Accounting for specific alleles can be necessary to prevent the loss of significant biodiversity and ecosystem services.
Abstract
The delineation of conservation units (CUs) is a challenging issue that has profound implications for minimizing the loss of biodiversity and ecosystem services. CU delineation typically seeks to prioritize evolutionary significance, and genetic methods play a pivotal role in the delineation process by quantifying overall differentiation between populations. Although CUs that primarily reflect overall genetic differentiation do protect adaptive differences between distant populations, they do not necessarily protect adaptive variation within highly connected populations. Advances in genomic methodology facilitate the characterization of adaptive genetic variation, but the potential utility of this information for CU delineation is unclear. We use genomic methods to investigate the evolutionary basis of premature migration in Pacific salmon, a complex behavioral and physiological phenotype that exists within highly connected populations and has experienced severe declines. Strikingly, we find that premature migration is associated with the same single locus across multiple populations in each of two different species. Patterns of variation at this locus suggest that the premature migration alleles arose from a single evolutionary event within each species and were subsequently spread to distant populations through straying and positive selection. Our results reveal that complex adaptive variation can depend on rare mutational events at a single locus, demonstrate that CUs reflecting overall genetic differentiation can fail to protect evolutionarily significant variation that has substantial ecological and societal benefits, and suggest that a supplemental framework for protecting specific adaptive variation will sometimes be necessary to prevent the loss of significant biodiversity and ecosystem services.
INTRODUCTION
Invaluable economic, ecological, and cultural benefits are being lost worldwide as biodiversity decreases due to human actions (1–3). Legislation that provides a framework to protect unique species and population segments below the species level exists in many countries throughout the world (4, 5). Protection is achieved by assessing the health of a defined conservation unit (CU), and if the unit is at risk, attempts are made to preserve/restore critical habitat and restrict stressors until the risk is eliminated. Assessing risk and developing a protection strategy is not possible without first establishing unit boundaries. Because the number of units that can be effectively managed is resource-limited (6), the delineation of units should be strategic and should prioritize evolutionary significance (4, 7–11). Several criteria, such as genetic and ecological exchangeability (10), have been proposed for assessing evolutionary significance for CU delineation, but directly evaluating these criteria in natural populations is difficult (5).
Genetic methods play a pivotal role in the process of delineating CUs (10, 12). To this end, genetic data from different regions of the genome are combined to produce measurements of overall genetic differentiation between populations. These measurements represent typical regions of the genome and serve as a proxy for evolutionary significance (13, 14). However, because most genomic regions are primarily influenced by gene flow and genetic drift as opposed to selection, these measurements may fail to account for important adaptive differences between populations (12). Recent advances in genetic methodology facilitate the identification and evolutionary analysis of adaptively important loci (15–22) and provide an alternative way to assess evolutionary significance, but the utility of these loci for CU delineation is unclear and disputed (12, 23–27).
Pacific salmon (Oncorhynchus spp.) provide a unique opportunity to investigate the application of genetic tools to the conservation of biodiversity below the species level (4, 6, 28–30). Despite extensive conservation efforts, Pacific salmon have been extirpated from almost 40% of their historical range in the contiguous United States, and many remaining populations have experienced marked declines and face increasing challenges from climate change (31–35). Reintroduction attempts of extirpated populations are largely unsuccessful because precise natal homing across highly heterogeneous environments has resulted in divergent selection and abundant local adaptation (19, 36–38). Thus, maintaining existing stocks is critical for preserving the species themselves as well as the communities and ecosystems that rely on their presence (39). Genetic methods have been used extensively in delineating CUs in Pacific salmon [referred to as evolutionarily significant units (ESUs) or distinct population segments (DPSs) depending on the species] and, as a consequence of patterns of gene flow, have resulted in units that primarily reflect geography (40–43). Although current ESUs and DPSs certainly protect adaptive differences between distant populations, adaptations within highly connected populations are not necessarily protected (10, 34). However, the evolutionary significance of these adaptations and the potential long-term consequences of not independently protecting them are poorly understood.
Perhaps the most recognized example of differential adaptation within highly connected populations of Pacific salmon is variation in adult migration timing (also called run timing) (44–46). In contrast to typical adult salmon that mature sexually before freshwater migration, premature migrating individuals have a complex behavioral and physiological adaptation that allows them to access distinct habitats, distributing ocean-derived nutrients higher into watersheds, and spawn earlier in the season (46). Because of their distinct migration time and high fat content (47), premature migrating populations also provide additional, more-coveted, and culturally important harvest opportunities (48). For example, indigenous peoples in the Klamath Basin in northern California celebrated the return of premature migrating salmon with ceremonies that progressed upriver with the salmon migration (49).
Premature migrating populations have suffered grossly disproportionate impacts from human actions, such as dam building, mining, and logging, because of their extended time in freshwater and reliance on headwater habitat (14, 34, 40, 42, 46, 50, 51). With few exceptions (for example, some interior Columbia Basin locations), genetic analyses find little differentiation between proximate premature and mature migrating populations (13, 52–59), and as a result, they are generally grouped into the same ESU or DPS (40, 42). Therefore, despite the extirpation or substantial decline of premature migrating populations, the ESUs or DPSs to which they belong usually retain relatively healthy mature migrating populations and thus have low extinction risk overall (14, 40, 42). Here, we investigate the genetic and evolutionary basis of premature migration to explore potential consequences of not independently protecting this beneficial adaptation as well as the utility of genomics for informing conservation.
RESULTS
Initial genomic analysis consistent with current steelhead DPS delineations
Dramatic examples of premature migration are observed in coastal (noninterior) populations of steelhead (anadromous rainbow trout; Oncorhynchus mykiss) and Chinook salmon (Oncorhynchus tshawytscha). In these populations, premature migrating individuals (called summer steelhead or spring Chinook) use receding spring flows during freshwater migration to reach upstream habitat before hostile summer conditions in the lower watershed, hold for several months in deep cool pools while their gametes mature, then spawn at similar times to mature migrating individuals that have just entered freshwater (44, 46). We began our investigation by compiling a set of 148 steelhead samples from five coastal locations across four DPSs in California and Oregon (Fig. 1A). Four of the locations (Eel, New, Siletz, and North Umpqua) represent the few remaining watersheds with significant wild premature migrating populations. The fifth location, Scott, contains only mature migrating individuals. Our sampling focused as much as possible on individuals that could be confidently categorized as premature or mature migrating based on collection date and location (Fig. 1B and table S1).
Fig. 1. Genetic structure of premature and mature migrating steelhead populations.
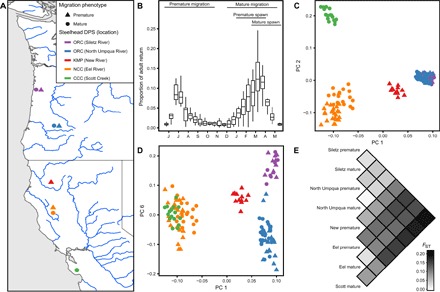
(A) Map of steelhead sample locations and migration phenotypes; color indicates location, and shape indicates migration phenotype. (B) Bimonthly proportion of annual adult steelhead return over Winchester Dam on the North Umpqua River (2003 to 2013); horizontal bars depict migration and spawn timing of premature and mature migrating populations. (C and D) Principal component analysis (PCA) and (E) pairwise FST estimates using genome-wide single-nucleotide polymorphism (SNP) data.
To collect high-resolution genomic information from these samples, we prepared individually barcoded restriction site associated DNA (RAD) libraries, sequenced them using paired-end Illumina technology, and aligned the sequence reads to a recent draft of the rainbow trout genome (tables S1 and S2) (60). We then used a probabilistic framework to discover SNPs and genotype them in each individual (61). A total of 9,864,960 genomic positions were interrogated in at least 50% of individuals, and 615,958 SNPs (that is, segregating sites) were identified (P < 10−6). Of these SNPs, 215,345 had one genotype posterior greater than 0.8 in at least 50% of individuals. Population structure characterization and genome-wide analyses in nonmodel organisms are typically carried out with far fewer SNPs (62). We conclude that the sequence data obtained are appropriate for genome-wide measurements and high-resolution analyses of specific genomic regions.
To characterize the genetic structure of these populations, we performed PCA and estimated pairwise FST using genome-wide genotype data (63). The first two PCs revealed four distinct groups corresponding to the four current DPSs (Fig. 1C). Siletz and North Umpqua, which are two different locations within the Oregon Coast DPS, did not break into distinct groups until PC6 (Fig. 1D), indicating relatively low genetic differentiation between distinct locations within a DPS. In all cases, individuals with different migration phenotypes from the same location were in the same group. The pairwise FST estimates also revealed strong genetic differentiation between locations but little differentiation between migration phenotypes from the same location (Fig. 1E). The mean pairwise FST between migration groups from the same location was 0.032 (range, 0.018 to 0.039; n = 3), whereas the mean between groups from different locations was 0.125 (range, 0.049 to 0.205; n = 25). The combination of this genetic structure and observations of hybridization between premature and mature migrating individuals (53) suggests higher rates of gene flow between different migration groups from the same location than between groups from different locations. Thus, as found in previous analyses, the overall genetic structure among steelhead populations is predominantly influenced by geography, as opposed to migration phenotype. We conclude that measurements of overall genetic differentiation from genome-wide SNP data are consistent with current steelhead DPS delineations.
Premature migrating steelhead explained by a single allelic evolutionary event at a single locus
To identify genomic loci associated with premature migration, we performed association mapping of migration category. We used a likelihood ratio test (64) with λ correction for population stratification (65) to compare 181,954 SNPs between migration categories in North Umpqua and found 14 SNPs that were significant (Bonferroni-corrected α level: P < 0.05). Strikingly, all of these SNPs were located within a 211,251–base pair (bp) region (568,978 to 780,229) on a single 1.95-Mb scaffold (Fig. 2A; fig. S1, A and B; and table S3). Furthermore, when this analysis was repeated with Eel individuals using 170,678 SNPs, we obtained a similar pattern of association (Fig. 2B; fig. S1, C and D; and table S3). The strongest associated SNPs in both sample locations were flanking two restriction sites approximately 50 kb apart and located just upstream and within a gene identified as GREB1L (Fig. 2C; see Discussion for more information on GREB1L). The strength of these associations was unexpected given the phenotypic complexity of premature migration and the relatively low number of samples analyzed. We conclude that the same single locus is strongly associated with migration phenotype in at least two DPSs.
Fig. 2. Genetic and evolutionary basis of premature migration in steelhead.
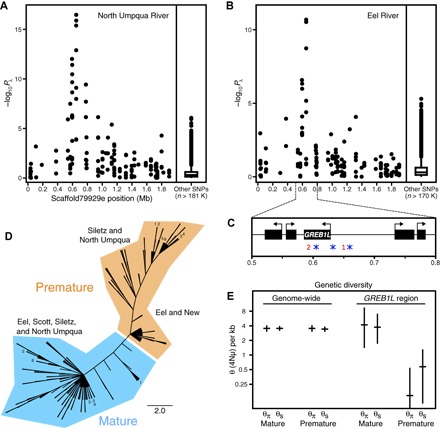
Association mapping of migration category in (A) North Umpqua River and (B) Eel River steelhead. (C) Gene annotation of region with strong association; red numbers indicate genomic positions of two restriction sites flanked by strongest associated SNPs, and blue asterisks indicate positions of amplicon sequencing. (D) Phylogenetic tree depicting maximum parsimony of phased amplicon sequences from all individuals; branch lengths, with the exception of terminal tips, reflect nucleotide differences between haplotypes; numbers identify individuals with one haplotype in each migration category clade. (E) Genome-wide and GREB1L region diversity estimates in North Umpqua for each migration category with 95% confidence intervals from coalescent simulations.
To investigate the evolutionary history of this locus, we sequenced three amplicons, each of approximately 500 bp, from the GREB1L region in all individuals from all populations (Fig. 2C and tables S1, S4, and S5) and used these sequences to construct a haplotype tree based on parsimony (66). Strikingly, the tree contained two distinct monophyletic groups corresponding to migration phenotype (Fig. 2D). For 123 of 129 individuals, both haplotypes separated into the appropriate migration category clade. The remaining six individuals (four Siletz and two North Umpqua samples originally classified as mature migrating) had one haplotype in each migration category clade (Fig. 2D), suggesting heterozygosity at the causative polymorphism(s). Furthermore, although there was little differentiation within the mature migration clade, premature migration haplotypes from Siletz and North Umpqua were more divergent from the mature migration clade than those from Eel and New (Fig. 2D; see Discussion for more information on heterozygotes and differentiation within the premature clade). The overall tree topology is inconsistent with premature migration alleles originating from independent evolutionary events in different locations because separate mutational events would be expected to occur on different haplotype backgrounds and result in premature migration alleles having a polyphyletic origin (15). We conclude that there is a nearly complete association between variation at this locus and migration category and that the premature migration alleles from all locations arose from a single evolutionary event.
To examine the evolutionary mechanisms leading to the dispersal of the premature migration allele as well as reconcile the difference between patterns of variation at the GREB1L locus and overall genetic structure, we summarized patterns of genetic variation using two estimators of θ (4Nμ). One estimator is based on average pairwise differences (θπ) (67), and the other is based on the number of segregating sites (θS) (68). When genome-wide data were used, both estimators produced similar θ values for each migration category (Fig. 2E). The GREB1L region of mature migrating individuals also produced θ values similar to the genome-wide analysis. However, premature migrating individuals from North Umpqua had strikingly lower θ values (Fig. 2E) and a significantly skewed site frequency spectrum (SFS) (Tajima’s D = −2.08; P = 0.001) (69) indicative of strong, recent positive selection in the GREB1L region. Premature migrating individuals from Eel also had reduced θ values in the GREB1L region (premature: θπ/kb = 2.48, θS/kb = 2.67; mature: θπ/kb = 3.59, θS/kb = 4.00), but the SFS was not significantly skewed, consistent with an older selection event. Although both demography and selection can reduce nucleotide diversity and skew the SFS, this pattern is specific to the GREB1L region as opposed to genome-wide, implicating selection as the cause. Furthermore, the combination of a stronger signature of selection and a more divergent sequence pattern in the northern premature migration haplotypes is consistent with a northward movement of the premature migration allele. We conclude that, upon entering new locations via straying, positive selection allowed the premature migration allele to persist despite ongoing hybridization with local mature migrating populations.
Premature migrating Chinook also explained by a single allelic evolutionary event in GREB1L region
To broaden our investigation into premature migration, we compiled a set of 250 Chinook samples from nine locations across five ESUs in California, Oregon, and Washington (Fig. 3A). Similar to steelhead, our sampling focused as much as possible on individuals that could be confidently categorized as premature or mature migrating based on collection time and location (table S6). We then prepared individually barcoded RAD libraries, sequenced them using paired-end Illumina technology, and aligned the sequence reads to the same rainbow trout reference assembly used above (tables S6 and S7). No reference genome is available for Chinook, and rainbow trout, which diverged from Chinook approximately 10 to 15 million years ago (70, 71), is the closest relative with a draft genome assembly. With the methods described above, a total of 3,910,009 genomic positions were interrogated in at least 50% of individuals and 301,562 SNPs were identified (P < 10−6). Of these SNPs, 55,797 had one genotype posterior greater than 0.8 in at least 50% of individuals. Although the alignment success was lower and subsequent SNP discovery and genotyping produced fewer SNPs compared to steelhead, the large number of SNPs discovered and genotyped should still be adequate for downstream analysis.
Fig. 3. Genetic and evolutionary basis of premature migration in Chinook.
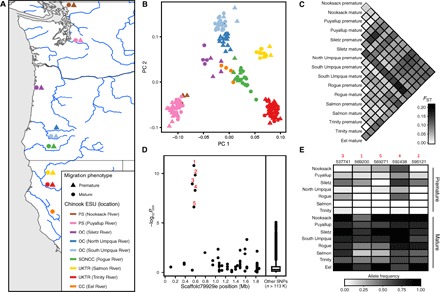
(A) Map of Chinook sample locations and migration phenotypes; color indicates location, and shape indicates migration category. (B) PCA and (C) pairwise FST estimates using genome-wide SNP data. (D) Association mapping of migration category in Chinook; red numbers indicate significant SNPs. (E) Allele frequency shift at significant SNPs between premature and mature migrating populations. Black numbers indicate SNP position on scaffold79929e.
To characterize the genetic structure of these populations, we performed PCA and estimated pairwise FST using the genotype information described above. The first two PCs revealed four groups: the largest group contained all coastal ESUs, the second contained the two Puget Sound ESU locations, and the last two groups corresponded to the two locations within the Upper Klamath–Trinity Rivers ESU and were only differentiated by the second axis (Fig. 3B). In all cases, individuals from the same location but with different migration phenotypes were in the same group, and locations within groups became differentiated as additional PCs were examined. The mean pairwise FST between migration categories from the same location was 0.037 (range, 0.009 to 0.093; n = 7), and the mean between groups from different locations was 0.097 (range, 0.021 to 0.199; n = 113) (Fig. 3C). Thus, similar to what we found in steelhead, the overall genetic structure is strongly influenced by geography, as opposed to migration phenotype. We conclude that measurements of overall genetic differentiation from genome-wide SNP data are consistent with current Chinook ESUs.
To investigate the genetic architecture and evolutionary basis of premature migration in Chinook, we conducted association mapping with 114,036 SNPs using a generalized linear framework with covariate correction for population stratification (65, 72). Strikingly, we again found a single significant peak of association (Bonferroni-corrected α level: P < 0.05) that contained five SNPs within 57,380 bp (537,741 to 595,121) in the same GREB1L region identified in steelhead (Fig. 3D and table S8). We next examined allele frequencies at these five SNPs and found a strong and consistent shift between all premature and mature migrating populations independent of location (Fig. 3E). Thus, despite having a lower genomic resolution and fewer samples per location, these results demonstrate that the GREB1L region is also the primary locus associated with premature migration in Chinook. Furthermore, the shift of allele frequencies in the same direction between premature and mature migrating populations across all locations is inconsistent with the premature migration alleles in Chinook being a product of multiple independent evolutionary events. Although the genomic region was consistent between species, the SNPs identified in Chinook were distinct from those in steelhead (tables S3 and S8). That is, the premature and mature migrating Chinook haplotypes are more similar to each other than to either of the steelhead haplotypes and vice versa, suggesting independent allelic evolutionary events in each species. We conclude that the same evolutionary mechanism used in steelhead, with a single allelic evolutionary event in the GREB1L region that subsequently spread to different locations, also explains premature migration in Chinook.
DISCUSSION
Our association analysis across multiple populations in each of two different species, as well as an independent analysis on Klickitat River steelhead (73), suggests that either the function or the regulation of GREB1L is modified in premature migrating individuals. Both GREB1L and its paralog GREB1 are ubiquitous in and highly conserved across vertebrates. Although GREB1 is known to encode a nuclear hormone receptor coactivator (74) and has been implicated in diverse biological processes (75–80), relatively little is known about GREB1L. However, a recent study found that GREB1L is differentially regulated by feeding and fasting in AgRP (agouti-related protein) neurons of the hypothalamic arcuate nucleus in mice (81). The strength of the associations, as well as the known role of AgRP neurons in modulating diverse behavior and metabolic processes such as foraging and fat storage (81, 82), provides evidence for and an explanation of how the complex premature migration phenotype could be controlled by this single locus. An alternative explanation is that the GREB1L region only influences a subset of the phenotypic components of premature migration and that other important loci were not identified because of technical or biological reasons. Regardless, our results indicate that an appropriate genotype at this locus is necessary for successful premature migration.
Given that premature migration alleles at this locus are critical for premature migration, our results on the evolutionary history of these alleles provide important insights into the potential for premature migration to persist during declines and reemerge if lost. Finding that the same locus is associated with premature migration in both steelhead and Chinook indicates that genetic mechanisms capable of producing this phenotype are very limited. Although some loci can be predisposed to functionally equivalent mutations in relatively short evolutionary time scales (83, 84), this does not appear to be the case with the GREB1L region. In predisposed loci, several independent mutations with the same phenotypic effect are observed in different populations of a single species (83, 84). In contrast, our survey of many populations revealed only one evolutionary event that produced a premature migration allele in each species despite the 10 to 15 million years since they diverged (70, 71). Regardless of whether or not additional allelic evolutionary events have occurred (for example, in the interior Columbia Basin), our finding that a broad array of populations shares alleles from a single evolutionary event suggests that mutational events that create new premature migration alleles are rare. Thus, if current premature migration alleles are lost, new premature migration alleles and the phenotype they promote cannot be expected to reevolve in time frames relevant to conservation planning (for example, tens to hundreds of years).
The rarity of mutational events that produce premature migration alleles at this locus highlights the importance of existing premature migration alleles. Unlike alleles with a small effect on phenotype, alleles with a large effect on phenotype are expected to be rapidly lost from a population when there is strong selection against the phenotype they promote (85). An important exception to this is when an allele is recessive and therefore masked in the heterozygous state (15, 85). Thus, the inheritance pattern of the GREB1L locus has critical implications for the persistence of premature migration alleles during declines of the premature migration phenotype. Although our sampling focused on migration peaks (Fig. 1B) and was not designed to investigate the migration phenotype of heterozygotes, the recently published Klickitat data (73) included samples collected outside the migration peaks. Strikingly, a reanalysis of these data suggests that the same haplotype is associated with premature migration (Fig. 4A and table S3) and that heterozygotes display an intermediate phenotype (Fig. 4B and fig. S2). This explains the high frequency of heterozygotes in our Siletz mature migrating samples (4 of 10), which were collected before the peak of mature migration and far upstream in the watershed (table S1). Thus, the premature migration allele does not appear to be masked in the heterozygous state and cannot be expected to be maintained as standing variation in populations that lack the premature migration phenotype.
Fig. 4. Inheritance pattern of GREB1L locus.
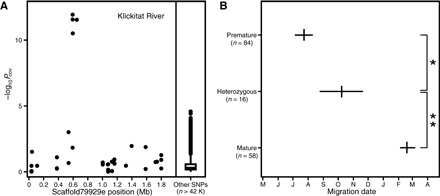
(A) Association mapping of migration date in Klickitat River steelhead. (B) Mean migration date and 95% confidence interval of the mean in Klickitat River steelhead categorized as homozygous for the premature migration allele, heterozygous, and homozygous mature. *P = 0.00574; **P = 2.95 × 10−5.
Two additional lines of evidence suggest that the premature migration allele will not be maintained as standing variation in mature migrating populations. First, the combination of the strong bimodal phenotypic distribution that is usually observed (for example, Fig. 1B) and the ecology of premature migration (see Introduction) (44, 46) suggests a general pattern of disruptive selection against individuals with an intermediate phenotype (for example, heterozygotes). Although heterozygotes are expected to be produced by hybridization in locations where both migration categories exist (for example, we observed two heterozygotes in North Umpqua, which has the lowest genetic differentiation between migration groups; Fig. 1E), their presence does not suggest that the premature migration allele will be maintained by mature migrating populations. Second, the genetic differentiation between premature migration haplotypes from California and Oregon steelhead (Fig. 2D) indicates that, unlike mature migration alleles, premature migration alleles are not freely moving across this area. This result reveals that mature migrating populations do not act as an influential source or conduit of premature migration alleles despite being abundant and broadly distributed. Therefore, premature migrating populations appear ultimately necessary for both the maintenance and spread of these alleles.
Previously, studies revealing that overall genetic structure among populations of steelhead and Chinook primarily reflects geography (as opposed to migration phenotype) suggested that premature migration evolved independently in many locations within each species (13, 54, 59). This implied that premature migration is evolutionarily replaceable over time frames relevant to conservation planning (13) and is not an important component in the evolutionary legacy of the species (14). Although these interpretations were logical given the data available at that time, our results demonstrate that the evolution was not independent in each location but instead relied on preexisting genetic variation. Thus, although evolving the premature migration phenotype in new locations could be rapid if robust premature migrating populations are present in proximate locations, the widespread extirpation and decline of premature migrating populations (14, 34, 40, 42, 46, 50, 51) has greatly diminished the potential restoration and expansion (for example, into new habitats that become available with climate change) of premature migration across at least a substantial proportion of the range for both species (19).
Future work characterizing the distribution of premature migration alleles would improve our understanding of the extent to which the potential restoration and expansion of the premature migration phenotype has been diminished. For example, testing for the presence of premature migration alleles in locations where the phenotype has recently been extirpated would reveal how quickly these alleles are lost and potential restoration options. One possibility is that some heterozygotes still exist in these locations and could be used to restore the premature migration phenotype. The alternative is that the premature migration allele has already been lost and restoration of the phenotype would require introducing the allele from an outside population. Regardless, the results presented here will serve as a foundation for future work to determine optimal strategies for the conservation and restoration of premature migrating populations. Additionally, given the complex premature migration phenotype and evolutionary importance of premature migration alleles, future work that provides mechanistic insight into the GREB1L locus [for example, identifying the causative polymorphism(s) and characterizing expression profiles] could have important implications for areas ranging from conservation to biomedicine.
The combination of three key results from this study has broad conservation implications, which highlight the utility of genomics for informing conservation. First, we present an example of how a single allele at a single locus can have economic, ecological, and cultural importance. Second, we show that mutations producing an important allele can be very rare from an evolutionary perspective, suggesting that the allele will not readily reevolve if lost. Last, we observe that patterns of significant adaptive allelic variation can be completely opposite from patterns of overall genetic differentiation. Together, our results demonstrate that CUs reflecting overall genetic differentiation can fail to protect evolutionarily significant variation that has substantial ecological and societal benefits, and suggest that a supplemental framework for protecting specific adaptive variation will sometimes be necessary to prevent the loss of significant biodiversity and ecosystem services.
MATERIALS AND METHODS
Sample collection and molecular biology
Fin clips were taken from live adults or post-spawn carcasses (tables S1 and S6), dried on Whatman qualitative filter paper (grade 1), and stored at room temperature. DNA was extracted with either the DNeasy Blood and Tissue Kit (Qiagen) or a magnetic bead–based protocol (22) and quantified using Quant-iT PicoGreen dsDNA Reagent (Thermo Fisher Scientific) with an FLx800 Fluorescence Reader (BioTek Instruments).
SbfI RAD libraries were prepared with well and plate (when applicable) barcodes using either the traditional or new RAD protocol (22) and sequenced with paired-end 100-bp reads on an Illumina HiSeq 2500 (tables S2 and S7). In some cases, the same sample was included in multiple libraries to improve sequencing coverage.
For amplicon sequencing, genomic DNA extractions were rearrayed into 96-well plates and diluted 1:40 with low TE buffer (pH 8.0; 10 mM tris-HCl and 0.1 mM EDTA). Two microliters of this diluted sample was used as polymerase chain reaction (PCR) template for each of the three amplicons in the GREB1L region (Fig. 2 and table S4). Multiple forward primers were synthesized for each amplicon. Each forward primer contained a partial Illumina adapter sequence, a unique inline plate barcode, and the amplicon-specific sequence (tables S4 and S5). Initial PCRs were performed in 96-well plates using OneTaq DNA polymerase (New England Biolabs) at the recommended conditions with an annealing temperature of 61°C and 35 cycles. These reaction plates were then combined into a single plate that preserved the well locations. The pooled PCR products were cleaned with Ampure XP beads (Beckman Coulter), and a second round of PCR with eight cycles was performed to add the remaining Illumina adapter sequence and a unique TruSeq barcode to each well (tables S4 and S5). From each final PCR, 2 μl was removed, pooled, and purified with Ampure XP beads. The final amplicon library was sequenced with paired-end 300-bp reads on an Illumina MiSeq.
RAD analysis
RAD sequencing data were demultiplexed by requiring a perfect barcode and partial restriction site match (22). Sequences were aligned to a slightly modified version of a recent rainbow trout genome assembly (see scaffold79929e assembly and annotation) (60) using the backtrack algorithm of Burrows-Wheeler Aligner (BWA) (86) with default parameters. SAMtools (87) was used to sort, filter for proper pairs, remove PCR duplicates, and index binary alignment map (BAM) files (tables S2 and S7). In cases where the same sample was sequenced in multiple libraries, BAM files from the same sample were merged before indexing using SAMtools (tables S1, S2, S6, and S7).
Additional BAM file sets were generated to account for technical variation among samples. To minimize variation associated with the two distinct library preparation protocols used in Chinook (table S7) (22), we generated a set of single-end BAM files for Chinook that contained only trimmed reads from the restriction site end of the RAD fragments. To prepare these files, we trimmed these reads to 75 bp from the 3′ end after removing 5 bp from the 5′ end. Next, paired-end alignments were performed and processed as above. Last, reads from the variable end of RAD fragments were removed (table S7). To remove variation associated with variable sequencing depth, we generated a set of subsampled BAM files by using SAMtools to randomly sample approximately 120,000 alignments from paired-end BAM files for steelhead and approximately 60,000 alignments from single-end BAM files for Chinook. Subsampling to a lower number of alignments allows more individuals to be included in the analysis. We determined the optimal alignment numbers for subsampling by testing a variety of thresholds and determining the minimum before which the sample groupings started to become dispersed in PCA.
All RAD analyses were performed using Analysis of Next Generation Sequencing Data (ANGSD) (61) with a minimum mapping quality score (minMapQ) of 10, a minimum base quality score (minQ) of 20, and the SAMtools genotype likelihood model (GL 1) (88). Unless otherwise noted, samples with less alignments than required for subsampling were excluded (tables S1 and S6), and only sites represented in at least 50% of the included samples (minInd) were used.
PCA and association mapping were performed by identifying polymorphic sites (SNP_pval 1e-6), inferring major and minor alleles (doMajorMinor 1) (72), estimating allele frequencies (doMaf 2) (64), and retaining SNPs with a minor allele frequency of at least 0.05 (minMaf). For PCA, subsampled BAM files were used and genotype posterior probabilities were calculated with a uniform prior (doPost 2). The ngsCovar (89) function implemented in ngsTools (63) was used to calculate a covariance matrix from called genotypes. For association mapping, paired-end BAM files were used with two distinct tests. The frequency test with known major and minor alleles (doAsso 1) implements a likelihood ratio test using read counts (64). This test has good statistical power even with lower coverage data but does not allow the inclusion of covariates to correct for population stratification. The score test (doAsso 2) uses a generalized linear framework on posterior genotype probabilities (72). This test allows the inclusion of covariates to correct for population stratification but has less statistical power than the frequency test. For the Umpqua and Eel steelhead associations, the frequency test with λ correction for population stratification (65) was used because there were relatively few samples and a weak population structure. λ is the ratio of observed and expected median χ2 values and used to correct the observed χ2 values before converting them to P values (fig. S1, A and C, and table S3) (65). For the Chinook association, the score test with covariate correction for population stratification was used because there were many samples and a complex population structure (fig. S1E). The positions of each sample along the first 15 PCs were used as covariates.
Genome-wide FST between population pairs was estimated by first estimating an SFS for each population (doSaf) (90) using paired-end BAM files for steelhead and single-end BAM files for Chinook. Two-dimensional SFS and global FST (weighted) between each population pair were then estimated using realSFS (61).
To calculate Watterson’s θ (68), Tajima’s θ (67), and Tajima’s D (69), we used SFS that were estimated as described above as priors (pest) with paired-end BAM files to calculate each statistic for each site (doThetas), which were averaged to obtain a single value for each statistic (91). The analysis was restricted to 565,000 to 785,000 bp of scaffold79929e for the GREB1L region analysis.
The coalescent simulation program ms (92) was used to determine 95% confidence intervals for the θ estimates from 10,000 simulations under a neutral demographic model. The input number of chromosomes was equal to the number of individuals used to calculate the θ statistics. For genome-wide confidence intervals, 100 independent loci and an input θ of 1, which is the approximate θ of a single RAD tag, were used. For the GREB1L region confidence intervals, a single locus and the empirical θ estimates were used. The significance of the empirical Tajima’s D value was evaluated by generating a Tajima’s D distribution from 10,000 ms simulations under a neutral demographic model. A single locus and the average between empirical values of Watterson’s and Tajima’s θ values in the GREB1L region were used. A Tajima’s D distribution was also generated using the extremes of the θ confidence intervals, and the empirical value remained significant.
Allele frequencies were estimated (doMaf 1) (64) for the significant Chinook SNPs in each population that had at least four individuals with enough alignments for subsampling. Paired-end BAM files were used with the reference genome assembly as the prespecified major allele (doMajorMinor 4). Because some populations had low sample sizes, all samples were included regardless of alignment number.
Amplicon analysis
Amplicon sequence data were demultiplexed by requiring perfect barcode and primer matches. Sequences were aligned to the reference genome assembly described above using the BWA-SW algorithm (93) with default parameters, and SAMtools was used to sort, filter for proper pairs, and index BAM files (table S5).
Phylogenetic analysis was performed on samples in which two or more amplicons had at least 20 alignments (tables S1 and S5). Genotypes for all sites were called using ANGSD with the SAMtools genotype likelihood model, a uniform prior, and a posterior cutoff of 0.8. The genotype output file was parsed and converted into biallelic consensus sequences, with an IUPAC (International Union of Pure and Applied Chemistry) nucleotide code denoting heterozygous positions. These consensus sequences were input into fastPHASE (94) to produce 1000 output files that each contained two phased haplotype sequences per individual. Default parameters were used except that a distinct subpopulation label was specified for each of the five locations and base calls with a posterior of less than 0.8 were converted to Ns (unknown bases). Parsimony trees were then constructed from each fastPHASE output, and a consensus tree was called using PHYLIP (66).
In the initial phylogenetic analysis, one sample from the Eel River that was originally classified as premature migrating clustered in the mature migration clade (table S1). A PCA specific to the Eel River placed this sample at an intermediate position between mature migrating and premature migrating sample groups. Furthermore, this was the only Eel River sample that was homozygous for a haplotype on chromosome Omy05 associated with residency (20). Examination of the original sampling information revealed that this fish was much smaller than others and collected upstream from the main premature steelhead holding area (56), suggesting that it was a resident trout as opposed to an anadromous steelhead. Therefore, this sample was removed, and the analysis was rerun.
Scaffold79929e assembly and annotation
Our initial RAD analysis was aligned against a published reference genome assembly (60) and identified highly associated SNPs on three independent scaffolds. Given the state of the assembly, the sizes of the scaffolds with highly associated SNPs, and the positions of the highly associated SNPs on the scaffolds, we hypothesized that these scaffolds might be physically linked despite not being connected in the current assembly. We aligned four large-insert mate-pair libraries to the published assembly to look for linkages and estimate the distance between linked scaffolds (table S9). A perfect sequence match was required, and alignments to regions with high coverage were discarded. The resulting alignments from all libraries strongly supported a linear assembly with a total size of 1,949,089 bp that included the three associated scaffolds as well as four others (tables S9 and S10). This assembled scaffold was named scaffold79929e (e for extended) and added to the published assembly, and the seven independent scaffolds that composed it were removed to create the modified reference assembly used in this study.
Scaffold79929e was annotated with MAKER (95) using rainbow trout and Atlantic salmon (Salmo salar) EST (expressed sequence tag) sequences from the NCBI (National Center for Biotechnology Information) database, the UniProt/Swiss-Prot database for protein homology, a rainbow trout repeat library (60) for masking, AUGUSTUS (human) and SNAP (mamiso) gene predictors, a maximum intron size of 20,000 bp for evidence alignments, and otherwise default parameters.
Klickitat steelhead analysis
Single-end RAD data from 237 Klickitat River steelhead samples (73) were aligned to the modified rainbow trout genome as described above. SAMtools (87) was used to remove unaligned reads, sort, index, and randomly subsample BAM files to 500,000 reads to reduce the effect of PCR duplicates (96). All subsequent analyses were performed on subsampled BAM files using ANGSD (61).
Association mapping was performed using the score test (doAsso 2), with the migration date at Lyle Falls (May 1 set to day 1) (73) as a quantitative proxy for the premature migration phenotype (yQuant) because more direct measures (for example, gonadal maturation and body fat content at freshwater entry) were not available (this information is difficult to obtain and may require lethal sampling). The positions of each sample along the first nine PCs were used as covariates to correct for population stratification (fig. S1F). The PCA used to generate covariates was performed as described above.
Genotype data from the four associated SNPs were used to categorize individuals as homozygous for the mature migration allele, heterozygous, or homozygous premature. Genotypes were called (doGeno 4) with a uniform prior (doPost 2) and a posterior probability cutoff of 0.8 (postCutoff 0.8). Seven hundred fifty-one of 948 genotypes passed this cutoff. Two SNPs were flanking sites on the same RAD tag, had near-perfect consistency between genotype calls, and were treated as a single genotype for categorization. For an individual to be categorized as homozygous or heterozygous, all called genotypes were required to be in agreement and at least two of the three genotypes must have been called. A total of 158 samples passed these requirements, whereas 51 failed because less than two genotypes were called and 28 failed because of disagreement between called genotypes.
Migration date means were calculated with May 1 set to day 1 because it is an approximate date for the beginning of premature migration at Lyle Falls (73). Confidence intervals of the means were calculated by bootstrapping with 1000 replicates. The significance of differences in mean migration date between genotype categories was evaluated with Welch’s t test. May 1 is somewhat arbitrary, and a subset of premature migrating individuals likely ascends Lyle Falls before this date (fig. S2). Furthermore, some individuals may enter freshwater then hold below Lyle Falls for an extended period before ascending to spawn. In either of these scenarios, individuals would be assigned a migration date indicative of mature migration, even though they were premature migrating. With the available information, we cannot be sure which individuals migrated under these scenarios. However, setting May 1 to day 1 is a conservative approach that, if anything, should underestimate the significance of the differences between mean migration dates for each genotype (Fig. 4B and fig. S2).
Acknowledgments
We thank C. Liu, J. Saxon, P. Tronquet, the National Marine Fisheries Service, the Oregon Department of Fish and Wildlife, and the California Department of Fish and Wildlife for assistance with sample acquisition as well as F. Allendorf, E. Anderson, J. Ashander, G. Coop, and R. Waples for valuable comments on an earlier version of the manuscript. M.R.M. thanks C. Doe, Y. Palti, and G. Thorgaard for invaluable support and encouragement. This work used the Vincent J. Coates Genomics Sequencing Laboratory at the University of California, Berkeley, supported by NIH S10 Instrumentation grants S10RR029668 and S10RR027303. Funding: This study was funded by the Department of Animal Science and College of Agricultural and Environmental Sciences at the University of California, Davis. Author contributions: Conceptualization: D.J.P., S.M.O., T.Q.T., and M.R.M.; resources: S.M.O., T.J.H., A.P.S., and M.R.M.; investigation: D.J.P., S.M.O., T.Q.T., O.A.A., H.S.L., I.K.S., and M.R.M.; formal analysis: D.J.P., T.Q.T., and M.R.M.; visualization: D.J.P., T.Q.T., and M.R.M.; writing of original draft: D.J.P. and M.R.M.; writing, review, and editing: D.J.P., S.M.O., T.Q.T., O.A.A., H.S.L., I.K.S., T.J.H., A.P.S., and M.R.M.; funding acquisition: S.M.O. and M.R.M.; supervision: M.R.M. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All sequence data generated in this study are available at the NCBI Sequence Read Archive with identifier SRP101883. Scripts used to execute analysis software are available at https://github.com/djprince/premature-migration-2017. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/8/e1603198/DC1
fig. S1. Observed versus expected statistics for association mapping of migration phenotype.
fig. S2. Migration date distribution of Klickitat River steelhead at Lyle Falls with weekly binning.
table S1. Steelhead samples.
table S2. Steelhead RAD sequence.
table S3. Steelhead migration associated SNPs.
table S4. Steelhead amplicon primers.
table S5. Steelhead amplicon sequence.
table S6. Chinook samples.
table S7. Chinook RAD sequence.
table S8. Chinook migration associated SNPs.
table S9. Scaffold links.
table S10. Scaffold79929e assembly.
REFERENCES AND NOTES
- 1.Millennium Ecosystem Assessment, Ecosystems and Human Well-Being (Island Press, 2005). [Google Scholar]
- 2.Krauss J., Bommarco R., Guardiola M., Heikkinen R. K., Helm A., Kuussaari M., Lindborg R., Ockinger E., Pärtel M., Pino J., Pöyry J., Raatikainen K. M., Sang A., Stefanescu C., Teder T., Zobel M., Steffan-Dewenter I., Habitat fragmentation causes immediate and time-delayed biodiversity loss at different trophic levels. Ecol. Lett. 13, 597–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pimm S. L., Jenkins C. N., Abell R., Brooks T. M., Gittleman J. L., Joppa L. N., Raven P. H., Roberts C. M., Sexton J. O., The biodiversity of species and their rates of extinction, distribution, and protection. Science 344, 1246752 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Waples R. S., Pacific salmon, Oncorhynchus spp., and the definition of “species” under the Endangered Species Act. Mar. Fish. Rev. 53, 11–22 (1991). [Google Scholar]
- 5.F. W. Allendorf, G. H. Luikart, S. N. Aitken, Conservation and the Genetics of Populations (Wiley-Blackwell, ed. 2, 2012). [Google Scholar]
- 6.Waples R. S., Evolutionarily significant units, distinct population segments, and the endangered species act: Reply to Pennock and Dimmick. Conserv. Biol. 12, 718–721 (1998). [Google Scholar]
- 7.Ryder O. A., Species conservation and systematics: The dilemma of subspecies. Trends Ecol. Evol. 1, 9–10 (1986). [DOI] [PubMed] [Google Scholar]
- 8.Dizon A. E., Lockyer C., Perrin W. F., Demaster D. P., Sisson J., Rethinking the stock concept: A phylogeographic approach. Conserv. Biol. 6, 24–36 (1992). [Google Scholar]
- 9.Moritz C., Defining ‘evolutionarily significant units’ for conservation. Trends Ecol. Evol. 9, 373–375 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Crandall K. A., Bininda-Emonds O. R. P., Mace G. M., Wayne R. K., Considering evolutionary processes in conservation biology. Trends Ecol. Evol. 15, 290–295 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Fraser D. J., Bernatchez L., Adaptive evolutionary conservation: Towards a unified concept for defining conservation units. Mol. Ecol. 10, 2741–2752 (2001). [PubMed] [Google Scholar]
- 12.Funk W. C., McKay J. K., Hohenlohe P. A., Allendorf F. W., Harnessing genomics for delineating conservation units. Trends Ecol. Evol. 27, 489–496 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waples R. S., Teel D. J., Myers J. M., Marshall A. R., Life-history divergence in Chinook salmon: Historic contingency and parallel evolution. Evol. Int. J. Org. Evol. 58, 386–403 (2004). [PubMed] [Google Scholar]
- 14.T. H. Williams, Upper Klamath and Trinity River Chinook Salmon Biological Review Team Report (U.S. Department of Commerce, 2013). [Google Scholar]
- 15.Colosimo P. F., Hosemann K. E., Balabhadra S., Villarreal G. Jr., Dickson M., Grimwood J., Schmutz J., Myers R. M., Schluter D., Kingsley D. M., Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307, 1928–1933 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Miller M. R., Dunham J. P., Amores A., Cresko W. A., Johnson E. A., Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 17, 240–248 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baird N. A., Etter P. D., Atwood T. S., Currey M. C., Shiver A. L., Lewis Z. A., Selker E. U., Cresko W. A., Johnson E. A., Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLOS ONE 3, e3376 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohenlohe P. A., Bassham S., Etter P. D., Stiffler N., Johnson E. A., Cresko W. A., Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLOS Genet. 6, e1000862 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller M. R., Brunelli J. P., Wheeler P. A., Liu S., Rexroad C. E. III, Palti Y., Doe C. Q., Thorgaard G. H., A conserved haplotype controls parallel adaptation in geographically distant salmonid populations. Mol. Ecol. 21, 237–249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearse D. E., Miller M. R., Abadía-Cardoso A., Garza J. C., Rapid parallel evolution of standing variation in a single, complex, genomic region is associated with life history in steelhead/rainbow trout. Proc. Biol. Sci. 281, 20140012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barson N. J., Aykanat T., Hindar K., Baranski M., Bolstad G. H., Fiske P., Jacq C., Jensen A. J., Johnston S. E., Karlsson S., Kent M., Moen T., Niemelä E., Nome T., Næsje T. F., Orell P., Romakkaniemi A., Sægrov H., Urdal K., Erkinaro J., Lien S., Primmer C. R., Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature 528, 405–408 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Ali O. A., O’Rourke S. M., Amish S. J., Meek M. H., Luikart G., Jeffres C., Miller M. R., RAD Capture (Rapture): Flexible and efficient sequence-based genotyping. Genetics 202, 389–400 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allendorf F. W., Hohenlohe P. A., Luikart G., Genomics and the future of conservation genetics. Nat. Rev. Genet. 11, 697–709 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Shafer A. B. A., Wolf J. B. W., Alves P. C., Bergström L., Bruford M. W., Brännström I., Colling G., Dalén L., De Meester L., Ekblom R., Fawcett K. D., Fior S., Hajibabaei M., Hill J. A., Hoezel A. R., Höglund J., Jensen E. L., Krause J., Kristensen T. N., Krützen M., McKay J. K., Norman A. J., Ogden R., Österling E. M., Ouborg N. J., Piccolo J., Popović D., Primmer C. R., Reed F. A., Roumet M., Salmona J., Schenekar T., Schwartz M. K., Segelbacher G., Senn H., Thaulow J., Valtonen M., Veale A., Vergeer P., Vijay N., Vilà C., Weissensteiner M., Wennerström L., Wheat C. W., Zieliński P., Genomics and the challenging translation into conservation practice. Trends Ecol. Evol. 30, 78–87 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Shafer A. B. A., Wolf J. B. W., Alves P. C., Bergström L., Colling G., Dalén L., De Meester L., Ekblom R., Fior S., Hajibabaei M., Hoezel A. R., Hoglund J., Jensen E. L., Krützen M., Norman A. J., Österling E. M., Ouborg N. J., Piccolo J., Primmer C. R., Reed F. A., Roumet M., Salmona J., Schwartz M. K., Segelbacher G., Thaulow J., Valtonen M., Vergeer P., Weissensteiner M., Wheat C. W., Vilà C., Zieliński P., Reply to Garner et al. Trends Ecol. Evol. 31, 83–84 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Garner B. A., Hand B. K., Amish S. J., Bernatchez L., Foster J. T., Miller K. M., Morin P. A., Narum S. R., O’Brien S. J., Roffler G., Templin W. D., Sunnucks P., Strait J., Warheit K. I., Seamons T. R., Wenburg J., Olsen J., Luikart G., Genomics in conservation: Case studies and bridging the gap between data and application. Trends Ecol. Evol. 31, 81–83 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Pearse D. E., Saving the spandrels? Adaptive genomic variation in conservation and fisheries management. J. Fish Biol. 89, 2697–2716 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Allendorf F. W., Bayles D., Bottom D. L., Currens K. P., Frissell C. A., Hankin D., Lichatowich J. A., Nehlsen W., Trotter P. C., Williams T. H., Prioritizing Pacific salmon stocks for conservation. Conserv. Biol. 11, 140–152 (1997). [Google Scholar]
- 29.Pennock D. S., Dimmick W. W., Critique of the evolutionarily significant unit as a definition for “distinct population segments” under the U.S. Endangered Species Act. Conserv. Biol. 11, 611–619 (1997). [Google Scholar]
- 30.Wainwright T. C., Waples R. S., Prioritizing Pacific salmon stocks for conservation: Response to Allendorf et al. Conserv. Biol. 12, 1144–1147 (1998). [Google Scholar]
- 31.Levin P., Schiewe M., Preserving salmon biodiversity. Am. Sci. 89, 220–227 (2001). [Google Scholar]
- 32.Augerot X., Atlas of Pacific salmon. Integr. Comp. Biol. 45, 952 (2005). [Google Scholar]
- 33.Gustafson R. G., Waples R. S., Myers J. M., Weitkamp L. A., Bryant G. J., Johnson O. W., Hard J. J., Pacific salmon extinctions: Quantifying lost and remaining diversity. Conserv. Biol. 21, 1009–1020 (2007). [DOI] [PubMed] [Google Scholar]
- 34.P. B. Moyle, J. A. Israel, S. E. Purdy, Salmon, Steelhead, and Trout in California: Status of an Emblematic Fauna (Center for Watershed Sciences, University of California, Davis, 2008). [Google Scholar]
- 35.Muñoz N. J., Farrell A. P., Heath J. W., Neff B. D., Adaptive potential of a Pacific salmon challenged by climate change. Nat. Clim. Change 5, 163–166 (2015). [Google Scholar]
- 36.Taylor E. B., A review of local adaptation in Salmonidac, with particular reference to Pacific and Atlantic salmon. Aquaculture 98, 185–207 (1991). [Google Scholar]
- 37.Fraser D. J., Jones M. W., McParland T. L., Hutchings J. A., Loss of historical immigration and the unsuccessful rehabilitation of extirpated salmon populations. Conserv. Genet. 8, 527–546 (2006). [Google Scholar]
- 38.Anderson J. H., Pess G. R., Carmichael R. W., Ford M. J., Cooney T. D., Baldwin C. M., McClure M. M., Planning Pacific salmon and steelhead reintroductions aimed at long-term viability and recovery. N. Am. J. Fish. Manag. 34, 72–93 (2014). [Google Scholar]
- 39.Committee on Protection and Management of Pacific Northwest Anadromous Salmonids, National Research Council, Division on Earth and Life Studies, Commission on Life Sciences, Upstream: Salmon and Society in the Pacific Northwest (National Academies Press, 1996). [Google Scholar]
- 40.P. J. Busby, Status Review of West Coast Steelhead from Washington, Idaho, Oregon, and California (U.S. Department of Commerce, 1996). [Google Scholar]
- 41.U.S. Fish Wildlife Service, National Oceanic and Atmospheric Administration , Policy regarding the recognition of distinct vertebrate population segments under the Endangered Species Act. Fed. Regist. 61, 4722–4725 (1996). [Google Scholar]
- 42.J. M. Myers, Status Review of Chinook Salmon from Washington, Idaho, Oregon, and California (U.S. Department of Commerce, 1998). [Google Scholar]
- 43.National Oceanic and Atmospheric Administration , Endangered and threatened species: Final listing determinations for 10 distinct population segments of west coast steelhead. Fed. Regist. 71, 834–862 (2006). [Google Scholar]
- 44.P. B. Moyle, Inland Fishes of California (University of California Press, Berkeley, ed. 1, 2002). [Google Scholar]
- 45.T. Quinn, The Behavior and Ecology of Pacific Salmon and Trout (University of Washington Press, 2005). [Google Scholar]
- 46.Quinn T. P., McGinnity P., Reed T. E., The paradox of “premature migration” by adult anadromous salmonid fishes: Patterns and hypotheses. Can. J. Fish. Aquat. Sci. 73, 1015–1030 (2016). [Google Scholar]
- 47.Hearsey J. W., Kinziger A. P., Diversity in sympatric Chinook salmon runs: Timing, relative fat content and maturation. Environ. Biol. Fishes 98, 413–423 (2014). [Google Scholar]
- 48.A. F. McEvoy, The Fisherman’s Problem: Ecology and Law in the California Fisheries, 1850-1980 (Cambridge Univ. Press, 1990). [Google Scholar]
- 49.Swezey S. L., Heizer R. F., Ritual management of salmonid fish resources in California. J. Calif. Anthropol. 4, 6–29 (1977). [Google Scholar]
- 50.Kareiva P., Marvier M., McClure M., Recovery and management options for spring/summer Chinook salmon in the Columbia River Basin. Science 290, 977–979 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Committee on Endangered and Threatened Fishes in the Klamath River Basin, Board on Environmental Studies and Toxicology, Division on Earth and Life Studies, National Research Council (2004), Endangered and Threatened Fishes in the Klamath River Basin: Causes of Decline and Strategies for Recovery (National Academies Press, 2004). [Google Scholar]
- 52.F. W. Allendorf, “Genetic variability in a species possessing extensive gene duplication: Genetic interpretation of duplicate loci and examination of genetic variation in populations of rainbow trout,” thesis, University of Washington (1975). [Google Scholar]
- 53.Chilcote M. W., Crawford B. A., Leider S. A., A genetic comparison of sympatric populations of summer and winter steelheads. Trans. Am. Fish. Soc. 109, 203–206 (1980). [Google Scholar]
- 54.Thorgaard G. H., Chromosomal differences among rainbow trout populations. Copeia 1983, 650–662 (1983). [Google Scholar]
- 55.Nielsen J. L., Fountain M. C., Microsatellite diversity in sympatric reproductive ecotypes of Pacific steelhead (Oncorhynchus mykiss) from the Middle Fork Eel River, California. Ecol. Freshw. Fish. 8, 159–168 (1999). [Google Scholar]
- 56.A. J. Clemento III, “Subpopulation structure of steelhead trout (Oncorhynchus mykiss) in the Middle Fork Eel River as determined by microsatellite DNA polymorphisms,” thesis, Humboldt State University (2006). [Google Scholar]
- 57.Kinziger A. P., Hellmair M., Hankin D. G., Garza J. C., Contemporary population structure in Klamath River basin Chinook salmon revealed by analysis of microsatellite genetic data. Trans. Am. Fish. Soc. 142, 1347–1357 (2013). [Google Scholar]
- 58.Brieuc M. S. O., Ono K., Drinan D. P., Naish K. A., Integration of Random Forest with population-based outlier analyses provides insight on the genomic basis and evolution of run timing in Chinook salmon (Oncorhynchus tshawytscha). Mol. Ecol. 24, 2729–2746 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Arciniega M., Clemento A. J., Miller M. R., Peterson M., Garza J. C., Pearse D. E., Parallel evolution of the summer steelhead ecotype in multiple populations from Oregon and Northern California. Conserv. Genet. 17, 165–175 (2016). [Google Scholar]
- 60.Berthelot C., Brunet F., Chalopin D., Juanchich A., Bernard M., Noël B., Bento P., Da Silva C., Labadie K., Alberti A., Aury J.-M., Louis A., Dehais P., Bardou P., Montfort J., Klopp C., Cabau C., Gaspin C., Thorgaard G. H., Boussaha M., Quillet E., Guyomard R., Galiana D., Bobe J., Volff J.-N., Genêt C., Wincker P., Jaillon O., Roest Crollius H., Guiguen Y., The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 5, 3657 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Narum S. R., Buerkle C. A., Davey J. W., Miller M. R., Hohenlohe P. A., Genotyping-by-sequencing in ecological and conservation genomics. Mol. Ecol. 22, 2841–2847 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fumagalli M., Vieira F. G., Linderoth T., Nielsen R., ngsTools: Methods for population genetics analyses from next-generation sequencing data. Bioinformatics 30, 1486–1487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S. Y., Lohmueller K. E., Albrechtsen A., Li Y., Korneliussen T., Tian G., Grarup N., Jiang T., Andersen G., Witte D., Jorgensen T., Hansen T., Pedersen O., Wang J., Nielsen R., Estimation of allele frequency and association mapping using next-generation sequencing data. BMC Bioinformatics 12, 231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price A. L., Zaitlen N. A., Reich D., Patterson N., New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 11, 459–463 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Felsenstein J., PHYLIP-phylogeny inference package (version 3.2). Cladistics 5, 164–166 (1989). [Google Scholar]
- 67.Tajima F., Evolutionary relationship of DNA sequences in finite populations. Genetics 105, 437–460 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watterson G. A., On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7, 256–276 (1975). [DOI] [PubMed] [Google Scholar]
- 69.Tajima F., Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crête-Lafrenière A., Weir L. K., Bernatchez L., Framing the Salmonidae family phylogenetic portrait: A more complete picture from increased taxon sampling. PLOS ONE 7, e46662 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhivotovsky L. A., Genetic history of salmonid fishes of the genus Oncorhynchus. Russ. J. Genet. 51, 491–505 (2015). [PubMed] [Google Scholar]
- 72.Skotte L., Korneliussen T. S., Albrechtsen A., Association testing for next-generation sequencing data using score statistics. Genet. Epidemiol. 36, 430–437 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Hess J. E., Zendt J. S., Matala A. R., Narum S. R., Genetic basis of adult migration timing in anadromous steelhead discovered through multivariate association testing. Proc. Biol. Sci. 283, 20153064 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohammed H., D’Santos C., Serandour A. A., Ali H. R., Brown G. D., Atkins A., Rueda O. M., Holmes K. A., Theodorou V., Robinson J. L. L., Zwart W., Saadi A., Ross-Innes C. S., Chin S.-F., Menon S., Stingl J., Palmieri C., Caldas C., Carroll J. S., Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 3, 342–349 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ghosh M. G., Thompson D. A., Weigel R. J., PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 60, 6367–6375 (2000). [PubMed] [Google Scholar]
- 76.Rae J. M., Johnson M. D., Cordero K. E., Scheys J. O., Larios J. M., Gottardis M. M., Pienta K. J., Lippman M. E., GREB1 is a novel androgen-regulated gene required for prostate cancer growth. Prostate 66, 886–894 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Pellegrini C., Gori I., Achtari C., Hornung D., Chardonnens E., Wunder D., Fiche M., Canny G. O., The expression of estrogen receptors as well as GREB1, c-MYC, and cyclin D1, estrogen-regulated genes implicated in proliferation, is increased in peritoneal endometriosis. Fertil. Steril. 98, 1200–1208 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Hodgkinson K. M., Vanderhyden B. C., Consideration of GREB1 as a potential therapeutic target for hormone-responsive or endocrine-resistant cancers. Expert Opin. Ther. Targets 18, 1065–1076 (2014). [DOI] [PubMed] [Google Scholar]
- 79.Hegarty K. G., Drummond F. J., Daly M., Shanahan F., Molloy M. G., GREB1 genetic variants are associated with bone mineral density in Caucasians. J. Bone Miner. Metab. 2017, 1–11 (2017). [DOI] [PubMed] [Google Scholar]
- 80.MacManes M. D., Austin S. H., Lang A. S., Booth A., Farrar V., Calisi R. M., Widespread patterns of sexually dimorphic gene expression in an avian hypothalamic–pituitary–gonadal (HPG) axis. Sci. Rep. 7, 45125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Henry F. E., Sugino K., Tozer A., Branco T., Sternson S. M., Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. eLife 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dietrich M. O., Zimmer M. R., Bober J., Horvath T. L., Hypothalamic Agrp neurons drive stereotypic behaviors beyond feeding. Cell 160, 1222–1232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cockram J., Mackay I. J., O’Sullivan D. M., The role of double-stranded break repair in the creation of phenotypic diversity at cereal VRN1 loci. Genetics 177, 2535–2539 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chan Y. F., Marks M. E., Jones F. C., Villarreal G. Jr., Shapiro M. D., Brady S. D., Southwick A. M., Absher D. M., Grimwood J., Schmutz J., Myers R. M., Petrov D., Jónsson B., Schluter D., Bell M. A., Kingsley D. M., Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.B. Charlesworth, D. Charlesworth, Elements of Evolutionary Genetics (W. H. Freeman, ed. 1, 2010). [Google Scholar]
- 86.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li H., A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fumagalli M., Vieira F. G., Korneliussen T. S., Linderoth T., Huerta-Sánchez E., Albrechtsen A., Nielsen R., Quantifying population genetic differentiation from next-generation sequencing data. Genetics 195, 979–992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nielsen R., Korneliussen T., Albrechtsen A., Li Y., Wang J., SNP calling, genotype calling, and sample allele frequency estimation from New-Generation Sequencing data. PLOS ONE 7, e37558 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Korneliussen T. S., Moltke I., Albrechtsen A., Nielsen R., Calculation of Tajima’s D and other neutrality test statistics from low depth next-generation sequencing data. BMC Bioinformatics 14, 289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hudson R. R., Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18, 337–338 (2002). [DOI] [PubMed] [Google Scholar]
- 93.Li H., Durbin R., Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scheet P., Stephens M., A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78, 629–644 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cantarel B. L., Korf I., Robb S. M. C., Parra G., Ross E., Moore B., Holt C., Sánchez Alvarado A., Yandell M., MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Andrews K. R., Hohenlohe P. A., Miller M. R., Hand B. K., Seeb J. E., Luikart G., Trade-offs and utility of alternative RADseq methods: Reply to Puritz et al. Mol. Ecol. 23, 5943–5946 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/8/e1603198/DC1
fig. S1. Observed versus expected statistics for association mapping of migration phenotype.
fig. S2. Migration date distribution of Klickitat River steelhead at Lyle Falls with weekly binning.
table S1. Steelhead samples.
table S2. Steelhead RAD sequence.
table S3. Steelhead migration associated SNPs.
table S4. Steelhead amplicon primers.
table S5. Steelhead amplicon sequence.
table S6. Chinook samples.
table S7. Chinook RAD sequence.
table S8. Chinook migration associated SNPs.
table S9. Scaffold links.
table S10. Scaffold79929e assembly.


