precedence for an auxiliary [4Fe–4S] cluster of Cp1 Fe–Fe hydrogenase, as well as in the hydrogenase cofactor maturase HydF.18–20 Additionally, in BioB, IscU and MitoNEET nitrogen coordination is utilized to bind transient [Fe–S] clusters.21 Nitrogen coordination to the cluster would provide a convenient path for cluster release through a simple protonation of the nitrogen ligand and release of NifB-co to NifEN or NifX.
Like NifB from M. acetivorans and M. thermoautotrophicum,14 MiNifB produces more SAH than AdoH in turnover assays (a 3:2 ratio). We note that SAM cleavage to AdoH has never been observed to be in excess of SAH in NifB suggesting a single H abstraction followed by oxidation/deprotonation events to remove the remaining H from the transferred methyl to form carbide. Interestingly, we observed a shift in the g3 feature at 352 mT of the WT EPR spectrum in the presence of SAM (Figure 2B), but no indication of binding of SAM to the RS cluster in the 3P-ESEEM, suggesting the existence of additional steps in the reaction mechanism to form NifB-co. In the mechanism that has been proposed for NifB,10,22 SAM transfers a methyl group to one of the clusters that will be fused to form NifB-co (Scheme S1). We suggest the observed perturbation to WT NifB EPR spectrum in the presence of SAM is due to methyl transfer to one of the auxiliary [Fe–S] clusters, poised for NifB-co formation, thus ensuring that, when 5′dAdo• is formed, the site of hydrogen abstraction is present preventing unwanted side reactions from the highly reactive primary carbon radical.
Supplementary Material
Acknowledgments
We thank Juan Fontecilla for providing pRSFIscMetKDuet-1, and Etienne Mulliez and Phanélie Perche-Letuvée for providing 15N-SAM and 13CD3-SAM. This work was supported by the European Research Council Starting Grant 205442 (to L.M.R.), MINECO Grant BIO2014-59131-R (to L.M.R.), FPI Fellowship BES-2010-038322 (to A.S.), and National Institutes of Health Grant GM-104543 (to R.D.B.).
Footnotes
Supporting Information: The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b03329.
Materials and Methods (PDF)
Notes: The authors declare no competing financial interest.
References
- 1.Broderick JB, Duffus BR, Duschene KS, Shepard EM. Chem Rev. 2014;114:4229–4317. doi: 10.1021/cr4004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frey PA, Hegeman AD, Ruzicka F. J Crit Rev Biochem Mol Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 3.Curatti L, Ludden PW, Rubio LM. Proc Natl Acad Sci USA. 2006;103:5297–5301. doi: 10.1073/pnas.0601115103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spatzal T, Aksoyoglu M, Zhang L, Andrade SL, Schleicher E, Weber S, Rees DC, Einsle O. Science. 2011;334:940. doi: 10.1126/science.1214025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen RM, Chatterjee R, Ludden PW, Shah VK. J Biol Chem. 1995;270:26890–26896. doi: 10.1074/jbc.270.45.26890. [DOI] [PubMed] [Google Scholar]
- 6.George SJ, Igarashi RY, Xiao Y, Hernandez JA, Demuez M, Zhao D, Yoda Y, Ludden PW, Rubio LM, Cramer SP. J Am Chem Soc. 2008;130:5673–5680. doi: 10.1021/ja0755358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah VK, Allen JR, Spangler NJ, Ludden PW. J Biol Chem. 1994;269:1154–1158. [PubMed] [Google Scholar]
- 8.Zhao D, Curatti L, Rubio LM. J Biol Chem. 2007;282:37016–37025. doi: 10.1074/jbc.M708097200. [DOI] [PubMed] [Google Scholar]
- 9.Curatti L, Hernandez JA, Igarashi RY, Soboh B, Zhao D, Rubio LM. Proc Natl Acad Sci USA. 2007;104:17626–17631. doi: 10.1073/pnas.0703050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiig JA, Hu Y, Lee CC, Ribbe MW. Science. 2012;337:1672–1675. doi: 10.1126/science.1224603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiig JA, Hu Y, Ribbe MW. Proc Natl Acad Sci USA. 2011;108:8623–8627. doi: 10.1073/pnas.1102773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arragain S, Scandurra AA, Jimenez-Vicente E, Echavarri-Erasun C, Rubio LM. XI European Nitrogen Fixation Conference. Tenerife, Spain: 2014. [Google Scholar]
- 13.Echavarri-Erasun C, Arragain S, Scandurra AA, Rubio LM. Methods Mol Biol. 2014;1122:19–31. doi: 10.1007/978-1-62703-794-5_3. [DOI] [PubMed] [Google Scholar]
- 14.Fay AW, Wiig JA, Lee CC, Hu Y. Proc Natl Acad Sci USA. 2015;112:14829–14833. doi: 10.1073/pnas.1510409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bandyopadhyay S, Chandramouli K, Johnson MK. Biochem Soc Trans. 2008;36:1112–1119. doi: 10.1042/BST0361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith AD, Jameson GN, Dos Santos PC, Agar JN, Naik S, Krebs C, Frazzon J, Dean DR, Huynh BH, Johnson MK. Biochemistry. 2005;44:12955–12969. doi: 10.1021/bi051257i. [DOI] [PubMed] [Google Scholar]
- 17.Iron-Sulfur Proteins. Vol. 4 John Wiley-Interscience; New York: 1982. [Google Scholar]
- 18.Cammack R, MacMillan F. In: Metals in Biology: Applications of High-Resolution EPR to Metalloenzymes. Hanson G, Berliner L, editors. Vol. 29. Springer; New York: 2010. pp. 11–44. [Google Scholar]
- 19.Berto P, Di Valentin M, Cendron L, Vallese F, Albertini M, Salvadori E, Giacometti GM, Carbonera D, Costantini P. Biochim Biophys Acta, Bioenerg. 2012;1817:2149–2157. doi: 10.1016/j.bbabio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 21.Bak DW, Elliott SJ. Curr Opin Chem Biol. 2014;19:50–58. doi: 10.1016/j.cbpa.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Wiig JA, Hu Y, Ribbe MW. Nat Commun. 2015;6:8034. doi: 10.1038/ncomms9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


