Abstract
Rare hereditary disorders provide unequivocal evidence of the importance of genes in human disease pathogenesis. Familial syndromes that predispose to osteosarcomagenesis are invaluable in understanding the underlying genetics of this malignancy. Recently, patient-derived pluripotent stem cells (iPSCs) have been successfully utilized to model Li-Fraumeni syndrome (LFS)-associated bone malignancy, demonstrating that iPSCs can serve as an in vitro disease model to elucidate osteosarcoma etiology. Here, we provide an overview of osteosarcoma predisposition syndromes and review recently established iPSC disease models for these familial syndromes. Merging molecular information gathered from these models with the current knowledge of osteosarcoma biology will help us gain a deeper understanding of the pathological mechanisms underlying osteosarcomagenesis and potentially aid in the development of future patient therapies.
Keywords: Cancer Etiology, Familial Cancer Syndrome, Induced Pluripotent Stem Cell, Osteosarcoma
Population at Risk for Osteosarcoma
Bone cancer is one of the most common primary malignancies in children and adolescents. Osteosarcoma comprises almost 60% of the common histological subtypes of bone sarcoma. It most frequently arises in the epiphysis (see Glossary) of long bones [1]. Standard treatment for osteosarcoma consists of surgery and chemotherapy. While the five-year survival rate of non-metastatic disease hovers at approximately 70%, metastatic disease -- most often to the lungs -- is associated with survival rates of 15% to 30% [1]. Osteosarcoma predominantly affects children and adolescents between the ages of 5 and 20 years, as well as adults in their seventies, with approximately 400 new pediatric cases diagnosed in the US annually [1].
Despite advances in surgery and multi-agent chemotherapy, lack of understanding of the molecular mechanisms of osteosarcomagenesis has prevented significant improvement in the survival of patients over the past 40 years. This malignancy makes osteosarcoma one of the leading causes of cancer mortality among children and adolescents. Therefore, elucidation of individual osteosarcoma-associated gene functions to explore the possible pathological mechanisms involved in osteosarcoma initiation, development and progression is critical for future osteosarcoma detection and treatment. People with certain rare hereditary genetic disorders are particularly at high risk for developing osteosarcoma. These includes patients with Li-Fraumeni syndrome (LFS), hereditary retinoblastoma (RB), Rothmund-Thomson syndrome (RTS), RAPADILINO syndrome (RAPA), Werner syndrome (WS), Bloom syndrome (BS), Diamond-Blackfan anemia (DBA), and Paget’s disease of bone (PDB) (see Boxes 1–5). Importantly, alterations in these genetic disease-associated genes have also been identified in samples of human sporadic osteosarcoma, accounting for the vast majority of osteosarcoma cases in the general population. As new information has emerged, so has the possibility of modeling osteosarcoma biology, and one approach has included the use of patient-derived pluripotent stem cells (iPSCs), as in the case of Li-Fraumeni syndrome (LFS)-associated bone malignancy. Indeed, investigations into inherited osteosarcoma-associated gene functions, coupled to iPSC modeling strategies will undoubtedly provide valuable insight into better understanding bone tumor initiation, development, progression and treatment.
Box 1. Li-Fraumeni syndrome (LFS).
LFS is a rare hereditary autosomal-dominant cancer disease first identified by Drs. FP Li and JF Fraumeni Jr. in 1969 [127]. They studied four families with soft-tissue sarcomas and found a high rate of familial tumor aggregation that could not be explained by chance alone. They further described this disease as a genetically heterogeneous inherited syndrome of pediatric soft-tissue sarcomas, breast cancers as well as other neoplasms in young adults [127]. Its autosomal dominant inheritance pattern was confirmed in 1988 [128]. Patients with LFS are likely to develop a primary cancer diagnosis by age twenty and secondary malignancies are common. Germline mutations in p53 were found to be responsible for most cases of LFS in 1990 [129]. In contrast to other inherited cancer syndromes largely characterized by tissue and site specificity, LFS patients present with a variety of early-onset primary tumors, including soft tissue sarcomas and osteosarcomas, breast cancers, brain tumors, leukemias, and adrenocortical carcinomas [128]. The clinical criteria for classic LFS is based on meeting the following three criteria: (1) a sarcoma diagnosed before the age of 45 years, (2) a first-degree relative with any cancer before the age of 45 years, and (3) a first- or second-degree relative with any cancer before the age of 45 years or a sarcoma at any age. In addition, some patients who do not meet the classic LFS criteria nevertheless harbor germline TP53 mutations, which led to the development of the less stringent Chompret criteria to identify families at risk for LFS and thus eligible for TP53 testing. [130]. LFS-associated germline p53 mutations are relatively rare, occurring in 1 in 20,000 individuals [131]. Over 300 distinct p53 mutations causing LFS have been identified (http://p53.free.fr/Database/p53_cancer_db.html). Approximately 70% of these mutations occur in the DNA binding domain encoded by exons 5–8; over 75% are missense mutations and usually generate a truncated p53 protein (http://p53.free.fr/Database/p53_cancer_db.html). The top mutational hotspots include R175, Y220, G245, R248, R273, R282 and R337 [22]. All of these TP53 mutations except R337H-- a founder mutation in Southern and Southeastern Brazilian populations --is widely found in sporadic cancers as well [22]. Commonly, mutations in p53 not only abolish p53 normal function but have also been associated with gain of oncogenic function [22, 30, 31].
Box 5. Diamond-Blackfan Anemia (DBA) and Paget’s Disease of Bone (PDB).
DBA is a rare inherited disease with an incidence of around 1:100,000 to 1:200,000 live births characterized by red blood cell aplasia. Affected patients frequently have congenital anomalies and display an increased risk of cancer development [158]. About half (~52.9%) of DBA patients carry heterozygous ribosomal protein mutations, and up to 9 ribosomal protein genes are reported to be involved in DBA [158]. The most commonly identified mutated gene in DBA is ribosomal protein S19 (RPS19), located at 19q13.2 and accounting for approximately 25% of DBA [158]. The next most common cause (~7%) is a mutation of RPL5, which is associated with a relatively high incidence of developmental anomalies compared with other forms of DBA [96, 158]. The specific pathogenesis of DBA is still unclear, although several hypotheses have been offered to explain the anemia phenotype. Due to the insufficiency of ribosomes for translation, some have proposed that ribosomal defects interfere predominantly with the development of highly proliferated cell populations, such as erythrocytes [159]. Others have suggested that disruption of the heme exporter FLVCR1 in RPS19-mutated DBA patients may lead to heme toxicity and apoptosis of erythroid progenitors [160]. In addition, ribosomal protein haploinsufficiency leading to a reduced translation of GATA1, a critical hematopoietic transcription factor for erythrocyte maturation, has been found in DBA patients [161]. Finally, a DBA zebrafish model reveals that defects in ribosome synthesis accelerate apoptotic death of hematopoietic progenitors due to p53 activation [92]. DBA patients have an unexpectedly high incidence of osteosarcoma, with 6 affected patients among 608 in the DBA Registry of North America [162, 163]. To date, there is no defined explanation for the high incidence of osteosarcoma in DBA patients.
PDB, first described by James Paget in 1876, is a focal disorder of bone turnover causing enlarged and disorganized bones [164]. The disease affects 1–2% of the general population and is more common in the elderly and in males [164]. Approximately 1 in 650 PDB patients develop osteosarcoma, a 30-fold increased incidence compared with the general population over 40 years [164]. The number, size and the sensitivity to growth factors of osteoclasts in PDB is increased, resulting in abnormal bone homeostasis and development of skeletal deformities [165]. This rapid bone turnover due to increases in both osteoclastogenesis and osteoblastogenesis may be the cause of increased incidence of osteosarcoma [164]. This turnover hypothesis may also explain why adolescents are particularly prone to development of osteosarcoma due to increased osteoblastogenesis between the ages of 10 and 19 years. The most frequent germline mutated gene of both inherited and sporadic Paget’s disease is SQSTM1 [165], a gene involved in the RANK signaling pathway and in autophagy. The similarity of cytokine profiles between osteoimmunological diseases (e.g., osteoporosis) and PDB as well as evidence of increased IFN-mediated signaling in PDB [166] suggests that PDB may be a potential osteoimmunological disorder.
Genetics of Osteosarcoma
Osteosarcoma frequently carries gross genomic mutations and rearrangements including chromosomal translocations [2–4]. Chromothripsis-- the presence of thousands of clustered chromosomal rearrangements -- is observed in 25% of clinical osteosarcoma human samples compared to 2–3% of cancers overall, such as chronic lymphocytic leukemia (CLL) [4]. Chromosomal translocations and mutations may juxtapose proto-oncogenes with constitutively active promoters, cause deletion of tumor suppressor genes, or produce chimeric oncogenes (e.g., PMP22-ELOVL5) [2–5]. Germline and somatic genome sequencing efforts have revealed potential pathological mechanisms involved in both osteosarcoma and syndromes with a genetic predisposition to osteosarcoma. For instance, The St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project compared the results of whole genome sequencing (WGS) from osteosarcoma specimens with matched germline DNA from affected patients and identified high rates of structural variations (SVs) and copy number alterations (CNAs) but low rates of single nucleotide variations (SNVs) in osteosarcoma tumors [2], indicating that chromosomal lesions by SVs and CNAs, rather than SNVs, are the main mechanism of recurrent mutations in osteosarcoma. Consistent with this, a study of genomic alterations in pediatric cancers indicated that osteosarcomas exhibit the highest frequency of SVs among all pediatric cancers [6]. The genes TP53, RB1, ATRX and DLG2 are altered via SVs and/or SNVs with high frequency in osteosarcoma [2]. Inactivation of the tumor suppressor p53 from translocation into the first intron of the TP53 gene has been detected in 9 out of 19 patient osteosarcoma tumors [2]. Although SNVs in the osteosarcoma genome are relatively uncommon, both SVs and SNVs can result in inactivating mutations in the p53 pathway, a feature found in ~95% of osteosarcomas [2]. Of note, kataegis, a pattern of localized hypermutations caused by SNVs, has also been widely found throughout the human osteosarcoma genome [2]. These results offer insight into novel genes that may contribute to the molecular pathogenesis of osteosarcoma and emphasize the value of comprehensive WGS on investigating the genetic features of osteosarcoma.
Hundreds of genomic rearrangements have been identified in osteosarcomas using genomic and transcriptomic analysis, including recurrent rearrangements of TP53, RB1, MDM2 and CDKN2A as well as PMP22-ELOVL5 gene fusions [2–5]. The most frequent TP53 rearrangements (e.g., TP53-VAV1, TP53-EMR1, TP53-PPRAD and TP53-KPNA3) resulted in the inactivation of p53 in osteosarcoma, explaining how TP53 gene function can be consistently disrupted in osteosarcoma despite the low observed prevalence of TP53 mutations in sporadic osteosarcomas, as shown in traditional mutation analyses [5, 7]. A study of transcriptome analysis on untreated clinical osteosarcoma samples revealed that two other osteosarcoma-specific fusion gene products, LRP1-SNRNP25 and KCNMB4-CCND3, were associated with osteosarcoma cell motility [8] and that a TP53-KPNA3 translocation was associated with chemotherapy resistance and metastasis [7]. These findings have not been validated in large studies since osteosarcoma-associated gene fusions are not as common as other sarcoma-associated gene fusions (e.g., EWSR1-FLI1 in Ewing sarcoma [9]). Therefore, further studies will be required to validate the role of these fusion genes in osteosarcomagenesis and to identify potential therapeutic targets.
Cellular Origins of Osteosarcoma
There are two primary competing hypotheses regarding the cellular origin of osteosarcoma, the mesenchymal stem cell (MSC) origin hypothesis and the osteoblast origin hypothesis [10–13]. The MSC hypothesis proposes that a mutation-carrying MSC will give rise to osteosarcoma [11, 13]. A high frequency of pathogenic variants in the TP53 and RB1 tumor suppressor genes and the c-MYC and RAS oncogenes is found in genomic studies of human osteosarcoma [2, 3]. Moreover, transformed human MSCs engineered to deplete RB1 and overexpress c-MYC -- a combination observed in patients with poor survival-- acquire malignant osteosarcoma-like properties. These MSCs express osteosarcoma markers CD99, ALP, osteonectin, and osteocalcin (also known as bone gamma-carboxyglutamic acid-containing protein (BGLAP)) and form lung and liver metastases in immunocompromised mice, suggesting that MSCs constitute the cellular origin of osteosarcomas [14].
In contrast, the osteoblast origin hypothesis suggests that osteosarcoma arises from defective differentiation of osteoblast-committed cells (Figure 1). This hypothesis stems from studies of MSCs derived from Trp53-mutant mice showing that a Trp53 mutation might result in early osteogenesis but impedes final maturation from osteoblast precursors into mature osteoblasts, which is evaluated by the expression of early and intermediate osteogenic marker osteopontin, rather than the terminal osteogenic marker osteocalcin [12]. Moreover, during osteogenic differentiation, depletion of Trp53 or both Trp53 and Rb1 in murine bone marrow-derived MSCs (BM-MSCs) -- but not adipose-derived MSCs (ASCs) -- induced the formation of osteosarcoma-like tumors [15]. Both undifferentiated BM-MSCs and ASCs developed leiomyosarcoma-like tumors but not osteosarcoma. This finding emphasizes that osteogenic differentiation of MSCs is critical for osteosarcoma development [15]. Induced pluripotent stem cell (iPSC)--derived osteoblasts but not MSCs obtained from LFS patients maintained in vitro and in vivo tumorigenesis, as evidenced by anchorage-independent growth (AIG) assays and xenotransplantation in immunocompromised nude mice, respectively [10]. This suggests that osteoblasts rather than MSCs are the cells of origin of osteosarcoma. Supporting this notion, RUNX2 and WNT signaling pathways, essential for osteogenic differentiation, have been found to be disrupted in human osteosarcoma samples, demonstrating loss of RUNX2 transcriptional activity and nuclear accumulation of β-Catenin, and thus, that osteosarcoma development might entail differentiation defects [16, 17]. In addition, activation of the intracellular domain of Notch1 in osteoblast-specific conditional Notch Intracellular Domain (NICD) transgenic mice, was shown to promote immature osteoblast proliferation, and was sufficient to induce osteosarcomagenesis [18].
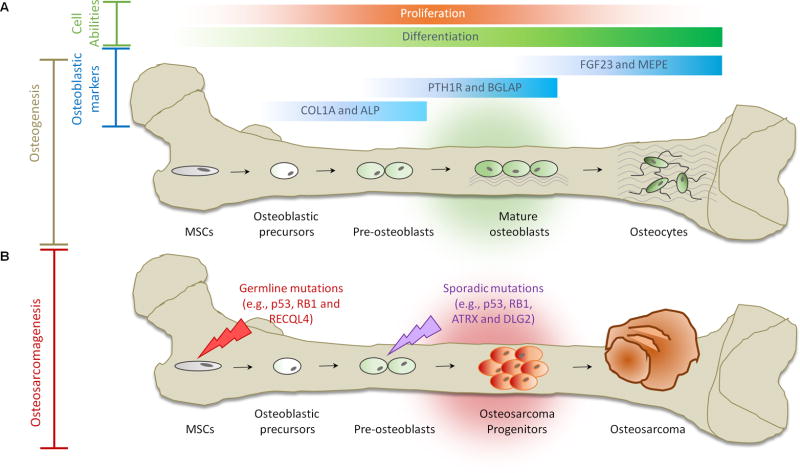
Figure 1. Osteogenesis and Osteosarcomagenesis.
(A) Initiation of osteogenic differentiation from MSCs. MSCs are multipotent bone marrow cells that are capable of differentiating to bone (osteoblast/osteocyte), fat (adipocyte) and cartilage (chondrocyte) tissues. Osteogenic differentiation is a tightly regulated process involving various signal transduction pathways (e.g., BMP and WNT), transcriptional regulators (e.g., p53, ZEB1, RUNX2 and ZNF521) and cell cycle controllers (e.g., RB1). Gene expression continuously changes through distinct osteogenic differentiation stages. COL1A and ALP are markers for osteoblastic progenitors and preosteoblasts. PTH1R and BGLAP serve as markers for mature osteoblasts. FGF23 and MEPE are markers for osteocytes. (B) Defects in osteogenesis lead to osteosarcomagenesis. Genetic alterations (e.g., germline mutations in p53, RB1 and RECQL4) probably interfere with the normal osteogenic process, resulting in incompletely differentiated osteoblasts or osteocytes in bone. These defects disrupt the balance between proliferation and differentiation and may cause a group of cells to display uncontrolled cell proliferation. Osteosarcoma progenitors may arise from these cells and expand to form osteosarcoma.
These hypotheses might be at least partially reconciled if the mutation-carrying MSCs indirectly result in osteosarcoma by potentiating the generation of osteoblasts with defective differentiation. Alternatively, given the variability across osteosarcoma tumor samples, both MSCs and osteoblasts might contribute to osteosarcomagenesis. Conditionally-disrupted TP53 and RB1 in murine MSCs, pre-osteoblasts and mature osteoblasts are all reported to develop into osteosarcoma [19]. Finally, osteosarcoma may arise from mature osteoblasts and osteocytes. For instance, the osteocyte marker dentin matrix acidic phosphoprotein 1 (DMP1) is increased in patient osteosarcoma samples, and SV40-immortalized mouse osteocyte cell lines can engraft as tumors in mice via either subcutaneous or intratibial injection [20]. Therefore osteocytes might also constitute an osteosarcoma progenitor cell type. Taken together, although there is still a debate regarding the cellular origins of osteosarcoma, specific genetic alterations may represent key factors in driving the development of osteosarcoma across cell types.
Molecular Mechanisms Involved in Syndrome-Associated Osteosarcomas
Hereditary genetic disorders associated with predisposition to osteosarcoma are relatively rare. However, studies of these diseases have led to important insights that generalize to the broader osteosarcoma population. Here, we discuss the most recent findings on eight genetic diseases that predispose to the development of osteosarcoma, collectively informing our understanding of the underlying molecular determinants: LFS (Box 1), RB (Box 2), RTS and RAPA (Box 3), WS and BS (Box 4), as well as DBA and PDB (Box 5) (Figure 2).
Box 2. Hereditary retinoblastoma (RB).
RB, caused by autosomal dominant mutations in the RB1 tumor suppressor gene, is a rare cancer of the retina typically found in children with approximately 300 cases in the US and 5–10,000 cases worldwide reported annually [132]. The RB autosomal dominant inheritance pattern suggests that one copy of the RB mutation in each cell is sufficient to increase patient RB risk [132]. Patients with bilateral RB are likely to acquire RB1 germline mutations. In contrast, patients with unilateral retinoblastoma usually harbor RB1 sporadic mutations. Clinical presentation suggests the incidence of unilateral retinoblastoma is 1.5 fold higher than that of bilateral retinoblastoma [133]. Although RB is one of the most treatable pediatric cancers with 95% 5-year actuarial survival rates in the US [134], RB patients are prone to develop a second cancer, including sarcomas, melanomas and brain cancers [135]. Osteosarcoma is the leading cause of death in RB survivors [136]. Receiving radiation therapy and systemic chemotherapy are important risk factors for developing osteosarcoma in RB patients [137].
One study indicated that among 848 radiation therapy-treated RB patients, 188 patients developed second cancers, 70 of which were osteosarcoma [136], consistent with other reported rates [135]. Somatic mutations of RB1 are also found in 30–75% of sporadic osteosarcomas [116, 122, 138]. Most of these osteosarcomas have been found to consist of poorly differentiated cells based on clinical histological data [139], implying that fully functional RB1 might be critical for normal osteoblast development [16, 43]. Interestingly, conditional osteoblast-specific Rb1 knockout mice exhibit defects in bone development and have increased osteoprogenitors in calvaria [140]; these osteoprogenitors might differentiate into immature osteoblasts and subsequently develop into a malignant osteosarcoma following additional cellular events (e.g., gain of somatic mutations and/or dysregulation of signaling pathways). This suggests that Rb1/RB1 might be able to control early osteoblast progression, and its dysregulation may increase the risk of osteosarcoma development; however, further studies will be required to validate this concept
Box 3. Rothmund-Thomson syndrome (RTS) and RAPADILINO syndrome (RAPA).
RTS is an autosomal recessive genetic disease characterized by a distinct skin rash called poikiloderma that develops between the age of three and six months [60]. The common clinical findings in RTS patients include poikiloderma, small stature, skeletal dysplasia, radial ray defect, sparse scalp hair, sparse brows/lashes, cataracts, skin cancer and osteosarcoma [141]. Recently, RTS has been categorized into two types, Type I and Type II, based on the absence (Type I) or presence (Type II) of mutations in the RECQL4 gene [142]. Notably, osteosarcoma is noted in 32% RTS II patients but not in RTS I patients [142]. RECQL4 is a member of the RECQ DNA helicase family, which also includes the WRN and BLM proteins that are associated with WS and BS, respectively[143]. The majority of patients with Type II RTS are compound heterozygous for RECQL4 mutations, and most of these mutations are predicted to cause truncated protein. Genotype-phenotype correlations have shown that Type II RTS patients are at much higher risk for skeletal defects and osteosarcoma compared to Type I RTS patients [60, 142, 144, 145].
RAPA is an autosomal recessive congenital disorder first reported in 1989 in the Finnish population [146]. RAPADILINO is an acronym for the disease features: RA stands for RAdial hypoplasia/aplasia, PA stands for PAtellar and Palate abnormalities, DI stands for DIarrhea and DIslocated joints, LI stands for LIttle size and LImb malformation, and NO stands for slender NOse and NOrmal intelligence [146]. Although RAPA patients have mutations in the same gene, RECQL4, as RTS patients, they lack poikiloderma which is the hallmark feature of RTS [147]. However, like RTS, RAPA patients also show a high incidence of osteosarcoma and lymphoma, with 2 and 4 cases, respectively, reported within a cohort of 15 RAPA patients [148].
Box 4. Werner syndrome (WS) and Bloom syndrome (BS).
WS is a rare autosomal recessive genetic disorder characterized by features of accelerated aging after puberty including premature loss of skin elasticity, gray hair, cataracts, diabetes, and cardiovascular disease [114]. Mutations in the DNA helicase WRN gene are associated with the development of WS [143]. To date, 83 different mutations spanning the WRN gene have been identified in WS patients [65]. Comprehensive genetic investigation of WS patients revealed a higher disease incidence among the Japanese [149]. WS patients are at a higher risk of rare cancers, including soft-tissue sarcoma, osteosarcoma, melanoma, meningioma, thyroid carcinoma, and gastric carcinoma [149]. In a case series of 158 patients with WS from 1996, 12 cases of osteosarcoma were reported [149].
First described by David Bloom in 1954, BS is a rare autosomal recessive disorder. BS is characterized by growth retardation, short stature, malar hypoplasia, hypo- and hyperpigmentation, immune deficiency, infertility, and occasional mild mental retardation, butterfly rash [150, 151], and is caused by mutations in the BLM gene [77]. Sixty-four different mutations of BLM have been reported up-to-date [77]. Among these 64 mutations, 19 could be found in more than one patient [77]. The Ashkenazi Jewish population exhibits a higher incidence of BS [152], with around 1% of Ashkenazi Jews being carriers who harbor a heterozygous frameshift mutation of BLM [153]. Patients with BS are predisposed to all cancers seen in the general population (such as leukemia, breast and colon cancers), but the onset of disease is much earlier than normal with an average of diagnosis of cancer at 27 year old [154]. Two cases of osteosarcoma were reported among 100 cases of cancers found in 168 BS patients in 1997 [155]. Although the incidence of osteosarcoma in BS is not as high as other syndromes predisposing to osteosarcoma, it still far exceeds the expected rate in the general population [156]. BLM functions in preventing recombination by disrupting recombination intermediates in vitro [157]. Human BS cells exhibit increased spontaneous chromosomal breaks and increased sister chromatid exchanges (SCEs), and these cytogenetic features are used clinically to help establish the diagnosis of BS [154].
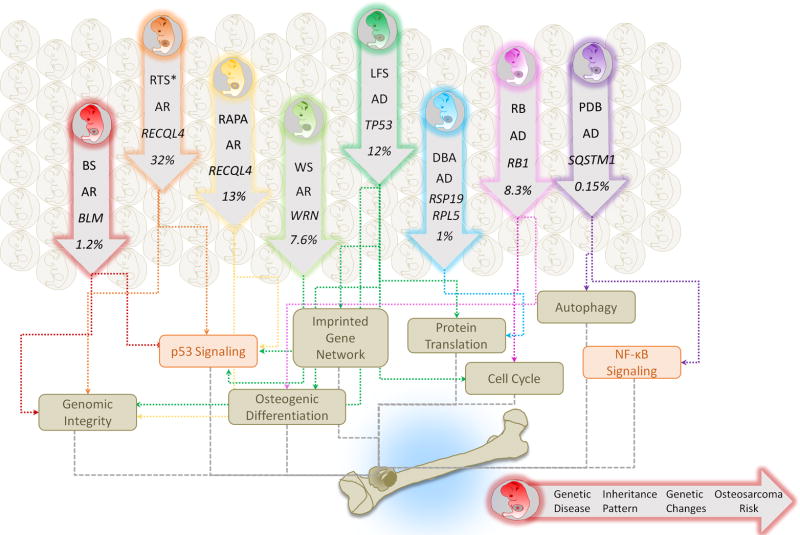
Figure 2. Familial Syndromes and Osteosarcoma.
A cluster of familial syndromes predispose patients to the development of osteosarcomas and are of relevance to understanding the underlying genetics of these tumors. These include LFS, RB, RTS, RAPA, WS, BS, DBA, and PDB. Each inherited syndrome harbors distinct gene mutations but shares a cancer predisposition to osteosarcoma. AD, autosomal dominant; AR, autosomal recessive. The dysregulation of variant biological processes (e.g., imprinted gene network, osteogenic differentiation, genomic integrity, protein translation, cell cycle and autophagy) and signaling (e.g., p53 and NF-κB) contributes to the syndrome-associated osteosarcomagenesis. The syndrome-associated risk of osteosarcoma is stated as percentage of patients with disorder developing osteosarcoma. *RTS indicates RTS Type II.
p53 in LFS
The tumor suppressor p53 is a transcription factor that binds DNA as a tetramer and inhibits tumorigenesis by regulating genes involved in cell cycle, DNA damage responses, apoptosis and metabolism [21]. As a transcription factor, p53 exerts its regulatory effects on thousands of genes regulating hallmarks of cancer [21, 22], including angiogenesis, metabolism, cell cycle, apoptosis, autophagy, metastasis and immune surveillance. Various studies have recently demonstrated that p53 plays additional roles in regulating embryonic and somatic stem cell differentiation and that defects in the p53 signaling pathway lead to impaired cellular differentiation [23, 24]. Impaired differentiation may in turn facilitate the development of several types of cancer such as osteosarcoma [25]. While p53 is traditionally thought to primarily promote cellular differentiation, studies of the role of p53 in bone development suggest that p53 might play a negative regulator role by attenuating osteoblast differentiation [26, 27]. Somatic mutations in the TP53 tumor suppressor gene are one of the most frequently noted alterations in almost every type of cancer, found in up to 70% of specimens (http://p53.free.fr/Database/p53_cancer_db.html). The hot-spot sporadic mutations are similar to germline mutations with the exception of the R337 mutation [22].
p53 has been shown to suppress tumor angiogenesis and proliferation in osteosarcoma by inhibiting the PI3K/AKT/mTOR pathway [28], further supporting that detrimental mutations in the TP53 gene can affect other pathways involved in cancer. The PI3K-AKT-mTOR pathway has been identified as an osteosarcoma driver by a Sleeping Beauty (SB) transposon-based forward genetic screen which introduces mutations into the genome [29]. Importantly, most LFS mouse models recapitulate human osteosarcoma susceptibility [30–33]. Specifically, introduction of mutant p53(R172H) into mice could lead to formation of osteosarcoma [30]. Functional inactivation of the tumor suppressor function of transactivation (TA) forms of p63 and p73 resulted in increased cell-transforming activity and reinitiation of DNA synthesis [30]. In addition, a LFS iPSC disease model of osteosarcoma revealed that impaired H19-mediated osteoblastic differentiation and tumor suppression were involved in mutant p53-associated oncogenic ability [10].
Gain-of-function mutations in p53 are thought to participate in multiple pathological activities. For instance, mutant p53 is capable of cooperating with the SWI/SNF chromatin remodeling complex to regulate tumor angiogenesis in 2D and 3D cultures of MDA-468 breast cancer cell lines [34]. It also interacts with ETS2 to bind and upregulate chromatin regulatory proteins including MLL1, MLL2 and MOZ to promote human breast cancer cell proliferation [35]. In addition to regulating chromatin function, mutant p53 associates with SREBP to abnormally upregulate the mevalonate pathway, resulting in disruption of normal acinar structures in human breast cancers [36]. Mutant p53 also upregulates PDGFRβ by inhibiting the p73/NF-Y complex leading to further invasion and metastasis in a pancreatic cancer mouse model [37]. The effects of these gain-of-function mutations have not yet been characterized in LFS-associated osteosarcomagenesis, and systematic studies of mutant p53 will be important in understanding LFS-associated osteosarcoma etiology.
RB1 in RB
The RB gene family, also known as pocket proteins, includes three members, RB1 (pRB or p105), RBL1 (p107) and RBL2 (p130) [38]. RB1 located at human chromosome 13q14, encoding the 110-kDa RB protein (RB1) which mainly participates in negative regulation of cell cycle progression [39]. In the G1 phase, RB1 is active and binds to E2F transcription factors, inhibiting the expression of both cell cycle and apoptotic genes [39]. During the G1-S phase transition, RB1 is inactivated via phosphorylation by CDKs. Phosphorylated RB1 releases E2Fs, allowing transcription and cell proliferation to proceed [39]. When RB1 is lost or inactivated by hyperphosphorylation, as commonly occurs in cancer, the cell maintains high expression levels of cell cycle genes [39]. However, only when RB1 is lost, whether by gene deletion, mutation or cleavage by caspases, can apoptotic genes be turned on [39]. In such cases and across species, the abrogation of the p53 proapoptotic pathway is required to protect cells from apoptosis [39]. Although expression of genes downstream of E2Fs leads to tumors, the lack of E2Fs also stimulates tumorigenesis in various species [40]. This finding not only implies the essential role of E2F in maintaining cell homeostasis, but also highlights the complexity of the RB-E2F pathway in cell proliferation.
RB1 also plays roles in the progression and metastasis of cancer by regulating angiogenesis and the epithelial-mesenchymal transition (EMT) [41]. Moreover, the self-renewal and differentiation properties of embryonic stem cells (ESCs)/cancer stem cells are controlled by the RB-E2F pathway [41]. Across different systems, RB1 can regulate differentiation by controlling lineage-specific transcription factors involved in erythrogenesis, myogenesis, cardiogenesis, adipogenesis and osteogenesis, [41, 42]. In addition, RB1 can interact with RUNX2 and peroxisome proliferator-activated receptor γ subunit (PPAR-γ) to respectively control osteogenesis and adipogenesis in MSCs, as shown for different species [16, 43]. And, the expression levels of RB1 can direct the cell fate of human MSCs toward either osteoblast/osteocyte or adipocyte lineages [16]. RB1 expression induces osteogenic lineage differentiation of mouse MSCs, which can differentiate into adipocytes in its absence [44]. Taken together, the role of RB1 in regulating osteoblast differentiation may explain the high incidence of osteosarcoma development in RB patients due to loss of RB1.
RECQL4 in RTS and RAPA
RECQL4 belongs to a member of the RecQ DNA helicase family. Its function has been demonstrated in initiation of DNA replication, DNA damage repair, and maintenance of the integrity of telomere and mitochondrial DNA [45]. The N-terminus (1~200 aa) of human RECQL4 shares homology with yeast Sld2 protein which is important for initiation of DNA replication [46, 47]. Human RECQL4 interacts with DNA replication licensing factor MCM10 to mediate the formation of the CMG (Cdc45; Mcm2–7; GINS) replication complex [48–50]. Since replication stress causes chromosomal instability in human cells [51], mutations in RECQL4 could cause replication stress leading to genome instability. In addition, RECQL4 directly participates in DNA damage repair, including nucleotide excision repair (NER) for UV DNA damage, base excision repair (BER) for oxidative DNA damage, and DNA double strand break repair (DSBR) through homologous recombination (HR) and non-homologous end-joining (NHEJ) pathways [45]. RECQL4 co-localizes and interacts with xeroderma pigmentosum group A (XPA) protein which is required for NER, and UV damage to H1299 and HeLa cells has resulted in increased co-immunoprecipitation intensity and co-localization between RECQL4 and XPA [52]. In response to H2O2 induced oxidative stress, RECQL4 was demonstrated to co-localize with and stimulate the biochemical activities of apurinic/apyrimidinic endodeoxyribonuclease 1 (APE1), DNA polymerase β, and flap structure-specific endonuclease 1 (FEN1), several key factors in the BER pathway, indicating that RECQL4 plays a role in BER in mammalian cells [53]. RECQL4 has also been shown to play a role in NHEJ-dependent DSBR by interacting with and stimulating the activity of the Ku heterodimer, an important member of NHEJ pathway [54]. RECQL4 interacts with p53 and masks the p53 nuclear localization signal, which in turn facilitates p53 mitochondrial localization in untreated normal human fibroblasts [55], providing a new regulatory mechanism of p53 activity. In mitochondrial nucleoids, the RECQL4-p53 complex physically interacts with mitochondrial DNA polymerase (PolyA/B2) in human fibroblasts and potentiates its binding to the mitochondrial DNA (mtDNA) control region (D-loop), as demonstrated by electrophoretic mobility shift assays [56]. In addition, RECQL4 can be recruited to laser-induced double strand breaks (DSB) by the MRE11 nuclease in the human osteosarcoma U2OS cell line, and is required for 5’ end resection of DSB, the initial step of HR-dependent DSBR [57]. In mammalian cells, RECQL4 also associates with RAD51, a key protein involved in homologous recombination (HR); thus, the defect in RECQL4 function is expected to result in defective HR associated genomic instability [58]. Supporting this idea, karyotype analyses in cells from Recql4-deficient mice show increased aneuploidy and premature centromere separation [59]. Therefore, it is possible that defective HR-induced genomic instability might contribute to initiating osteosarcoma development in RTS patients with RECQL4 mutations. Indeed, although the molecular mechanism of osteosarcomagenesis in RTS remains unclear, genomic instability due to mutations in RECQL4 has been implicated in disease development. For instance, RECQL4 is also mutated in RAPA, which also presents an increased risk of osteosarcomagenesis[60].
While the mouse global knockout of Recql4 (targeting exon 5 to 8) causes embryonic lethality [61], inactivation of Recql4 in skeletal tissues has recapitulated some features of RTS, including skeletal abnormalities and small stature [62]. Cells from such Recql4 conditional mutant mice display elevated p53 activity [55, 62], and the skeletal phenotypes in these mice can be partially rescued by p53 inactivation [62]. Furthermore, mice that lack Recql4 in osteoblast progenitor cells demonstrate a decrease in mineral apposition rate and bone formation rate [63] but no increase in osteosarcoma. These results imply that osteosarcoma susceptibility is most likely triggered by mutant, not null, alleles of RECQL4 in RTS patients.
WRN in WS
The human WRN protein contains a conserved 3’ to 5’ helicase domain as well as a RecQ helicase conserved (RQC) region. The helicase, RNAse D, C-terminal conserved (HRDC) region in WRN is also shared by BLM. WRN is the only RecQ helicase member to also possess 3’to 5’ DNA exonucleolytic activity [64]. WRN can also unwind unusual G quadruplex (G4) DNA structures and four-way junctions [65]. WRN phosphorylation at S1133 by CDK1 favors WRN-DNA replication helicase/nuclease 2 (DNA2)-dependent long-range DNA resection and promotes homologous recombination in human fibroblasts [66]. WRN can also interact with HDAC1 and 2 to promote replication restart following replication stress in mammalian cells [67]. Although loss of WRN in WS may directly lead to cancer development through increased chromosome instability [68], its overall function in cancer is not clear. WRN has been reported to be up-regulated in several types of cancers. The ability of WRN to stabilize chromosomes (e.g., base excision repair, proofreading, resolution of G4 for proper DNA transcription and telomere stabilization) may benefit cancer cells with higher rates of proliferation and associated errors [69]. In addition to maintaining genomic integrity, WRN can interact with the carboxyl terminus of p53 through its carboxyl-terminal region [70]. Moreover, overexpression of WRN in cancer can enhance the transcriptional activity of p53 which leads to increased p21WAF1 protein expression in mammalian cells [70]. p38 MAPK activation leads to accelerated cell senescence in WS fibroblasts, implying a potential therapeutic role for a p38 inhibitor in WS [71, 72]. Because vitamin C supplementation increased the life span of WRN-mutated mice and C. elegans [73, 74], vitamin C has been reported to be a potential new treatment for WS [75, 76]. However, the effect of these compounds on WS-associated osteosarcoma and/or osteosarcoma with defects in WRN-associated signaling has not yet been explored.
BLM in BS
BLM belongs to the RecQ family of DNA helicases. Numerous mutations in human BLM causing premature stop codons have been shown to lead to BS [77]. Like WRN, BLM contains a helicase domain, a RecQ-C-terminal (RecQ-Ct; RQC) domain, a HRDC domain and a C-terminal NLS [78]. In addition to ATP-dependent 3’to 5’ DNA helicase activities, BLM can interact with topoisomerase III alpha (TOP3A) and stimulate torsional stress relief during DNA unwinding [79]. BLM and TOP3A cooperatively resolve the double Holliday junction (dHJ) and prevent sister-chromatid exchanges (SCEs) [80] with help from RecQ mediated genome instability 1 (RMI1) and RecQ mediated genome instability 2 (RMI2). Using biochemical assays, DNA replication helicase/nuclease 2 (DNA2) has been shown to form a complex with either WRN or BLM and promote DNA repair by facilitating dsDNA degradation [81]. Additionally, BLM is involved in chromosome segregation and telomere maintenance in human cells [82, 83]. Like WRN, BLM also interacts with the C-terminal domain of p53 and can facilitate p53-induced apoptosis [82]. p53 can help the localization of BLM to PML nuclear bodies in mammalian cells [84]. Despite the strong structural similarity between WRN and BLM, only BLM-deficient cells exhibit increased spontaneous chromosomal breaks (BS>WS>WT), sister chromatid exchanges (SCEs) and HR-mediated genomic instability [85], and we speculate that accumulation of genome alterations may induce BS-associated malignancies. In contrast, WS cells are characterized by reduced HR repair and enhanced mutations and chromosomal aberrations (e.g., variegated translocation mosaicism and large chromosomal deletions) [85]. BLM also interacts with several components of the Fanconi Anemia (FA) gene complex [85]. FA comprises a collection of diseases characterized by mutations in FA genes leading to genomic instability and increased cancer risk. Indeed, BLM can promote FA group D2 (FANCD2) protein activation through its motif VI, indicating that BLM might partially function through FANCD2 in DNA replication [86]. This connection across multiple syndromes highlights the importance of chromosomal instability in cancer [86].
Ribosomal Proteins (RPs) in DBA
DBA is a disorder of ribosome function [87]. RPs are required for normal translation in all cells, and expression of about a quarter of the RPs varies greatly across human tissue types [88]. Hematopoietic cells have among the most heterogeneous RP expression, potentially explaining why DBA has primarily hematologic manifestations [89]. Upregulation of p53 function is commonly found upon dysregulation of ribosome biogenesis caused by ribosomal abnormalities. Some RPs (e.g., RPL5, RPL11, RPS7, and RPS26) can interact with MDM2 and inhibit E3 ubiquitination of p53, leading to p53 accumulation in both cell lines and mouse model systems [90, 91]. Other RPs (e.g., RPS19) have no direct interactions with MDM2/p53, but their mutation nonetheless increases p53 levels in zebrafish [92]. In general, defects in ribosome synthesis accelerate apoptotic cell death of hematopoietic progenitors due to p53 activation [93]. Furthermore, ribosomal insufficiency caused by RPS19 and RPS24 mutations can result in G0/G1 arrest and G2/M reduction, respectively, suggesting that cell proliferation defects might contribute to the decreased number of erythroid precursors observed in DBA [94]. The dysregulation of specific RPs has also been noted in cancers [89]. For instance, RPL5 heterozygous mutations or deletions are found in 11% of glioblastomas, 28% of melanomas and 34% of breast cancer patients [95]. Besides being the most common RP mutation in cancer, RPL5 mutations also lead to craniofacial anomalies [96]. RPS19, the most frequently mutated gene in DBA, is upregulated in several cancers and causes tumor-associated immunosuppression via the complement C5a receptor 1 protein [97, 98]. Because of the diverse functions of RPs, the precise mechanisms by which altered ribosome biogenesis in DBA may lead to cancer remains unknown. However, the association between DBA and osteosarcoma suggests an underappreciated role for ribosomal synthesis in regulating and preventing cancer.
p62/SQSTM1 in PDB
p62 (also known as SQSTM1) encoded by the SQSTM1 gene is a potent regulator of cell signaling, triggering NF-κB activation, adipogenesis, mTORC1 activation, apoptosis and selective autophagy through interactions mediated by its different functional domains [99]. When the supply of amino acids is abundant, p62 interacts with regulatory-associated protein of mTOR (RPTOR, also known as RAPTOR) and RAG C/D and leads to p62 translocation into lysosomes, activating mTORC1 signaling [100]. p62 functions by recognizing and binding to selectively ubiquitinated macromolecules: p62 translocates with the ubiquitinated cargo to the lysosome, where it is phosphorylated by mTORC1 at Ser349 [101]. Phosphorylated p62 then causes NRF2 release from KEAP1 and NRF2 stabilization, after which NRF2 can translocate to the nucleus, turning on downstream anti-oxidative genes or forming autophagosome [101]. Interactions between p62 and TRAF6 promote K63 polyubiquitination of mTORC1, which also facilitates mTORC1 activation in both mammalian cells and mouse model systems [102].
Most p62 mutations found in PDB patients occur in the autophagy- and ubiquitin-associated domain [103]. Studies have suggested that these mutations enhance the sensitivity of RANKL and accelerate PDB patient osteoclast formation through NF-κB activation [104]. This signaling pathway might dysregulate the homeostatic cross-talk between osteoclasts and osteoblasts and promote bone malignancy in PDB due to abnormally-enhanced osteoclast formation. In addition, accumulation of p62 in epithelial cells, as occurs in pancreatitis and liver degenerative diseases, has been reported to promote tumor initiation and progression [99]. By contrast, p62 is commonly downregulated in tumor stromal fibroblasts; the reduced p62 level decreases mTORC1 activity and c-Myc levels, ultimately increasing oxidative stress within the tumor microenvironment and promoting cancer progression [99]. It is unknown which of these processes, including autophagy, hyperactivation of RANKL-NF-κB signaling, and changes in the tumor microenvironment contribute to PDB-associated osteosarcoma. Thus, further studies will be required to robustly define these associations.
Current Advances in Osteosarcoma Biology Using iPSC Models
In vitro modeling of human disease has recently become possible with iPSC methodologies [105] (Figure 3A). Numerous laboratories have demonstrated that PSCs (ESCs and iPSCs, Box 6) overcome many limitations of other model systems and can serve as a relevant model system to study the etiologies of cancer including osteosarcoma (in LFS [10], WS [106, 107], DBA[108, 109] and RB [110]) (Figure 3B), brain tumors [111, 112], and leukemia [113].
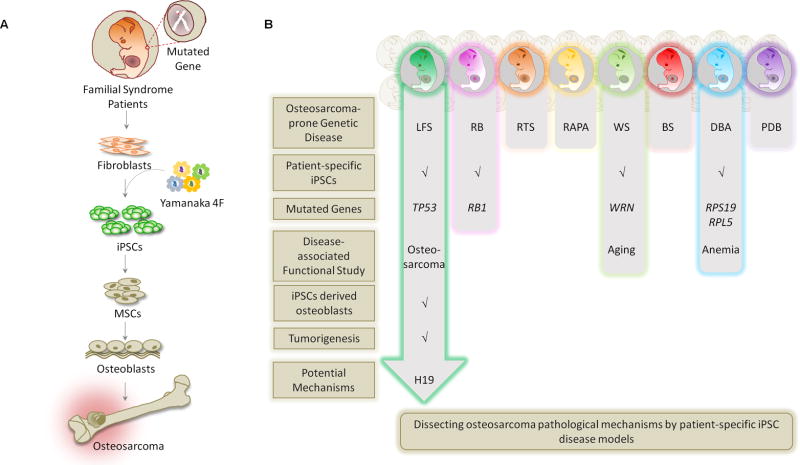
Figure 3. Patient-Derived iPSC Disease Models for Osteosarcoma.
(A) Patient-derived iPSCs are used to model human familial cancer syndromes and reveal a role of the mutant gene(s) in disease development. To apply iPSC methodology to study genetic disease-associated bone malignancy, patient fibroblasts are biopsied from skin and then reprogrammed to iPSCs by the four "Yamanaka factors" (OCT4, SOX2, KLF4 and c-MYC). iPSCs are then differentiated into MSCs, and then further, to osteoblasts. These iPSC-derived osteoblasts can be examined for osteoblast differentiation defects and tumorigenic ability. Systematic comparison of the genome/transcriptome/interactome between mutant and wild-type osteoblasts can further elucidate pathological mechanisms. (B) Current progress of applying LFS, RB, RTS, RAPA, WS, BS, DBA and PDB patient-derived iPSCs to model disease etiology and dissect disease-associated osteosarcomas.
Box 6. Modeling diseases by patient-derived iPSCs and ESCs.
Characterized by the ability to self-renew indefinitely and differentiate into all cell lineages of an organism similar to ESCs, iPSCs provide a powerful and unlimited source of cells to generate differentiated cells that can be used to elucidate disease pathogenesis, facilitate drug discovery and development, and provide crucial understanding that will be required to provide personalized healthcare [125]. Although iPSCs have been used for several years to model diseases such as neurodegeneration, mental retardation, heart disease, and metabolism disorders [125], they have only recently been used for cancer research [126].
iPSCs not only provide unlimited and consistent cell resources but also enhance the applicability of findings by permitting studies within a human model system. Correcting disease mutations in iPSCs by cutting-edge genome editing methodologies (e.g. ZNFs, TALENs, and CRISPR/Cas9) can generate a perfect isogenic control for disease modeling. However, the behavior of each iPSC line must be understood in the context of its genetic background and disease-related specific mutation. A panel of iPSCs, each carrying a distinct genetic alteration in a distinct background, would most generally address the central pathological mechanisms involved in disease development from an affected patient population. However, limited patient samples and high cost of iPSC generation may restrict this approach. The incorporation of engineered ESCs may help compensate for this limitation by controlling for genetic background. Various disease-associated mutations can be engineered into the same ESC line, permitting comparison across different disease mutations to understand central disease pathogenesis. Both patient-derived iPSCs and ESCs will aid our understanding of disease pathogenesis and complement existing animal models.
Malignancy in the LFS iPSC disease model
Osteosarcoma is one of the main cancer types seen in LFS patients. A LFS disease model established from patient-derived iPSCs delineated the pathological mechanisms caused by heterozygous p53 (G245D) mutation in osteosarcoma [10]. LFS iPSC-derived osteoblasts recapitulate clinical osteosarcoma features including defective osteoblastic differentiation and gain of tumorigenic ability [10]. Gene expression patterns of LFS osteoblasts are similar to those of tumor samples obtained from primary osteosarcoma patients, and these tumorigenic features strongly correlate with shorter tumor recurrence times and poorer patient survival rates [10].
The numerous chromosomal alterations and rearrangements present in osteosarcoma make analyses of the initial steps of osteosarcoma development particularly challenging in most model systems; however, LFS-derived osteoblasts lack cytogenetic rearrangements and therefore permit the study of early tumor initiation without interference from other gene alterations [10]. Analyses of the global transcriptome in LFS osteoblasts identified impaired expression of the long noncoding RNA H19; functional studies concluded that H19 is an essential gene for normal osteogenesis and tumor suppression and acts by regulating bone development-associated genes and the imprinted gene network (IGN) [10]. This LFS iPSC disease model highlights a previously unappreciated role of p53 in regulating the human IGN that culminates in osteogenic differentiation defects and osteosarcomagenesis [10].
Premature aging in the WS iPSC disease model
Premature cellular aging, telomere dysfunction and early cell senescence are commonly found in WS patient mesenchymal-derived tissues [114]. One study reprogrammed WS patient fibroblasts to iPSCs and then differentiated these iPSCs to MSCs [107]. Premature senescence was observed in WS MSCs, and a role of telomerase in mediating these defects was suggested [107]. In parallel, another study revealed the effects of long-term culture on WS iPSCs and documented that WS iPSCs maintain normal self-renewal and differentiation abilities throughout two years of long-term culture, indicating that somatic reprogramming might suppress premature senescence in WS [106]. The WS iPSC disease model may help further explore the link between aging and senescence in cancer development including osteosarcomagenesis.
Anemia in the DBA iPSC disease model
Erythroid hypoplasia is one of the key features of DBA patients. DBA iPSCs from affected patient fibroblasts harboring either heterozygous RPS19 (Q126X) or RPL5 (R23X) mutations have been established [109]. In comparison with wild-type controls, both RPS19-mutated iPSCs and RPL5-mutated iPSCs have been found to exhibit defective ribosome biosynthesis and globally impaired erythroid progenitor differentiation [109]. These findings suggest that the DBA iPSC model might help explore the pathological role of dysregulation of ribosome biosynthesis in developing anemia [109]. The same conclusion was also made by another study using distinct DBA patient iPSCs harboring either RPS19 (R94X) or RPL5 (Y16X) [108]. Here, an unbiased chemical screen was performed on DBA iPSC-derived hematopoietic progenitors and the autophagy inducer SMER28 was identified as a potential compound that might enhance erythropoiesis and ameliorate the defect in erythroid differentiation [108].
Although DBA iPSCs have not been used to elucidate the role of ribosome biosynthesis in the pathogenesis of osteosarcoma, many of these earlier findings in anemia may translate to osteosarcoma due to the essential role of ribosome synthesis in cellular homeostasis. It will be valuable to explore whether (1) osteoblasts and hematopoietic progenitors in DBA patients share a similar differentiation defect due to heterozygous null mutations in ribosome proteins; (2) defective autophagy in DBA iPSC-derived osteoblasts contributes to osteosarcomagenesis; (3) SMER28 can be applied to treat and/or prevent DBA-associated osteosarcoma; and (4) defective ribosome biosynthesis is a general mechanism leading to osteosarcomagenesis. Because dysregulation of ribosome protein function (e.g., RPL5 and RPL11) influences the p53/MDM2 axis [115], this disease model may also help clarify the complex signaling networks in osteosarcoma.
Early studies of the RB iPSC disease model
iPSCs were recently generated from a RB patient carrying a heterozygous RB1 (S888A) mutation [110]. Although RB iPSCs expressed pluripotency markers and were capable of differentiation to all germ layers in vivo, no pathological and/or mechanistic investigations were conducted in this study [110]. Given the presence of RB1 alterations in 30–75% of sporadic osteosarcoma [116] and lack of success thus far in recapitulating the osteosarcoma phenotype in RB mouse models[117–119], RB iPSC-derived osteoblasts have the potential to substantially improve our understanding of the role of RB1 in osteosarcomagenesis.
Remaining iPSC disease models
RTS, RAPA, BS and PDB patient-specific iPSC disease models have not been yet established. Consequently, the generation of these disease models through iPSC approaches will be of substantial value in investigation of the global picture of the molecular mechanisms involved in osteosarcoma development. Comparison among the dysregulated gene profiles and signaling networks found in these new lines, as well as those described above, will be crucial to understanding the etiology of osteosarcoma.
Concluding Remarks
Multiple inherited syndromes have been linked to osteosarcoma predisposition. Although attempts at studying the genetics of these diseases have provided fruitful results, our understanding of the role of these genes in development of osteosarcoma is still lacking. There have been attempts to model these genetic diseases with various animal model systems, but few are capable of fully capturing the cancer phenotype. Currently, LFS mice carrying R172H and R270H (R175H and R273H in human) as well as humanized p53 knock-in (HUPKI) mice harboring R175H, R248W, R248Q and R273H are capable of modeling much of the broad cancer spectrum found in humans and highlight the importance of gain-of-function activity in p53 [30–32, 120, 121]. Unfortunately, another HUPKI mouse harboring G245S, one of the most commonly inherited familial p53 mutations, shows similar tumor onset and survival compared with the p53-null HUPK mice and fails to recapitulate its dominant-negative function in vivo [32]. Moreover, although children with germline RB1 mutations are likely to experience bilateral multifocal RB and increased risk of osteosarcoma [122], mice with a similar disruption of Rb1 do not develop either retinoblastoma or osteosarcoma [117–119]. RTS patients carrying RECQL4 mutations commonly develop osteosarcoma, but deletion of Recql4 in mice only conveys some RTS clinical features, such as skeletal abnormalities, but not osteosarcoma [62, 123]. These results highlight the limitations of using mice to model known genetic risk factors for bone malignancy and emphasize differences across species. This lack of osteosarcoma phenotype in most current mouse models suggests the importance of using other complementary models to study genetic predisposition to osteosarcoma.
Progress in precise gene-editing methodologies including Zinc finger nuclease (ZFNs), Transcription activator-like effector nucleases (TALENs) and clustered, regularly interspaced, short palindromic repeat/Cas9 (CRISPR/Cas9) will aid in providing an alternative means for introduction of a genetic disease trait into human ESCs (hESCs) and wild-type iPSCs, particularly when patient samples are difficult to acquire [124]. With these technologies, osteosarcoma-associated gene alterations (gene deletion, amplification, mutation and gene fusion) can be easily and arbitrarily introduced into hESCs and wild-type iPSCs to permit the study of their role in osteosarcomagenesis.
While researchers have found applications for PSCs in a broad range of human genetic diseases with either Mendelian or complex inheritance patterns [125], the application of PSCs to cancer research remains in its infancy. Still, the work of groups that have begun to apply iPSCs to phenocopy cancer, to explore pathological mechanisms, and to screen potential therapeutic compounds [126] highlights the potential of using human PSCs in medical research. These models of cancer pathogenesis overcome previous limitations related to patient sample availability or inapplicability of results from animal models or cell lines with inappropriate genetic backgrounds due to species differences. Modeling genetic disorders with an osteosarcoma predisposition has been successfully demonstrated using the LFS iPSC model [10]. We anticipate significant developments stemming from other hereditary PSC disease models in the coming years (see Outsatnding Questions and Box 7). A careful study of the diverse mechanisms of osteosarcomagenesis in these cell lines will help delineate the pathological mechanisms and reveal potential opportunities for treatment of osteosarcoma in both genetic and sporadic cases.
Box 7. Clinician’s Corner.
Patient-derived iPSCs offer substantial potential for clinical application. A clearer understanding of the stepwise genetic changes that occur upon osteosarcomagenesis and progression can be utilized to perform better and earlier diagnosis. In addition, PSCs offer a powerful platform for drug discovery and development, aiding in the discovery of targeted treatments for these genetic diseases.
The genome in osteosarcoma exhibits more alterations than almost all other cancers. Combining WGS and whole exon sequencing (WES) will help reveal the complex genomic evolution of osteosarcoma and point us towards the identity of osteosarcoma-initiating cells.
Patient-derived iPSC disease models can be applied for modeling disease development, elucidating pathological mechanisms, screening effective compounds to treat and/or prevent patient illness, and test drug toxicity. In the future, it may be possible to apply these osteosarcoma predisposition iPSC models in in vitro clinical trials to identify potential compounds targeting osteosarcoma. The panels of identified chemicals could be applied for the first-line screening and suggest personalized therapies for affected patients.
Trends box.
Osteosarcoma can be derived from undifferentiated/dedifferentiated MSCs and osteoblast-committed cells with differentiation defects.
Other than mutations, genomic rearrangements are also involved in osteosarcomagenesis, which cany be ignored by traditional mutation analysis.
Osteosarcoma-specific fusion genes can offers potential therapeutic targets for further osteosarcoma treatment.
Insights gained from osteosarcoma-prone diseases highlight numerous interesting concepts linked to cancer development, including differentiation control, tumor-associated immunosuppression and autophagy.
Several osteosarcoma-prone iPSC disease models have been established, including LF, RB, WS, and DBA. These systems provide a new platform for modeling and investigating the pathogenesis of osteosarcoma.
Outstanding Questions.
How do deficiencies in genes with ostensibly similar functions (e.g., RECQL4, BLM, and WRN) result in different patient phenotypes? Why do all of these phenotypes include osteosarcoma?
What is the relationship between these osteosarcoma-prone genetic diseases? Do these distinct genetic disorders share common genome/transcriptome/interactome profiles that may promote initiation and progression of osteosarcoma?
Can investigation of these osteosarcoma-prone genetic diseases aid in the development of new strategies to treat sporadic osteosarcoma?
Can differentiation defects constitute a trigger to initiate osteosarcoma development? What are the pathological mechanisms (e.g., epigenetic regulation and defect on autophagy) involved in osteogenesis defects?
Can patient-derived iPSCs comprehensively recapitulate human diseases in the absence of a physiological microenvironment (e.g. inside human bodies)?
Acknowledgments
We sincerely apologize to authors whose work we could not include due to space limitations. D.-F.L. is the CPRIT scholar in Cancer Research and supported by NIH Pathway to Independence Award R00 CA181496 and CPRIT Award RR160019. L.LW is supported by NIAMS AR059063, BCM IDDRC P30HD024064 and The Rolanette and Berdon Lawrence Bone Disease Program of Texas.
Glossary Box
- Epiphysis
bony tissue at the end of a long bone. Before bone growth completes, it is separated from the bone shaft by the growth plate cartilage. After that, it is connected with the bone shaft by the ossification of growth plate cartilage.
- Induced pluripotent stem cells (iPSCs)
By introducing defined factors (e.g., the "Yamanaka factors" OCT4, SOX2, KLF4 and c-MYC), fully-differentiated somatic cells can be reprogrammed into pluripotent stem cells and gain full differentiation abilities.
- Chromothripsis
Extensive chromosomal rearrangements that occur in one or a few chromosomes. This chromosomal patchwork pattern leads to genomic chaos.
- Whole genome sequencing (WGS)
The process of analyzing the complete DNA sequence of an organism. It provides comprehensive genetic information and can be used to identify all variations from a reference genome.
- Structural variations (SVs)
Variation in a DNA region >1kB arising from insertions, deletions, duplications, copy-number alterations, inversions and translocations that change the structure of the affected region.
- Copy number alterations (CNAs)
Some genes are duplicated or deleted in the genome. A difference of duplicated number in a repeated genome area defines its copy number alteration.
- Single nucleotide variations (SNVs)
Alterations in the DNA sequence comprising only a single nucleotide change.
- Kataegis
A large number of mutations located at a certain genomic positions rather than spread throughout the genome. Katagegis (Greek for thunderstorm), represents the nature of clustering of this mutational thunderstorm. Chromosomal rearrangements are also involved in these regions.
- Leiomyosarcoma-like tumors
tumors bearing histological features of leiomyosarcoma during in vivo tumor formation. Leiomyosarcoma is a soft-tissue sarcoma arising from smooth muscle cells.
- Epithelial-mesenchymal transition (EMT)
The transition of a cancer cell from an epithelial to a mesenchymal morphology, allowing for the movement of the cancer cell into lymph and blood vessels, thereby promoting metastasis. In order to accomplish this, several genes, such as E-cadherin, SNAIL and TWIST are alternatively regulated.
- Embryonic stem cells (ESCs)
Cells isolated from the inner cell mass of the blastocyst at the preimplantation stage and cultured. These cells can differentiate into all adult lineages (pluripotency) or proliferate indefinitely without differentiation (self-renewal), based on the environmental conditions.
- Nucleotide excision repair (NER)
A DNA repair mechanism that eliminates DNA lesions induced by ultraviolet light (UV).
- Base excision repair (BER)
A process that removes DNA base lesions induced by oxidation, deamination and alkylation.
- DNA double strand break repair (DSBR)
The process that repairs DNA with breaks in both strands.
- Homologous recombination (HR)
The exchange of DNA sequences between homologous DNA strands.
- Non-homologous end-joining (NHEJ)
The pathway to repair double strand breaks by direct ligation of two broken DNA strands. It is an error-prone process.
- Mitochondrial nucleoids
The complexes formed by mitochondrial DNA (mtDNA) and proteins within mitochondria.
- Holliday junction (HJ)
A DNA structure joined by four double-stranded DNA strands during homologous recombination. Named after Robin Holliday.
- Sister-chromatid exchange (SCE)
The breaking and rejoining of DNA sequences between two sister chromatids of one chromosome during DNA replication.
- PML nuclear bodies
Spherical structures found in the nuclear matrix and generally composed exclusively of proteins. They play important roles in transcription, apoptosis, and the DNA-damage response.
- Variegated translocation mosaicism
A cytogenetic characteristic of WS fibroblasts in which chromosomal rearrangements in cell lines isolated from an individual patient demonstrate a clonal effect.
- Fanconi Anemia (FA)
A genetic disease that causes bone marrow failure to produce new blood cells and increased risk of certain types of cancer.
- Autophagy
A normal destructive mechanism of the cell to clean up dysfunctional components and recycle usable materials.
- Autophagosome
A double-layer membranous structure forming during the process of autophagy. By fusing with the lysosome, the cell can clear unnecessary or dysfunctional components.
- Imprinted gene network (IGN)
A specific group of imprinted genes with correlated expression. These genes co-regulate each other’s expression during embryonic growth. The IGN may control a complex regulatory network in order to induce rapid but controlled developmental processes.
- Zinc finger nuclease (ZFNs)
This genome editing methodology works by introducing FokI restriction enzyme-conjugated zinc finger proteins. Two zinc finger proteins, each targeted to a specific strand of DNA in opposite directions, work together to define the targeting site. The FokI restriction enzyme domains, brought together by the zinc-finger domains, function only as a dimer, allowing sequence and orientation specificity to generate a double strand break and facilitate homologous recombination.
- Transcription activator-like effector nucleases (TALENs)
highly accurate genome editing methodology that combines the FokI restriction enzyme with transcription activator-like effectors (TALEs). TALEs are derived from Xanthomonas bacteria and are built from highly conserved 33–34 amino acid sequences, each of which can recognize a unique base pair. Like ZFNs, target recognition brings FokI to a specific location and permits induction of a double stranded break.
- Clustered, regularly interspaced, short palindromic repeat/Cas9 (CRISPR/Cas9)
genome editing methodology built from guide RNA(gRNA)-conjugated DNA nuclease or nickase. The CRISPR/Cas9 system borrows from the bacterial immune system that defends against foreign genetic elements to produce CRISPR RNA(crRNA) based on foreign RNA material. The CRISPR/Cas9 system is constituted by a guide gRNA for recognition, and Cas9 for cleavage. The CAS crRNA complex can target and cut DNA at an arbitrary site based on base-pairing to complementary RNA. Currently, this system is the most convenient methodology for genome editing, although the more accurate but limited paired CRISPR/Cas9 nickase system is also available to target genomic loci. Cas9 will not successfully bind and cleave target DNA regions unless they contain the 5′NGG3′ protospacer adjacent motifs (PAM) sequence.
- Calvaria
(p1, of calvarium). The upper portion of the skull comprised of the occipital, frontal and parietal bones that cover the cranial cavity containing the brain, excluding the jaw and facial regions.
References
- 1.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Chen X, et al. Recurrent somatic structural variations contribute to turnorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7:104–112. doi: 10.1016/j.celrep.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kansara M, et al. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 4.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorenz S, et al. Unscrambling the genomic chaos of osteosarcoma reveals extensive transcript fusion, recurrent rearrangements and frequent novel TP53 aberrations. Oncotarget. 2016;7:5273–5288. doi: 10.18632/oncotarget.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Downing JR, et al. The Pediatric Cancer Genome Project. Nat Genet. 2012;44:619–622. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen KS, et al. A novel TP53-KPNA3 translocation defines a de novo treatment-resistant clone in osteosarcoma. Cold Spring Harb Mol Case Stud. 2016;2:a000992. doi: 10.1101/mcs.a000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, et al. Recurrent LRP1-SNRNP25 and KCNMB4-CCND3 fusion genes promote tumor cell motility in human osteosarcoma. J Hematol Oncol. 2014;7:76. doi: 10.1186/s13045-014-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu J, et al. The histogenesis of Ewing sarcoma. Cancer Rep Rev. 2017;1:1–2. doi: 10.15761/CRR.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee DF, et al. Modeling familial cancer with induced pluripotent stem cells. Cell. 2015;161:240–254. doi: 10.1016/j.cell.2015.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez R, et al. Modeling sarcomagenesis using multipotent mesenchymal stem cells. Cell Res. 2012;22:62–77. doi: 10.1038/cr.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tataria M, et al. Absence of the p53 tumor suppressor gene promotes osteogenesis in mesenchymal stem cells. J Pediatr Surg. 2006;41:624–632. doi: 10.1016/j.jpedsurg.2005.12.001. discussion 624–632. [DOI] [PubMed] [Google Scholar]
- 13.Xiao W, et al. Mesenchymal stem cell transformation and sarcoma genesis. Clin Sarcoma Res. 2013;3:10. doi: 10.1186/2045-3329-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang JY, et al. Generation of Osteosarcomas from a Combination of Rb Silencing and c-Myc Overexpression in Human Mesenchymal Stem Cells. Stem Cells Transl Med. 2017;6:512–526. doi: 10.5966/sctm.2015-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubio R, et al. The differentiation stage of p53-Rb-deficient bone marrow mesenchymal stem cells imposes the phenotype of in vivo sarcoma development. Oncogene. 2013;32:4970–4980. doi: 10.1038/onc.2012.507. [DOI] [PubMed] [Google Scholar]
- 16.Thomas DM, et al. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J Cell Biol. 2004;167:925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haydon RC, et al. Cytoplasmic and/or nuclear accumulation of the beta-catenin protein is a frequent event in human osteosarcoma. Int J Cancer. 2002;102:338–342. doi: 10.1002/ijc.10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao J, et al. Notch activation as a driver of osteogenic sarcoma. Cancer Cell. 2014;26:390–401. doi: 10.1016/j.ccr.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quist T, et al. The impact of osteoblastic differentiation on osteosarcomagenesis in the mouse. Oncogene. 2015;34:4278–4284. doi: 10.1038/onc.2014.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sottnik JL, et al. Osteocytes serve as a progenitor cell of osteosarcoma. J Cell Biochem. 2014;115:1420–1429. doi: 10.1002/jcb.24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 22.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee DF, et al. Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell. 2012;11:179–194. doi: 10.1016/j.stem.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molchadsky A, et al. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis. 2010;31:1501–1508. doi: 10.1093/carcin/bgq101. [DOI] [PubMed] [Google Scholar]
- 25.Tang N, et al. Osteosarcoma development and stem cell differentiation. Clin Orthop Relat Res. 2008;466:2114–2130. doi: 10.1007/s11999-008-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, et al. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J Cell Biol. 2006;172:115–125. doi: 10.1083/jcb.200507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lengner CJ, et al. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J Cell Biol. 2006;172:909–921. doi: 10.1083/jcb.200508130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song R, et al. P53 suppresses cell proliferation, metastasis, and angiogenesis of osteosarcoma through inhibition of the PI3K/AKT/mTOR pathway. Int J Surg. 2015;20:80–87. doi: 10.1016/j.ijsu.2015.04.050. [DOI] [PubMed] [Google Scholar]
- 29.Moriarity BS, et al. A Sleeping Beauty forward genetic screen identifies new genes and pathways driving osteosarcoma development and metastasis. Nat Genet. 2015;47:615–624. doi: 10.1038/ng.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Hanel W, et al. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20:898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wijnhoven SW, et al. Mice expressing a mammary gland-specific R270H mutation in the p53 tumor suppressor gene mimic human breast cancer development. Cancer Res. 2005;65:8166–8173. doi: 10.1158/0008-5472.CAN-05-1650. [DOI] [PubMed] [Google Scholar]
- 34.Pfister NT, et al. Mutant p53 cooperates with the SWI/SNF chromatin remodeling complex to regulate VEGFR2 in breast cancer cells. Genes Dev. 2015;29:1298–1315. doi: 10.1101/gad.263202.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu J, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525:206–211. doi: 10.1038/nature15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freed-Pastor WA, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weissmueller S, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell. 2014;157:382–394. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grana X, et al. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene. 1998;17:3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- 39.Indovina P, et al. RB1 dual role in proliferation and apoptosis: cell fate control and implications for cancer therapy. Oncotarget. 2015;6:17873–17890. doi: 10.18632/oncotarget.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamasaki L, et al. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 41.Schaal C, et al. The Rb-E2F transcriptional regulatory pathway in tumor angiogenesis and metastasis. Adv Cancer Res. 2014;121:147–182. doi: 10.1016/B978-0-12-800249-0.00004-4. [DOI] [PubMed] [Google Scholar]
- 42.Sage J. The retinoblastoma tumor suppressor and stem cell biology. Genes Dev. 2012;26:1409–1420. doi: 10.1101/gad.193730.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fajas L, et al. The retinoblastoma-histone deacetylase 3 complex inhibits PPARgamma and adipocyte differentiation. Dev Cell. 2002;3:903–910. doi: 10.1016/s1534-5807(02)00360-x. [DOI] [PubMed] [Google Scholar]
- 44.Calo E, et al. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010;466:1110–1114. doi: 10.1038/nature09264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu L, et al. Aging in Rothmund-Thomson syndrome and related RECQL4 genetic disorders. Ageing Res Rev. 2017;33:30–35. doi: 10.1016/j.arr.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Sangrithi MN, et al. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 47.Matsuno K, et al. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kliszczak M, et al. Interaction of RECQ4 and MCM10 is important for efficient DNA replication origin firing in human cells. Oncotarget. 2015;6:40464–40479. doi: 10.18632/oncotarget.6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Im JS, et al. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc Natl Acad Sci U S A. 2009;106:15628–15632. doi: 10.1073/pnas.0908039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu X, et al. MCM10 mediates RECQ4 association with MCM2–7 helicase complex during DNA replication. EMBO J. 2009;28:3005–3014. doi: 10.1038/emboj.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lamm N, et al. Genomic Instability in Human Pluripotent Stem Cells Arises from Replicative Stress and Chromosome Condensation Defects. Cell Stem Cell. 2016;18:253–261. doi: 10.1016/j.stem.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 52.Fan W, Luo J. RecQ4 facilitates UV light-induced DNA damage repair through interaction with nucleotide excision repair factor xeroderma pigmentosum group A (XPA) J Biol Chem. 2008;283:29037–29044. doi: 10.1074/jbc.M801928200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schurman SH, et al. Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum Mol Genet. 2009;18:3470–3483. doi: 10.1093/hmg/ddp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shamanna RA, et al. RECQ helicase RECQL4 participates in non-homologous end joining and interacts with the Ku complex. Carcinogenesis. 2014;35:2415–2424. doi: 10.1093/carcin/bgu137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De S, et al. RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. J Cell Sci. 2012;125:2509–2522. doi: 10.1242/jcs.101501. [DOI] [PubMed] [Google Scholar]
- 56.Gupta S, et al. RECQL4 and p53 potentiate the activity of polymerase gamma and maintain the integrity of the human mitochondrial genome. Carcinogenesis. 2014;35:34–45. doi: 10.1093/carcin/bgt315. [DOI] [PubMed] [Google Scholar]
- 57.Lu H, et al. RECQL4 Promotes DNA End Resection in Repair of DNA Double-Strand Breaks. Cell Rep. 2016;16:161–173. doi: 10.1016/j.celrep.2016.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petkovic M, et al. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- 59.Mann MB, et al. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum Mol Genet. 2005;14:813–825. doi: 10.1093/hmg/ddi075. [DOI] [PubMed] [Google Scholar]
- 60.Lu L, et al. RECQ DNA helicases and osteosarcoma. Adv Exp Med Biol. 2014;804:129–145. doi: 10.1007/978-3-319-04843-7_7. [DOI] [PubMed] [Google Scholar]
- 61.Ichikawa K, et al. [Preparation of the gene targeted knockout mice for human premature aging diseases, Werner syndrome, and Rothmund-Thomson syndrome caused by the mutation of DNA helicases] Nihon Yakurigaku Zasshi. 2002;119:219–226. doi: 10.1254/fpj.119.219. [DOI] [PubMed] [Google Scholar]
- 62.Lu L, et al. RECQL4 Regulates p53 Function In Vivo During Skeletogenesis. J Bone Miner Res. 2015;30:1077–1089. doi: 10.1002/jbmr.2436. [DOI] [PubMed] [Google Scholar]
- 63.Ng AJ, et al. The DNA helicase recql4 is required for normal osteoblast expansion and osteosarcoma formation. PLoS Genet. 2015;11:e1005160. doi: 10.1371/journal.pgen.1005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang S, et al. The premature ageing syndrome protein, WRN, is a 3'-->5' exonuclease. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yokote K, et al. WRN Mutation Update: Mutation Spectrum, Patient Registries, and Translational Prospects. Hum Mutat. 2017;38:7–15. doi: 10.1002/humu.23128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palermo V, et al. CDK1 phosphorylates WRN at collapsed replication forks. Nat Commun. 2016;7:12880. doi: 10.1038/ncomms12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kehrli K, et al. Class I Histone Deacetylase HDAC1 and WRN RECQ Helicase Contribute Additively to Protect Replication Forks upon Hydroxyurea-induced Arrest. J Biol Chem. 2016;291:24487–24503. doi: 10.1074/jbc.M115.708594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lauper JM, et al. Spectrum and risk of neoplasia in Werner syndrome: a systematic review. PLoS One. 2013;8:e59709. doi: 10.1371/journal.pone.0059709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orlovetskie N, et al. Targeted inhibition of WRN helicase, replication stress and cancer. Biochim Biophys Acta. 2017;1867:42–48. doi: 10.1016/j.bbcan.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 70.Blander G, et al. Physical and functional interaction between p53 and the Werner's syndrome protein. J Biol Chem. 1999;274:29463–29469. doi: 10.1074/jbc.274.41.29463. [DOI] [PubMed] [Google Scholar]
- 71.Davis T, et al. Evaluating the Role of p38 MAPK in the Accelerated Cell Senescence of Werner Syndrome Fibroblasts. Pharmaceuticals (Basel) 2016:9. doi: 10.3390/ph9020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bagley MC, et al. Use of p38 MAPK Inhibitors for the Treatment of Werner Syndrome. Pharmaceuticals (Basel) 2010;3:1842–1872. doi: 10.3390/ph3061842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dallaire A, et al. Expression profile of Caenorhabditis elegans mutant for the Werner syndrome gene ortholog reveals the impact of vitamin C on development to increase life span. BMC Genomics. 2014;15:940. doi: 10.1186/1471-2164-15-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Massip L, et al. Vitamin C restores healthy aging in a mouse model for Werner syndrome. FASEB J. 2010;24:158–172. doi: 10.1096/fj.09-137133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y, et al. Vitamin C alleviates aging defects in a stem cell model for Werner syndrome. Protein Cell. 2016;7:478–488. doi: 10.1007/s13238-016-0278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aumailley L, et al. Impact of vitamin C on the cardiometabolic and inflammatory profiles of mice lacking a functional Werner syndrome protein helicase. Exp Gerontol. 2015;72:192–203. doi: 10.1016/j.exger.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 77.German J, et al. Syndrome-causing mutations of the BLM gene in persons in the Bloom's Syndrome Registry. Hum Mutat. 2007;28:743–753. doi: 10.1002/humu.20501. [DOI] [PubMed] [Google Scholar]
- 78.Killoran MP, Keck JL. Sit down, relax and unwind: structural insights into RecQ helicase mechanisms. Nucleic Acids Res. 2006;34:4098–4105. doi: 10.1093/nar/gkl538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu L, Hickson ID. The Bloom's syndrome helicase stimulates the activity of human topoisomerase IIIalpha. Nucleic Acids Res. 2002;30:4823–4829. doi: 10.1093/nar/gkf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 81.Pinto C, et al. Human DNA2 possesses a cryptic DNA unwinding activity that functionally integrates with BLM or WRN helicases. Elife. 2016:5. doi: 10.7554/eLife.18574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chan KL, et al. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rezazadeh S. On BLM helicase in recombination-mediated telomere maintenance. Mol Biol Rep. 2013;40:3049–3064. doi: 10.1007/s11033-012-2379-0. [DOI] [PubMed] [Google Scholar]
- 84.Wang XW, et al. Functional interaction of p53 and BLM DNA helicase in apoptosis. J Biol Chem. 2001;276:32948–32955. doi: 10.1074/jbc.M103298200. [DOI] [PubMed] [Google Scholar]
- 85.de Renty C, Ellis NA. Bloom's syndrome: Why not premature aging?: A comparison of the BLM and WRN helicases. Ageing Res Rev. 2017;33:36–51. doi: 10.1016/j.arr.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Panneerselvam J, et al. BLM promotes the activation of Fanconi Anemia signaling pathway. Oncotarget. 2016;7:32351–32361. doi: 10.18632/oncotarget.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Armistead J, Triggs-Raine B. Diverse diseases from a ubiquitous process: the ribosomopathy paradox. FEBS Lett. 2014;588:1491–1500. doi: 10.1016/j.febslet.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 88.Bortoluzzi S, et al. Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics. 2001;17:1152–1157. doi: 10.1093/bioinformatics/17.12.1152. [DOI] [PubMed] [Google Scholar]
- 89.Guimaraes JC, Zavolan M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 2016;17:236. doi: 10.1186/s13059-016-1104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cui D, et al. The ribosomal protein S26 regulates p53 activity in response to DNA damage. Oncogene. 2014;33:2225–2235. doi: 10.1038/onc.2013.170. [DOI] [PubMed] [Google Scholar]
- 91.Macias E, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Danilova N, et al. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 93.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Badhai J, et al. Ribosomal protein S19 and S24 insufficiency cause distinct cell cycle defects in Diamond-Blackfan anemia. Biochim Biophys Acta. 2009;1792:1036–1042. doi: 10.1016/j.bbadis.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fancello L, et al. The ribosomal protein gene RPL5 is a haploinsufficient tumor suppressor in multiple cancer types. Oncotarget. 2017 doi: 10.18632/oncotarget.14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gazda HT, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kondoh N, et al. Differential expression of S19 ribosomal protein, laminin-binding protein, and human lymphocyte antigen class I messenger RNAs associated with colon carcinoma progression and differentiation. Cancer Res. 1992;52:791–796. [PubMed] [Google Scholar]
- 98.Markiewski MM, et al. The Ribosomal Protein S19 Suppresses Antitumor Immune Responses via the Complement C5a Receptor 1. J Immunol. 2017 doi: 10.4049/jimmunol.1602057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moscat J, et al. p62 in Cancer: Signaling Adaptor Beyond Autophagy. Cell. 2016;167:606–609. doi: 10.1016/j.cell.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duran A, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Katsuragi Y, et al. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282:4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- 102.Linares JF, et al. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell. 2013;51:283–296. doi: 10.1016/j.molcel.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rea SL, et al. New insights into the role of sequestosome 1/p62 mutant proteins in the pathogenesis of Paget's disease of bone. Endocr Rev. 2013;34:501–524. doi: 10.1210/er.2012-1034. [DOI] [PubMed] [Google Scholar]
- 104.Chamoux E, et al. The p62 P392L mutation linked to Paget's disease induces activation of human osteoclasts. Mol Endocrinol. 2009;23:1668–1680. doi: 10.1210/me.2009-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 106.Shimamoto A, et al. Reprogramming suppresses premature senescence phenotypes of Werner syndrome cells and maintains chromosomal stability over long-term culture. PLoS One. 2014;9:e112900. doi: 10.1371/journal.pone.0112900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cheung HH, et al. Telomerase protects werner syndrome lineage-specific stem cells from premature aging. Stem Cell Reports. 2014;2:534–546. doi: 10.1016/j.stemcr.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Doulatov S, et al. Drug discovery for Diamond-Blackfan anemia using reprogrammed hematopoietic progenitors. Sci Transl Med. 2017:9. doi: 10.1126/scitranslmed.aah5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Garcon L, et al. Ribosomal and hematopoietic defects in induced pluripotent stem cells derived from Diamond Blackfan anemia patients. Blood. 2013;122:912–921. doi: 10.1182/blood-2013-01-478321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zeng S, et al. Generation of induced pluripotent stem cells (iPSCs) from a retinoblastoma patient carrying a c.2663G>A mutation in RB1 gene. Stem Cell Res. 2016;17:208–211. doi: 10.1016/j.scr.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 111.Duan S, et al. PTEN deficiency reprogrammes human neural stem cells towards a glioblastoma stem cell-like phenotype. Nat Commun. 2015;6:10068. doi: 10.1038/ncomms10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Funato K, et al. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 2014;346:1529–1533. doi: 10.1126/science.1253799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mulero-Navarro S, et al. Myeloid Dysregulation in a Human Induced Pluripotent Stem Cell Model of PTPN11-Associated Juvenile Myelomonocytic Leukemia. Cell Rep. 2015;13:504–515. doi: 10.1016/j.celrep.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ostler EL, et al. A model for the phenotypic presentation of Werner's syndrome. Exp Gerontol. 2002;37:285–292. doi: 10.1016/s0531-5565(01)00194-2. [DOI] [PubMed] [Google Scholar]
- 115.Deisenroth C, et al. The Evolution of the Ribosomal Protein-MDM2-p53 Pathway. Cold Spring Harb Perspect Med. 2016:6. doi: 10.1101/cshperspect.a026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gebhardt MC. Molecular biology of sarcomas. Orthop Clin North Am. 1996;27:421–429. [PubMed] [Google Scholar]
- 117.Clarke AR, et al. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 118.Jacks T, et al. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 119.Lee EY, et al. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 120.Liu DP, et al. A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene. 2010;29:949–956. doi: 10.1038/onc.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Song H, et al. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 122.Ottaviani G, Jaffe N. The etiology of osteosarcoma. Cancer Treat Res. 2009;152:15–32. doi: 10.1007/978-1-4419-0284-9_2. [DOI] [PubMed] [Google Scholar]
- 123.Hoki Y, et al. Growth retardation and skin abnormalities of the Recql4-deficient mouse. Hum Mol Genet. 2003;12:2293–2299. doi: 10.1093/hmg/ddg254. [DOI] [PubMed] [Google Scholar]
- 124.Hockemeyer D, Jaenisch R. Induced Pluripotent Stem Cells Meet Genome Editing. Cell Stem Cell. 2016;18:573–586. doi: 10.1016/j.stem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Grskovic M, et al. Induced pluripotent stem cells--opportunities for disease modelling and drug discovery. Nat Rev Drug Discov. 2011;10:915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- 126.Gingold J, et al. Modeling Cancer with Pluripotent Stem Cells. Trends Cancer. 2016;2:485–494. doi: 10.1016/j.trecan.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer, and other neoplasms. A familial syndrome? Ann Intern Med. 1969;71:747–752. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- 128.Li FP, et al. A cancer family syndrome in twenty-four kindreds. Cancer Res. 1988;48:5358–5362. [PubMed] [Google Scholar]
- 129.Malkin D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 130.Tinat J, et al. 2009 version of the Chompret criteria for Li Fraumeni syndrome. J Clin Oncol. 2009;27:e108–109. doi: 10.1200/JCO.2009.22.7967. author reply e110. [DOI] [PubMed] [Google Scholar]
- 131.Gonzalez KD, et al. Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol. 2009;27:1250–1256. doi: 10.1200/JCO.2008.16.6959. [DOI] [PubMed] [Google Scholar]
- 132.Dyer MA. Lessons from Retinoblastoma: Implications for Cancer, Development, Evolution, and Regenerative Medicine. Trends Mol Med. 2016;22:863–876. doi: 10.1016/j.molmed.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yun J, et al. Epidemiology and Rb1 gene of retinoblastoma. Int J Ophthalmol. 2011;4:103–109. doi: 10.3980/j.issn.2222-3959.2011.01.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Broaddus E, et al. Survival with retinoblastoma in the USA: 1975–2004. Br J Ophthalmol. 2009;93:24–21. doi: 10.1136/bjo.2008.143842. [DOI] [PubMed] [Google Scholar]
- 135.Yu CL, et al. Cause-specific mortality in long-term survivors of retinoblastoma. J Natl Cancer Inst. 2009;101:581–591. doi: 10.1093/jnci/djp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wong FL, et al. Cancer incidence after retinoblastoma. Radiation dose and sarcoma risk. JAMA. 1997;278:1262–1267. doi: 10.1001/jama.278.15.1262. [DOI] [PubMed] [Google Scholar]
- 137.Temming P, et al. Incidence of second cancers after radiotherapy and systemic chemotherapy in heritable retinoblastoma survivors: A report from the German reference center. Pediatr Blood Cancer. 2017;64:71–80. doi: 10.1002/pbc.26193. [DOI] [PubMed] [Google Scholar]
- 138.Toguchida J, et al. Preferential mutation of paternally derived RB gene as the initial event in sporadic osteosarcoma. Nature. 1989;338:156–158. doi: 10.1038/338156a0. [DOI] [PubMed] [Google Scholar]
- 139.Unni KK, Dahlin DC. Osteosarcoma: pathology and classification. Semin Roentgenol. 1989;24:143–152. doi: 10.1016/0037-198x(89)90010-2. [DOI] [PubMed] [Google Scholar]
- 140.Gutierrez GM, et al. Impaired bone development and increased mesenchymal progenitor cells in calvaria of RB1−/− mice. Proc Natl Acad Sci U S A. 2008;105:18402–18407. doi: 10.1073/pnas.0805925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang LL, et al. Absence of RECQL4 mutations in poikiloderma with neutropenia in Navajo and non-Navajo patients. Am J Med Genet A. 2003;118A:299–301. doi: 10.1002/ajmg.a.10057. [DOI] [PubMed] [Google Scholar]
- 142.Wang LL, et al. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J Natl Cancer Inst. 2003;95:669–674. doi: 10.1093/jnci/95.9.669. [DOI] [PubMed] [Google Scholar]
- 143.Oshima J, et al. Werner Syndrome. In: Pagon RA, et al., editors. GeneReviews(R) 1993. [Google Scholar]
- 144.Mehollin-Ray AR, et al. Radiographic abnormalities in Rothmund-Thomson syndrome and genotype-phenotype correlation with RECQL4 mutation status. AJR Am J Roentgenol. 2008;191:W62–66. doi: 10.2214/AJR.07.3619. [DOI] [PubMed] [Google Scholar]
- 145.Wang LL, et al. Clinical manifestations in a cohort of 41 Rothmund-Thomson syndrome patients. Am J Med Genet. 2001;102:11–17. doi: 10.1002/1096-8628(20010722)102:1<11::aid-ajmg1413>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 146.Kaariainen H, et al. RAPADILINO syndrome with radial and patellar aplasia/hypoplasia as main manifestations. Am J Med Genet. 1989;33:346–351. doi: 10.1002/ajmg.1320330312. [DOI] [PubMed] [Google Scholar]
- 147.Siitonen HA, et al. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum Mol Genet. 2003;12:2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- 148.Siitonen HA, et al. The mutation spectrum in RECQL4 diseases. Eur J Hum Genet. 2009;17:151–158. doi: 10.1038/ejhg.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Goto M, et al. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- 150.Bloom D. Congenital telangiectatic erythema resembling lupus erythematosus in dwarfs; probably a syndrome entity. AMA Am J Dis Child. 1954;88:754–758. [PubMed] [Google Scholar]
- 151.Ellis NA, et al. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 152.German J, et al. Bloom's syndrome. VI. The disorder in Israel and an estimation of the gene frequency in the Ashkenazim. Am J Hum Genet. 1977;29:553–562. [PMC free article] [PubMed] [Google Scholar]
- 153.Li L, et al. Carrier frequency of the Bloom syndrome blmAsh mutation in the Ashkenazi Jewish population. Mol Genet Metab. 1998;64:286–290. doi: 10.1006/mgme.1998.2733. [DOI] [PubMed] [Google Scholar]
- 154.Cunniff C, et al. Bloom's Syndrome: Clinical Spectrum, Molecular Pathogenesis, and Cancer Predisposition. Mol Syndromol. 2017;8:4–23. doi: 10.1159/000452082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.German J. Bloom's syndrome. XX. The first 100 cancers. Cancer Genet Cytogenet. 1997;93:100–106. doi: 10.1016/s0165-4608(96)00336-6. [DOI] [PubMed] [Google Scholar]
- 156.Savage SA, Mirabello L. Using epidemiology and genomics to understand osteosarcoma etiology. Sarcoma. 2011;2011:548151. doi: 10.1155/2011/548151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Karow JK, et al. The Bloom's syndrome gene product promotes branch migration of holliday junctions. Proc Natl Acad Sci U S A. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vlachos A, et al. Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol. 2008;142:859–876. doi: 10.1111/j.1365-2141.2008.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dianzani I, Loreni F. Diamond-Blackfan anemia: a ribosomal puzzle. Haematologica. 2008;93:1601–1604. doi: 10.3324/haematol.2008.000513. [DOI] [PubMed] [Google Scholar]
- 160.Rey MA, et al. Enhanced alternative splicing of the FLVCR1 gene in Diamond Blackfan anemia disrupts FLVCR1 expression and function that are critical for erythropoiesis. Haematologica. 2008;93:1617–1626. doi: 10.3324/haematol.13359. [DOI] [PubMed] [Google Scholar]
- 161.Ludwig LS, et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med. 2014;20:748–753. doi: 10.1038/nm.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vlachos A, et al. Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood. 2012;119:3815–3819. doi: 10.1182/blood-2011-08-375972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lipton JM, et al. Osteogenic sarcoma associated with Diamond-Blackfan anemia: a report from the Diamond-Blackfan Anemia Registry. J Pediatr Hematol Oncol. 2001;23:39–44. doi: 10.1097/00043426-200101000-00009. [DOI] [PubMed] [Google Scholar]
- 164.Hansen MF, et al. Osteosarcoma in Paget's disease of bone. J Bone Miner Res. 2006;21(Suppl 2):P58–63. doi: 10.1359/jbmr.06s211. [DOI] [PubMed] [Google Scholar]
- 165.Roodman GD, Windle JJ. Paget disease of bone. J Clin Invest. 2005;115:200–208. doi: 10.1172/JCI24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Numan MS, et al. Paget's disease of bone: an osteoimmunological disorder? Drug Des Devel Ther. 2015;9:4695–4707. doi: 10.2147/DDDT.S88845. [DOI] [PMC free article] [PubMed] [Google Scholar]