Abstract
Beyond protein synthesis and autophagy, emerging evidence has implicated mTORC1 in regulating protein folding and proteasomal degradation as well, highlighting its prominent role in cellular proteome homeostasis or proteostasis. In addition to growth signals, mTORC1 senses and responds to a wide array of stresses, including energetic/metabolic stress, genotoxic stress, oxidative stress, osmotic stress, ER stress, proteotoxic stress, and psychological stress. Whereas growth signals unanimously stimulate mTORC1, stresses exert complex impacts on mTORC1, most of which are repressive. mTORC1 suppression, as a generic adaptive strategy, empowers cell survival under various stressful conditions. In this essay, we provide an overview of the emerging role of mTORC1 in proteostasis, the distinct molecular mechanisms through which mTORC1 reacts to diverse stresses, and the schemes exploited by cancer cells to circumvent stress-induced mTORC1 suppression. Hence, acting as a stress sensor, mTORC1 intimately couples stresses to cellular proteostasis.
Keywords: mTORC1, proteostasis, stress sensor, SAPK/JNK, HSF1
Introduction
As one of the fundamental building blocks of cells, proteins are vital to life. Unsurprisingly, the cellular proteome is subjected to tight and dynamic regulation at the points of protein synthesis, folding and degradation. As a consequence, the cellular proteome exists in dynamic equilibrium: a delicate state referred to as proteome homeostasis or proteostasis [1]. Congruent with its paramount importance, a wealth of evidence has implicated proteostasis in a broad range of pathophysiological conditions in humans [1].
mTORC1 governs proteostasis
One key component of this homeostatic network is protein synthesis. Among the various pathways governing protein production inside cells is mechanistic target of rapamycin (mTOR) signaling. First discovered in 1991 in yeasts [2], mTOR is a Ser/Thr kinase that belongs to the phosphoinositide kinase-related family of protein kinases (PIKKs) [3]. After two decades of intense studies, a great deal of knowledge has been gleaned about mTOR. Inside cells, mTOR is assembled into two distinct multi-component protein complexes: mTOR complex 1 (mTORC1) and complex 2 (mTORC2) [3]. mTORC1 comprises mTOR, regulatory associated protein of mTOR (RAPTOR), mammalian lethal with SEC13 protein 8 (mLST8 or GβL), proline-rich AK transforming (AKT) substrate 40kDa (PRAS40), DEP domain containing mTOR-interacting protein (DEPTOR), and newly identified telomere maintenance 2 (TEL2) and TEL2 interacting protein 1 (TTI1) [3,4]. While mTOR is the core kinase, RAPTOR is proposed to function as a scaffold protein that recruits the substrates for phosphorylation by mTOR [5]. The RAPTOR-mTOR interaction can be either loosened or tightened depending on the stimuli. For instance, whereas rapamycin disrupts this interaction, amino acid deprivation strengthens it [6]. Therefore, the precise impacts of RAPTOR on mTOR may be complex: at the very least, they remain to be fully elucidated. mLST8/GβL, by interacting with mTOR’s kinase domain and stabilizing the RAPTOR-mTOR interaction, positively regulates mTORC1 [7]. However, this model is challenged by the observation that RAPTOR-mTOR interactions are not impaired in mice deficient in mLST8/GβL [8]. In contrast to RAPTOR, the AKT substrate PRAS40 is generally believed to be a negative regulator of mTORC1 [9]. Through direct interactions with RAPTOR, PRAS40 blocks the binding of substrates to mTOR [10, 11]. The phosphorylation of PRAS40 at Thr246 by AKT causes its dissociation from RAPTOR [10], thereby relieving the suppression. Like PRAS40, DEPTOR also negatively impacts mTORC1 by binding to the FRAP-ATM-TTRAP (FAT) domain of mTOR directly [12].
mTORC2, like mTORC1, also contains mTOR, mLST8/GβL, DEPTOR, TEL2 and TTI1. In addition to these common subunits, mTORC2 consists of three unique components additionally—namely rapamycin-insensitive companion of mTOR (RICTOR), mammalian stress-activated MAP kinase-interacting protein 1 (mSIN1), and protein observed with RICTOR (PROTOR) [3]. While RICTOR is necessary for the assembly of mTORC2 [13], mSIN1 plays a critical role in maintaining the RICTOR-mTOR interaction [14]. Congruently, both RICTOR and mSIN1 positively impact mTORC2 activity. By contrast, PROTOR is dispensable for mTORC2 assembly and appears only to affect the phosphorylation of selective substrates of mTORC2 in a tissue-specific manner [15]. Sharply contrasting with its dispensability for the RAPTOR-mTOR interaction, in vivo studies confirm the necessity of mLST8/GβL for the RICTOR-mTOR interaction [8]. Similar to its impact on mTORC1, DEPTOR negatively regulates mTORC2 as well [12].
mTORC1 and mTORC2, despite sharing the very same core catalytic subunit, differ greatly in substrate recognition. The prominent substrates for mTORC1 include eukaryotic translation initiation factor 4E–binding protein 1 (eIF4E–BP1) [16], ribosomal protein S6 kinase 1 (S6K1) [16], unc-51 like autophagy activating kinase 1 (ULK1) [17], autophagy related 13 (ATG13) [18,19], phosphatidic acid phosphatase LIPIN1 (LPIN1 or PAP1) [20], growth factor receptor bound protein 10 (GRB10) [21], and cytoplasmic linker protein 170 (CLIP-170) [22]. Accordingly, mTORC1 controls protein translation, autophagy and lipogenesis [3]. By contrast, mTORC2 phosphorylates AKT [23], protein kinase C (PKC) [13], and serum/glucocorticoid regulated kinase 1 (SGK1) [24,25], thereby controlling, cell survival, cytoskeleton organization, and stress responses [3].
A plethora of evidence has unequivocally pinpointed a crucial role of mTORC1 in governing protein translation via two key substrates--eIF4E–BP1 and S6K1 [3,16]. The mTORC1-mediated Thr389 phosphorylation leads to dissociation of S6K1 from the eukaryotic initiation factor complex 3 (eIF3) [26], followed by the phosphoinositide-dependent kinase 1 (PDK1)-mediated S6K1 Thr229 phosphorylation [27]. Upon activation, S6K1 phosphorylates ribosomal protein S6 (RPS6) to stimulate ribosomal biogenesis [28], eukaryotic elongation factor 2 kinase (eEF2K) to de-repress translational elongation [29], as well as eIF4B to heighten translational initiation [26,30]. Following S6K1 dissociation, mTORC1 accesses eIF4E–BP1, which interacts with eIF4E at the 5’ cap structures of mRNAs [31]. The mTORC1-mediated Thr37/46 and Ser65/Thr70 phosphorylation releases eIF4E–BP1 from eIF4E, thereby enabling the assembly of eIF4F complex that will recruit the 40S ribosomal subunits to the cap structures to initiate translation [31]. In aggregate, mTORC1 is a key player in controlling protein synthesis.
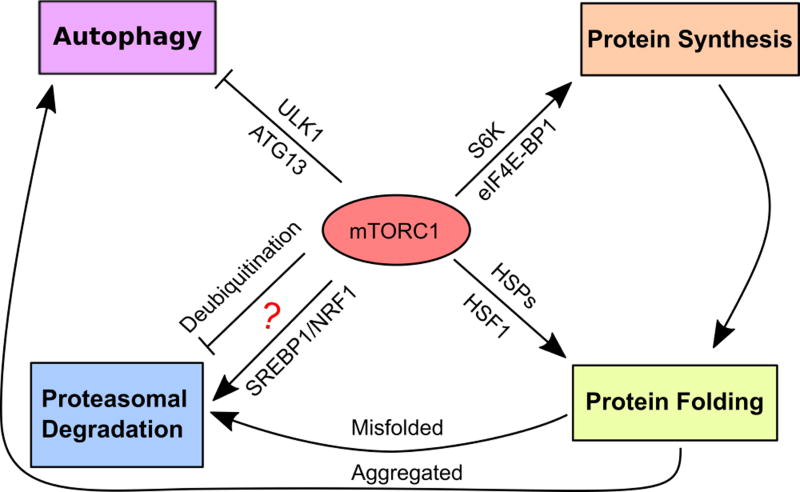
Beyond protein production, mTORC1 also regulates other aspects of proteostasis. As an intracellular lysosome-based self-degradation mechanism, autophagy impacts proteostasis through clearance of aggregated proteins [32]. By phosphorylating ULK1 and ATG13[17–19], two key components of the autophagy initiation complex, mTORC1 suppresses autophagy. Mechanistically, the mTORC1-mediated Ser757 phosphorylation impedes the activating phosphorylation of ULK1 at Ser317 and Ser777 by the metabolic sensor AMP-activated protein kinase (AMPK) [17]. Recently, mTORC1 has further been implicated in regulating proteasomal degradation, another key protein-quality control mechanism. It was reported that mTORC1 activation stimulates proteasome biogenesis via the nuclear factor erythroid-derived 2-related factor 1 (NRF1)-mediated transcription of proteasome subunit genes [33], suggesting coordinated protein synthesis and degradation. However, this was challenged by the observation that mTORC1 inhibition promotes proteasomal degradation via enhanced protein ubiquitination [34]. Apparently, more thorough studies are warranted to delineate how mTORC1 affects the ubiquitin-proteasome system (UPS). In addition to protein degradation, mTORC1 also influences cellular chaperoning capacity under proteotoxic stress. It has been widely recognized that cells cope well with proteotoxic stress by mobilizing a potent transcriptional program named the heat-shock, or proteotoxic, stress response (HSR/PSR) [35]. The most notable feature of this stress response is rapid induction of heat-shock proteins (HSPs) or molecular chaperones [35]. Under proteotoxic stress, HSPs are essential to cellular proteostasis by safeguarding proper protein folding, resolving protein misfolding, and promoting the ubiquitination and degradation of damaged proteins that are beyond repair [35]. Of note, under proteotoxic stress mTORC1 is required for robust translation of both heat shock factor 1 (HSF1) and highly transcribed HSP mRNAs [36]. Thus, mTORC1 contributes to cellular proteostasis through translational augmentation of the HSR/PSR [36]. Collectively, these findings highlight an important role of mTORC1 in protein-quality control, beyond its well-known impact on protein-quantity control (Fig. 1).
Figure 1. A key role of mTORC1 in proteostasis.
While mTORC1 is widely recognized to control protein synthesis and autophagy, emerging evidence has implicated mTORC1 in regulating protein folding and proteasomal degradation as well. Thus, mTORC1 impacts all aspects of cellular proteostasis. The lines with an arrow head denote positive regulations. The T-bar lines denote negative or inhibitory regulations. The question mark denotes the controversial roles of mTORC1 in regulating proteasomal degradation. SREBP1: sterol regulatory element-binding protein 1.
Diverse extracellular and intracellular cues regulate mTORC1
In light of its biological importance, unsurprisingly, mTORC1 is under stringent and complex regulations. The Ras-homolog expressed in brain (RheB), a small GTPase that physically interacts with and activates mTORC1 [37], is an additional immediate regulator of mTORC1. Furthermore, RheB is directly inhibited by the tuberous sclerosis complex 1 and 2 (TSC1/2) [38,39], well-known tumor suppressor complexes. Recently, a third core subunit of this complex, TBC1 domain family member 7 (TBC1D7), has been identified [40]. TSC1-TSC2-TBC1D7 complexes act as GTPase-activating proteins (GAP) that inhibit RheB activity by converting RheB-GTP into RheB-GDP [38,39]. In this way, mTORC1 is critically regulated by the TSC-RheB cascade.
mTORC1 senses growth factors and nutrients
Two major signals that stimulate mTORC1 are growth factors, including insulin and insulin-like growth factor 1 (insulin/IGF-1), and amino acids [3]. In response to growth stimulation, the phosphoinositide 3-kinase (PI3K)/AKT and rat sarcoma small GTPase/rapidly accelerated fibrosarcoma oncogene /MAP-ERK kinase/extracellular signal-regulated kinase/90KDa ribosomal protein S6 kinase (RAS/RAF/MEK/ERK/RSK) signaling pathways become activated. Subsequently, AKT phosphorylates TSC2 at Ser939 and Thr1462, leading to inactivation of the TSC complex [41,42]. The ERK-mediated Ser540/664 and RSK1-mediated Ser1798 phosphorylation of TSC2 achieve the similar effects [43,44].
Despite the well-known fact that amino acids, especially leucine, are an essential activator of mTORC1 [45], the underlying molecular mechanisms remain elusive until recently. Studies have now revealed that on the lysosomal surface a large protein complex, which at least comprises vacuolar H+-ATPase (v-ATPase), RAGULATOR (a pentameric complex consisting of LAMTOR1–5), and Ras-related GTP-binding protein (Rag) small GTPases, mediates the amino acid-sensing of mTORC1 [45]. Within this complex, v-ATPase physically interacts with RAGULATOR, a guanine nucleotide exchange factor (GEF) for RagA/B GTPases [46,47]. In response to amino acid stimulation, v-ATPase activates RAGULATOR [46,47], which may involve amino acid-dependent changes in v-ATPase assembly [48]. Subsequently, the GTP-loaded RagA/B that is heterodimerized with the GDP-loaded RagC/D recruit inactive mTORC1 from the cytosol to the lysosomal surface, where it becomes activated by RheB [49,50]. Sestrin2 was identified recently as an intracellular sensor for leucine. Binding of leucine causes dissociation of Sestrin2 from GAP activity towards Rags 2 (GATOR2), a protein complex positively regulating mTORC1 by inhibiting GATOR1 [51], another protein complex that acts as a GAP for RagA/B [52]. Moreover, two arginine sensors have also been identified. One is solute carrier family 38 member 9 (SLC38A9), a lysosomal transmembrane protein that interacts with both Rag GTPases and RAGULATOR [53]. The other is cellular arginine sensor for mTORC1 (CASTOR1), a protein physically interacting with and inhibiting GATOR2 [54]. Binding of arginine to CASTOR1 causes its dissociation from GATOR2, thus activating GATOR2 and relieving the suppression of RagA/B-GTP by GATOR1 [54].
mTORC1 senses a diversity of stresses
In addition to growth factors and amino acids, mTORC1 also senses and responds to a large variety of stresses. Under this scenario, mTORC1 becomes suppressed, likely reflecting a general cellular strategy to adapt to stressful conditions. Accumulating studies have started unraveling the molecular regulations of mTORC1 by stresses.
mTORC1 is suppressed by energetic/metabolic stress
Cellular energetic stress is hallmarked by the decline of ATP and/or increase of AMP levels inside cells. As a consequence, AMPK, a key cellular metabolic sensor, becomes mobilized [55]. Among the numerous AMPK substrates are TSC2 and RAPTOR. While the AMPK-mediated Ser1387 (Ser1345 in the rat counterpart) phosphorylation activates TSC2 [56], phosphorylation of RAPTOR at Ser722 and Ser792 by AMPK induces the binding of 14-3-3 proteins, leading to RAPTOR inhibition [57]. Furthermore, independently of AMPK, energy depletion suppresses mTORC1 via the p38 mitogen-activated protein kinase (p38 MAPK) signaling. Following 2-deoxy-D-glucose treatment, p38 regulated/activated kinase (PRAK) directly phosphorylates RheB at Ser130, a modification that impairs the GTP/GDP binding of RheB and leads to mTORC1 inactivation [58]. In addition, under energetic stress p38β phosphorylates forkhead box class O (FOXO) transcription factors, which induce BCL2 interacting protein 3 (BNIP3) expression [59]. BNIP3, in turn, interacts with and inactivates RheB, leading to mTORC1 suppression [59]. Moreover, energetic stress can also suppress mTORC1 via TEL2-TTI1-TTI2 (TTT)-RUVBL1/2 [60], an AAA+ ATPase-containing protein complex that is implicated in regulating PIKKs [61]. Depletion of glucose and glutamine causes disassembly of the mTORC1-interacting TTT-RUVBL1/2 complex, impeding mTORC1 dimerization and lysosomal translocation [60]. Thus, via multiple distinct pathways energetic stress suppresses mTORC1.
mTORC1 is both suppressed and stimulated by oxidative stress
Imbalance between cellular reactive oxygen species (ROS) and antioxidants leads to oxidative stress, which has been implicated in a wide array of human pathophysiological conditions, including aging, diabetes, atherosclerosis, neurodegenerative disorders, and cancer [62].
Elevated ROS can activate ataxia telangiectasia mutated (ATM) [63], a key tumor suppressor sensing DNA damage [64]. In turn, ATM suppresses mTORC1 via the liver kinase B1 (LKB1)-AMPK-TSC2 signaling cascade [63]. In addition, under arsenite-induced oxidative stress sperm associated antigen 5 (SPAG5 or ASTRIN), a protein regulating mitotic spindles, sequesters RAPTOR apart from mTORC1 to stress granules (SGs), thereby suppressing mTORC1 [65]. SGs are cytoplasmic aggregates of messenger ribonucleoproteins (mRNPs) that develop under conditions wherein translation is inhibited, such as diverse stresses [66]. Moreover, nemo-like kinase (NLK) has also been shown to play a role in suppressing mTORC1 under oxidative stress [67]. Through direct phosphorylation of RAPTOR at Ser863, NLK disrupts RAPTOR-Rag GTPase interactions, thereby impeding mTORC1’s lysosomal translocation and activation [67].
However, oxidants have also been reported to stimulate mTORC1. For example, the cysteine oxidant phenylarsine oxide activates RheB and thereby mobilizes mTORC1 [68]. Moreover, arsenite activates p38β, which leads to mTORC1 activation via phosphorylating RAPTOR [69]. Thus, the impacts of oxidative stress on mTORC1 seem complex, which may, at least in part, be determined by the intensity and duration of stress.
mTORC1 is suppressed by genotoxic stress
Various DNA damages, caused by oxidation, hydrolysis, alkylation, nucleotide mismatch and radiation, elicit genotoxic stress inside cells. To repair the damage and maintain genome integrity, cells rely on the DNA damage response (DDR) [70].
ATM and ataxia telangiectasia and Rad3-related (ATR) are two key DNA-damage sensors and regulators of the DDR [71]. Like oxidative stress, genotoxic stress activates ATM, which could subsequently suppress mTORC1 through the LKB1-AMPK-TSC2 cascade [63]. Activated ATM can also mobilize tumor protein p53 (TP53), a well-known tumor suppressor, via Ser15 phosphorylation [72]. Interestingly, Sestrin1/2, two target genes of TP53, can either activate the AMPK–TSC2 cascade or inhibit the guanine nucleotide exchange of Rag GTPases, thereby causing mTORC1 suppression [73,74]. In addition, regulated in development and DNA damage responses 1 (REDD1), another TP53 target gene involved in the TP53-mediated apoptosis in response to DNA damage [75], acts as a TSC2 activator through either blockade of 14-3-3 protein binding to TSC2 or inactivation of AKT by recruiting PP2A, thereby suppressing mTORC1 [76,77]. The tumor suppressor phosphatase and tensin homolog (PTEN), whose expression is also induced by TP53 [78], can also suppress mTORC1 by inhibiting PI3K/AKT signaling [79], a well-established signaling pathway that stimulates mTORC1. Moreover, TP53 activation also stimulates TSC2 expression [80]. Taken together, all these studies indicate mTORC1 suppression as an important aspect of the cellular response to genotoxic stress.
mTORC1 is suppressed by hyperosmotic stress
Unequal osmolarity between intra- and extracellular environments causes flux of water into or out of cells, eliciting osmotic stress [81]. In particular, hyperosmotic stress is referred to as the condition where extracellular osmolarity is higher than intracellular one. Accordingly, cells shrink and become dehydrated, owing to loss of intracellular water, which triggers a broad range of molecular changes, including DNA and protein damage [81].
It has been reported that under hyperosmotic stress NLK, similarly to under oxidative stress, mediates mTORC1 suppression by phosphorylating RAPTOR at Ser863 to disrupt the RAPTOR-Rag interactions [67]. Moreover, hyperosmotic stress can also inactivate AKT, likely via a Calyculin-A-sensitive phosphatase, to promote TSC2 activation, thereby suppressing mTORC1 [82]. Interestingly, hyperosmotic stress was reported to cause sequestration of both mTORC1 and the dual specificity tyrosine-phosphorylation-regulated kinase 3 (DYRK3) into SGs [83]. During stress DYRK3 sequestration prevents dissolution of SGs and mTORC1 release [83]. However, following stress DYRK3 directly phosphorylates and inactivates PRAS40, an inhibitory component of mTORC1, thereby re-activating mTORC1 [83]. Of interest, emerging studies suggest SGs as an important stress-inducible compartment for mTORC1 suppression during stress. In contrast to hyperosmotic stress, little is known about the impacts of hypo-osmotic stress on mTORC1.
mTORC1 is both suppressed and stimulated by ER stress
The endoplasmic reticulum (ER) is a central organelle where secreted and membrane-bound proteins are sorted and processed. ER homeostasis is vital to the health of cells, especially those specialized in secretion. Perturbation of this homeostasis, due to either external insults or internal defects, inevitably causes accumulation of many misfolded and aggregated proteins inside the ER lumen, a severely adverse condition widely referred to as ER stress [84]. To counteract this type of stress, cells have evolved a defensive mechanism named the ER stress response or unfolded protein response (UPR) [84].
Protein kinase RNA-like endoplasmic reticulum kinase (PERK), inositol-requiring enzyme 1 (IRE1) and activating transcription factor 6 (ATF6) are the three principal ER-stress sensors that mediate the UPR [84]. Given the nature of ER stress, attenuation of protein translation is not surprising. Indeed, under ER stress, activated PERK phosphorylates eIF2α, a subunit of eIF2 that is essential for the ternary complex (TC) formation [85], at Ser51 [86]. This phosphorylation blocks TC formation and thereby inhibits the translation initiation. In stark contrast to this wellilluminated mechanism, whether and how ER stress impacts mTORC1 are much less clear. Nonetheless, it has been reported that PERK activation following ER stress leads to selective translation of ATF4, a transcription factor mediating the UPR, via the upstream open reading frame (uORF) mechanism [87]. In turn, ATF4 induces the expression of REDD1 and tribbles pseudokinase 3 (TRIB3) [88,89]. While REDD1 is an activator of TSC2, TRIB3 is a direct AKT inhibitor [90]. Thus, increased REDD1 and TRIB3 both lead to mTORC1 suppression. Importantly, cells deficient in either Tsc1 or Tsc2 are hypersensitive to ER stress [91], suggesting the necessity of mTORC1 suppression for surviving ER stress. Congruently, mTORC1 inhibition enhances cellular survival of ER stress, although it was ascribed to blockade of the IRE1-JNK signaling cascade [92]. Conflicting with the reported role of ATF4 in mediating mTORC1 suppression, it was reported that during ER stress ATF4, in cooperation with the C/EBP homologous protein (CHOP), promotes global protein synthesis via transcriptional regulation of target genes involved in protein translation [93]. Similarly, evidence also indicates that ER stress stimulates AKT in the acute phase, thereby leading to mTORC1 activation [92]. Given the translation inhibition already imposed by eIF2α phosphorylation, it remains possible that the initial mTORC1 activation may exert effects other than protein translation.
Despite these seemingly contradictory findings, it is generally agreed upon that attenuation of protein synthesis promotes survival of ER stress, at least during the acute phase; and, during the chronic phase protein synthesis resumes. In contrast to the evident role of eIF2α in attenuating protein translation under ER stress, the implication of mTORC1 remains obscure.
mTORC1 is suppressed by proteotoxic stress
While there are considerable amounts of secreted and transmembrane proteins inside cells, however, a large proportion of the entire cellular proteome are consisted of proteins that are not processed in the ER. Instead, they mature and reside in the cytosol and/or nuclei. Molecular chaperones or HSPs, the counterparts of ER chaperones such as BiP/GRP78, are responsible for the proper folding, transportation, assembly, as well as degradation of these cytosolic and nuclear proteins, thereby playing a pivotal role in preserving cellular proteostasis [1,94]. Disruption of this equilibrium will provoke proteotoxic stress and, in turn, mobilize the powerful HSR/PSR [35]. Similar to the UPR, the HSR/PSR is also a transcriptional program that is primarily governed by HSF1 [95,96].
It has been long recognized that classic proteotoxic stressors, such as heat shock, causes global inhibition of protein synthesis. Again, this has been mainly ascribed to eIF2α phosphorylation [97]. In light of the importance of mTORC1 in controlling translation, surprisingly, little is known about whether and how mTORC1 responds to proteotoxic stress. It was reported that HSPs, in particular HSP90, help to assemble mTORC1 [98]. Owing to the titration by elevated levels of misfolded proteins under heat stress, the mTORC1-chaperoning activity of HSP90 is diminished, leading to disassembly and suppression of mTORC1 [98]. Given the abundance of cellular HSP90, this mechanism likely takes effect under chronic or intense proteotoxic stress where HSP90’s availability becomes a limiting factor. By contrast, it remains unclear how mTORC1 reacts to proteotoxic stress acutely.
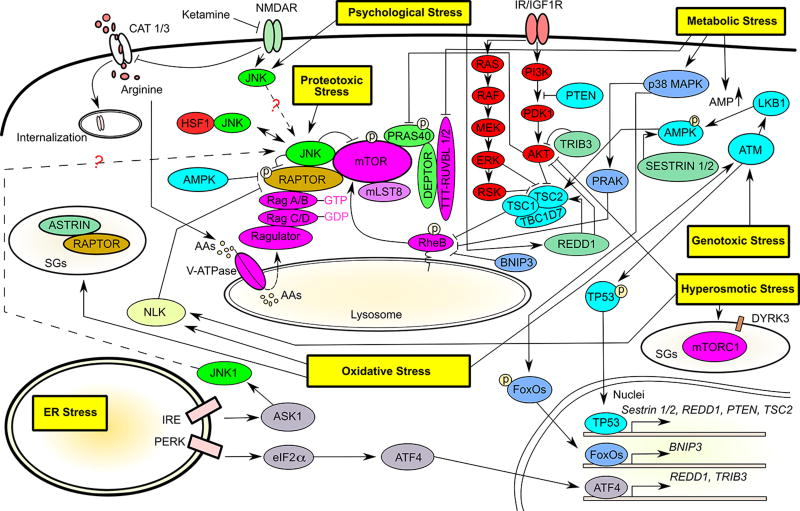
Prompt attenuation of general protein translation would be a logical reaction of cells to adapt to and survive proteotoxic stress. In line with this notion, a recent study revealed that diverse proteotoxic stressors, including heat shock and proteasome inhibitors, potently suppress mTORC1 and its mediated protein translation [36]. Of note, this is an acute response. For example, following proteasomal inhibition, mTORC1 activity begins to decline within 10 minutes, accompanied by concurrent Stress-activated protein kinase/c-Jun NH(2)-terminal kinase (SAPK/JNK) activation [36]. Mechanistically, proteotoxic stressors with distinct modes of action universally trigger JNK activation, which is both necessary and sufficient for mTORC1 suppression [36]. Of particular interest, JNK physically interacts with and phosphorylates mTORC1 [36], intimately integrating proteotoxic stress with protein translational control. Strikingly, JNK associates with mTORC1 in a constitutive manner and proteotoxic stress does not enhance this physical interaction, rather triggers JNK activation [36]. Following activation, JNK phosphorylates both RAPTOR at Ser863 and mTOR at Ser567, respectively, leading to mTORC1 disassembly [36]. This disassembly is partial, selectively excluding mTOR and its positive regulator mLST8/GβL from the complex but retaining JNK-RAPTOR-PRAS40 interactions [36]. Collectively, these new findings support the concept that mTORC1 is poised to respond to proteotoxic stress promptly with JNK as an integral sensor (Fig. 2).
Figure 2. mTORC1 senses diverse stresses.
The detailed molecular regulators are described in the main text. The question marks denote unexplored regulations. The objects in red represent oncogenes or pro-oncogenic factors; the objects in cyan represent tumor suppressors; and the objects in light green represent negative regulators of mTORC1. Within the mTORC1, the positive regulators are marked in purple and the negative regulators are marked in bright green. AAs: amino acids. AMP: Adenosine monophosphate. ASK1: apoptosis signal-regulating kinase. GTP: guanosine 5’-triphosphate. GDP: guanosine diphosphate. IR/IGF1R: insulin receptor/insulin-like growth factor 1 receptor. P: phosphorylation. PDK1: phosphoinositide-dependent kinase 1. SGs: stress granules.
Suppression of general protein translation, albeit beneficial to proteotoxic-stress adaptation by lessening overall proteomic burden, also creates a dilemma. Marked elevation of HSP expression is vital to surviving proteotoxic stress and, ultimately, restoring cellular proteostasis. Uncontrolled translation inhibition would diminish HSP production, dampening the HSR/PSR and exacerbating cell death. Thus, cells need to develop a means to prevent profound translational inhibition during proteotoxic stress. Indeed, JNK activation by proteotoxic stress is tightly modulated by HSF1, the master regulator of the HSR/PSR. Through physical interactions, HSF1 sequesters JNK away from mTORC1 [36], thereby ensuring attenuation, but not deep repression, of protein translation. Of great interest, HSF1 exerts this effect independently of its canonical transcriptional action [36]. This mode of action highlights that the abundance of HSF1 proteins governs the severity of mTORC1 suppression by proteotoxic stress. Beyond the stress response, importantly, this regulation has profound impacts on normal cellular and organismal growth. Cells and mice deficient for Hsf1 display elevated JNK activation, resulting in suppressed mTORC1 signaling and protein translation, and, ultimately, diminished cell and body size [36].
In aggregate, acting like a rheostat, HSF1 elegantly attunes mTORC1’s response to proteotoxic stress. Importantly, HSF1, through regulations of both mTORC1 and the HSR/PSR, intimately coordinates the protein quantity- and quality-control machineries. Thereby, a healthy cellular proteostasis is safeguarded.
mTORC1 is suppressed by psychological stress
It has been well established that protein translation is key to synaptic plasticity, learning and memory [99]. Compared to the various physical stresses discussed above, the knowledge of whether and how psychological stress impacts mTORC1 is scarce. Nonetheless, emerging studies start shedding light on this issue.
In rats, chronic unpredictable stress (CUS) causes mTORC1 suppression through induction of REDD1, a negative mTORC1 regulator that activates TSC2, in the prefrontal cortex region (PFC) [100]. Importantly, Redd1 deficiency markedly reverses the mTORC1 suppression, synaptic loss, and depressive behavior induced by CUS in rats [100]. Supporting a causative role of mTORC1 suppression in depressive disorders, ketamine, a N-methyl-D-aspartic acid receptor (NMDAR) antagonist that is currently on clinical trials for treating major depressive disorder (MDD), activates mTORC1 in the PFC of rats, which underlies its rapid antidepressant effects [101].
Interestingly, acute immobilization stress can induce JNK activation in the hippocampus, causing deficits in associative learning [102]. Moreover, JNK is also activated by NMDAR [103], and NMDAR activation leads to mTORC1 suppression [104]. Although this suppression has been ascribed to impaired neuronal arginine uptake, owing to depletion of cell surface cationic amino acid transporters 1/3 (CAT1/3) [104], it remains elusive whether JNK activation also contributes to mTORC1 suppression in this context (Fig. 2). In further support of a causative role of JNK activation in anxiety and depression, in mice Jnk1 deletion promotes hippocampal neurogenesis, alleviating anxiety and depressive-like behaviors [105]. Taken together, further efforts are warranted to fully elucidate the roles of mTORC1 and JNK in psychological stress, which is crucial to our better understanding of anxiety and depression, and may further open new therapeutic avenues.
mTORC1 suppression: a common theme of stress adaptation
Under ER or proteotoxic stress, cells mitigate protein synthesis to reduce proteomic input, a key strategy enabling more efficient proteomic repair and proteostasis restoration. Then, why does mTORC1 suppression also occur under numerous other types of stress?
Interplay between different cellular stresses may be one simplified explanation. For instance, hyperosmotic stress can induce protein misfolding as well, which likely signals mTORC1 suppression. Alternatively, osmotic stress may affect mTORC1 assembly directly. Oxidative stress can also lead to protein oxidation, a type of protein damage that likely elicits proteotoxic stress and subsequent mTORC1 suppression.
Another simplified explanation is energy conservation. To adapt to energetic stress, it is imperative for cells to minimize anabolic processes, among which protein synthesis consumes a large amount of cellular ATP. Attenuation of protein synthesis would markedly diminish ATP consumption, empowering cells to restore energy homeostasis. Why does genotoxic stress also suppress mTORC1? One of the plausible explanations is oxidative stress, as genotoxic stressors, including DNA alkylating agent, UV and ionizing radiation, frequently induce intracellular ROS [106–108]. High levels of ROS will cause oxidative stress, which leads to mTORC1 suppression. Another possibility is general transcriptional arrest. It has been known that DNA damage causes transcriptional arrest, through either hyperphosphorylation of RNA polymerase II, which prevents its association with the pre-initiation complex [109] or sequestration of TATA-binding protein (TBP) to damaged DNA, which limits its participation in transcription [110]. With reduced mRNA templates, it would be uneconomical to sustain high-rate protein translation.
Cancer cells elude stress-induced mTORC1 suppression
Unambiguous evidence has demonstrated the critical role of mTORC1 in promoting tumorigenesis. However, cancer cells also experience various stresses constantly from within and without. Inevitably, this constitutively stressed state would restrain robust mTORC1 activation. To eliminate or mitigate this constraint, cancer cells adopt diverse strategies. One of them is dampening the negative regulations of mTORC1. Inactivating mutations in tumor suppressor genes, including PTEN [111], the negative regulator of PI3K/AKT signaling, LKB1 [112], an upstream activator of AMPK, and TSC1/2 [113], render mTORC1 constitutively active and alleviate stress-induced suppression. Similarly, inactivation of the JNK signaling cascade also occurs in human cancers. Mutations in MAP2K4/MKK4 and MAP2K7/MKK7 [114,115], two key upstream kinases of JNK, impair JNK activation, in line with their tumor-suppressive functions [116,117]. Moreover, mutations in MAPK8/JNK1 and MAPK9/JNK2 are also identified [114], although the functional consequences remain to be elucidated. Alternatively, cancer cells can alleviate the JNK-mediated mTORC1 suppression through physical sequestration of JNK, as elevated HSF1 expression is widespread in human cancers [94]. Contrary to HSF1 overexpression, in most human cancers the expression of DEPTOR, a negative regulatory component of mTORC1, is markedly reduced [12].
In contrast to the de-repressing strategy, activating mutations of positive upstream regulators of mTORC1, including PI3KCA, AKT, RAS, RAF, MEK, and RheB [118–121], result in constitutive mTORC1 activation. Furthermore, mutations in mLST8/GβL have also been identified in human cancers (COSMIC/http://cancer.sanger.ac.uk), although their functional consequences remain unaddressed. Moreover, cancer cells could select for activating mutations within mTOR kinase itself. A number of mTOR-activating mutations have been uncovered in human cancers, some of which result in diminished DEPTOR binding and partial resistance of mTORC1 to nutrient deprivation, and resistance to the REDD1-mediated suppression [122,123]. Therefore, at least some of these activating mutations are expected to render mTORC1 resilient to suppression by other stresses as well.
Conclusions and Perspectives
mTORC1: a generic stress sensor?
It is evident that mTORC1 responds to diverse stresses via distinct mechanisms, some of which are indirect and require nascent transcription and/or translation. Likely, these mechanisms operate in the late-phase or chronic mTORC1 response to stress.
JNK is known to be activated by numerous stresses, including oxidative, osmotic, energetic, genotoxic, and ER stress [124–128]. Given its constitutive association with mTORC1 [36], it is tempting to speculate that mTORC1 senses a wide variety of stress signals immediately via JNK. This configuration is expected to empower mTORC1 to react to stress in a generic and prompt fashion. With the initial work focusing on proteotoxic stress only, further studies are required to confirm this notion in the context of other stresses.
Cellular stress: a double-edged sword in tumorigenesis?
Upon acute stress, reduction in global protein, lipid, and RNA synthesis and arrest of cell cycle progression represent a collective adaptive response enhancing survival and promoting damage repair. These systemic changes are incompatible with the anabolic lifestyle required to sustain malignancy. Thus, acute cellular stress likely antagonizes malignant growth.
Paradoxically, chronic cellular stress may promote malignancy, partly due to the selection pressure that drives the emergence of genetic and/or epigenetic alterations helping them both survive and resist stresses. This is supported by the occurrence of mutations rendering cancer cells refractory to stress-induced mTORC1 suppression [122,123]. In this way, chronic cellular stress may hasten tumor progression and cause the emergence of aggressive malignant phenotypes, analogous to the accelerated evolution by stressful environments [129,130]. This notion, albeit speculative, may have key implications. Chemo- and radiotherapies largely provoke genotoxic stress in cancer cells. Theoretically, these therapies should be administered to elicit acute and drastic cellular stresses. By doing so, cancer cells will be less likely to adapt, survive, and evolve.
Acknowledgments
The authors sincerely apologize to those whose work could not be cited in this review because of space limitations. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Abbreviations
- ER
endoplasmic reticulum
- HSF1
heat shock factor 1
- HSR/PSR
heat-shock response/proteotoxic stress response
- mTORC 1/2
mechanistic target of rapamycin complex 1/2
- Proteostasis
proteome homeostasis
- ROS
reactive oxygen species
- SAPK/JNK
Stress-activated protein kinase/c-Jun NH(2)-terminal kinase
- UPR
unfolded protein response
Footnotes
CONFLICTS OF INTEREST
The authors have declared no conflicts of interest.
References
- 1.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–9. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–9. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 3.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaizuka T, Hara T, Oshiro N, Kikkawa U, et al. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem. 2010;285:20109–16. doi: 10.1074/jbc.M110.121699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hara K, Maruki Y, Long X, Yoshino K, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–89. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 6.Kim DH, Sarbassov DD, Ali SM, King JE, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, Sarbassov DD, Ali SM, Latek RR, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 8.Guertin DA, Stevens DM, Thoreen CC, Burds AA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–71. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, et al. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–23. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 10.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–15. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Thedieck K, Polak P, Kim ML, Molle KD, et al. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS One. 2007;2:e1217. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson TR, Laplante M, Thoreen CC, Sancak Y, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–86. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarbassov DD, Ali SM, Kim DH, Guertin DA, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 14.Frias MA, Thoreen CC, Jaffe JD, Schroder W, et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16:1865–70. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Pearce LR, Sommer EM, Sakamoto K, Wullschleger S, et al. Protor-1 is required for efficient mTORC2-mediated activation of SGK1 in the kidney. Biochem J. 2011;436:169–79. doi: 10.1042/BJ20102103. [DOI] [PubMed] [Google Scholar]
- 16.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–18. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung CH, Jun CB, Ro SH, Kim YM, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganley IG, Lam du H, Wang J, Ding X, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson TR, Sengupta SS, Harris TE, Carmack AE, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–20. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu PP, Kang SA, Rameseder J, Zhang Y, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–22. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi JH, Bertram PG, Drenan R, Carvalho J, et al. The FKBP12-rapamycin-associated protein (FRAP) is a CLIP-170 kinase. EMBO Rep. 2002;3:988–94. doi: 10.1093/embo-reports/kvf197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–85. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 25.Yan L, Mieulet V, Lamb RF. mTORC2 is the hydrophobic motif kinase for SGK1. Biochem J. 2008;416:e19–21. doi: 10.1042/BJ20082202. [DOI] [PubMed] [Google Scholar]
- 26.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–80. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 27.Frodin M, Antal TL, Dummler BA, Jensen CJ, et al. A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J. 2002;21:5396–407. doi: 10.1093/emboj/cdf551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–8. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Li W, Williams M, Terada N, et al. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–9. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raught B, Peiretti F, Gingras AC, Livingstone M, et al. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004;23:1761–9. doi: 10.1038/sj.emboj.7600193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–26. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 32.Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci. 2015;16:345–57. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513:440–3. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J, Zhai B, Gygi SP, Goldberg AL. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci U S A. 2015;112:15790–7. doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morimoto RI. The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb Symp Quant Biol. 2011;76:91–9. doi: 10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- 36.Su KH, Cao J, Tang Z, Dai S, et al. HSF1 critically attunes proteotoxic stress sensing by mTORC1 to combat stress and promote growth. Nat Cell Biol. 2016;18:527–39. doi: 10.1038/ncb3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, et al. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–13. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 38.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E–BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–66. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 39.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–34. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dibble CC, Elis W, Menon S, Qin W, et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47:535–46. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inoki K, Li Y, Zhu T, Wu J, et al. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–57. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 42.Cai SL, Tee AR, Short JD, Bergeron JM, et al. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006;173:279–89. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, et al. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–93. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 44.Roux PP, Ballif BA, Anjum R, Gygi SP, et al. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci U S A. 2004;101:13489–94. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol. 2013;14:133–9. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zoncu R, Bar-Peled L, Efeyan A, Wang S, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–83. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stransky LA, Forgac M. Amino Acid Availability Modulates Vacuolar H+-ATPase Assembly. J Biol Chem. 2015;290:27360–9. doi: 10.1074/jbc.M115.659128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolfson RL, Chantranupong L, Saxton RA, Shen K, et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–8. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–6. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S, Tsun ZY, Wolfson RL, Shen K, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–94. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chantranupong L, Scaria SM, Saxton RA, Gygi MP, et al. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell. 2016;165:153–64. doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–62. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–90. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 57.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–26. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng M, Wang YH, Wu XN, Wu SQ, et al. Inactivation of Rheb by PRAK-mediated phosphorylation is essential for energy-depletion-induced suppression of mTORC1. Nat Cell Biol. 2011;13:263–72. doi: 10.1038/ncb2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin A, Yao J, Zhuang L, Wang D, et al. The FoxO-BNIP3 axis exerts a unique regulation of mTORC1 and cell survival under energy stress. Oncogene. 2014;33:3183–94. doi: 10.1038/onc.2013.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim SG, Hoffman GR, Poulogiannis G, Buel GR, et al. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol Cell. 2013;49:172–85. doi: 10.1016/j.molcel.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24:1939–50. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lobo V, Patil A, Phatak A, Chandra N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn Rev. 2010;4:118–26. doi: 10.4103/0973-7847.70902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alexander A, Cai SL, Kim J, Nanez A, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci U S A. 2010;107:4153–8. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 65.Thedieck K, Holzwarth B, Prentzell MT, Boehlke C, et al. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell. 2013;154:859–74. doi: 10.1016/j.cell.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 66.Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim Biophys Acta. 2015;1849:861–70. doi: 10.1016/j.bbagrm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuan HX, Wang Z, Yu FX, Li F, et al. NLK phosphorylates Raptor to mediate stress-induced mTORC1 inhibition. Genes Dev. 2015;29:2362–76. doi: 10.1101/gad.265116.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshida S, Hong S, Suzuki T, Nada S, et al. Redox regulates mammalian target of rapamycin complex 1 (mTORC1) activity by modulating the TSC1/TSC2-Rheb GTPase pathway. J Biol Chem. 2011;286:32651–60. doi: 10.1074/jbc.M111.238014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu XN, Wang XK, Wu SQ, Lu J, et al. Phosphorylation of Raptor by p38beta participates in arsenite-induced mammalian target of rapamycin complex 1 (mTORC1) activation. J Biol Chem. 2011;286:31501–11. doi: 10.1074/jbc.M111.233122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hakem R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 2008;27:589–605. doi: 10.1038/emboj.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marechal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Banin S, Moyal L, Shieh S, Taya Y, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–7. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 73.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–60. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng M, Yin N, Li MO. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell. 2014;159:122–33. doi: 10.1016/j.cell.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, et al. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 76.DeYoung MP, Horak P, Sofer A, Sgroi D, et al. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–51. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dennis MD, Coleman CS, Berg A, Jefferson LS, et al. REDD1 enhances protein phosphatase 2A–mediated dephosphorylation of Akt to repress mTORC1 signaling. Sci Signal. 2014;7:ra68. doi: 10.1126/scisignal.2005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stambolic V, MacPherson D, Sas D, Lin Y, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–25. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 79.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–96. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 80.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–9. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burg MB, Ferraris JD, Dmitrieva NI. Cellular response to hyperosmotic stresses. Physiol Rev. 2007;87:1441–74. doi: 10.1152/physrev.00056.2006. [DOI] [PubMed] [Google Scholar]
- 82.Plescher M, Teleman AA, Demetriades C. TSC2 mediates hyperosmotic stress-induced inactivation of mTORC1. Sci Rep. 2015;5:13828. doi: 10.1038/srep13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, et al. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 84.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 85.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–27. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–4. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 87.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:11269–74. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Whitney ML, Jefferson LS, Kimball SR. ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem Biophys Res Commun. 2009;379:451–5. doi: 10.1016/j.bbrc.2008.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ohoka N, Yoshii S, Hattori T, Onozaki K, et al. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24:1243–55. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–7. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- 91.Kang YJ, Lu MK, Guan KL. The TSC1 and TSC2 tumor suppressors are required for proper ER stress response and protect cells from ER stress-induced apoptosis. Cell Death Differ. 2011;18:133–44. doi: 10.1038/cdd.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kato H, Nakajima S, Saito Y, Takahashi S, et al. mTORC1 serves ER stress-triggered apoptosis via selective activation of the IRE1-JNK pathway. Cell Death Differ. 2012;19:310–20. doi: 10.1038/cdd.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Han J, Back SH, Hur J, Lin YH, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–90. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dai C, Sampson SB. HSF1: Guardian of Proteostasis in Cancer. Trends Cell Biol. 2016;26:17–28. doi: 10.1016/j.tcb.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vihervaara A, Sistonen L. HSF1 at a glance. J Cell Sci. 2014;127:261–6. doi: 10.1242/jcs.132605. [DOI] [PubMed] [Google Scholar]
- 96.Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J Biol Chem. 2005;280:33097–100. doi: 10.1074/jbc.R500010200. [DOI] [PubMed] [Google Scholar]
- 97.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–27. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 98.Qian SB, Zhang X, Sun J, Bennink JR, et al. mTORC1 links protein quality and quantity control by sensing chaperone availability. J Biol Chem. 2010;285:27385–95. doi: 10.1074/jbc.M110.120295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- 100.Ota KT, Liu RJ, Voleti B, Maldonado-Aviles JG, et al. REDD1 is essential for stress-induced synaptic loss and depressive behavior. Nat Med. 2014;20:531–5. doi: 10.1038/nm.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li N, Lee B, Liu RJ, Banasr M, et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science. 2010;329:959–64. doi: 10.1126/science.1190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sherrin T, Blank T, Hippel C, Rayner M, et al. Hippocampal c-Jun-N-terminal kinases serve as negative regulators of associative learning. J Neurosci. 2010;30:13348–61. doi: 10.1523/JNEUROSCI.3492-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Centeno C, Repici M, Chatton JY, Riederer BM, et al. Role of the JNK pathway in NMDA-mediated excitotoxicity of cortical neurons. Cell Death Differ. 2007;14:240–53. doi: 10.1038/sj.cdd.4401988. [DOI] [PubMed] [Google Scholar]
- 104.Huang Y, Kang BN, Tian J, Liu Y, et al. The cationic amino acid transporters CAT1 and CAT3 mediate NMDA receptor activation-dependent changes in elaboration of neuronal processes via the mammalian target of rapamycin mTOR pathway. J Neurosci. 2007;27:449–58. doi: 10.1523/JNEUROSCI.4489-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mohammad H, Marchisella F, Ortega-Martinez S, Hollos P, et al. JNK1 controls adult hippocampal neurogenesis and imposes cell-autonomous control of anxiety behaviour from the neurogenic niche. Mol Psychiatry. 2016 doi: 10.1038/mp.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rowe LA, Degtyareva N, Doetsch PW. DNA damage-induced reactive oxygen species (ROS) stress response in Saccharomyces cerevisiae. Free Radic Biol Med. 2008;45:1167–77. doi: 10.1016/j.freeradbiomed.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kang MA, So EY, Simons AL, Spitz DR, et al. DNA damage induces reactive oxygen species generation through the H2AX-Nox1/Rac1 pathway. Cell Death Dis. 2012;3:e249. doi: 10.1038/cddis.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yamamori T, Yasui H, Yamazumi M, Wada Y, et al. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radic Biol Med. 2012;53:260–70. doi: 10.1016/j.freeradbiomed.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 109.Rockx DA, Mason R, van Hoffen A, Barton MC, et al. UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. Proc Natl Acad Sci U S A. 2000;97:10503–8. doi: 10.1073/pnas.180169797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vichi P, Coin F, Renaud JP, Vermeulen W, et al. Cisplatin- and UV-damaged DNA lure the basal transcription factor TFIID/TBP. EMBO J. 1997;16:7444–56. doi: 10.1093/emboj/16.24.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sanchez-Cespedes M. A role for LKB1 gene in human cancer beyond the Peutz-Jeghers syndrome. Oncogene. 2007;26:7825–32. doi: 10.1038/sj.onc.1210594. [DOI] [PubMed] [Google Scholar]
- 113.Orlova KA, Crino PB. The tuberous sclerosis complex. Ann N Y Acad Sci. 2010;1184:87–105. doi: 10.1111/j.1749-6632.2009.05117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Parsons DW, Wang TL, Samuels Y, Bardelli A, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 115.Greenman C, Stephens P, Smith R, Dalgliesh GL, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ahn YH, Yang Y, Gibbons DL, Creighton CJ, et al. Map2k4 functions as a tumor suppressor in lung adenocarcinoma and inhibits tumor cell invasion by decreasing peroxisome proliferator-activated receptor gamma2 expression. Mol Cell Biol. 2011;31:4270–85. doi: 10.1128/MCB.05562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schramek D, Kotsinas A, Meixner A, Wada T, et al. The stress kinase MKK7 couples oncogenic stress to p53 stability and tumor suppression. Nat Genet. 2011;43:212–9. doi: 10.1038/ng.767. [DOI] [PubMed] [Google Scholar]
- 118.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:103–19. doi: 10.1517/14728222.2011.645805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nikolaev SI, Sotiriou SK, Pateras IS, Santoni F, et al. A single-nucleotide substitution mutator phenotype revealed by exome sequencing of human colon adenomas. Cancer Res. 2012;72:6279–89. doi: 10.1158/0008-5472.CAN-12-3869. [DOI] [PubMed] [Google Scholar]
- 121.Ghosh AP, Marshall CB, Coric T, Shim EH, et al. Point mutations of the mTOR-RHEB pathway in renal cell carcinoma. Oncotarget. 2015;6:17895–910. doi: 10.18632/oncotarget.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Grabiner BC, Nardi V, Birsoy K, Possemato R, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–63. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu J, Pham CG, Albanese SK, Dong Y, et al. Mechanistically distinct cancer-associated mTOR activation clusters predict sensitivity to rapamycin. J Clin Invest. 2016;126:3526–40. doi: 10.1172/JCI86120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Benhar M, Engelberg D, Levitzki A. ROS, stress-activated kinases and stress signaling in cancer. EMBO Rep. 2002;3:420–5. doi: 10.1093/embo-reports/kvf094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rosette C, Karin M. Ultraviolet light and osmotic stress: activation of the JNK cascade through multiple growth factor and cytokine receptors. Science. 1996;274:1194–7. doi: 10.1126/science.274.5290.1194. [DOI] [PubMed] [Google Scholar]
- 126.Gehart H, Kumpf S, Ittner A, Ricci R. MAPK signalling in cellular metabolism: stress or wellness? EMBO Rep. 2010;11:834–40. doi: 10.1038/embor.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Picco V, Pages G. Linking JNK Activity to the DNA Damage Response. Genes Cancer. 2013;4:360–8. doi: 10.1177/1947601913486347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–90. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hoffmann AA, Hercus MJ. Environmental stress as an evolutionary force. Bioscience. 2000;50:217–26. [Google Scholar]
- 130.Wright BE. Stress-directed adaptive mutations and evolution. Mol Microbiol. 2004;52:643–50. doi: 10.1111/j.1365-2958.2004.04012.x. [DOI] [PubMed] [Google Scholar]