Abstract
Phytoplasmas are cell wall-less plant pathogenic bacteria responsible for major crop losses throughout the world. In grapevine they cause grapevine yellows, a detrimental disease associated with a variety of symptoms. The high economic impact of this disease has sparked considerable interest among researchers to understand molecular mechanisms related to pathogenesis. Increasing evidence exist that a class of small non-coding endogenous RNAs, known as microRNAs (miRNAs), play an important role in post-transcriptional gene regulation during plant development and responses to biotic and abiotic stresses. Thus, we aimed to dissect complex high-throughput small RNA sequencing data for the genome-wide identification of known and novel differentially expressed miRNAs, using read libraries constructed from healthy and phytoplasma-infected Chardonnay leaf material. Furthermore, we utilised computational resources to predict putative miRNA targets to explore the involvement of possible pathogen response pathways. We identified multiple known miRNA sequence variants (isomiRs), likely generated through post-transcriptional modifications. Sequences of 13 known, canonical miRNAs were shown to be differentially expressed. A total of 175 novel miRNA precursor sequences, each derived from a unique genomic location, were predicted, of which 23 were differentially expressed. A homology search revealed that some of these novel miRNAs shared high sequence similarity with conserved miRNAs from other plant species, as well as known grapevine miRNAs. The relative expression of randomly selected known and novel miRNAs was determined with real-time RT-qPCR analysis, thereby validating the trend of expression seen in the normalised small RNA sequencing read count data. Among the putative miRNA targets, we identified genes involved in plant morphology, hormone signalling, nutrient homeostasis, as well as plant stress. Our results may assist in understanding the role that miRNA pathways play during plant pathogenesis, and may be crucial in understanding disease symptom development in aster yellows phytoplasma-infected grapevines.
Introduction
Phytoplasmas are known to infect hundreds of plant species worldwide and are responsible for devastating yield losses of many economically important crops, fruit trees, and ornamental plants [1]. They are obligate cell wall-less bacterial pathogens (class Mollicutes), and rely on plants and homopterous phloem-sucking insects for biological dispersal. In plants, they are mainly restricted to the phloem tissue where they can move and multiply through the sieve tube elements [2].
The aster yellows (AY) phytoplasma group (16SrI, subgroup A and B) represents the most diverse and widespread phytoplasma group and is also known as ‘Candidatus Phytoplasma asteris’ [3]. AY phytoplasma-infection can cause a severe disease in grapevine (Vitis vinifera L.), known as grapevine yellows (GY). Phytoplasma-like symptoms have been observed in South African vineyards since 2006, and were later shown to be caused by AY phytoplasma (16SrI-B) [4]. Transmission experiments conducted on vineyards in the vicinity of Vredendal (Western Cape) suggested that Mgenia fuscovaria (Hemiptera: Cicadellidae) is a vector of AY phytoplasma in South Africa [5]. GY disease incidence in the same region was monitored for different cultivars (Chenin blanc, Shiraz, Chardonnay, Cabernet Franc, Sauvignon blanc, Pinotage and Colombar), and revealed that Chardonnay is especially susceptible, based on a GY increase from 0.5% to 7.5% in two years in a single vineyard [6]. Typical symptoms caused by GY disease include discolouration and necrosis of leaf veins and laminae, downward curling of leaves, abnormal leaf shape and size, incomplete lignification, stunting and necrosis of shoots, flower abortion and berry withering. These symptoms eventually lead to reduced plant vitality and fruit yield that may hold devastating consequences for the wine and table grape industries [1,7]. Currently, the only available control strategies include early eradication of infected crops, early eradication of infected source plants (weed control), and chemical control of vectors through regular insecticide treatments [8].
V. vinifera is one of the most important fruit and/or beverage crops in the world and, like all land plants, grapevines have to develop various mechanisms at a physiological and molecular level in order to cope with their ever-changing environment. Significant progress has been made to understand plant-pathogen interactions and the multiple gene regulatory mechanisms they invoke during plant defence responses. The recent successful, axenic cultivation of phytoplasmas [9] will allow direct in planta investigation of molecular interactions postulated to exist between phytoplasmas and their plant and insect vectors. In addition, high-throughput transcriptome analysis of next-generation sequencing (NGS) and microarray data, as well as proteomics, have served as valuable approaches for gaining new insights into physiological, biochemical and molecular mechanisms underlying phytoplasma disease symptom development in grapevine and other plant species [10–16].
Increasing evidence has shown that a class of small non-coding endogenous RNAs known as microRNAs (miRNAs), play a major role in post-transcriptional gene regulation during plant development and plant responses to biotic and abiotic stresses [17,18]. Mature miRNAs are typically 19 to 24 nt in length and originate from miRNA (MIR) genes that are transcribed by RNA Polymerase II. These transcripts, known as primary miRNAs (pri-miRNA), form imperfect fold-back hairpins that are cleaved by RNase III-like Dicer 1 (DCL1) to produce miRNA precursors (pre-miRNA). Each pre-miRNA contains one or more short intermediate complementary miRNA/miRNA* duplexes. These duplexes are then cleaved by DCL1 from the stem region and processed inside the nucleus to be exported to the cytoplasm where the leading miRNA is incorporated into the RNA-induced silencing complex (RISC). When associated with the RISC, guided binding of the miRNA to its complementary target mRNA(s) or non-coding trans-acting siRNA (TAS) transcript(s) occurs. This facilitates either translational inhibition or degradation of target mRNA(s), or slicing of TAS transcripts that lead to generation of trans-acting siRNAs (tasiRNAs). Target degradation occurs through endonucleolytic cleavage by the RISC core protein ARGONAUTE 1 (AGO1) [19–21].
It has been suggested that the miRNA pathway contributes to pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), which refers to a basal defence response upon recognition of certain pathogenic elements, such as flagellin [22]. The bacterial PAMP peptide flg22 causes induced expression of the Arabidopsis miR393, which was the first miRNA identified to play a role in plant PTI. Overexpression of miR393 caused down-regulation of auxin receptor mRNAs, including transport inhibitor response 1 (TIR1), through degradation, which caused increased resistance to virulent Pseudomonas syringae pv. tomato (Pst) DC3000 [23].
The availability of two draft V. vinifera cv. ‘Pinot noir’ genome sequences obtained from NGS projects [24,25] has enabled rapid discovery of miRNAs that further supports efforts to explore small RNA (sRNA)-based regulatory networks in grapevine. The use of computational analyses of high-throughput sequencing and microarray data, followed by experimental validation, have been used to identify highly conserved miRNAs, some of which play important roles in grapevine development [26,27]. To date, 186 mature grapevine miRNA sequences from 47 different miRNA families have been deposited in miRBase v21 [28].
This study is the first to utilise a bioinformatics pipeline to dissect complex high-throughput sRNA sequencing (sRNA-seq) data in order to identify miRNAs that are differentially expressed in V. vinifera cv. ‘Chardonnay’ in response to AY phytoplasma-infection. Furthermore, we used computational resources for the in silico prediction and annotation of putative miRNA targets to explore the involvement of possible pathogen response pathways. Understanding sRNA-mediated gene regulation is crucial to expanding our knowledge of gene regulatory pathways involved in different stress-regulated physiological processes. Our results provide insight into miRNA-mediated pathogenesis in V. vinifera and may shed light on disease control strategies for molecular breeding in the future.
Materials and methods
Plant material
We visually selected and tagged 50 symptomatic and 50 asymptomatic V. vinifera cv. ‘Chardonnay’ plants in a 7-year-old vineyard in the Olifants River Valley (Western Cape) (Fig 1). The vineyard was part of a high disease incidence area mapped by the Agricultural Product Inspection Services (APIS) of the Department of Agriculture, Forestry and Fisheries (DAFF). Permission was granted by the owner to conduct the study on his farm, Daltana. During the peak summer season, whole leaf material, including the blade and petiole, were collected from each plant, immediately flash frozen in liquid nitrogen, transported on dry ice and stored at -80°C until use. RNA was extracted using a modified CTAB method [29], while genomic DNA was extracted using a NucleoSpin® Plant II kit (Macherey-Nagel; Düren, Germany). Phytoplasma infection was confirmed by a nested-PCR procedure, specifically amplifying a region of the phytoplasma 16S rDNA. The first PCR round was performed using a universal primer pair R16mF2/mR1, followed by a second PCR with the R16F2n/R2 primer pair [30]. Afterwards samples were screened for the most prevalent grapevine viruses, including Grapevine leafroll-associated virus 3 (GLRaV-3), Grapevine virus A (GVA), Grapevine virus E (GVE), and Grapevine rupestris stempitting-associated virus (GRSPaV), using two-step RT-PCR assays. Primer sequences for virus screening were obtained from previous publications (S1 File). Results from these diagnostics were used to select material, free from these viruses, from three AY phytoplasma-infected, and three healthy plants for further experiments.
Fig 1.
Vitis vinifera cv. ‘Chardonnay’ with asymptomatic leaves (A), and leaves showing typical aster yellows (AY) disease symptoms (B).
Total RNA extraction and sRNA-seq
Large-scale RNA extractions were carried out on one gram of plant material for each of the six experimental plants using PureLink® Plant RNA Reagent (Thermo Fisher Scientific, Waltham, Massachusetts, United States), according to the manufacturer’s protocol, with an additional phenol-chloroform extraction step when further purification was necessary. Total RNA was quantified on a NanoDrop ND-1000, while RNA integrity was assessed using a Plant RNA Nano Assay using an Agilent 2100 Bioanalyzer. Ten micrograms of total RNA from each plant were sent to Fasteris SA (Plan-les-Ouates, Switzerland) for sRNA-seq. The six sRNA libraries were constructed using the TruSeq® Small RNA Library Prep Kit protocol (Illumina, San Diego, California, USA), followed by sRNA-seq on an Illumina HiSeq2000 platform (https://support.illumina.com).
sRNA bioinformatic analysis
After sRNA-seq, high-quality, adapter-trimmed sequence data was received from the service provider in Illumina-fastq format. FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used as a tool to visualise different quality control measurements. In order to confirm RT-PCR results of the virus screening, we produced de novo assemblies with the 18 to 26 nt sRNA reads of each sample, using Velvet v1.1 [31]. The resulting contigs were compared against the NCBI database using nucleotide BLAST [32].
The unique (non-redundant) 18 to 26 nt sequences with accompanying copy numbers, across all six libraries (representing the six biological samples), were submitted to miRanalyzer [33] (http://bioinfo5.ugr.es/miRanalyzer/miRanalyzer.php) for known miRNA analysis, allowing one mismatch. All reads that mapped to other non-coding RNAs (ncRNAs) in RFam (http://www.sanger.ac.uk/science/tools/rfam) and RepBase (http://www.girinst.org/repbase/) were removed, and the remaining reads were mapped against the canonical grapevine miRNA (vvi-miRNA) sequences deposited in miRBase v21. Mapped read counts for libraries obtained from the phytoplasma-infected group were compared to those from the healthy (control) group using the DESeq v2 package for differential expression analysis [34] (http://bioconductor.org/packages/release/bioc/html/DESeq.html).
For novel miRNA predictions, sRNA library files of the 18 to 26 nt reads, from all six libraries, were grouped into a single file that served as input for sRNAbench v0.9 [35] (http://bioinfo5.ugr.es/srnatoolbox/srnabench), and Shortstack v0.4.1 [36], using the default parameters of the respective packages. sRNAbench was also used for the discovery of sequence variants of known miRNAs, also known as miRNA isoforms (isomiRs). The V. vinifera (cv. ‘Pinot noir’; PN40024) 12x coverage genome assembly (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/) served as the reference sequence to which the sRNA reads were mapped [24]. Importantly, primary criteria described by Meyers et al. [37] for duplex-forming precursors (pre-miRNAs) are used by both programs. These include that (1) the miRNA and miRNA* are derived from opposite arms within the stem region to form a duplex with two 3’-nucleotide overhangs; (2) extensive base-pairing exist between the miRNA and the other arm of the hairpin, which includes the miRNA*; and (3) asymmetric bulges are minimal in size and frequency, especially within the miRNA/miRNA* duplex.
The Unified Nucleic Acid Folding (UNAFold) software was used to calculate the minimum folding free energy (MFE; ΔG) of novel pre-miRNA sequences [38] (http://mfold.rna.albany.edu/). In an effort to find more comprehensive evidence that miRNAs differ from other RNAs, Zhang et al. [39] described a statistical method incorporating pre-miRNA folding free energies, base pairing, nucleotide composition, and other characteristics. This method was defined by two criteria known as the adjusted minimum folding free energy (AMFE) and the minimal folding free energy index (MFEI). The AMFE and MFEI were calculated using the following equations:
Precursor sequences were analysed in RNAfold to view their stem-loop secondary structures [40]. Novel mature miRNA sequences were compared against the miRBase v21 database using BLASTn v2.2.29+ [32,41] (http://www.ncbi.nlm.nih.gov/books/NBK1763/) for the identification of miRNA homologs. Only the top BLAST results, with an identity of ≥90%, zero gaps and not more than two mismatches (over a seed region of 18 nt), were regarded as homologs. For each resulting BLAST hit, we compared the associated precursor sequence against miRBase with the miRBase BLASTn tool, using less stringent parameters, to identify homologous pre-miRNA sequences.
The number of sRNA reads that aligned to novel mature miRNA sequences present in all six libraries were obtained with Bowtie v1.0.1 [42] and a customised shell script. The resulting count data were analysed in DESeq v2 to obtain differentially expressed novel miRNAs. Only log2-fold changes with an adjusted p-value of ≤ 0.05 were considered significant.
Validation of miRNA expression by real-time RT-qPCR
Stem-loop reverse transcription quantitative PCR (RT-qPCR) assays were performed according to the methods of Chen et al. [43] to validate the DESeq differential expression results. High-quality total RNA was prepared as described above. For each miRNA a 20 μl reverse transcription reaction was prepared containing 100 U of Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA), 20 U of RiboLock RNase inhibitor (Thermo Scientific, Waltham, Massachusetts, United States), 4 μl first-strand buffer (5x), 5 mM DTT, 500 nM dNTPs and 1 μl miRNA-specific stem-loop RT primer (10 μM) and 1.2 μg total RNA. Cycling conditions were as follows: 30 min at 16°C, 60 cycles at 30°C for 30 s, 42°C for 30 s, and 50°C for 1 s, heat inactivation for 5 min at 85°C, and cooling at 4°C. qPCR was performed using the Universal ProbeLibrary (UPL) probe assay with UPL probe #21 (Roche Diagnostics, Basel, Switzerland). Each 10 μl reaction mixture was prepared in triplicate and contained 1 μl cDNA, 5 μl FastStart TaqMan® Probe Master (2x) (Roche Diagnostics, Basel, Switzerland), 0.5 μl miRNA-specific forward primer (10 μM), 0.5 μl universal reverse primer (10 μM), 0.1 μl UPL probe (10 μM), and nuclease-free water. A control reaction, without cDNA template, was included for each miRNA. Based on previous results from geNorm analysis (qBasePLUS v2.0, Biogazelle, Ghent, Belgium) [44], miR167a was chosen as internal control to normalise miRNA expression levels (data not shown). PCR amplification was performed in an Applied Biosystems 7900HT Fast Real-Time PCR System, in which the baseline and threshold cycles (Ct) were automatically determined with SDS v2.3 software. Cycling conditions were as follows: 95°C for 5 min, 45 cycles at 95°C for 10 s and 60°C for 1 min. Relative miRNA expression analysis was performed using qBasePLUS v2.0 software (Biogazelle, Ghent, Belgium).
miRNA target prediction and functional annotation
Potential targets of differentially expressed miRNAs were predicted using the psRNAtarget analysis server [45] (http://plantgrn.noble.org/psRNATarget/), with default parameters which included a threshold cut-off of 3.0 for low false-positive prediction, a complementarity scoring length of 20 bp, and the energy required for target accessibility equal to 25 kcal/mole. The collection of annotated transcript sequences of the V. vinifera (PN40024) 12x assembly was used for the miRNA target search (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/). Predicted targets for both the known and novel differentially expressed miRNAs were functionally annotated using Blast2GO v2.2.7 [46]. This was done by using NCBI BLASTx to find homologous sequences, a mapping step to retrieve gene ontology (GO) terms associated with BLAST hits (http://geneontology.org/page/go-database), and assigning functional attributes to each query sequence in terms of biological processes, cellular components and molecular functions, in a species-independent manner. Afterwards a combined graph was generated using a GO sequence similarity level of 3 and an annotation cut-off value of 7.
Results and discussion
Plant material
According to the diagnostic PCR screening results (data not shown), 19 out of the 50 plants that were visually tagged as ‘healthy’ were AY phytoplasma-positive, while 32 out of the 50 plants that were visually tagged as phytoplasma-infected were confirmed positive for AY phytoplasma. The remaining 31 ‘healthy’ (no phytoplasma-infection) and 32 phytoplasma-infected candidate plants were subjected to further virus screening. All plants that tested positive, following the virus-screening, were eliminated from the study. BLAST results for the de novo assembled contigs also confirmed the absence of any prevalent grapevine viruses (data not shown). Our final test groups consisted of three phytoplasma and virus-free Chardonnay plants for the control group (h55, h85, h89), and three AY phytoplasma-infected, but virus-free, Chardonnay plants for our experimental group (p73, p93, p99).
sRNA-seq
To investigate miRNA expression profiles in response to GY disease, individual sRNA libraries were constructed from RNA extracted from pooled leaf material of the six plants. High-quality, adapter-trimmed reads were generated from the respective sRNA-seq libraries and the number of reads are displayed in Table 1.
Table 1. Summary of total small RNA reads.
| Small RNA library type | ||
|---|---|---|
| Total high-quality reads | Healthy | AY |
| p73 | N/A | 10,893,265 |
| p93 | N/A | 10,476,093 |
| p99 | N/A | 10,511,436 |
| h55 | 10,878,402 | N/A |
| h85 | 12,424,487 | N/A |
| h89 | 11,510,533 | N/A |
| All | 34,813,422 | 31,880,794 |
| 18–26 nt | 26,474,279 | 24,314,330 |
| 18–26 nt: unique | 6,388,422 | 5,726,632 |
| 18–26 nt: mapped | 22,515,584 | 20,782,176 |
H: Health (control) sample group
AY: AY phytoplasma-infected sample group
N/A: Not applicable
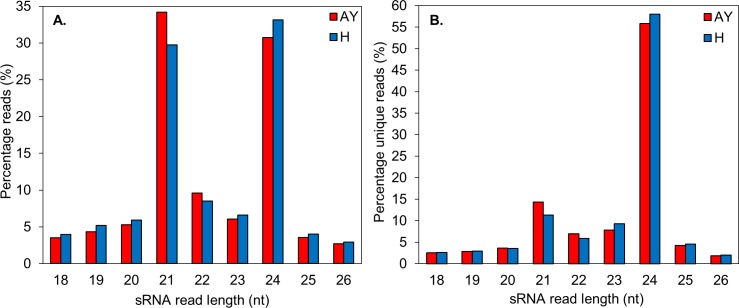
Analysis of the size distribution of sRNA sequences in the 18 to 26 nt range showed the most abundant sequences to be between 21 and 24 nt in length, with sizes 21 nt and 24 nt as the major classes (Fig 2). These results were consistent with those of other grapevine cultivars, as well as Arabidopsis, Citrus trifoliate, Oryza sativa, Eugenia uniflora, and Glycine max [26, 47–51]. The library generated from the phytoplasma-infected samples indicated that 21 nt sRNAs were more abundant (34.2%) than those in the library obtained from the healthy plant samples (29.7%). A similar profile was observed for Mexican lime infected with ‘Candidatus Phytoplasma aurantifolia’ [52]. The 24 nt sRNAs, however, were more abundant in the library from the healthy plant samples (33.2%) compared to the library from the phytoplasma-infected samples (30.7%). This observation points to differences in complexity between the two pools of sRNAs that may infer an underlying miRNA-mediated regulatory response triggered by biotic stress. The unique (non-redundant) 21 nt reads were also more abundant in the phytoplasma-infected samples. Their length is characteristic of canonical miRNAs, and they possessed a high reads/unique reads ratio (Fig 2), reflecting their regulatory impact and abundance in plants. The 24 nt reads, which are predominantly repeat-associated siRNAs (rasiRNAs), exhibited the highest sequence diversity, consistent with the origin of this size class (Fig 2B) [53].
Fig 2.
(A) The size distribution of the total 18 to 26 nt sRNA reads in the healthy (H) and AY phytoplasma-infected (AY) libraries. (B) The size distribution of the total 18 to 26 nt unique sRNA reads in the healthy (H) and AY phytoplasma-infected (AY) libraries.
Identification of known miRNAs and their sequence variants
We used sRNAbench v0.9 to detect both the canonical vvi-miRNA sequences (from miRBase), and all isomiRs present in the pooled phytoplasma-infected and healthy (control) read data, respectively. The different sequences were classified and are presented in a simple table output (S2 File). IsomiRs are defined as different sequence variants of known miRNAs that may arise from post-transcriptional modifications and alternative processing [54]. IsomiR types included those reads having non-template additional nucleotides (where the read sequence starts and ends at the same position as the canonical sequence in the pre-microRNA, but shows sequence variation), “flush fitting” length variants (where the read sequence always starts or ends at the same position as the canonical sequence but a terminal trimming or extension is evident), and multiple length variants (where the read sequence does not coincide with either the 3’ or 5’ terminal nucleotides of the canonical sequence). Those reads that contained the same 5’ terminal nucleotides as the canonical vvi-miR166b sequence, but showed divergence of length in their 3’ terminal extension, as a result of alternative DCL1 cleavage, were the dominantly expressed isomiRs in the ‘healthy libraries’ (S2 File). In the case of the ‘AY phytoplasma-infected libraries’, those reads that contained the same 3’ terminal nucleotides as the canonical vvi-miR166e-5p sequence, but showed divergence of length in their 5’ terminal extension, as a result of alternative DCL1 cleavage, were the most dominantly expressed isomiRs (S2 File).The mechanism by which miRNA heterogeneity arises has been extensively reviewed. Different findings have suggested that multiple isomiRs that arose from a single miRNA locus are not randomly generated artefacts, but rather generated in vivo through biological relevant processes. Consequently, such sequence variations may drastically alter miRNA association with their targets, and also influence miRNA stability during Argonaut (AGO)-RISC loading [55–57].
The vvi-miR166 family showed the highest levels of expression, but had no significant difference in terms of the total normalised read counts between the two different library types (S2 File). Vvi-miR166b and its isomiRs constituted ~40% of the total normalised read counts in both library types. This high level of vvi-miR166 expression was also seen in a previous study where it was the most dominantly expressed miRNA family in all assayed grapevine tissues [26]. A degradome sequencing approach revealed that vvi-miR166b regulates a Class III homeodomain leucine zipper (HDZIP-III) transcription factor which is involved in secondary cell wall biosynthesis [25,58,59]. Direct evidence from the identification and analysis of corresponding activation tagged mutants has implicated the regulatory involvement of miR165/166 in leaf and vascular morphogenesis [60,61].
Differential expression analysis of known miRNAs
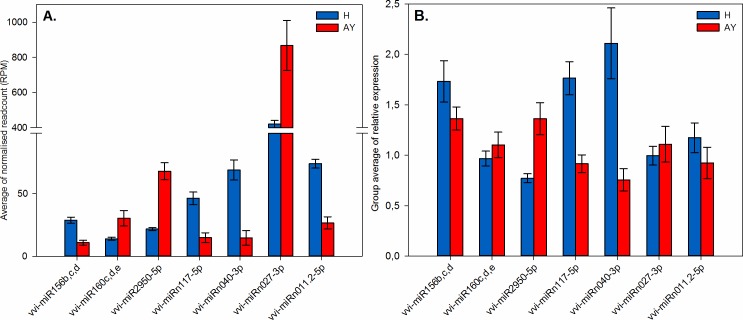
Comparative profiling, with DESeq v2, between the healthy (control) and AY phytoplasma-infected samples was used to determine the differential expression of known miRNAs in the AY phytoplasma-infected material. Based on false discovery rate (FDR) for multiple testing, we encountered seven significantly differentially expressed known vvi-miRNA families that had log2-fold changes with adjusted p-values (q) ≤ 0.05. (Fig 3, Table 2).
Fig 3. Bar charts displaying profiles of differentially expressed vvi-miRNAs (q ≤ 0.05) in healthy (H) and AY phytoplasma-infected (AY) samples that were further validated.
Vertical bars indicate the standard error (SE) of the mean. (A) Average normalised read counts of vvi-miRNAs. Group averages were given in terms of the average of reads per million mapped reads (RPM) for three biological replicates. (B) Relative expression analysis with real-time RT-qPCR, confirming expression profiles of vvi-miRNAs. Each bar represents the average of three biological replicates with three technical replicates.
Table 2. List of significantly differentially expressed known vvi-miRNAs.
| Kown miRNA | Sequence (5'-3') | Length (nt) | Avg of normalised read counts† | DESeq results (H vs AY) | |||
|---|---|---|---|---|---|---|---|
| H | AY | log2FC | p-value | Adj. p-value (q-value) | |||
| vvi-miR156b,c,d¶ | UGACAGAAGAGAGUGAGCAC | 20 | 29.29 | 11.06 | -1.18 | 0.0011 | 0.0102 |
| vvi-miR159c | UUUGGAUUGAAGGGAGCUCUA | 21 | 3869.83 | 8860.93 | 1.15 | 3.89E-05 | 0.0007 |
| vvi-miR399g | UGCCAAAGGAGAUUUGCCCCU | 21 | 463.24 | 103.94 | -2.02 | 3.89E-05 | 0.0007 |
| vvi-miR171a,c,d,I,j | UGAUUGAGCCGUGCCAAUAUC | 21 | 20.56 | 37.71 | 0.87 | 0.0006 | 0.0071 |
| vvi-miR172d | UGAGAAUCUUGAUGAUGCUGCAU | 23 | 243.48 | 736.45 | 1.42 | 0.0007 | 0.0075 |
| vvi-miR160c,d,e¶ | UGCCUGGCUCCCUGUAUGCCA | 21 | 14.23 | 30.9 | 1.00 | 0.0060 | 0.0477 |
| vvi-miR2950-5p¶ | UUCCAUCUCUUGCACACUGGA | 21 | 22.15 | 69.15 | 1.59 | 3.26E-10 | 2.35E-08 |
H: Healthy sample group
AY: AY phytoplasma-infected sample group
¶Validated using real-time RT-qPCR
†Average of reads per million mapped reads (RPM) between three biological replicates
An additional nine known miRNAs from seven families, had log2-fold changes with significant p-values (p ≤ 0.05), which indicate they may be of biological importance (S3 File). A total of eight miRNA families, viz. vvi-miR159c, vvi-miR160c-e, vvi-miR171acdij, vvi-miR172d, vvi-miR2950-5p, vvi-miR319bcef, vvi-miR3627-5p, and vvi-miR395a-m, were up-regulated, and five, viz. vvi-miR156bcd, vvi-miR3629(a-3p, b-3p, c-5p), vvi-miR3638-5p, vvi-miR399aheg, vvi-miR479, were down-regulated (Table 2, S3 File). The differential expression of conserved miRNA families (vvi-miR156, miR159, vvi-miR160, vvi-miR171, vvi-miR172, vvi-miR319), known to be involved in different aspects of plant development [18], make these potential candidates that play a role in the interactions leading to symptoms associated with GY.
Novel miRNA prediction and differential expression analysis
The pooled sRNA reads from all six libraries served as input for sRNAbench v0.9 and Shortstack v0.4.1 for predicting novel miRNAs. These sRNA sequences were aligned to the V. vinifera (PN40024) 12x assembled genome sequence to identify loci that may harbour potential pre-miRNA sequences, based on secondary structure and read distribution. Known V. vinifera pre-miRNA chromosomal locations found in miRBase v21 were flagged during each analysis to obtain unique precursor sequences that did not match these loci. Secondary fold structures were viewed using RNAfold and all miRNA precursors displayed appropriate stem-loop hairpin secondary structures (data not shown). Based on structural criteria described by Meyers et al. [37], these miRNAs can be regarded as authentic candidates that adhere to biogenesis and expression criteria for confident miRNA annotation.
In total, 175 novel pre-miRNA sequences were predicted, each derived from a unique genome location (S4 File). Three of the pre-miRNAs were predicted with both prediction pipelines. We also identified multiple pre-miRNAs that produce mature miRNAs with similar sequences, e.g. vvi-miRn024a to vvi-miRn024c.These miRNAs can be considered members of the same miRNA family (S4 File). Likewise, vvi-miRn019a to vvi-miRn019g represent a larger family of duplicated miRNA paralogs with identical precursor and mature miRNA sequences (S4 File).
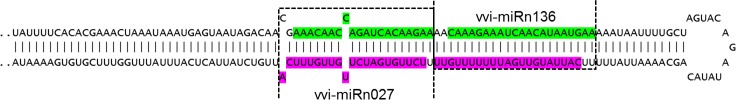
Pre-vvi-miRn027, predicted with Shortstack, may serve as an example of a large precursor that could give rise to two different miRNA duplexes since the sRNAbench-predicted pre-vvi-miRn136 falls within its location (Fig 4; S4 File). Precursor sequences ranged from 54 nt to 742 nt while mature miRNA sequences ranged from 20 nt to 25 nt in length, the majority being 21 nt. Most mature miRNA sequences started with an uracil at the first position, corroborating data described by Baumberger and Baulcombe [62] that showed a preferential association of the AGO1 protein with sRNAs containing a 5’-terminal uracil. This may indicate an important characteristic for miRNA biogenesis through recognition of miRNA duplexes by RISC.
Fig 4. An example of a novel pre-miRNA hairpin structure that may give rise to two different miRNA duplexes (See S4 File).
The sequences highlighted in green and magenta represents the 5’ and 3’ mature miRNA sequences, respectively.
Zhang et al. [39], implemented a criterion to better distinguish miRNAs from other sRNAs, known as MFEI which incorporates MFE, sequence length and GC content. The Unified Nucleic Acid Folding (UNAFold) software was used to calculate the MFE. MFE for predicted novel pre-miRNAs ranged from -13.2 kcal/mol to -428.9 kcal/mol and the MFEI ranged from -0.47 to -2.51. The majority (>94%) of the novel Shortstack-predicted pre-miRNAs possessed MFEI-values in accordance with the expected value (≥-0.85), while only 39% of sRNAbench-predicted pre-miRNAs had strong negative MFEI-values of more than -0.85 (S4 File).
Each novel pre-miRNA sequence, as well as its most abundant mature miRNA sequence, was subjected to a homology-based search against miRBase using BLASTn. Results indicated that some of the newly identified pre-miRNAs have either precursor and/or mature sequences homologous to conserved miRNAs from other plant species, including grapevine miRNAs. Vvi-miRn025a and vvi-miRn025b, for example, share high homology with vvi-miR171 (S4 File). We identified 17 novel vvi-miRNAs belonging to 13 miRNA families that can be classified as newly-identified members of known miRNAs, based on homology to known plant miRNAs for both precursor and mature sequences (S4 File).
Differential expression analysis revealed that 10 novel miRNAs were significantly up-regulated, while 13 novel miRNAs were significantly down-regulated in sRNA libraries prepared from AY phytoplasma-infected leafmaterial (Table 3). We used a Perl script provided by Shen et al. [63] to generate hairpin structures of these differentially expressed novel miRNAs (S1 Fig). These images demonstrated complementary 5’ and 3’ mature miRNA sequences, each within a duplex that was possibly DCL1-derived.
Table 3. List of significantly differentially expressed novel vvi-miRNAs.
| Novel miRNA | Sequence (5'-3') | Length (nt) | Avg of normalised read counts† | DESeq results (H vs AY) | |||
|---|---|---|---|---|---|---|---|
| H | AY | log2FC | p-value | Adj. p-value (q-value) | |||
| vvi-miRn010.2-3p (miR529_new) | GCUGUACCCUCUCUCUUCCCC | 21 | 8.71 | 2.20 | -1.58 | 3.88E-07 | 5.39E-05 |
| vvi-miRn025b/n025a-3p (miR171_new) | UGAUUGAGCCGUGCCAAUAUC | 21 | 20.56 | 37.71 | 0.93 | 4.40E-07 | 5.39E-05 |
| vvi-miRn011.2-5p¶ (miR391_new) | AGGAGAGAUGACGCCGUCGCC | 21 | 75.28 | 27.16 | -1.23 | 9.60E-07 | 7.84E-05 |
| vvi-miRn133-5p | AGACUGGUAGAAAGAUUUAUA | 21 | 19.36 | 2.73 | -1.78 | 7.27E-06 | 4.45E-04 |
| vvi-miRn140-3p | UCACCUUGUUGAGUGCCCGGU | 21 | 6.48 | 1.59 | -1.50 | 1.29E-05 | 6.31E-04 |
| vvi-miRn040-3p¶ | UGGGUUCAAAGUAGACAAUAUUUA | 24 | 70.10 | 14.94 | -1.57 | 2.09E-05 | 8.53E-04 |
| vvi-miRn131-3p (miR399_new) | UGCCAAAGGAGAUUUGCCCCG | 21 | 2.73 | 0.53 | -1.56 | 4.22E-05 | 1.48E-03 |
| vvi-miRn117-5p¶ | UGGACCCUCAUGACUUUAAAAUGC | 24 | 47.07 | 15.09 | -1.29 | 6.07E-05 | 1.86E-03 |
| vvi-miRn139-3p | GGGGGCUGACCUGUUGAAGAG | 21 | 21.50 | 8.60 | -1.04 | 0.0002 | 0.0045 |
| vvi-miRn150-5p | UUUUUCAUGGUCUGAUUGAGC | 21 | 15.97 | 36.25 | 1.11 | 0.0002 | 0.0045 |
| vvi-miRn022b-5p (miR1446_new) | UCUGAACUCUCUCCCUCAUUGGC | 23 | 0.76 | 2.45 | 1.35 | 0.0002 | 0.0045 |
| vvi-miRn008.1-3p (miR169_new) | AGGCAGUCACCUUGGCUAACU | 21 | 3.72 | 1.17 | -1.22 | 0.0004 | 0.0081 |
| vvi-miRn147-5p | UGGUGAACCAAAUAACUCUGG | 21 | 33.29 | 63.81 | 0.93 | 0.0009 | 0.0174 |
| vvi-miRn027-3p¶ | UCUUGUGAUCUUGUUGUUUCA | 21 | 420.78 | 867.56 | 0.99 | 0.0010 | 0.0174 |
| vvi-miRn115-3p | AGGAAUGUGCUUCUUGGCAUA | 21 | 6.45 | 1.84 | -1.19 | 0.0016 | 0.0261 |
| vvi-miRn070-3p | UAAGGACUAAAUUGGUAGACC | 21 | 1.92 | 4.03 | 0.97 | 0.0022 | 0.0334 |
| vvi-miRn089-5p | UACACAUGUAGUGCCAUCAUAUGA | 24 | 53.03 | 16.67 | -1.13 | 0.0025 | 0.0365 |
| vvi-miRn007.1-3p | UGAUAUUAGCAGCUGAGAACA | 21 | 7.19 | 3.71 | -0.76 | 0.0032 | 0.0386 |
| vvi-miRn003-5p | UUACACAGAGAGAUGACGGUGG | 22 | 24.78 | 53.83 | 0.98 | 0.0031 | 0.0386 |
| vvi-miRn051-5p | AGAGACCACCUAGUCCUGUUAAGA | 24 | 31.20 | 19.81 | -0.52 | 0.0029 | 0.0386 |
| vvi-miRn129-5p | UUUUGGAACUAGAGUGCUUGC | 21 | 1.34 | 2.89 | 0.98 | 0.0035 | 0.0410 |
| vvi-miRn137 -5p | CAACAAUCUAAAUGAAACAUAGA | 23 | 3.40 | 6.61 | 0.90 | 0.0043 | 0.0478 |
| vvi-miRn022a-5p (miR1446_new) | UCUGAACUCUCUCCCUCAUGGC | 22 | 8.36 | 15.35 | 0.85 | 0.0047 | 0.0497 |
H: Healthy sample group
AY: AY phytoplasma-infected sample group
¶Validated using real-time RT-qPCR
†Average of reads per million mapped reads (RPM) between three biological replicates
Known miRNA homologs are given in brackets
Validation of miRNA expression profiles by real-time RT-qPCR
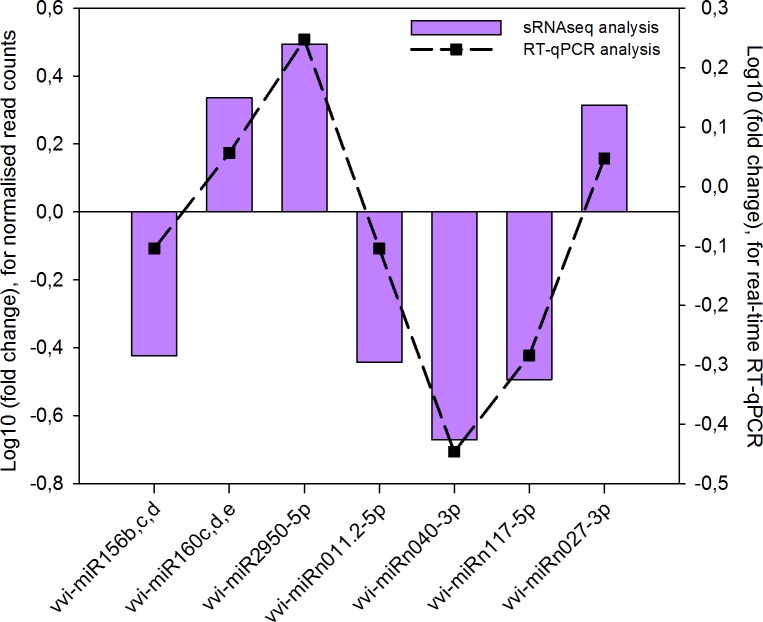
A stem-loop RT-qPCR assay was applied to verify the results for the miRNA differential expression analysis. Primers sequences are listed in the S5 File. The relative expression of seven significantly differentially expressed miRNAs, three known (viz. miR156bcd, miR160cde, and miR2950-5p) and four novel (viz. miRn011.2-5p, miRn040-3p, miRn117-5p, and miRn027-3p), was measured in healthy and phytoplasma-infected leaves using real-time RT-qPCR analysis (Fig 3B). We were also able to validate the expression of less significant known miRNAs (q ≤ 0.15) (viz. miR319e, miR399e, and miR479) (S3 File). This suggests that modulation of these miRNAs may hold biological importance. Although the non-conserved miRNAs (viz. vvi-miR479 and vvi-miR2950), and certain novel miRNAs were present at low levels, they were detected using real-time RT-qPCR. The trend of expression obtained from the RT-qPCR analysis was consistent with the average normalised read abundance observed in the sRNA-seq data (Fig 5; S3 File). Since the expression of the novel miRNA candidates were confirmed using real-time RT-qPCR they can be tentatively classified as authentic miRNAs. The use of stable and robust degradome data, however, will provide us with more concrete evidence to confirm these results [64].
Fig 5. Correlation graph comparing average normalised read counts with real-time RT-qPCR results, thereby confirming vvi-miRNA expression patterns.
Identification of putative targets for differentially expressed known miRNAs
Over the past decade, increasing evidence have demonstrated how miRNAs can play an important role in modulating gene expression during plant-microorganism interactions [65]. It is important to consider miRNA target identification and validation in order to elucidate the biological functions of miRNAs. Multiple ‘Pinot noir’ target mRNAs have been identified for known miRNAs using a high-throughput degradome sequencing approach [26]. To gain further insight into the function of the differentially expressed miRNAs found in this study, we performed a complementary-based search with psRNAtarget to search for putative target-binding sites found in grapevine mRNAs. We adopted strict parameters, which provided perfect or near-perfect complementarity between a miRNA and its target, suggesting DCL1-cleavage or translational inhibition of miRNA-targeted mRNAs [66,67] (S6 File).
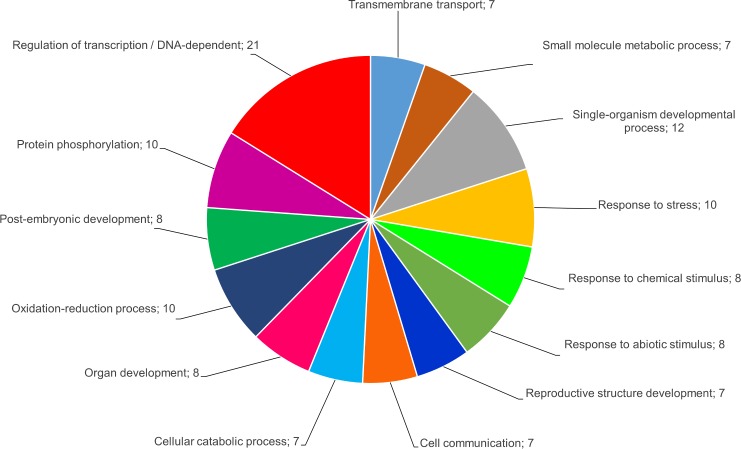
In order to obtain a holistic view of biological pathways possibly influenced by miRNA-mediated regulation, in silico predicted targets for both the differentially expressed known and novel miRNAs were functionally annotated using Blast2GO v2.2.7. After GO analysis we found 71 functionally annotated putative targets for 15 of the known miRNAs and 54 functionally annotated putative targets for 17 of the novel miRNAs. For some of the targets, however, functional attributes could not be assigned, using default parameters within Blast2GO. Detailed annotation results are provided in the S6 File. A combined graph was generated and depicted different categories in which the targets grouped in terms of biological processes (Fig 6).
Fig 6. A combined graph depicting the main categories of putative vvi-miRNA targets grouped in terms of biological processes (GO level 3; annotation cut-off = 7.0).
There were 14 categories of which the four major processes included transcriptional regulation, developmental processes, response to stress, and metabolic processes that included phosphoregulation and oxidoreductase activity. This suggested that the differentially expressed miRNAs are involved in a broad range of physiological functions. Putative targets of conserved miRNA families, such as miR156, miR159, miR171 and miR399, identified in this study, correspond to targets found in numerous plant species, including several grapevine cultivars, while the predicted functions of these targets were also similar with previous findings [26,27,58,60,68–70] (S6 File). Recent studies have revealed that several of these miRNA targets share common roles in the crosstalk between signalling pathways modulated by both biotic and abiotic stresses [71]. Our results revealed that some of these target genes encode transcription factors, including squamosa-promoter binding protein (SPB)-box, MYB, NAC-domain, Scarecrow-like/GRAS-domain, AP2, HDZIP-III and bHLH transcription factors, previously reported for grapevine and other plant species [72–75].
Phytoplasma-infection may cause miRNA-mediated changes in plant morphology and architecture
Plant morphological changes can be attributed to changes in the expression of certain transcription factors, as well as regulatory changes at a post-transcriptional and epigenetic level. The miR156/157 family, which is highly conserved in plants, can target numerous members of the SBP-box genes in V. vinifera. Evidence has shown that changes in the expression levels of these genes play a role in phase transition and reproductive development [76,77]. Studies on Arabidopsis and rice showed that cleavage of squamosa-promoter binding-like (SPL) proteins, due to miR156 overexpression, give rise to plants that are smaller, show delayed flowering and loss of apical dominance, initiate growth of more leaves with shorter plastochrons (in Arabidopsis) and causes reduced panicle size (in rice) [78,79]. Likewise, miR156-overexpression in poplar (Populus spp.) caused an increase in leaf size and leaf initiation rate, and reduced apical dominance [80]. The modification of leaf morphology due to regulation of SBP-box transcripts by miR156 overexpression was demonstrated in phytoplasma-infected Mexican lime trees, mulberry, and red date [52,81,82]. Expression analysis in this study, however, revealed a significant decrease in abundance of certain vvi-miR156 members in the phytoplasma-infected samples, which cannot be explained at this point.
It has been shown that down-regulation of miR156 results in an increase in SPLs that promote juvenile to adult phase transition and flowering through activation of miR172 and MADS box genes in Arabidopsis [83,84]. The Arabidopsis AtSPL9, can positively regulate the expression of miR172, demonstrating the presence of a miR156-AtSPL9-miR172 regulatory cascade [85]. It was proposed that the miR156-SPL-miR172 regulatory pathway was activated in mulberry in response to phytoplasma infection [81]. Higher levels of miR172 associated with viral pathogenesis in tomato leaf curl disease and grapevine leafroll disease have also been reported [86,87]. The APETALA2/Ethylene-responsive transcription factor (AP2/ERF)-like mRNA was identified as a possible target of vvi-miR172 in our study. The interaction between miR172 and AP2/ERF-like targets is well conserved and are known to be involved in transitions between developmental stages, regulating flowering time and specifying floral organ identity [88,89]. Differential expression of vvi-miR156 and vvi-miR172, leading to restricted phase transition, may lead to symptoms associated with GY such as abnormal leaf shape and size, as well as downward curling of leaves and flower abortion. It was suggested that expression changes of miR156 and miR172 may lead to development of green leaf-like structures instead of flowers, also referred to as phyllody, as well as flower sterility in phytoplasma-infected red date and mulberry [81,82].
Levels of vvi-miR159 and vvi-miR319 were also significantly higher in the AY phytoplasma-infected leaves, and in silico analysis predicted that they may target a GAMYB-like mRNA and a R2R3-MYB mRNA. Recent studies identified their association with plant disease during fungal and bacterial infection in Arabidopsis and Populus trichocarpa, respectively [90,91], and have experimentally validated their targets as being mRNAs encoding MYB transcription factors [92]. MYB genes constitutes a large and widespread gene family in plants (estimated at 279 members in grapevine, of which 108 belong to the R2R3 family), and are involved in a variety of plant-specific functions including primary and secondary metabolism, cell fate and differentiation, developmental processes and responses to biotic and abiotic stresses [93,94]. Consequently, altered expression of miR159 and miR319 may also contribute to deformation of grapevine leaves [82,86].
Differential miRNA expression may lead to modulated auxin signalling
Disease symptoms caused by certain pathogens have also been described as a result of interference with plant hormone signalling that lead to the disturbance of plant defence responses. Phytoplasma diseases have been classified as ‘auxonic diseases’ which refers to possible interactions with the auxin balance of the host [95]. Auxin, an important phytohormone, regulates many plant developmental processes, and its influence during pathogen resistance responses has been described [96]. A substantial increase of indole-3-acetic acid (IAA) was observed in phytoplasma-infected Mexican lime trees, possibly indicating susceptibility to the pathogen [52]. Certain proteins, known as virulence effectors, are secreted by pathogens during infection and are known to modulate hormone and signalling pathways by altering gene transcription levels. AY-WB effectors, SAP11 and TENGU, are known to be unloaded from the phloem sieve cells to the target cell nuclei where they interact and destabilise certain transcription factors, resulting in severe changes in leaf morphology and increased susceptibility to phytoplasma insect vectors [97–99]. Microarray analysis of transgenic Arabidopsis lines overexpressing TENGU demonstrated regulation of several auxin responsive genes and auxin efflux carrier genes. SAP11 destabilises CINCINNATA (CIN)-TEOSINTE BRANCHED1, CYCLOIDEA, PROLIFERATING CELL FACTOR (TCP) transcription factors 1 and 2, known to be regulated by miR319 in Arabidopsis, resulting in the suppression of Jasmonate (JA) production that create favourable conditions for insect vector proliferation [97–99].
A group of miRNAs can promote plant defence responses by coordinate regulation of hormone signalling pathways in response to pathogen attack. Among them, miR160, miR167, miR390 and miR393 contribute to PTI by regulating the expression of genes encoding different auxin response factors (ARFs) and auxin receptors involved in auxin signalling, thereby promoting inhibition of pathogen growth [90]. miR393 expression, induced by bacterial elicitor flg22, was the first shown to be implicated in the repression of auxin receptor genes in Arabidopsis [23]. Our results showed that vvi-miR160, which may target ARF mRNAs, was significantly up-regulated in the AY phytoplasma-infected leaves. ARF transcription factors are known to regulate auxin-inducible genes by binding to elements in their auxin-responsive promoters to either activate or repress transcription [100]. Other instances where miR160 accumulated during biotic stress response were demonstrated in clubroot-infected Brassica napus root [101], powdery mildew infection in wheat [102], and phytoplasma-infected mulberry [81].
Phytoplasma-responsive miRNA expression may play a role in nutrient homeostasis
The AY phytoplasma chromosome is extremely reduced and lacks many essential genes related to amino acid and fatty acid biosynthesis, the tricarboxylic acid cycle and oxidative phosphorylation. This suggested that phytoplasmas have evolved as intracellular parasites in nutrient-rich host environments and therefore possess multiple transporter genes in order to assimilate important mineral nutrients for their survival [103]. Several plant miRNAs have been reported for their role in nutrient homeostasis in response to deficiencies of phosphate, nitrogen, sulphur, and copper [104]. A few of these, including vvi-miR395 and vvi-miR399, were differentially expressed in the present study, possibly in response to AY phytoplasma-infection. miR395 is known to target members of the ATP-sulphurylase (ATPS) gene family and a low-affinity sulphate transporter gene SULTR2;1, both crucial for regulating sulphate homeostasis in Arabidopsis [105] (S6 File). The induction of miR395 levels leads to sulphate accumulation in the leaves due to increased translocation from the roots [106,107]. In this study miR395 was up-regulated in the AY phytoplasma-infected leaves, and may contribute to favourable conditions for pathogen growth. Alternatively, sulphur starvation can cause physiological imbalances, impaired plant growth, and reduced plasticity against environmental changes and pathogen attack [108]. The role of miR399 in the maintenance of phosphate homeostasis has been well characterised. It is involved in the regulation and allocation of inorganic phosphate (Pi) from the roots to the shoots as well as remobilisation from old to young leaves [109,110]. miR399 positively regulates Pi uptake and translocation by down-regulating PHO2, which encodes a ubiquitin-conjugating E2 enzyme, UBC24 [109,111]. PHO2, on the other hand, acts as a negative regulator by suppressing these activities when external Pi is ample, thereby preventing phosphate toxicity. Our results revealed significant down-regulation of vvi-miR399 in AY phytoplasma-infected leaves. Interestingly, lower levels of miR399 was also found in phytoplasma-infected material of Mexican lime trees, mulberry, and red date [52,81,82]. An adequate supply of Pi is required for optimal growth and reproduction due to its involvement in essential plant functions, including energy transfer, photosynthesis, enzyme regulation, metabolite transport and nucleic acid synthesis. Therefore, the down-regulation of miR399 may cause suppression of Pi uptake, further contributing to disease symptom development.
Identification of putative targets for differentially expressed novel miRNAs
In addition to the targets of known miRNA, we also predicted possible targets for the in silico predicted novel miRNAs that were significantly differentially expressed in the AY phytoplasma-infected leaves (S6 File). Some of these target mRNAs encode certain transcription factors, such as Scarecrow-like/GRAS-domain protein, TPR-like protein, MADS-box protein, bHLH-like protein, and a NAC-domain protein. We also identified targets that encode proteins involved in hydrolase activity, e.g. ARM repeat superfamily isoform 2-like protein, beta-fructofuranosidase, glucan endo-1,3-beta-glucosidase, and a calcineurin-like metallo-phosphoesterase. Receptor-like kinase (RLK) proteins that are involved in signal transduction, such as a G-type lectin S-receptor-like serine/threonine-protein kinase, a leucine-rich repeat (LRR) receptor EXS-like kinase, a disease resistance At3g14460-like protein, a RLK HSL1-like protein, and a LRR receptor-like serine/threonine At4g08850-like kinase were also identified.
Some signal transduction proteins are surface-located, transmembrane receptor molecules that are activated by external stimuli, such as plant hormones and pathogens. These, in turn, are sequentially transmitted to initiate complex downstream signalling pathways that induce PAMP -and effector-triggered immunity (PTI and ETI) and/or hypersensitive cell death resistance responses. The majority of these innate immune receptors are proteins that contain a nucleotide-binding site (NBS) and leucine-rich repeats (LRR) that are encoded by resistance (R) genes [112]. Another versatile function of certain miRNAs is targeting diverse members of NBS-LRRs which are then processed by RNA-dependent RNA polymerase 6 (RDR6) to dsRNA and then cleaved by DCL4 to produce phased, secondary siRNAs (phasiRNAs) [113]. This was demonstrated in resistant Solanum lycopersicum (tomato) and Gossypium raimondii (cotton) where the miR482-mediated silencing cascade was suppressed in pathogen-infected plants so that certain NBS-LRRs were up-regulated to confer resistance [114,115]. Interestingly, vvi-miRn027, which may target a disease resistance mRNA (GSVIVG01027229001), was severely increased in AY phytoplasma-infected leaves in comparison to the other novel miRNAs that might target RLK mRNAs. This miRNA and its putative target may serve as potential candidates in transient expression studies to investigate an underlying defence response to AY phytoplasma.
Furthermore, most of the other novel miRNAs were expressed at a lower abundance than that of conserved miRNAs and are likely to be grapevine-specific miRNAs, which may be classified into non-conserved miRNAs. It can be suspected that they are likely candidates involved in developmental, metabolic and transmembrane transport processes as proposed by the gene ontology results, but it would require additional experimental approaches to address these hypotheses.
Conclusions
In summary, our study employed different computational tools to provide the first report on the identification of differentially expressed miRNAs in grapevine leaves infected with AY phytoplasma. In addition to known vvi-miRNAs, we detected a large group of putative novel miRNAs by utilizing two different analysis pipelines. Some of the novel miRNAs shared a high degree of homology with other known plant miRNAs, and were therefore classified as newly-identified members of existing miRNA families. Further experimentation concerning the regulation of their target mRNA(s), however, would be required to confirm this.
Differential expression analysis was done via comparative miRNA profiling between sRNA libraries constructed from healthy control plants and plants diagnosed with AY phytoplasma, respectively. Changes in the expression of various miRNAs were clearly observed in the diseased group, possibly modulated in response to biotic stress. The relative expression of certain known and novel miRNAs was determined with real-time RT-qPCR analysis, thereby demonstrating a similar trend in expression regarding the normalised sRNA read data. There is increasing evidence for the involvement of miRNAs in plant-microorganism interaction and how they mediate gene expression related to pathogenesis.
In order to identify potential miRNA targets, we applied a simple complementary-based, in silico approach with psRNAtarget. This method relies on perfect or near-perfect complementarity of plant miRNAs with their target(s), known to facilitate gene regulation through mRNA cleavage or translational inhibition. To further validate grapevine-specific miRNAs and the mRNAs they target would require the use of stable and robust degradome sequencing data that would assist in the elucidation of different modes of regulation in a tissue-specific and developmental stage-specific manner. Target mRNAs regulated by translational inhibition, however, would be undetectable in degradome data. Furthermore, high-throughput gene expression profiling techniques such as microarray-hybridisation analysis and RNAseq/transcriptome analysis would allow us to observe expression levels of miRNAs and their anti-correlated target mRNAs.
The miRNA expression patterns observed in AY phytoplasma-infected grapevine leaves, followed by putative miRNA target description and annotation, led us to believe that our results were compatible with evidence of perturbations found in other pathogen-infected plants. Putative miRNA target predictions indicated the involvement of miRNA pathways that may influence plant development and morphology either directly or by auxin imbalance. We also identified targets involved in nutrient homeostasis, as well as a few important novel miRNA targets involved in signal transduction, which may hold the key to activating pathogen-resistance pathways in grapevine. Taken together, our findings suggest some hypothetical associations between miRNAs and certain physiological changes that may be crucial in understanding disease symptom development in AY phytoplasma-infected grapevines. Further investigations of these miRNA-mediated pathways may shed new light on the roles and mechanisms of miRNAs in plant pathogenesis.
Supporting information
(PDF)
(XLSX)
(XLSX)
(PDF)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
This work is based upon the research supported by the National Research Foundation (NRF) of South Africa. The grant holder (UID 78073) acknowledges that opinions, findings and conclusions or recommendations expressed in any publication generated by the NRF-supported research are those of the author(s) and the NRF accepts no liability whatsoever in this regard. Furthermore, the authors are also very appreciative towards the Technology Innovation Agency (TIA) for financial support for the duration of this study. We would also like to express our gratitude to Anelda van der Walt, Marike Visser and Beatrix Coetzee (Department of Genetics, Stellenbosch University), as well as Charl Möller (IT, Stellenbosch University), for their generous contributions and/or technical assistance regarding the computational work.
Data Availability
All sequence data have been submitted to the Sequence Read Archive (SRA) of the NCBI (http://www.ncbi.nlm.nih.gov/) as BioProject number PRJNA385718. BioSample accessions: SAMN06908861, SAMN06908862, SAMN06908863, SAMN06908864, SAMN06908865, SAMN06908866.
Funding Statement
This work is based upon the research supported by the National Research Foundation of South Africa (grand number UID78073), and the Technology Innovation Agency (PlantBio project number PB109/08). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lee I-M, Davis RE, Gundersen-Rindal DE. Phytoplasma: Phytopathogenic Mollicutes 1. Annu Rev Microbiol. 2000; 54: 221–255. doi: 10.1146/annurev.micro.54.1.221 [DOI] [PubMed] [Google Scholar]
- 2.Hogenhout SA, Oshima K, Ammar el-D, Kakizawa S, Kingdom HN, Namba S. Phytoplasmas: bacteria that manipulate plants and insects. Mol Plant Pathol. 2008; 4:403–23. doi: 10.1111/j.1364-3703.2008.00472.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee IM. 'Candidatus Phytoplasma asteris', a novel phytoplasma taxon associated with aster yellows and related diseases. Int J Syst Evol Microbiol. 2004; 54: 1037–1048. doi: 10.1099/ijs.0.02843-0 [DOI] [PubMed] [Google Scholar]
- 4.Engelbrecht M, Joubert J, Burger JT. First report of aster yellows phytoplasma in grapevines in South Africa. Plant Dis. 2010; 94: 373–373. doi: 10.1094/pdis-94-3-0373a [DOI] [PubMed] [Google Scholar]
- 5.Krüger K, De Klerk A, Douglas-Smit N, Joubert J, Pietersen G, Stiller M. Aster yellows phytoplasma in grapevines: identification of vectors in South Africa . Bulletin of Insectology 2011; 64: S137–S1387. [Google Scholar]
- 6.Carstens R, Petersen Y, Stephan D, Burger JT. Current status of Aster yellows disease in infected vineyards in the Vredendal grape producing area of South Africa. Phytopathogenic Mollicutes 2011; 1(2): 83–85. doi: 10.5958/j.2249-4669.1.2.014 [Google Scholar]
- 7.Belli G, Bianco PA, Conti M. Grapevine yellows in Italy: past, present and future. J. Plant Pathol. 2010; 92: 303–326. [Google Scholar]
- 8.Maixner M. Grapevine yellows—current developments and unsolved questions. In: Proceedings of the 15th meeting of the international council for the study of virus and virus-like diseases of the grapevine (ICVG), Stellenbosch, South Africa; 2006. pp.86-88.
- 9.Contaldo N, Bertaccini A, Paltrinieri S, Windsor HM, Windsor GD. Axenic culture of plant pathogenic phytoplasmas. Phytopathol Mediterr. 2012; 51: 607–617. doi: https://doi.org/10.14601/Phytopathol_Mediterr-11773 [Google Scholar]
- 10.Hren M, Nikolić P, Rotter A, Blejec A, Terrier N, Ravnikar M, et al. 'Bois noir' phytoplasma induces significant reprogramming of the leaf transcriptome in the field grown grapevine. BMC Genomics. 2009; 10: 460 doi: 10.1186/1471-2164-10-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albertazzi G, Milc J, Caffagni A, Francia E, Roncaglia E, Ferrari F, et al. Gene expression in grapevine cultivars in response to Bois Noir phytoplasma infection. Plant Sci. 2009; 176: 792–804. doi: 10.1016/j.plantsci.2009.03.001 [Google Scholar]
- 12.Margaria P, Palmano S. Response of the Vitis vinifera L. cv. ‘Nebbiolo’ proteome to Flavescence dorée phytoplasma infection. Proteomics. 2010; 11: 212–224. doi: 10.1002/pmic.201000409 [DOI] [PubMed] [Google Scholar]
- 13.Mou H-Q, Lu J, Zhu S-F, Lin C-L, Tian G-Z, Xu X, et al. Transcriptomic analysis of Paulownia infected by Paulownia ‘Witches'-Broom phytoplasma. PLoS ONE. 2013; 8: e77217 doi: 10.1371/journal.pone.0077217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margaria P, Abbà S, Palmano S. Novel aspects of grapevine response to phytoplasma infection investigated by a proteomic and phospho-proteomic approach with data integration into functional networks. BMC Genomics. 2013; 14: 38 doi: 10.1186/1471-2164-14-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monavarfeshani A, Mirzaei M, Sarhadi E, Amirkhani A, Khayam Nekouei M, Haynes PA, et al. Shotgun proteomic analysis of the Mexican lime tree infected with “Candidatus Phytoplasma aurantifolia”. J Proteome Res. 2013; 12: 785–795. doi: 10.1021/pr300865t [DOI] [PubMed] [Google Scholar]
- 16.Liu LYD, Tseng HI, Lin CP, Lin YY, Huang YH, Huang CK, et al. High-Throughput transcriptome analysis of the leafy flower transition of Catharanthus roseus induced by Peanut Witches'-Broom phytoplasma infection. Plant Cell Physiol. 2014; 55: 942–957. doi: 10.1093/pcp/pcu029 [DOI] [PubMed] [Google Scholar]
- 17.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol. 2009; 60: 485–510. doi: 10.1146/annurev.arplant.043008.092111 [DOI] [PubMed] [Google Scholar]
- 18.Sunkar R, Li Y-F, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012; 17: 196–203. doi: 10.1016/j.tplants.2012.01.010 [DOI] [PubMed] [Google Scholar]
- 19.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006; 20: 759–771. doi: 10.1101/gad.1410506 [DOI] [PubMed] [Google Scholar]
- 20.Budak H, Akpinar BA. Plant miRNAs: biogenesis, organization and origins. Funct Integr Genomics. 2015; 15: 523–31. doi: 10.1007/s10142-015-0451-2 [DOI] [PubMed] [Google Scholar]
- 21.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005; 121: 207–221. doi: 10.1016/j.cell.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 22.Padmanabhan C, Zhang X, Jin H. Host small RNAs are big contributors to plant innate immunity. Curr Opin Plant Biol. 2009; 12: 465–472. doi: 10.1016/j.pbi.2009.06.005 [DOI] [PubMed] [Google Scholar]
- 23.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006; 312: 436–439. doi: 10.1126/science.aae0382 [DOI] [PubMed] [Google Scholar]
- 24.Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007; 449: 463–467. doi: 10.1038/nature06148 [DOI] [PubMed] [Google Scholar]
- 25.Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE. 2007; 2: e1326 doi: 10.1371/journal.pone.0001326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, et al. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 2010; 62:960–76. doi: 10.1111/j.0960-7412.2010.04208.x [DOI] [PubMed] [Google Scholar]
- 27.Mica E, Piccolo V, Delledonne M, Ferrarini A, Pezzotti M, Casati C, et al. Correction: High throughput approaches reveal splicing of primary microRNA transcripts and tissue specific expression of mature microRNAs in Vitis vinifera. BMC Genomics. 2010; 11: 109 doi: 10.1186/1471-2164-11-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011; 39: D152–D157. doi: 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White EJ, Venter M, Hiten NF, Burger JT. Modified Cetyltrimethylammonium bromide method improves robustness and versatility: The benchmark for plant RNA extraction. Biotechnol J. 2008; 3: 1424–1428. doi: 10.1002/biot.200800207 [DOI] [PubMed] [Google Scholar]
- 30.Gundersen DE, Lee IM. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol Mediterr. 1996; 35: 144–151. [Google Scholar]
- 31.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008; 18:821–9. doi: 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990; 215: 403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 33.Hackenberg M, Rodriguez-Ezpeleta N, Aransay AM. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res. 2011; 39: W132–W138. doi: 10.1093/nar/gkr247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010; 11: R106 doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barturen G, Rueda A, Hamberg M, Alganza A, Lebron R, Kotsyfakis M, et al. sRNAbench: profiling of small RNAs and its sequence variants in single or multi-species high-throughput experiments. Methods in Next Generation Sequencing. 2014; 1 doi: 10.2478/mngs-2014-0001 [Google Scholar]
- 36.Axtell MJ. ShortStack: Comprehensive annotation and quantification of small RNA genes. RNA. 2013; 19: 740–751. doi: 10.1261/rna.035279.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, et al. Criteria for annotation of plant microRNAs. Plant Cell. 2008; 20: 3186–3190. doi: 10.1105/tpc.108.064311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008; 453: 3–31. doi: 10.1007/978-1-60327-429-6_1 [DOI] [PubMed] [Google Scholar]
- 39.Zhang BH, Pan XP, Cox SB, Cobb GP, Anderson TA. Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci. 2006; 63: 246–254. doi: 10.1007/s00018-005-5467-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA Websuite. Nucleic Acids Res. 2008; 36: W70–W74. doi: 10.1093/nar/gkn188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10: 421 doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005; 33: e179–e179. doi: 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellemans J MG, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007; 8: R19 doi: 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011; 39: W155–W159. doi: 10.1093/nar/gkr319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conesa A, Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008; 2008: 1–12. doi: 10.1155/2008/619832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006; 20: 3407–3425. doi: 10.1101/gad.1476406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, et al. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008; 18: 1456–1465. doi: 10.1101/gr.075572.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song C, Wang C, Zhang C, Korir N, Yu H, Ma Z, et al. Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genomics. 2010; 11: 431 doi: 10.1186/1471-2164-11-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song Q-X, Liu Y-F, Hu X-Y, Zhang W-K, Ma B, Chen S-Y, et al. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 2011; 11: 5 doi: 10.1186/1471-2229-11-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guzman F, Almerão MP, Körbes AP, Loss-Morais G, Margis R. Identification of microRNAs from Eugenia uniflora by high-throughput sequencing and bioinformatics analysis. PLoS ONE. 2012; 7: e49811 doi: 10.1371/journal.pone.0049811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ehya F, Monavarfeshani A, Mohseni Fard E, Karimi Farsad L, Khayam Nekouei M, Mardi M, et al. Phytoplasma-responsive microRNAs modulate hormonal, nutritional, and stress signalling pathways in Mexican lime trees. PLoS ONE. 2013; 8: e66372 doi: 10.1371/journal.pone.0066372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lelandais-Brière C, Sorin C, Declerck M, Benslimane A, Crespi M, Hartmann C. Small RNA diversity in plants and its impact in development. Curr Genomics. 2010; 11:14–23. doi: 10.2174/138920210790217918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 2009; 37: 2461–2470. doi: 10.1093/nar/gkp093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chugh P, Dittmer DP. Potential pitfalls in microRNA profiling. Wiley Interdiscip Rev RNA. 2012; 3: 601–616. doi: 10.1002/wrna.1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013; 14: 475–488. doi: 10.1038/nrm3611 [DOI] [PubMed] [Google Scholar]
- 57.Guo L, Chen F. A challenge for miRNA: multiple isomiRs in miRNAomics. Gene. 2014; 544: 1–7. doi: 10.1016/j.gene.2014.04.039 [DOI] [PubMed] [Google Scholar]
- 58.Carra A, Mica E, Gambino G, Pindo M, Moser C, Pè ME, et al. Cloning and characterization of small non-coding RNAs from grape. Plant J. 2009; 59: 750–763. doi: 10.1111/j.1365-313X.2009.03906.x [DOI] [PubMed] [Google Scholar]
- 59.Du Q, Wang H. The role of HD-ZIP III transcription factors and miR165/166 in vascular development and secondary cell wall formation. Plant Signal Behav. 2015; 10:e1078955 doi: 10.1080/15592324.2015.1078955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun G. MicroRNAs and their diverse functions in plants. Plant Mol Biol. 2012; 80: 17–36. doi: 10.1007/s11103-011-9817-6 [DOI] [PubMed] [Google Scholar]
- 61.Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, et al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 2005; 42: 84–94. doi: 10.1111/j.1365-313X.2005.02354.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005; 102: 11928–11933. doi: 10.1073/pnas.0505461102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen W, Chen M, Wei G, Li Y. MicroRNA prediction using a fixed-order Markov model based on the secondary structure pattern. PLoS ONE. 2012; 7: e48236 doi: 10.1371/journal.pone.0048236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.German MA, Pillay M, Jeong D-H, Hetawal A, Luo S, Janardhanan P, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotech. 2008; 26: 941–946. doi: 10.1038/nbt1417 [DOI] [PubMed] [Google Scholar]
- 65.Khraiwesh B, Zhu JK, Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta. 2012; 1819:137–48. doi: 10.1016/j.bbagrm.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005; 8: 517–527. doi: 10.1016/j.devcel.2005.01.018 [DOI] [PubMed] [Google Scholar]
- 67.Carrington JC, Ambros V. Role of MicroRNAs in plant and animal development. Science. 2003; 301: 336–338. doi: 10.1126/science.1085242 [DOI] [PubMed] [Google Scholar]
- 68.Wang C, Wang X, Kibet NK, Song C, Zhang C, Li X, et al. Deep sequencing of grapevine flower and berry short RNA library for discovery of novel microRNAs and validation of precise sequences of grapevine microRNAs deposited in miRBase. Physiol Plant. 2011a; 143: 64–81. doi: 10.1111/j.1399-3054.2011.01481.x [DOI] [PubMed] [Google Scholar]
- 69.Wang C, Leng X, Zhang Y, Kayesh E, Sun X. Transcriptome-wide analysis of dynamic variations in regulation modes of grapevine microRNAs on their target genes during grapevine development. Plant Mol Biol. 2014; 84 doi: 10.1007/s11103-013-0132-2 [DOI] [PubMed] [Google Scholar]
- 70.Belli Kullan J, Lopes Paim Pinto D, Bertolini E, Fasoli M, Zenoni S, Tornielli GB, et al. miRVine: a microRNA expression atlas of grapevine based on small RNA sequencing. BMC Genomics. 2015; 16: 1–23. doi: 10.1186/1471-2164-16-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Curaba J, Singh MB, Bhalla PL. miRNAs in the crosstalk between phytohormone signalling pathways. J Exp Bot. 2014. doi: 10.1093/jxb/eru002 [DOI] [PubMed] [Google Scholar]
- 72.Kantar M, Lucas SJ, Budak H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta. 2011; 233: 471–84. doi: 10.1007/s00425-010-1309-4 [DOI] [PubMed] [Google Scholar]
- 73.Gonzalez-Ibeas D, Blanca J, Donaire L, Saladié M, Mascarell-Creus A, Cano-Delgado A, et al. Analysis of the melon (Cucumis melo) small RNAome by high-throughput pyrosequencing. BMC Genomics. 2011; 12: 393 doi: 10.1186/1471-2164-12-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Colaiacovo M, Subacchi A, Bagnaresi P, Lamontanara A, Cattivelli L, Faccioli P. A computational-based update on microRNAs and their targets in barley (Hordeum vulgare L.). BMC Genomics. 2010; 11: 595 doi: 10.1186/1471-2164-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ozhuner E, Eldem V, Ipek A, Okay S, Sakcali S, Zhang B, et al. Boron stress responsive MicroRNAs and their targets in barley. PLoS ONE. 2013; 8: e59543 doi: 10.1371/journal.pone.0059543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hou H, Li J, Gao M, Singer SD, Wang H, Mao L, et al. Genomic organization, phylogenetic comparison and differential expression of the SBP-Box family genes in grape. PLoS ONE. 2013; 8: e59358 doi: 10.1371/journal.pone.0059358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huijser P, Schmid M. The control of developmental phase transitions in plants. Development. 2011; 138: 4117–4129. doi: 10.1242/dev.063511 [DOI] [PubMed] [Google Scholar]
- 78.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008; 67: 183–195. doi: 10.1007/s11103-008-9310-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie K, Wu C, Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006; 142: 280–293. doi: 10.1104/pp.106.084475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J-W, Park M, Wang L-J, Koo Y, Chen X-Y, Weigel D. MiRNA control of vegetative phase change in trees. PLoS Genet. 2011. b; 7: e1002012 doi: 10.1371/journal.pgen.1002012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gai Y-P, Li Y-Q, Guo F-Y, Yuan C-Z, Mo Y-Y, Zhang H-L, et al. Analysis of phytoplasma-responsive sRNAs provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease. Sci Rep. 2014; 4: 5378 doi: 10.1038/srep05378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shao F, Zhang Q, Liu H, Lu S, Qiu D. Genome-wide identification and analysis of microRNAs involved in Witches’-Broom phytoplasma response in Ziziphus jujuba. PLoS ONE. 2016; 11: e0166099 doi: 10.1371/journal.pone.0166099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang J-W, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. 2008; 20: 1231–1243. doi: 10.1105/tpc.108.058180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu G, Park MY, Conway SR, Wang J-W, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009; 138: 750–759. doi: 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen X, Zhang Z, Liu D, Zhang K, Li A, Mao L. SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J Integr Plant Biol. 2010; 52: 946–951. doi: 10.1111/j.1744-7909.2010.00987.x [DOI] [PubMed] [Google Scholar]
- 86.Naqvi AR, Haq QM, Mukherjee SK. MicroRNA profiling of tomato leaf curl new delhi virus (tolcndv) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol J. 2010; 7: 1–16. doi: 10.1186/1743-422X-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alabi OJ, Zheng Y, Jagadeeswaran G, Sunkar R, Naidu RA. High-throughput sequence analysis of small RNAs in grapevine (Vitis vinifera L.) affected by grapevine leafroll disease. Mol Plant Pathol. 2012; 13: 1060–1076. doi: 10.1111/j.1364-3703.2012.00815.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen X. A MicroRNA as a Translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004; 303: 2022–2025. doi: 10.1126/science.1088060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu Q-H, Helliwell CA. Regulation of flowering time and floral patterning by miR172. J Exp Bot. 2011; 62: 487–495. doi: 10.1093/jxb/erq295 [DOI] [PubMed] [Google Scholar]
- 90.Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhou X, et al. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol. 2011; 75: 93–105. doi: 10.1007/s11103-010-9710-8 PubMed PMID: PMC3005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao J-P, Jiang X-L, Zhang B-Y, Su X-H. Involvement of microRNA-mediated gene expression regulation in the pathological development of Stem Canker Disease in Populus trichocarpa. PLoS ONE. 2012; 7: e44968 doi: 10.1371/journal.pone.0044968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Du H, Feng B-R, Yang S-S, Huang Y-B, Tang Y-X. The R2R3-MYB Transcription factor gene family in maize. PLoS ONE. 2012; 7: e37463 doi: 10.1371/journal.pone.0037463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Galbiati M, Matus JT, Francia P, Rusconi F, Cañón P, Medina C, et al. The grapevine guard cell-related VvMYB60 transcription factor is involved in the regulation of stomatal activity and is differentially expressed in response to ABA and osmotic stress. BMC Plant Biol. 2011; 11: 1–15. doi: 10.1186/1471-2229-11-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010; 15: 573–581. doi: 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- 95.Musetti R. Biochemical changes in plants infected by phytoplasmas. In: Weintraub PG, editor. Phytoplasmas: Genomes, plant hosts and vectors; 2010. pp. 138–146.
- 96.Kazan K, Manners JM. Linking development to defense: auxin in plant pathogen interactions. Trends Plant Sci. 2009; 14: 373–382. doi: 10.1016/j.tplants.2009.04.005 [DOI] [PubMed] [Google Scholar]
- 97.Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell. 2007; 13: 115–125. doi: 10.1016/j.devcel.2007.04.012 [DOI] [PubMed] [Google Scholar]
- 98.Hoshi A, Oshima K, Kakizawa S, Ishii Y, Ozeki J, Hashimoto M, et al. A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc Natl Acad Sci USA. 2009; 106: 6416–6421. doi: 10.1073/pnas.0813038106 PubMed PMID: PMC2669400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sugio A, MacLean AM, Kingdom HN, Grieve VM, Manimekalai R, Hogenhout SA. Diverse targets of phytoplasma effectors: from plant development to defense against insects. Annu Rev Phytopathol. 2011; 49: 175–195. doi: 10.1146/annurev-phyto-072910-095323 [DOI] [PubMed] [Google Scholar]
- 100.Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol. 2002; 49:373–85. . [PubMed] [Google Scholar]
- 101.Verma SS, Rahman MH, Deyholos MK, Basu U, Kav NNV. Differential expression of miRNAs in Brassica napus root following infection with Plasmodiophora brassicae. PLoS ONE. 2014; 9: e86648 doi: 10.1371/journal.pone.0086648 PubMed PMID: PMC3909011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, et al. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 2010; 10: 1–11. doi: 10.1186/1471-2229-10-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oshima K, Kakizawa S, Nishigawa H, Jung H-Y, Wei W, Suzuki S, et al. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat Genet. 2004; 36: 27–29. doi: 10.1038/ng1277 [DOI] [PubMed] [Google Scholar]
- 104.Shriram V, Kumar V, Devarumath RM, Khare TS, Wani SH. MicroRNAs As Potential targets for abiotic stress tolerance in plants. Front Plant Sci. 2016; 7: 817 doi: 10.3389/fpls.2016.00817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Matthewman CA, Kawashima CG, Húska D, Csorba T, Dalmay T, Kopriva S. miR395 is a general component of the sulfate assimilation regulatory network in Arabidopsis. FEBS Lett. 2012; 586: 3242–8. doi: 10.1016/j.febslet.2012.06.044 [DOI] [PubMed] [Google Scholar]
- 106.Liang G, Yang F, Yu D. MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 2010; 62: 1046–57. doi: 10.1111/j.1365-313X.2010.04216.x [DOI] [PubMed] [Google Scholar]
- 107.Kawashima CG, Matthewman CA, Huang S, Lee BR, Yoshimoto N, Koprivova A, et al. Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis. Plant J. 2011; 66: 863–76. doi: 10.1111/j.1365-313X.2011.04547.x [DOI] [PubMed] [Google Scholar]
- 108.Jagadeeswaran G, Li YF, and Sunkar R. Redox signaling mediates the expression of a sulfate-deprivation-inducible microRNA395 in Arabidopsis. Plant J. 2014; 77: 85–96. doi: 10.1111/tpj.12364 [DOI] [PubMed] [Google Scholar]
- 109.Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su C. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 2006; 18: 412–421. doi: 10.1105/tpc.105.038943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pant BD, Buhtz A, Kehr J, Scheible WR. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008; 53: 731–8. doi: 10.1111/j.1365-313X.2007.03363.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009; 151: 2120–32. doi: 10.1104/pp.109.147280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rafiqi M, Bernoux M, Ellis JG, Dodds PN. In the trenches of plant pathogen recognition: Role of NB-LRR proteins. Semin Cell Dev Biol. 2009; 20: 1017–1024. doi: 10.1016/j.semcdb.2009.04.010 [DOI] [PubMed] [Google Scholar]
- 113.Fei Q, Xia R, Meyers BC. Phased, Secondary, Small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013; 25: 2400–2415. doi: 10.1105/tpc.113.114652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shivaprasad PV, Chen H-M, Patel K, Bond DM, Santos BACM, Baulcombe DC. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012; 24: 859–874. doi: 10.1105/tpc.111.095380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhu Q-H, Fan L, Liu Y, Xu H, Llewellyn D, Wilson I. MiR482 Regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS ONE. 2014; 8: e84390 doi: 10.1371/journal.pone.0084390 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(XLSX)
(XLSX)
(PDF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All sequence data have been submitted to the Sequence Read Archive (SRA) of the NCBI (http://www.ncbi.nlm.nih.gov/) as BioProject number PRJNA385718. BioSample accessions: SAMN06908861, SAMN06908862, SAMN06908863, SAMN06908864, SAMN06908865, SAMN06908866.