Abstract
Theoretical approaches have long shaped neuroscience, but current needs for theory are elevated and prospects for advancement are bright. Advances in measuring and manipulating neurons demand new models and analyses to guide interpretation. Advances in theoretical neuroscience offer new insights into how signals evolve across areas and new approaches for connecting population activity with behavior. These advances point to a global understanding of brain function based on a hybrid of diverse approaches.
Theoretical approaches have a long history of contributing to neuroscience research, but never before has the need for them been so high nor the prospects for advancement so great. The explosion in technologies available for measuring and manipulating neurons has created a call for analysis techniques that are scalable to extremely large data sets, that take into account the heterogeneity of neurons, and that can predict and interpret the effects of complex manipulations of activity. The growing importance of theory is also driven by developments in theoretical neuroscience itself, advances that expand the reach of theoretical approaches and extend their ability to offer insight into long-standing puzzles. In the coming years, we will obtain enormous quantities of behavioral, recording (both electrical and optical), connectomic, gene expression and other forms of data. Obtaining deep understanding from this onslaught will require, in addition to the skillful and creative application of experimental technologies, substantial advances in data analysis methods and intense application of theoretic concepts and models.
The path from data to understanding requires three stages of analysis (Fig. 1). First, extracting relevant signals from the raw data: for example, image stabilization and isolation of regions-of-interest for imaging. Second, reducing large and complex data into forms that summarize the results in a more compact and understandable way. Third, using modeling to link the results to underlying mechanisms and overlying principles. The perspectives and reviews in this issue primarily address the third, and some of the second, stage, surveying new developments and modeling-based insights in topics ranging from understanding and interpreting network spiking activity1–3, exploring visual processing4 and memory5, and studying the representation and computation of probability6, to investigations of higher level cognition and mental illness7. In addition, major advances in the other stages of analysis have driven the entire program to evolve considerably in recent years. Two such advances and their accompanying challenges are described below.
Figure 1.
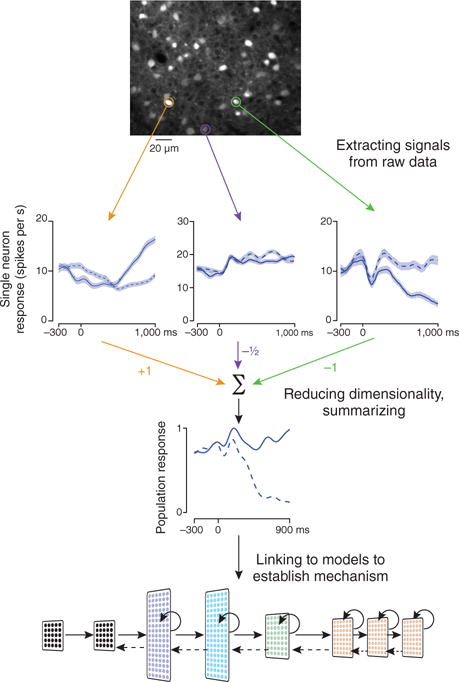
Multiple stages of analysis needed for neural data to Inform our understanding of neural mechanism. Top, extracting raw signals from data. Image shows neurons in mouse parietal cortex measured using a two-photon microscope (courtesy of F. Najafi). Regions of interest corresponding to single neurons are circled. Arrows point to schematized firing rates from the neurons, grouped by behavioral condition. Middle, dimensionality reduction is used to summarize responses of a large population. In the example, a weight is assigned to each neuron, and contributions are summed to extract a population response that correlates with behavioral conditions of interest. Bottom, models then link this reduced dimensionality representation of the data to mechanism. Adapted from ref. 4.
First, there are new challenges in data analysis driven by advances in our ability to simultaneously measure responses from many neurons. Specifically, it is not clear how to reduce large and complex data sets into forms that are understandable. Simply averaging responses of many neurons could obscure important signals: neural populations often have massive diversity in cell type8, projection target9 and response property10. One solution to this problem is to leverage methods that are naturally suited to multi-neuron measurements. Correlations across neurons, for example, can offer insight into the connectivity and state of a network1. Another solution to this problem is the use of machine-learning-based readouts and classifiers to interpret activity at the population level and relate it to behavior. Classifiers work by determining how best to combine neurons to extract differences in population activity across conditions (Fig. 1). This approach differs critically from traditional averaging in that neurons contribute in varying degrees, or even negatively (Fig. 1), to the extracted signal. Simple classifiers are becoming a standard tool for interpreting neural data10,11. More complex classifiers, such as those with multiple processing layers, are rapidly being developed in both neuroscience and industry12. One example is the hierarchical convolutional neural network (HCNN), which is based on ideas developed in studies of the visual system. As discussed in this issue, HCNNs can be used to interpret activity at various stages along the primate visual pathway during object recognition4.
Second, new challenges for data analysis arise from advances in experimentalists’ ability to manipulate neural activity with temporal, pathway and cell-type precision. The results of such manipulations can be difficult to interpret. For example, the neuronal heterogeneity described above implies that a population of neurons may encode a parameter through a mixture of firing rate increases and decreases. Thus, a pan-neuronal firing rate change from optogenetic stimulation or suppression could have mixed effects. This could occur even for manipulations of an identified class of cells because recent experimental findings indicate heterogeneity far beyond the currently identified categories13,14. Furthermore, as with more traditional methods of stimulation, propagation of activity to neighboring areas can create ‘off target’ effects that may erroneously suggest that an area has a causal role when, in fact, it does not15. Circuit-level modeling is a basic theoretical tool for dealing with these issues, but it must be extended to the multi-area level to address them more fully16.
Beyond the work described in this issue, what form might future models take? Work on the worm C. elegans provides insights that we expect will apply to other systems as well. In a study involving large-scale imaging of the C. elegans nervous system, a principal component–based description of neural activity was extracted, and patterns of activity across the entire neuronal population were correlated with behavioral states of the worm17. This analysis may ultimately lead to a model that provides an accurate, high-level description of neural activity and accounts for important aspects of worm behavior. The ‘units’ of this type of model are projections across a large neuronal population, not neurons themselves. Such a model could be criticized for leaving unexplained the responses of individual neurons; indeed, there is a temptation to require such models to dig down and explain fine-grained phenomena as well. As an alternative, we suggest a hybrid, rather than hierarchical, approach in which high-level and fine-grained models coexist, each explaining different features of the data. For example, in C. elegans the activity of a small number of modulatory neurons can alter the state transitions predicted by the high level model18, but these neurons are unlikely to be described well at the population level. The best approach may be to combine high-level and fine-grained components into a hybrid model that provides a more complete account of nervous system function than either component could by itself.
The perspectives and reviews in this issue embody this diversity of approach. Some of the articles in this issue start with basic, although simplified, biophysics and try to derive conclusions by assembling well-understood elements into less well-understood networks1–3,5. Others start with a general principle or idea of how a computation should be performed and work down from this ‘normative’ starting point to infer how functionality is achieved6,7. Yet another approach is to draw on analogies with another system, such as an HCNN4, even if it is quite different at the mechanistic level. Given the range of phenomena studied in neuroscience, demanding unity of viewpoint, methodology or depth of understanding, even in a single system, seems both unrealistic and counterproductive. Global understanding, when it comes, will likely take the form of highly diverse panels loosely stitched together into a patchwork quilt.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Contributor Information
Anne K Churchland, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA.
L F Abbott, Department of Neuroscience, and Department of Physiology and Cellular Biophysics, Columbia University College of Physicians and Surgeons, New York, New York, USA.
References
- 1.Doiron B, Litwin-Kumar A, Rosenbaum R, Ocker GK, Josić K. Nat Neurosci. 2016;19:383–393. doi: 10.1038/nn.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denève S, Machens CK. Nat Neurosci. 2016;19:375–382. doi: 10.1038/nn.4243. [DOI] [PubMed] [Google Scholar]
- 3.Abbott LF, DePasquale B, Memmesheimer RM. Nat Neurosci. 2016;19:350–355. doi: 10.1038/nn.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamins DLK, Dicarlo JJ. Nat Neurosci. 2016;19:356–365. doi: 10.1038/nn.4244. [DOI] [PubMed] [Google Scholar]
- 5.Fiete I, Chaudhuri R. Nat Neurosci. 2016;19:394–403. doi: 10.1038/nn.4237. [DOI] [PubMed] [Google Scholar]
- 6.Pouget A, Drugowitsch J, Kepecs A. Nat Neurosci. 2016;19:366–374. doi: 10.1038/nn.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huys QJM, Maia TV, Frank MJ. Nat Neurosci. 2016;19:404–413. doi: 10.1038/nn.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang ZJ, Zeng H. Annu Rev Neurosci. 2013;36:183–215. doi: 10.1146/annurev-neuro-062012-170307. [DOI] [PubMed] [Google Scholar]
- 9.Li N, Chen TW, Guo ZV, Gerfen CR, Svoboda K. Nature. 2015;519:51–56. doi: 10.1038/nature14178. [DOI] [PubMed] [Google Scholar]
- 10.Raposo D, Kaufman MT, Churchland AK. Nat Neurosci. 2014;17:1784–1792. doi: 10.1038/nn.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagan M, Urban LS, Wohl MP, Rust NC. Nat Neurosci. 2013;16:1132–1139. doi: 10.1038/nn.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeCun Y, Bengio Y, Hinton G. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 13.Tasic B, et al. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cembrowski MS, et al. Neuron. 2016 Jan 13; doi: 10.1016/j.neuron.2015.12.013. [DOI] [Google Scholar]
- 15.Otchy TM, et al. Nature. 2015;528:358–363. doi: 10.1038/nature16442. [DOI] [PubMed] [Google Scholar]
- 16.Chaudhuri R, Bernacchia A, Wang XJ. eLife. 2014;3:e01239. doi: 10.7554/eLife.01239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato S, et al. Cell. 2015;163:656–669. doi: 10.1016/j.cell.2015.09.034. [DOI] [PubMed] [Google Scholar]
- 18.Flavell SW, et al. Cell. 2013;154:1023–1035. doi: 10.1016/j.cell.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


