Abstract
The majority of Drosophila genes are expressed in a temperature-dependent manner, but the way in which small RNAs may contribute to this effect is completely unknown as we currently lack an idea of how small RNA transcriptomes change as a function of temperature. Applying high-throughput sequencing techniques complemented by quantitative real-time PCR experiments, we demonstrate that altered ambient temperature induces drastic but reversible changes in sequence composition and total abundance of both miRNA and piRNA populations. Further, mRNA sequencing reveals that the expression of miRNAs and their predicted target transcripts correlates inversely, suggesting that temperature-responsive miRNAs drive adaptation to different ambient temperatures on the transcriptome level. Finally, we demonstrate that shifts in temperature affect both primary and secondary piRNA pools, and the observed aberrations are consistent with altered expression levels of the involved Piwi-pathway factors. We further reason that enhanced ping–pong processing at 29°C is driven by dissolved RNA secondary structures at higher temperatures, uncovering target sites that are not accessible at low temperatures. Together, our results show that small RNAs are an important part of epigenetic regulatory mechanisms that ensure homeostasis and adaptation under fluctuating environmental conditions.
Keywords: small noncoding RNAs, gene expression, temperature adaptation, miRNA, piRNA, Drosophila ovaries
INTRODUCTION
Most homoio- and poikilothermal organisms are exposed to recurrently changing environmental conditions and evolution has realized manifold solutions that act to maintain physiological integrity, for example, along the day–night cycle or the change of seasons. Temperature is among the most important abiotic environmental parameters and affects all known physiological processes (Precht et al. 1973; Franks et al. 1990).
Prominent players in response to detrimental thermal stress are the evolutionary conserved transcription factor heat shock factor (HSF) and downstream acting heat shock proteins (HSP). Beyond HSF and HSP, several studies have demonstrated that effective adaptation to fluctuating ambient temperatures requires a physiological response in terms of altered gene expression in plant and animal species (Goldspink 1995; Sonna et al. 2002; Hannah et al. 2005; Voolstra et al. 2009; May et al. 2013; Chen et al. 2015). Temperature-sensitive structures such as DNA, RNA, proteins, and lipids are supposed to represent further layers in thermosensation within the physiological range. In fact, specialized histones were found to play a crucial role in in the perception of temperature (Kumar and Wigge 2010). However, despite these fascinating insights, our knowledge about the underlying mechanisms that trigger adaptation on the transcriptomic level is still far from being complete. Over the past decade, small noncoding (snc-) RNAs have emerged as important regulators of transcriptional and post-transcriptional gene regulation in virtually all eukaryotic organisms (Wilson and Doudna 2013). Linking temperature-dependent gene regulation with epigenetic mechanisms, i.e., the newly arising small RNA world, temperature-responsive micro (mi-) RNAs have been proposed as putative drivers of gene expression changes in Arabidopsis (May et al. 2013). However, in animals, these links remain very poorly understood and studies are still fragmentary (Bizuayehu et al. 2015).
In order to reveal putative connections between sncRNAs and temperature-dependent gene expression, we analyzed small RNA transcriptomes and global gene expression in Drosophila melanogaster flies kept at different temperatures. Applying high-throughput sequencing complemented by quantitative real-time PCR experiments, we scrutinized the expression levels of small noncoding RNAs in response to thermal gradients, and asked for changes of sequence composition and total abundance of both miRNA- and Piwi-interacting (pi-) RNA populations and the reversibility of putative changes. We further use mRNA-seq data to reveal if the expression of miRNAs and their predicted target transcripts correlates along these gradients, which could unveil the presence of regulatory circuits, triggered by temperature-responsive miRNAs that ensure epigenetic adaptation to different ambient temperatures.
RESULTS AND DISCUSSION
We chose Drosophila melanogaster as a model to study temperature-dependent sncRNA and gene expression in a poikilothermal taxon as they express all major classes of sncRNAs in the female germline. Two separate Drosophila populations were kept at 18°C and 29°C, respectively. After sampling ovaries for small RNA sequencing from 40 individuals per population (probes are referred to as “18°C #1” and “29°C #1” in the following, respectively), we switched the temperature for the remaining flies from 18°C to 29°C and from 29°C to 18°C, respectively (Fig. 1A). Then, we again sampled ovaries from 40 individuals per population (probes are referred to as 29°C #2 and 18°C #2 in the following, respectively). Total RNA was extracted from the obtained probes (two independent biological replicates per probe) and small RNAs were sequenced on an Illumina HiSeq 4000 platform. A detailed annotation of the obtained sequence data can be found in Supplemental Table S1.
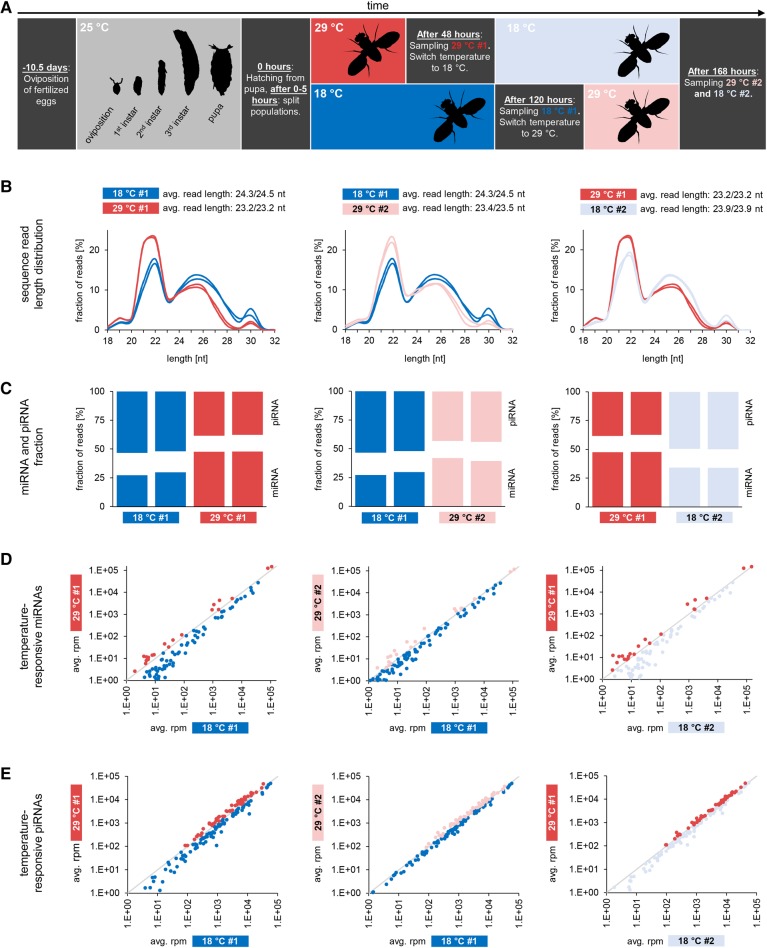
FIGURE 1.
Sequence length distribution and composition of small RNA transcriptomes from ovaries of flies kept at different temperatures. (A) Experimental setup. Colors used in this figure are used to denote different probes in all following figures (Figs. 2–4). (B) Comparison of sequence length distribution. Left plot compares sequence length distribution of probes obtained from two different fly populations, one kept at 18°C after hatching (blue, two replicates), the other kept at 29°C after hatching (red, two replicates). The middle plot compares sequence length distribution of probes obtained from one population, initially kept at 18°C (blue, two replicates) and then at 29°C (light red, two replicates). The right plot compares sequence length distribution of probes obtained from another population, initially kept at 29°C (red, two replicates) and then at 18°C (light blue, two replicates). (C) Abundance of miRNAs and piRNAs. Comparisons are as described above with two replicates per probe shown. miRNAs are more abundant under 29°C conditions whereas piRNAs are more abundant under 18°C conditions. (D) Temperature-responsive miRNAs. Comparisons are as described above. Dots in red (dark and light), as well as dots in blue (dark and light), refer to identical miRNAs across different probes in all three plots. (E) Temperature-responsive piRNAs, grouped according to the transposons from which they derive. Comparisons are as described above. Dots in red (dark and light), as well as dots in blue (dark and light), refer to identical miRNAs across different probes in all three plots.
Temperature-dependent changes of miRNA and piRNA abundance
Approximately 350 million prefiltered small RNA sequence reads (27–64 million per replicate) ranging from 18 to 44 nt in length were successfully mapped to the genome of Drosophila melanogaster (BDGP6). To get a first impression whether the expression of specific small RNA classes may change in a temperature-dependent manner, we compared the overall length profiles of mapped sequence reads. All probes show distinct peaks at 22 nt and 25–26 nt corresponding to the typical size of mi-/siRNAs and piRNAs, respectively. Remarkably, while replicates of one probe exhibit nearly identical length profiles, probes obtained from high-temperature housed flies clearly differ from those of low-temperature housed flies in that they exhibit a stronger 22-nt peak in relation to the 25–26-nt peak (Fig. 1B).
Indeed, sequence annotation of small RNAs for the different probes revealed that miRNAs are more abundant in flies that were kept at 29°C after hatching (29°C #1) compared to flies that were kept at 18°C after hatching (18°C #1). In line with this, miRNA abundance decreases upon a 29°C-to-18°C switch (29°C #1 versus 18°C #2) but increases upon an 18°C-to-29°C switch (18°C #1 versus 29°C #2, Fig. 1C; Supplemental Table S1).
To check our sequence data sets for the piRNA fraction, we focused on sequences that did not match any other class of noncoding RNA. These sequences show all hallmarks of genuine piRNAs, including length distribution, 5′-U bias, transposon content, and ping–pong signature, and correspond to piRNA sequences previously described based on immunoprecipitation experiments (Supplemental Fig. S1; Brennecke et al. 2007; Wang et al. 2015). Moreover, 7.5%–9% of putative piRNA reads (typical for genuine Drosophila piRNAs) map to known piRNA clusters (Supplemental Table S1). We will therefore bona fide refer to these sequences as piRNAs hereafter. In contrast to miRNAs, piRNAs are more abundant in flies that were kept at 18°C after hatching (18°C #1) compared to flies that were kept at 29°C after hatching (29°C #1). Accordingly, piRNA abundance decreases upon an 18°C-to-29°C switch (18°C #1 versus 29°C #2) and increases upon a 29°C-to-18°C switch (29°C #1 versus 18°C #2, Fig. 1C, detailed sequence annotation can be found in Supplemental Table S1).
The above results demonstrate that the different ratios of miRNA reads to piRNA reads in flies that were initially housed at 18°C and 29°C can be completely reversed by switching the temperature from 18°C to 29°C and from 29°C to 18°C, respectively.
Temperature-dependent expression changes of specific miRNAs and piRNAs
Considering the fact that high-temperature housing results in an increased abundance of miRNA reads compared to piRNA reads, we checked whether this is caused by a general up-regulation of miRNAs or whether alternatively distinct miRNAs behave differently upon changes in temperature. We therefore analyzed the expression of individual miRNAs and screened for expression changes that are consistent with changes in housing temperature. We found that 195 different miRNAs (including iso-miRs), accounting for 35%–40% of all miRNA reads in the different probes, exhibit temperature-dependent expression profiles (Fig. 1D). In order to focus on biological relevant expression changes, we rejected miRNAs with less than 1.1-fold expression change, finally retaining 129 temperature-responsive miRNAs (Supplemental Fig. S2A; Supplemental Table S2). Since small RNA-seq data can only give information on the relative abundance of a small RNA, we performed additional real-time (RT) quantitative PCR experiments on selected miRNAs which verify absolute expression changes, although the changes for miR-989-3p and miR-7-5p are rather moderate (Fig. 2). Interestingly, temperature-responsive miRNAs can be categorized into those that show higher abundance at hyper-optimal housing temperature (HOT = 29°C, n = 25, in the following referred to as HOT-miRNAs) and those that show higher abundance at near-optimal housing temperature (NOT = 18°C, n = 104, hereafter referred to as NOT-miRNAs; the prefixes HOT and NOT will be used analogously for piRNAs, genes, and transcripts hereafter). This heterogeneous picture suggests that temperature does not primarily affect elementary factors of the miRNA pathway, such as Dcr-1 or AGO1, and thus miRNA biogenesis per se, but more likely leads to a complex physiological–epigenetic response that is mirrored in a considerably modified miRNA expression profile.
FIGURE 2.
Quantitative real-time PCR on selected miRNAs. (A) Validation of miRNA-specific primers and primers for reference genes by PCR applied to a 2% agarose gel. Primers for miR-31a-5p and miR-988-5p were found to amplify the corresponding precursor, as shown by black arrows (these miRNAs were excluded from further analysis). (B) Expression of selected miRNAs at 18°C and 29°C normalized by five different nontemperature-dependent genes. Values for the condition with lower miRNA expression were set to one.
We next wanted to know if similar patterns could be observed for the piRNA fraction. Since piRNA pools are highly diverse, comprising tens of thousands of individual sequences, we grouped piRNAs according to their transposon targets and checked whether the expression of piRNAs that match to a specific transposon differs significantly when comparing high- and low-temperature housing. Indeed, expression of most transposon-derived piRNAs (57%–60% of transposon-derived piRNA reads) changes in a temperature-dependent manner (Fig. 1E), although the magnitude of change lags behind those observed for miRNAs (Supplemental Fig. S2B). As is the case for miRNAs, expression of distinct subfractions of piRNAs changes contrarily, with piRNA groups being consistently up-regulated or consistently down-regulated in high-temperature housed flies. Consequently, the composition of piRNA pools with respect to transposon target sequences changes in correlation with temperature. It is worth mentioning that reports on activation of Drosophila transposons at high temperature exist, though they yield no consistent overall picture (García Guerreiro 2012). In order to analyze a possible enrichment for specific transposon classes within the pool of HOT- or NOT-piRNAs, we grouped piRNAs matching to individual transposons according to major transposon classes into LINE, LTR, DNA, and other elements. However, we did not detect significant changes in either transposon composition or the total amount of transposon-derived piRNAs that correlate with changes in housing temperature when applying this general classification (Supplemental Fig. S1). Furthermore, the overall ratio of sense- and antisense-piRNAs and the amount of 1U/10A reads remain virtually unaffected (Supplemental Fig. S1). To check if altered expression of specific transposons can explain the modified piRNA target repertoire, we performed RNA-seq, probing ovaries of flies housed at 18°C and 29°C after hatching. Considering the entire transposon repertoire, we did not detect a correlation between transposon expression and the expression of corresponding piRNAs. While the expression of piRNAs derived from 115 transposons corresponds to expression changes of their source transposons, the opposite is true for 96 transposons (Pearson correlation coefficient, r = −0.058). Thus, we focused on transposons whose expression changes significantly in a temperature-dependent manner. We found that the expression of 27 individual transposons (significantly) depends on temperature, and our results confirm previous observations on heat-induced transcriptional activation of Copia elements (Strand and McDonald 1985). Moreover, piRNAs derived from transposons with higher expression at 29°C tend to exhibit higher abundance at 29°C, while the opposite is true for piRNAs derived from transposons with higher expression at 18°C (Supplemental Table S3). However, the difference between these groups does not reach significance (χ2 = 2.27, P = 0.13), and transposon/piRNA pairs that contradict the overall trend are present, implying that altered transposon activity cannot fully explain the altered piRNA target repertoire.
It is noteworthy that we also identified miRNAs and piRNAs whose expression depends on the developmental stage of the flies, rather than the ambient temperature (Supplemental Fig. S3; Supplemental Tables S4, S5). However, since the design of this study is clearly not optimized to analyze age-dependent miRNA- or piRNA expression changes, we prefer not to go further than having mentioned them, providing a reference for future studies.
Temperature-responsive miRNAs direct gene expression changes
A temperature-dependent expression of specific miRNAs has been previously reported, especially for organisms that depend on ambient temperature such as plants (Liu et al. 2008; Lee et al. 2010; May et al. 2013) or ectothermic animals (Bizuayehu et al. 2015). Further, miRNA expression changes upon shifts in temperature have also been reported for cultured human small airway epithelial cells (Potla et al. 2015). The existing studies also report on altered gene expression linked to differential miRNA expression. In view of the fact that the expression of most Drosophila genes changes in a temperature-dependent manner (Chen et al. 2015), we speculated that temperature-responsive miRNAs could represent the driving force for altered gene expression patterns in Drosophila ovaries. In order to clarify this issue, we compared global gene expression in ovaries of flies housed at 18°C and 29°C after hatching based on RNA-seq data. Searching for differentially expressed transcripts, we identified 4727 transcripts with significantly altered expression, comparing 18°C and 29°C conditions (Supplemental Table S6A–C). These transcripts correspond to 764 genes with higher expression under 29°C conditions (HOT-genes) and 926 genes with higher expression under 18°C conditions (NOT-genes, Fig. 3; Supplemental Table S6A–C). For 989 of the identified temperature-responsive genes, a linear temperature-dependent expression change has been previously reported based on whole-body gene expression data (Chen et al. 2015). Unsurprisingly, expression of the majority (n = 733) of these genes behaves similarly in ovaries compared to the whole body (Supplemental Fig. S4; Supplemental Table 6B).
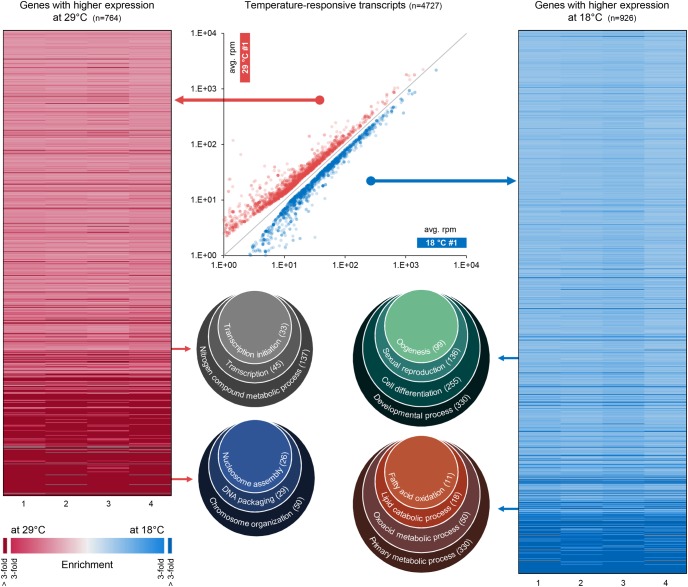
FIGURE 3.
Temperature-dependent gene expression changes. Differential expression analysis on the transcript level reveals 3671 transcripts with temperature-dependent expression (scatter plot in the middle). These transcripts correspond to 543 HOT-genes (left heatmap) and 496 NOT-genes (right heatmap). Each gene is represented with one line in one of the heatmaps. Heatmap columns numbered from 1 to 4 refer to four pairwise comparisons of mRNA expression in different probes and replicates (a and b): 1 = 18°C #1 a versus 29°C #1 a; 2 = 18°C #1 a versus 29°C #1 b; 3 = 18°C #1 b versus 29°C #1 a; 4 = 18°C #1 b versus 29°C #1 b. HOT-genes and NOT-genes are enriched for factors involved in different biological processes (Venn diagrams).
We performed Gene Ontology (GO) enrichment analysis for HOT and NOT genes separately and found that the two groups are enriched for genes involved in different biological processes (Fig. 3). HOT-genes are enriched for genes involved in transcription and chromatin organization, and down-regulation of these genes at 18°C might compensate for reduced chromatin accessibility at low temperatures (Chereji et al. 2016). On the other side, genes with higher expression at 18°C are enriched for genes involved in oogenesis and lipid metabolism, where the latter might reflect the need for increased energy release at low temperatures (Athenstaedt and Daum 2006; Arrese and Soulages 2010). Although the phenotypic consequences of this altered gene expression remain to be investigated, the overall picture suggests that the observed expression changes are part of an adaptive response to fluctuating temperature linking physiological and epigenetic adaptation.
We next turned to the question of in what way temperature-responsive miRNAs may contribute to the observed gene expression changes, and we predicted target sites for temperature-responsive miRNAs using miRanda (John et al. 2004). In a first attempt, we separately analyzed the expression of all transcripts that represent putative targets for a specific temperature-responsive miRNA. Applying this rather plain approach, we detected no significantly altered expression for the predicted target genes as a whole, comparing high- and low-temperature conditions. However, since transcripts typically comprise multiple putative miRNA binding sites, we next focused on the expression of those transcripts that are potentially targeted by either HOT- or NOT-miRNAs but not both. Indeed, we detected a significant (χ2 = 7.98, P = 0.0047) difference between these two groups of transcripts. Remarkably, transcripts targeted exclusively by NOT-miRNAs tend to exhibit higher expression at 29°C, while transcripts targeted exclusively by HOT-miRNAs tend to exhibit higher expression at 18°C (Fig. 4A). This fact is also clearly reflected by the summed up rpm and fpkm counts for transcripts of these two groups at different temperatures (Fig. 4B).
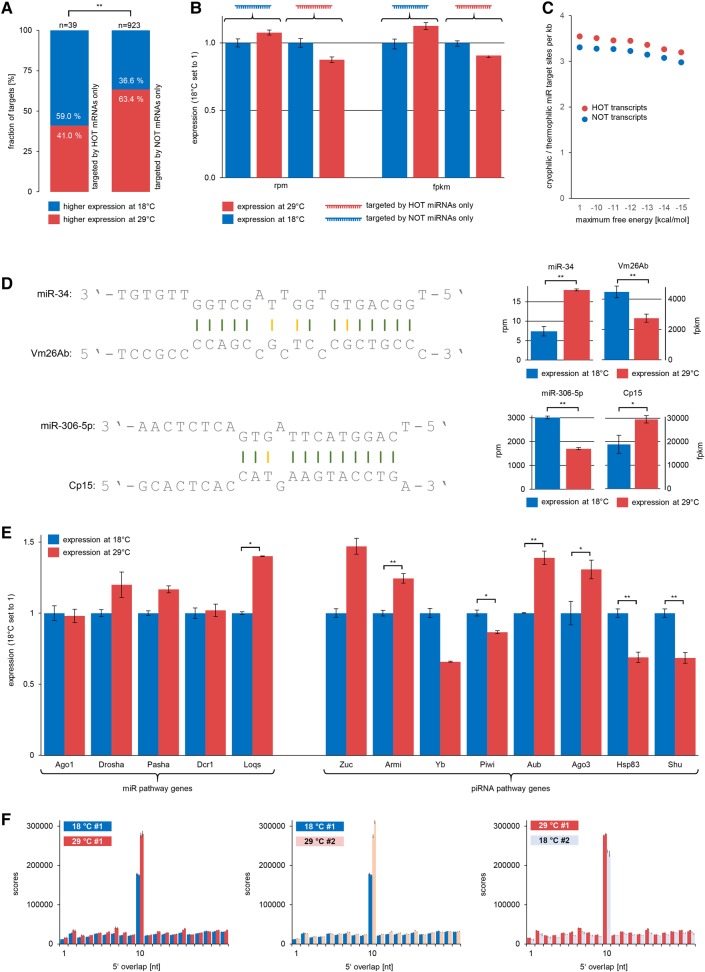
FIGURE 4.
Expression of temperature-responsive miRNAs and their targets; expression of small RNA pathway factors and ping–pong-amplification. Error bars refer to standard deviation across replicates. Asterisks refer to statistical significant P-values ([*] P ≤ 0.05; [**] P ≤ 0.01) according to edgeR differential expression analysis for genes or χ2 tests for miRNAs and comparison of gene groups in A. (A) Genes targeted only by HOT-miRNAs tend to be down-regulated at 29°C. Genes targeted only by NOT-miRNAs tend to be down-regulated at 18°C. (B) Total expression level of genes targeted only by HOT-miRNA compared to genes targeted only by NOT-miRNAs. Gene expression measured in rpm (left) and fpkm (right). Values for 18°C are set to one. (C) NOT-transcripts have more target sites for HOT-miRNAs (per kb transcript). HOT-transcripts have more target sites for NOT-miRNAs (per kb transcript). This is independent of the applied maximum free energy threshold for miRNA target site prediction. (D) miRNA target sites for Vm26Ab and Cp15. Gene expression and expression of targeting miRNAs correlates inversely. (E) Expression of small RNA pathway factors at different temperatures. (F) Average ping–pong signatures of 100 pseudoreplicate data sets from each probe, each comprising one million bootstrapped sequence reads. Sequences from 29°C probes consistently display a more pronounced ping–pong signature.
To gain further evidence for this inverse correlation, we focused on those genes whose expression changed significantly in a temperature-dependent manner and counted the number of predicted target sites for HOT- and NOT-miRNAs per kb transcript. In line with the above-mentioned finding, the ratio of NOT-to-HOT miRNA target sites is higher for genes that show higher expression at 29°C compared to genes that show higher expression at 18°C. Of note, this relation did not change when we modified the free energy thresholds for putative miRNA binding sites (Fig. 4C). To be more specific, we analyzed the expression and miRNA targeting of Vm26Ab (FBgn0003980) and Cp15 (FBgn0000355) in more detail (Fig. 4D). Vm26Ab is critical for eggshell formation and egg activation (Schüpbach and Wieschaus 1991; Tadros et al. 2003; Wu et al. 2010). Protein Cp15 is part of the oocyte-surrounding chorion (Waring and Mahowald 1979). Transcripts of both genes are among the most abundant temperature-responsive transcripts in the Drosophila ovary. Vm26Ab has a strong predicted target site for a HOT-miRNA, namely an isoform of miR-34 with more than 2.4-fold abundance at 29°C compared to 18°C. In line with this, the expression of Vm26Ab is reduced by 25% at 29°C compared to 18°C (Fig. 4D). Cp15 has binding sites for three different NOT-miRNAs, miR-306-5p and isoforms of miR-965 and miR-31b. As is the case for Vm26Ab and miR-34, expression levels of Cp15 and its targeting miRNAs correlate inversely (miR-965 and miR-31b not shown). Together, these results strongly suggest that temperature-dependent expression changes of specific genes that are critically involved in the reproduction process are at least in part directed by altered expression of temperature-responsive miRNAs. However, we must clearly stress that additional factors, whether on the transcriptional or post-transcriptional level, may determine the expression of a gene in dependency of the ambient temperature. The data at hand does not allow us to exactly quantify the amount of temperature-dependent regulation that is exerted by miRNAs compared to other factors. Since we further believe that this amount may differ significantly from gene to gene, disentangling these effects for genes of interest must be a subject for future studies.
Differential expression of essential small RNA pathway factors
Although gene expression on the protein level does not necessarily correlate with the expression of the according mRNA (Edfors et al. 2016), we did not want to miss the opportunity to use our RNA-seq data in order to get an impression regarding the expression patterns of essential miRNA and piRNA pathway factors to further substantiate the above-mentioned observations and inferences (Fig. 4E). Therefore, the following results should be considered with appropriate reservation.
The fact that specific miRNAs behave differently upon changes in ambient temperature leads us to speculate that temperature has no effect on the essential miRNA pathway factors, otherwise we would expect all miRNAs to be globally up- or down-regulated. Indeed, mRNA levels of Drosha, Pasha, DCR-1, and AGO1 do not show significant expression level differences. However, the mRNA level of LOQS, a double-stranded RNA binding protein that together with DCR-1 is involved in pre-miRNA processing (Förstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005), is significantly up-regulated under 29°C conditions. However, whether up-regulation of LOQS contributes to the dominance of miRNAs under 29°C conditions in consideration of unchanged DCR-1 and AGO1 expression remains disputable based on the available data. What is more, experiments using loqs knockout flies suggest that only a subset of miRNAs depend on LOQS (Liu et al. 2007), but importantly this subset does not correspond to those miRNAs that are more abundant under 29°C conditions, where LOQS shows higher expression levels.
Regarding piRNA pathway factors, we found that the mRNA level of Zucchini, the endonuclease that produces primary piRNAs (Ipsaro et al. 2012; Nishimasu et al. 2012), is tendentially up-regulated under 29°C conditions. Further, Armitage, a Piwi binding factor that is required for piRNA loading and correct localization of Piwi into Yb bodies (Saito et al. 2010), is significantly up-regulated on the mRNA level at 29°C. At first glance, this finding is surprising in light of reduced overall piRNA abundance in 29°C housed flies. However, we further noticed significant down-regulation of the mRNAs of Shutdown (Shu) and HSP83, two factors that are crucial for loading of piRNAs onto PIWI proteins and Shu-knockdown results in drastic reduction of both primary and secondary piRNAs (Preall et al. 2012). In addition, Yb, the eponymous factor for Yb bodies, which is crucial for piRNA loading onto Piwi (Saito et al. 2010), tends to be down-regulated on the mRNA level. Assuming that unloaded piRNAs are rapidly degraded, the expression levels of Yb, Shu, and HSP83 could represent a limiting factor for total piRNA population size. This would provide a plausible explanation for the reduced overall piRNA abundance in 29°C housed flies.
High-temperature housing results in increased ping–pong rates
Interestingly and in contrast to Yb, Shu, and HSP83, Aub and Ago3 are significantly up-regulated in 29°C housed flies. Aub and Ago3 represent the two PIWI proteins that participate in the ping–pong amplification loop and thus are essential for secondary piRNA biogenesis (Czech and Hannon 2016). Based on this observation, we checked whether we could detect a higher amount of ping–pong amplification under 29°C conditions, which would be mirrored by a more pronounced ping–pong signature. Since ping–pong scores increase nonlinearly with growing sequencing depth, we initially had to resample each sequence data set to simulate equal sequencing depth. We then calculated ping–pong scores for 100 pseudoreplicate data sets per original data set (18°C #1, 18°C #2, 29°C #1, 29°C #2, two replicates each, Fig. 4F). Remarkably and in line with the observation that Aub and Ago3 are up-regulated under 29°C conditions, piRNAs produce a more pronounced ping–pong signature when flies were housed at 29°C directly after hatching, compared to 18°C (Fig. 4F, left). Consistent with this, ping–pong signatures become more pronounced upon an 18°C-to-29°C switch (Fig. 4F, middle), while becoming less pronounced upon a 29°C-to-18°C switch (Fig. 4F, right). In addition to up-regulation of Aub and Ago3, higher ping–pong rates may also result from temperature-related effects on RNA secondary structure. RNA secondary structure is known to be a major determinant of si- and miRNAi efficiency (Shao et al. 2007; Kiryu et al. 2011). Further, findings from Tupaia belangeri suggest that piRNAs specifically target transposon transcripts at sites that do not form fold-back secondary structures since these sites remain accessible for ping–pong-mediated silencing (Rosenkranz et al. 2015a). To gain support for these hypotheses, we made the following consideration: Assuming higher expression of a few transposons to be responsible for the increased ping–pong rates, we would expect that the transposon composition of ping–pong piRNAs changes accordingly. Alternatively, if up-regulation of Aub and Ago3, and/or a lesser degree of RNA secondary structure, represent the main factors that drive ping–pong amplification, we would expect all transposon families to produce more ping–pong piRNAs, which would not result in an altered transposon composition. Therefore, we analyzed the transposon composition of ping–pong piRNAs at different temperatures and found no evidence that piRNAs from any transposon family are over- or underrepresented in the fraction of ping–pong piRNAs at 18°C or 29°C, respectively (Supplemental Table S7). We further checked ping–pong rates at loci with a minimum average ping–pong pair count of 1 (n = 37439), 5 (n = 14429), 10 (n = 9763), 50 (n = 3221), and 100 (n = 1810, based on bootstrap pseudoreplicate data sets) and found that more loci show increased ping–pong rates at 29°C compared to 18°C (Supplemental Table S7). Hence, we propose that heat-induced unfolding of secondary structures may expose formerly inaccessible target sites, thus explaining the more efficient ping–pong amplification which is in addition facilitated by concurrent up-regulation of Aub and Ago3. Assuming that a shift from 18°C to 29°C does not equally expose all putative piRNA targets having a smaller effect on very stable or already unfolded targets, altered RNA secondary structures could provide a plausible explanation for the slightly modified target repertoire of piRNAs (Fig. 1E; Supplemental Fig. S2B) which cannot be fully explained by altered transposon activity. In summary, these results demonstrate that higher temperatures induce an overall decrease of piRNAs, possibly in connection with down-regulation of Yb, Shu, and HSP83. At the same time, in line with up-regulation of Aub and Ago3 and the assumption that increasing temperature leads to more accessible RNA secondary structures, the fraction of secondary piRNAs within the total piRNA population increases, which results in more pronounced ping–pong signatures under 29°C conditions.
Concluding remarks
In Drosophila, temperature dependency of gene activity was reported as early as the 1910s, a time when the term gene still referred to not more than a conceptional idea (Driver 1931 and references therein). Focusing on the Drosophila female germline, this study provides evidence for a link between temperature-dependent gene expression and small RNA populations that considerably change along thermal gradients. We further argue that the overall pattern of differentially expressed miRNAs and genes mirrors important functional adaptations. Generally, the present study emphasizes the enormous influence that abiotic factors such as temperature can exert on regulatory networks involving epigenetic mechanisms mediated by sncRNA and mRNAs. This might be of special importance in considering assays that apply heat-shock induced conditional knockouts. We believe that it is worth keeping in mind these temperature-related phenomena when studying functional aspects of small RNA pathways in particular, considering the fact that these systems did not evolve under 25°C laboratory conditions. Otherwise, we might miss the most interesting regulatory mechanisms that exist and act to ensure homeostasis under fluctuating environmental conditions. Finally, considering the fact that the described adaptations on the transcriptomic level take place in the germline, we want to point out the possibility that phenotypic effects may also occur across generations, and may explain a number of phenomena observed in conjunction with different developmental and parental temperature (Huey et al. 1995; Zamudio et al. 1995; Gilchrist and Huey 2001).
MATERIALS AND METHODS
Fly culture
A Drosophila melanogaster population (strain: Oregon R, wild-type) was kept in culture tubes at 25°C for 48 h. We then removed the adult flies and allowed the eggs to continue developing at 25°C. Adult female flies were collected not later than 5 h after hatching of the first adult fly to ensure same age and virginity of the collected individuals. The collected female flies were split into two groups (∼150 individuals each) and transferred to new culture tubes that were kept at 18°C and 29°C, respectively. Since the speed of development is subject to temperature, we sampled ovaries from 29°C flies after 48 h and ovaries from 18°C flies after 120 h to ensure that both groups had reached identical developmental stages (Ashburner et al. 2011). For each probe, we pooled ovaries from 40 individuals. We refer to these probes as 18°C #1 and 29°C #1. After the first sampling procedure, we switched the temperature for the remaining flies from 18°C to 29°C and from 29°C to 18°C, respectively. A second sampling procedure was performed after a further 48 h for flies now kept at 29°C and after 120 h for flies now kept at 18°C; that is, flies in both populations reached the age of 168 h. We refer to the probes of the second sampling procedure as 18°C #2 and 29°C #2. The whole procedure was conducted twice in order to obtain two independent biological replicates (referred to as a and b) for each probe.
Small RNA-seq library construction and sequencing
Total RNA was extracted from pooled ovaries (probes 18°C #1, 18°C #2, 29°C #1, 29°C #2, two replicates each [a and b]) with TRI Reagent (Ambion) according to the manufacturer's instructions. rRNA was removed from total RNA extracts with Illumina's Ribo-Zero rRNA Removal Kit according to the manufacturer's instructions. rRNA-depleted RNA was then applied to a 10% urea-based denaturing polyacrylamide gel and run for 20 min at 1300 V and 60 W along with GeneRuler Ultra Low Range DNA Ladder (Thermo Scientific). The small RNA fraction ranging from ∼15 to 40 nt was excised from the gel and resolved in 30 µL nuclease-free water using Ultrafree-MC and Ultra-0.5 3K centrifugal devices (Amicon) according to the manufacturer's instructions. RNA concentration and integrity was tested with Agilent's 2100 Bioanalyzer system using the Agilent RNA 6000 Nano Kit. The NanoDrop 2000 system (Thermo Scientific) was used to quantify inorganic ions and test for polycarbonate contamination. Adaptor ligation was performed using Illumina's TruSeq Small RNA Sample Pre Kit according to the manufacturer's instructions. RNA with ligated adaptors was subject to reverse transcription using Invitrogen's Super Script II Kit. The resulting cDNA was PCR-amplified according to the following thermal cycling profile: 30″ at 98°C, [10″ at 98°C, 30″ at 60°C, 15″ at 72°C] × 11, 10′ at 72°C. DNA concentration of the libraries was tested with Agilent's 2100 Bioanalyzer system using the Agilent DNA 1000 Kit. Finally, eight libraries (four probes, two replicates each) were sequenced on one lane on an Illumina HiSeq 4000 system (single end, read length = 50). Sequence data were deposited at NCBI's Sequence Read Archive and can be accessed using the BioProject ID PRJNA350885.
Small RNA-seq data processing and annotation
Adapter-trimmed raw sequence reads ranging from 18 to 44 nt were collapsed to remove redundant sequence reads while storing the information on read counts using the program collapse. Subsequently, we removed low complexity reads using the program duster with default settings. Both programs are part of the NGS TOOLBOX (Rosenkranz et al. 2015b). We mapped the remaining sequences to the Drosophila melanogaster genome build BDGP6 with sRNAmapper requiring a perfect match for positions 1 to 18 and allowing one internal mismatch in the following part of the sequence. In addition, we allowed up to three nontemplate 3′ nucleotides in order to map sequences that were subject to post-transcriptional 3′ tailing (Roovers et al. 2015; Rosenkranz et al. 2015b). Successfully mapped sequences were annotated on the basis of available ncRNA data sets from the Ensembl database (release 85, Yates et al. 2016), miRBase (release 21, Kozomara and Griffiths-Jones 2014), GtRNAdb (Chan and Lowe 2009), and SILVA rRNA gene database (release 123.1, Quast et al. 2013). siRNAs were identified using Drosophila Ago2-IP data sets with SRA accession SRR060647 and SRR1042409 (Rozhkov et al. 2010; Abe et al. 2014). Further, the coordinates of mapped sequences were compared with RepeatMasker annotation to identify transposon-derived sequence reads and with known piRNA producing loci (Rosenkranz 2016) using in-house Perl scripts. Putative miRNA target sites were predicted with miRanda (John et al. 2004) applying a score threshold of 150 to reduce the number of false-positive predictions.
RNA-seq library construction and sequencing
Total RNA was extracted from pooled ovaries (probes 18°C #1, 29°C #1, two replicates each) with TRI Reagent (Ambion) according to the manufacturer's instructions. RNA concentration and purity of the probes was tested as described for small RNAs. mRNA libraries were prepared according to the TruSeq RNA Sample Prep Kit v2 (Illumina) and final DNA concentration of the libraries was tested as described above. Four libraries (two probes, two replicates each) were sequenced on an Illumina HiSeq 4000 system (paired end, read length = 100). Sequence data were deposited at NCBI's Sequence Read Archive and can be accessed using the BioProject ID PRJNA350885.
RNA-seq data processing and annotation
Gene expression analysis was performed using the miARma-seq analysis pipeline (Andrés-León et al. 2016). After initial quality control using FASTQC (v0.11.5), the paired-end sequence reads were mapped to the Drosophila melanogaster genome build BDGP6 using TopHat (v2.1.1, Kim et al. 2013) in connection with Bowtie 2 (v2.2.8, Langmead et al. 2009). Read counts were calculated using featureCounts (v1.5.0-p1) (Liao et al. 2014), differential expression analysis on the gene and transcript level was performed with edgeR (v3.12.1, Robinson et al. 2010). We used Gene Ontology database Released 2016-09-24 gene classifications to check whether temperature-responsive genes can be assigned to specific biological processes (Gene Ontology Consortium 2015). HOT- and NOT-genes were analyzed separately. The overrepresentation test was performed using a complete list of ∼11 thousand ovary-expressed genes as background and applying Bonferroni correction for multiple testing. The significance threshold was set to P ≤ 0.05. Transposon expression analysis was conducted with TEtranscripts (v1.5.1) (Jin et al. 2015) with standard parameters. Input files for TEtranscripts were created using STAR (v2.5.2b) (Dobin et al. 2013). In order to enhance the recovery of multimappers, the options -outFilterMultimapNmax and -winAnchorMultimapNmax were both set to 100. For conducting the differential expression analysis TEtranscripts makes use of DESeq (v1.22.1) (Anders and Huber 2010). By default, the DESeq standard normalization method is used and only features exhibiting a minimum read count of 1, a FDR ≤ 0.05 as well as a log2 fold-change are considered.
Identification of temperature-responsive miRNAs and piRNAs
In order to identify miRNAs whose expression depends on housing temperature, we screened our probes for miRNAs that showed consistently higher expression under high-temperature, or under low-temperature conditions. Instead of applying the general linear model (GLM) as we did for gene expression analysis, we conducted a more conservative customized procedure to make sure that a mi-/piRNA in question shows consistent up- or down-regulation in all pairwise comparisons, which is not the case when applying the GLM. Therefore, we defined HOT-miRNAs as miRNAs that are significantly enriched in the miRNA pools of flies kept at 29°C after hatching compared to miRNA pools of flies kept at 18°C after hatching. Further, the abundance of HOT-miRNAs had to increase significantly upon an 18°C-to-29°C switch, and had to decrease significantly upon a 29°C-to-18°C switch. Accordingly, NOT-miRNAs were defined as those miRNAs that behave contrarily. To test for compliance with each of these three requirements, we compared individual miRNA expression rates performing the following pairwise probe comparisons: 18°C #1 versus 29°C #1, 18°C #1 versus 29°C #2 and 29°C #1 versus 18°C #2. Since we used both biological replicates of each probe independently, 12 pairwise comparisons were performed in total, all of which had to reveal a consistent tendency. For each comparison, we calculated χ2 values from 2 × 2 contingency tables comprising sequence read counts of the miRNA in question and remainder miRNA read counts of the corresponding replicates. Since this test only considers miRNA reads, it is not affected by a possible global up- or down-regulation of other small RNA populations. The expression of a specific miRNA was designated as temperature-responsive if all 12 pairwise comparisons revealed coherent and significant (P ≤ 0.05) expression differences. P-values refer to the two-tailed probability of the χ2 distribution. A similar procedure was applied to piRNAs, with the difference that we did not examine single piRNA molecules, but rather groups of piRNAs that match to identical transposons.
RT-PCR of selected miRNAs
Total RNA from flies kept at 18°C or 29°C after hatching (probes 18°C #1 and 29°C #1, respectively) was split into two equal fractions (1 and 2). Fraction 1 was subject to reverse transcription with SuperScript IV (Invitrogen) using random hexamers according to the manufacturer's instructions. Fraction 2 was subject to poly(A) tailing using the A-Plus Poly(A) Polymerase Tailing Kit (Biozym) according to the manufacturer's instructions. Poly(A)-tailed RNA was then reversely transcribed with SuperScript IV using the RT-primer 5′-CGAATTCTAGAGCTCGAGGCAGGCGACATGT25VN-3′. We performed qPCR with a cDNA template obtained from fraction 1 (both probes) on reference genes whose expression does not depend on temperature according to our RNA-seq data (difference 18°C versus 29°C < 5%) and data from Chen et al. 2015 (adjusted P-value for differential expression >0.95: FBgn0032586, FBgn0026401, FBgn0036053, FBgn0035807, FBgn0010905). PCR targets were verified by Sanger sequencing. qPCR runs with a cDNA template obtained from fraction 2 (both probes) were conducted with miRNA-specific forward primers together with a universal reverse primer (5′-CGAATTCTAGAGCTCGAGGCAGG-3′). Prior to miRNA RT-PCR, we performed regular PCR on selected candidate miRNAs to exclude amplification of miRNA precursor molecules. Expression of miRNAs was normalized by expression of the reference genes. All experiments were performed using two independent biological replicates per condition and three technical replicates per RT-PCR. Primer sequences are available in Supplemental Table S8.
Cross-library comparison of ping–pong signatures
Analysis of ping–pong signatures based on detection of 5′ complementary sequence reads and sequencing depth critically impacts its result. However, score values for 5′ overlaps cannot be simply rescaled in an rpm-like fashion since this would not account for differences in the number of nonidentical sequences. To compare ping–pong signatures across data sets with different sequencing depth we performed a bootstrapping procedure focusing on reads that could not be annotated as any other small ncRNA, thus presumably representing the piRNA fraction. We built 100 pseudoreplicate data sets per original data set, each comprising one million sequence reads resulting in a comparable sequence diversity. Then, the ping–pong signature including a Z-score (Zhang et al. 2011) was calculated for each pseudoreplicate data set. Subsequently, we calculated the average score value and standard deviation per bp overlap from the 100 pseudoreplicate data sets to plot a comparable ping–pong signature for each original data set. Finally, we compared the average score for 10-bp overlap rather that the average Z-score, since the latter refers to the probability for ongoing ping–pong amplification (score for 10-bp overlap compared to the background) rather than its absolute magnitude.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Miguel Andrade from the Institute of Molecular Biology (IMB), Mainz, for helpful discussion and advice regarding miRNA target prediction. Further thanks go to Jean Wang from BGI for his obliging assistance in probe handling and NGS sequencing. Finally, we want to thank Julia Jehn and Julian Kiefer for their assistance in performing RT-PCR experiments. This project was supported by the Natural and Medical Sciences Research Center of the University Medicine Mainz (NMFZ) and the International PhD Programme coordinated by the Institute of Molecular Biology (IMB), Mainz, funded by the Boehringer Ingelheim Foundation.
Author contributions: I.F., C.B., and D.R. are responsible for the overall design of this study. I.F. performed all laboratory experiments. C.H. performed analysis of mRNA-seq data. I.F., L.W., D.G., and D.R. conducted computational analyses of sRNA sequence data including coding of individual Perl scripts. J.S. assisted in statistical analysis of gene expression. H.Z., I.F., L.W., D.G., and C.B. made valuable inputs during the preparation of the manuscript. D.R. wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.061119.117.
REFERENCES
- Abe M, Naqvi A, Hendriks GJ, Feltzin V, Zhu Y, Grigoriev A, Bonini NM. 2014. Impact of age-associated increase in 2′-O-methylation of miRNAs on aging and neurodegeneration in Drosophila. Genes Dev 28: 44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-León E, Núñez-Torres R, Rojas AM. 2016. miARma-Seq: a comprehensive tool for miRNA, mRNA and circRNA analysis. Sci Rep 6: 25749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Soulages JL. 2010. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol 55: 207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Golic KG, Hawley RS. 2011. Drosophila: a laboratory handbook, 2nd ed Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Athenstaedt K, Daum G. 2006. The life cycle of neutral lipids: synthesis, storage and degradation. Cell Mol Life Sci 63: 1355–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizuayehu TT, Johansen SD, Puvanendran V, Toften H, Babiak I. 2015. Temperature during early development has long-term effects on microRNA expression in Atlantic cod. BMC Genomics 16: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2009. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 37: D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Nolte V, Schlötterer C. 2015. Temperature-related reaction norms of gene expression: regulatory architecture and functional implications. Mol Biol Evol 32: 2393–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereji RV, Kan TW, Grudniewska MK, Romashchenko AV, Berezikov E, Zhimulev IF, Guryev V, Morozov AV, Moshkin YM. 2016. Genome-wide profiling of nucleosome sensitivity and chromatin accessibility in Drosophila melanogaster. Nucleic Acids Res 44: 1036–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. 2016. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci 41: 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver EC. 1931. Temperature and gene expression in Drosophila. J Exp Zool 59: 1–28. [Google Scholar]
- Edfors F, Danielsson F, Hallström BM, Käll L, Lundberg E, Pontén F, Forsström B, Uhlén M. 2016. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol Syst Biol 12: 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. 2005. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 3: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks F, Mathias SF, Hatley RH. 1990. Water, temperature and life. Philos Trans R Soc Lond B Biol Sci 326: 517–531. [DOI] [PubMed] [Google Scholar]
- García Guerreiro MP. 2012. What makes transposable elements move in the Drosophila genome? Heredity 108: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. 2015. Gene Ontology Consortium: going forward. Nucleic Acids Res 43: D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist GW, Huey RB. 2001. Parental and developmental temperature effects on the thermal dependence of fitness in Drosophila melanogaster. Evolution 55: 209–214. [DOI] [PubMed] [Google Scholar]
- Goldspink G. 1995. Adaptation of fish to different environmental temperature by qualitative and quantitative changes in gene expression. J Therm Biol 20: 167–174. [Google Scholar]
- Hannah MA, Heyer AG, Hincha DK. 2005. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet 1: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey RB, Wakefield T, Crill WD, Gilchrist GW. 1995. Within- and between-generation effects of temperature on early fecundity of Drosophila melanogaster. Heredity (Edinb) 74: 216–223. [DOI] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. 2012. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. 2005. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev 19: 1674–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Tam OH, Paniagua E, Hammell M. 2015. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31: 3593–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. 2004. Human MicroRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryu H, Terai G, Imamura O, Yoneyama H, Suzuki K, Asai K. 2011. A detailed investigation of accessibilities around target sites of siRNAs and miRNAs. Bioinformatics 27: 1788–1797. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. 2010. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Yoo SJ, Lee JH, Kim W, Yoo SK, Fitzgerald H, Carrington JC, Ahn JH. 2010. Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res 38: 3081–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Liu X, Park JK, Jiang F, Liu Y, McKearin D, Liu Q. 2007. Dicer-1, but not Loquacious, is critical for assembly of miRNA-induced silencing complexes. RNA 13: 2324–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. 2008. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14: 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, Liao W, Wu Y, Shuai B, McCombie WR, Zhang MQ, Liu QA. 2013. The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat Commun 4: 2145. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. 2012. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491: 284–287. [DOI] [PubMed] [Google Scholar]
- Potla R, Singh IS, Atamas SP, Hasday JD. 2015. Shifts in temperature within the physiologic range modify strand-specific expression of select human microRNAs. RNA 21: 1261–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preall JB, Czech B, Guzzardo PM, Muerdter F, Hannon GJ. 2012. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA 18: 1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Precht H, Christophersen J, Hensel H, Larcher W. 1973. Temperature and life. Springer, New York. [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers EF, Rosenkranz D, Mahdipour M, Han CT, He N, Chuva de Sousa Lopes SM, van der Westerlaken LAJ, Zischler H, Butter F, Roelen BAJ, et al. 2015. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep 10: 2069–2082. [DOI] [PubMed] [Google Scholar]
- Rosenkranz D. 2016. piRNA cluster database: a web resource for piRNA producing loci. Nucleic Acids Res 44: D223–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkranz D, Rudloff S, Bastuck K, Ketting RF, Zischler H. 2015a. Tupaia small RNAs provide insights into function and evolution of RNAi-based transposon defense in mammals. RNA 21: 911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkranz D, Han CT, Roovers EF, Zischler H, Ketting RF. 2015b. Piwi proteins and piRNAs in mammalian oocytes and early embryos: from sample to sequence. Genom Data 5: 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Aravin AA, Zelentsova ES, Schostak NG, Sachidanandam R, McCombie WR, Hannon GJ, Evgen'ev MB. 2010. Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. RNA 16: 1634–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC. 2005. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol 3: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E. 1991. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129: 1119–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Chan CY, Maliyekkel A, Lawrence CE, Roninson IB, Ding Y. 2007. Effect of target secondary structure on RNAi efficiency. RNA 13: 1631–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonna LA, Fujita J, Gaffin SL, Lilly CM. 2002. Invited review: effects of heat and cold stress on mammalian gene expression. J Appl Physiol (1985) 92: 1725–1742. [DOI] [PubMed] [Google Scholar]
- Strand DJ, McDonald JF. 1985. Copia is transcriptionally responsive to environmental stress. Nucleic Acids Res 13: 4401–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Houston SA, Bashirullah A, Cooperstock RL, Semotok JL, Reed BH, Lipshitz HD. 2003. Regulation of maternal transcript destabilization during egg activation in Drosophila. Genetics 164: 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voolstra CR, Schnetzer J, Peshkin L, Randall CJ, Szmant AM, Medina M. 2009. Effects of temperature on gene expression in embryos of the coral Montastraea faveolata. BMC Genomics 10: 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Han BW, Tipping C, Ge DT, Zhang Z, Weng Z, Zamore PD. 2015. Slicing and binding by Ago3 or Aub trigger Piwi-bound piRNA production by distinct mechanisms. Mol Cell 59: 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring GL, Mahowald AP. 1979. Identification and time of synthesis of chorion proteins in Drosophila melanogaster. Cell 16: 599–607. [DOI] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. 2013. Molecular mechanisms of RNA interference. Annu Rev Biophys 42: 217–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Manogaran AL, Beauchamp JM, Waring GL. 2010. Drosophila vitelline membrane assembly: a critical role for an evolutionarily conserved cysteine in the “VM domain” of sV23. Dev Biol 347: 360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, et al. 2016. Ensembl 2016. Nucleic Acids Res 44: D710–D716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio KR, Huey RB, Crill WD. 1995. Bigger isn't always better: body size, developmental and parental temperature and male territorial success in Drosophila melanogaster. Anim Behav 49: 671–677. [Google Scholar]
- Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, Zamore PD. 2011. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol Cell 44: 572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.