Under drought stress, lipid-anchored MfNACsa translocates to the nucleus by thioesterase MtAPT1-dependent de-S-palmitoylation, regulating GlyI gene expression and GSH/GSSG ratio.
Abstract
The plant-specific NAC (NAM, ATAF1/2, and CUC2) transcription factors (TFs) play a vital role in the response to drought stress. Here, we report a lipid-anchored NACsa TF in Medicago falcata. MfNACsa is an essential regulator of plant tolerance to drought stress, resulting in the differential expression of genes involved in oxidation reduction and lipid transport and localization. MfNACsa is associated with membranes under unstressed conditions and, more specifically, is targeted to the plasma membrane through S-palmitoylation. However, a Cys26-to-Ser mutation or inhibition of S-palmitoylation results in MfNACsa retention in the endoplasmic reticulum/Golgi. Under drought stress, MfNACsa translocates to the nucleus through de-S-palmitoylation mediated by the thioesterase MtAPT1, as coexpression of APT1 results in the nuclear translocation of MfNACsa, whereas mutation of the catalytic site of APT1 results in colocalization with MfNACsa and membrane retention of MfNACsa. Specifically, the nuclear MfNACsa binds the glyoxalase I (MtGlyl) promoter under drought stress, resulting in drought tolerance by maintaining the glutathione pool in a reduced state, and the process is dependent on the APT1-NACsa regulatory module. Our findings reveal a novel mechanism for the nuclear translocation of an S-palmitoylated NAC in response to stress.
INTRODUCTION
Among the environmental cues, drought stress and water deficits greatly affect plant growth and productivity, resulting in cellular energy depletion, redox imbalances, and oxidative damage. To tolerate stress, complex signaling pathways are triggered that regulate cellular homeostasis and promote survival (Golldack et al., 2014). Drought signal perception and transmission lead to the activation of transcriptional control. The NAM/ATAF1/2/CUC2 (NAC), AP2/EREBP, MYB/MYC, WRKY, and nuclear factor-Y transcription factors (TFs) are involved in the drought response and drought tolerance (Abe et al., 1997; Dietz et al., 2010; Krasensky and Jonak, 2012; Ambawat et al., 2013; Baldoni et al., 2015; Chen et al., 2015; Miao et al., 2015; Singh and Laxmi, 2015). NAC TFs are one of the largest families of plant-specific TFs, and most of these TFs exclusively localize to the nucleus to activate responses involved in reactive oxygen species homeostasis (Wu et al., 2012; You et al., 2014; Fang et al., 2015; Huang et al., 2015), osmolyte production (Jeong et al., 2010), and detoxification under drought stress (Tran et al., 2004; Hu et al., 2006). Moreover, some membrane-associated NAC members referred to as NTLs (NAC with transmembrane motif 1-like) are responsible for plasma membrane (PM) or endoplasmic reticulum (ER) membrane anchoring, which must be activated for nuclear translocation upon exposure to stress through the posttranscriptional regulation of alternate splicing (Kim et al., 2010), posttranslational regulation of intramembrane proteolysis (Kim et al., 2006; Kim et al., 2008; Yoon et al., 2008; Ng et al., 2013; Yang et al., 2014; Liang et al., 2015), or phosphorylation (Kim et al., 2012). However, it is unclear whether other modifications associated with the nuclear import of NAC TFs exist.
So far, only one article has reported a lipid-modified TF in animals (Eisenhaber et al., 2011). The NFAT5α (nuclear factor of activated T cells 5, isoform α) TF of Homo sapiens is transported to the PM via the ER and the Golgi due to both myristoylation and palmitoylation in the resting state. The nuclear import of NFAT5α upon high salt stress is not based on proteolytic processing. However, how the reversible palmitoylation mechanism causes NFAT5α to be released from the membranes under salt stress remains unclear. In this study, we identified an S-palmitoylated NAC TF in Medicago falcata that was associated with membranes in an uninduced state, and we examined the mechanism underlying the release of the MfNACsa protein from membranes.
In plants, the receptor-like kinases, ABC transporters, soluble NSF attachment protein receptor, ion channels, various heterotrimeric G-proteins, Ras-related small GTPases, and α-tubulin protein have been reported to be S-palmitoylated (Lavy and Yalovsky, 2006; Batistic et al., 2008; Hemsley et al., 2008, 2012; Sharma et al., 2008; Singaraja et al., 2009; Shipston, 2011; Agudo-Ibáñez et al., 2015). Most of these proteins participate in a range of signaling processes involving GTPase signaling, calcium perception, and the responses to pathogens (Hemsley and Grierson, 2008). However, the regulatory module whereby S-palmitoylated TFs perform their roles in plants is unknown. In this study, we found that MfNACsa is a positive regulator of drought tolerance. Thus, we asked whether the S-palmitoylation of MfNACsa is part of a signaling cascade that initiates the cellular reaction to drought stress.
M. falcata, a model candidate for studying abiotic stress-responsive mechanisms in legumes, grows in adverse environments (Zhang et al., 2011). PI502449, a diploid variety of M. falcata, is more tolerant to drought and cold stresses than the model legume Medicago truncatula (Miao et al., 2015). Thus, research on the regulatory mechanisms underlying drought stress in M. falcata PI502449 may provide information for the breeding of drought resistance in legumes. Here, we provide the first report of a lipid-anchored NAC TF that translocates to the nucleus under drought stress and directly or indirectly activates the expression of stress-, lipid transport-, and lipid localization-related genes in response to drought stress. We found that de-S-palmitoylation mediated by the thioesterase MtAPT1 is required for the nuclear translocation of MfNACsa. Moreover, the regulation of nuclear import also applies to other S-palmitoylated NAC TFs.
RESULTS
The MfNACsa Transcription Factor Is Associated with Membranes
In a previous study, the changes in the transcriptome of M. falcata cv PI502449 in response to dehydration, high salinity, or cold stresses were assessed to characterize the abiotic stress-responsive mechanism in legumes. A total of 62 NAC transcripts were found to be upregulated after dehydration stress (Miao et al., 2015). In this study, a candidate dehydration stress-responsive transcript referred to as MfNACsa (accession number KY673692) was selected for further analysis. To examine the role played by MfNACsa in response to dehydration stress, MfNACsa transcript levels were determined via quantitative real-time PCR (RT-qPCR) and normalized to MfEF1α expression. Dehydration stress significantly induced the expression (∼36-fold) of MfNACsa at 2 h compared with normal conditions (Supplemental Figure 1A). The transcriptional level of MfNACsa in the leaf was relatively higher than that in the root and stem, and the induction was also detected in these tissues after 2 h of dehydration (Supplemental Figure 1B), indicating that MfNACsa is involved in the dehydration response.
We next cloned full-length MfNACsa for further study. MfNACsa encodes a 293-amino acid protein belonging to the ATAF NAC domain-containing protein family, according to the classification of NACs (Ooka et al., 2003) (Supplemental Figures 1C and 2). The MfNACsa protein possesses an N-terminal NAC domain structure that is typical of NAC family members, which are subdivided into five subdomains (A to E) (Tran et al., 2004; Christianson et al., 2010), including a D subdomain harboring the predicted nuclear localization signal (NLS), and a C-terminal transcriptional activation region (TAR) containing a conserved EVQSEPKW motif that is only found in members of the ATAF group (Ooka et al., 2003; Mendes et al., 2013) (Supplemental Figures 1D and 3). To examine the transactivation activity of MfNACsa, a transient expression assay using a GAL4 DBD (DNA binding domain)-responsive reporter system in yeast cells was performed. Cells harboring the full-length MfNACsa and the positive control grew well in synthetic dropout (SD) medium lacking tryptophan, histidine, and adenine and showed α-galactosidase (α-Gal) activity (Supplemental Figure 1E, i). By contrast, the N-terminal region lacking the TAR and the negative control did not show any spot (Supplemental Figure 1E, ii), suggesting that MfNACsa exhibits transactivation activity in vitro.
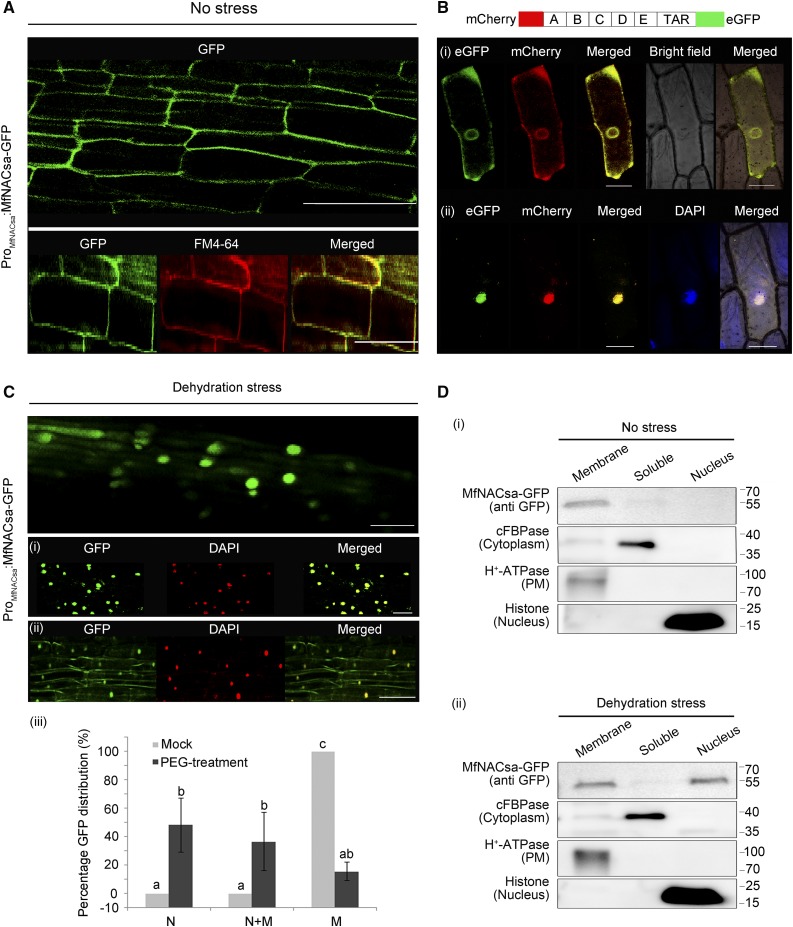
To determine the subcellular localization of the MfNACsa, we generated Pro35S:MfNACsa-GFP or ProMfNACsa:MfNACsa-GFP constructs that expressed MfNACsa from the constitutive cauliflower mosaic virus 35S promoter or the endogenous promoter up to –1185 bp upstream of the predicted transcription start site that had been amplified from M. falcata cv PI502449 genomic DNA using high-efficiency thermal asymmetric interlaced PCR (hiTAIL-PCR) (Liu and Chen, 2007). We introduced the two constructs into M. truncatula roots via Agrobacterium rhizogenes-mediated hairy root transformation. Surprisingly, confocal imaging revealed that MfNACsa-GFP protein driven by 35S or native promoter was not localized to the nucleus (Figure 1A). To identify the exact localization of MfNACsa, cellular fractionation of Nicotiana benthamiana leaf cells transiently expressing MfNACsa-GFP fusion protein driven by the native promoter was performed. After the soluble cytoplasm and insoluble membranes (a mixture of small membrane vesicles and fragments arising from the PM, tonoplast, ER, and Golgi apparatus) were separated, immunoblot analysis showed that MfNACsa was present only in the insoluble membrane fraction (Figure 1B, i). To determine whether MfNACsa was associated with the nucleus, we separated the cytoplasmic and nuclear components using ProMfNACsa:MfNACsa-GFP transient transgenic plants, and immunoblot results indicated that GFP-tagged MfNACsa was not located in either nuclear or cytoplasmic fraction (Figure 1B, ii).
Figure 1.
MfNACsa Is Associated with Membranes.
(A) Confocal imaging of transgenic M. truncatula hairy roots expressing MfNACsa-GFP fusion proteins driven by the constitutive CaMV 35S promoter (i) or the MfNACsa endogenous promoter (ii). GFP signal was detected with an Olympus FluoView FV1000 confocal laser scanning microscope. Bars = 50 μm.
(B) Cellular fractionation and immunoblot analysis of ProMfNACsa:MfNACsa-GFP protein in N. benthamiana leaves. Soluble and membrane proteins (i), and cytoplasmic and nuclear proteins were separated and subjected the immunoblot analysis (ii). H+-ATPase, Histone H3, and cFBPase were used as PM, nuclear, and cytosolic fraction markers, respectively.
(C) MfNACsa localizes to the PM and ER compartment in onion epidermal cells. Colocalization of MfNACsa-eGFP fusion proteins driven by CaMV35S promoter with the RFP-tagged PIP2A PM marker (i) or the HDEL ER marker (ii), and costaining with the endocytic dye FM4-64 (iii) or nuclear dye DAPI (iv). The signal patterns were observed at 488 nm (eGFP: green), 546 nm (RFP or FM4-64: magenta), or 358 nm (DAPI: blue) fluorescence excitation wavelength by confocal imaging (Olympus FluoView FV1000). Bars = 50 μm. The eGFP-labeled MfNACsa is green, RFP-labeled PIP2A or HDEL or FM4-64 dye is magenta, and merged image is yellow. The ICQs of MfNACsa-eGFP against PIP2A-RFP, RFP-HDEL, FM4-64 dye, and DAPI are 0.363, 0.341, 0.239 and –0.154, respectively.
Furthermore, the MfNACsa-eGFP fusion protein transiently expressed in onion epidermal cells colocalized with Arabidopsis thaliana PIP2A (Cutler et al., 2000), a PM aquaporin, with an intensity correlation quotient (ICQ) of 0.363; HDEL (Gomord et al., 1997), an ER retention motif, with an ICQ of 0.341; and the endosomal marker FM4-64 (Jennings et al., 2005; Rigal et al., 2015), a cellular membrane-specific lipophilic dye for tracing endocytosis and vesicle trafficking in living cells, with an ICQ of 0.239. However, the negative (−0.154) ICQ for the costaining of 4′,6-diamidino-2-phenylindole (DAPI) (Tarnowski et al., 1991), a dye employed for nuclear quantitation, showed that MfNACsa protein was not associated with the nucleus (Figure 1C; Supplemental Figure 4). To determine whether MfNACsa targeting could be influenced by the localization of the GFP reporter, a construct in which the N terminus of MfNACsa was tagged with GFP was designed. We observed costaining of GFP-MfNACsa with FM4-64 at the membrane, with an ICQ of 0.030, but this fusion was not associated with the nucleus according to the negative (–0.025) ICQ of DAPI against GFP-MfNACsa (Supplemental Figure 5). These data verified that MfNACsa localized to membranes.
MfNACsa Is Translocated to the Nucleus under Dehydration Stress
Considering that MfNACsa is a membrane-associated NAC TF, we investigated whether this protein translocates to the nucleus to function. To evaluate this hypothesis, we generated transgenic M. truncatula hairy roots that expressed MfNACsa-GFP driven by the native promoter to determine the subcellular localization of MfNACsa-GFP under dehydration stress. To scrutinize the fine localization of the MfNACsa-GFP fusion protein under normal conditions or dehydration stress, we used membrane or nuclear dyes to obtain insight into the exact subcellular localization patterns. A membrane distribution pattern of the MfNACsa protein was detected under normal conditions, which exhibited colocalized with FM4-64 dye for immediate staining (Figure 2A). However, the nuclear-localized MfNACsa-GFP protein was more abundant in PEG-treated hairy roots than in untreated roots. Colocalization with DAPI dye further confirmed the nuclear translocation of MfNACsa-GFP fusion proteins under 50% PEG-8000 treatment for 2 h (Figure 2B, i and ii). Furthermore, the distribution of cells expressing GFP in the nucleus, nucleus and membranes, or membranes revealed that the percentage of cells expressing GFP exclusively in the nucleus or in both membranes and nucleus was significantly increased following a 2-h treatment with PEG-8000 compared with mock treatment (nonparametric one-way ANOVA and Duncan multiple range test, P < 0.05) (Figure 2B, iii), suggesting that dehydration stress induced mobilization of MfNACsa for nuclear import.
Figure 2.
MfNACsa Is Translocated to the Nucleus under PEG-Imposed Dehydration Stress.
(A) Transgenic M. truncatula hairy roots expressing the MfNACsa-GFP fusion proteins driven by native promoter were stained with the membrane dye FM4-64 for 5 min under normal conditions (no stress), with an ICQ of 0.114.
(B) After hairy roots transiently expressing MfNACsa-GFP driven by the native promoter were treated with 50% PEG-8000 for 2 h (dehydration stress), the steady state locations of MfNACsa-GFP protein were imaged and stained with DAPI dye (pseudocolor, red; [i] and [ii]). Bars = 50 μm. The percentage of cells expressing GFP at the nucleus (N), nucleus and membranes (N+M), or membranes (M) in transgenic hairy root cells were counted respectively (iii). The data represent the mean and se of three independent replicates (n > 15) (nonparametric one-way ANOVA and Duncan multiple range test, P < 0.05).
(C) Nucleus translocation of the mCherry-MfNACsa-eGFP fusion protein under 10% PEG-8000 treatment in onion epidermal cells. The mCherry and eGFP signal patterns were observed at 0 (i) and 2 h (ii) after 10% PEG-8000 treatment by confocal imaging (Olympus FluoView FV1000). The same cells were imaged before and after treatment. Bars = 50 μm.
(D) Cellular fractionation and immunoblot detection of transiently expressed ProMfNACsa:MfNACsa-GFP in N. benthamiana leaves under normal conditions (i) or 30% PEG-8000 treatment for 2 h (ii). MfNACsa-GFP is shown. cFBPase, H+-ATPase, and Histone H3 mark the cytoplasm, PM, and nuclear fraction, respectively.
Furthermore, cellular fractionation and immunoblot analysis using ProMfNACsa:MfNACsa-GFP transient transgenic plants confirmed that MfNACsa was present only in the membrane fraction under normal conditions, whereas it was predominantly detected in membrane and nuclear components after 2 h of 30% PEG-8000 treatment (Figure 2D), implying that MfNACsa localization is affected by dehydration stress. Little cytoplasm-localized MfNACsa protein was detected, likely because MfNACsa protein driven by the native promoter was induced by dehydration stress, and resulted in intracellular membrane shuttling or nuclear translocation of MfNACsa in N. benthamiana cells.
To determine which subdomain was sufficient for the membrane localization of MfNACsa protein under normal conditions, truncated MfNACsa proteins were respectively fused with eGFP at the C-terminal region and transiently expressed in onion epidermal cells. Confocal imaging showed that the forms containing the A subdomain were associated with membrane (Supplemental Figure 6, i to vi), whereas the forms lacking the A subdomain and containing the NLS motif in the D subdomain were exclusively localized to the nucleus (Supplemental Figure 6, vii to ix), suggesting that the A subdomain at N terminus of MfNACsa were sufficient to mediate plasma membrane targeting.
Next, we questioned whether proteolytic cleavage in the A subdomain would release MfNACsa from the membrane into the nucleus during dehydration stress. It has been reported that ANAC017 was released to the nucleus upon cleavage of the C-terminal TM region (Ng et al., 2013). As previously described by Ng et al. (2013), a fusion protein consisting of full-length ANAC017 with RFP fused to the N-terminal region and GFP fused to the C-terminal region was constructed to observe the localization of ANAC017. The authors detected green fluorescence only in the ER, but the RFP-fused N terminus of ANAC017 was localized to the nucleus following proteolysis. In this study, we generated a similar construct consisting of full-length MfNACsa with mCherry fused to the N-terminal region and eGFP fused to the C-terminal region and transiently transformed the construct in onion cells to observe the localization of the N and C termini of MfNACsa. If proteolysis processing indeed occurred, the mCherry-fused N terminus of MfNACsa that contained the A subdomain would be attached to membrane, but the GFP-fused C terminus harboring the NLS would be localized to the nucleus. However, we found that mCherry and eGFP fluorescence signals were colocalized either under normal conditions or PEG treatment. Both fluorescence signals were associated with the membrane under normal conditions, but exclusively in the nucleus upon PEG treatment for 2 h (Figure 2C; Supplemental Figure 7). Therefore, we deduced that the nuclear-relocated MfNACsa protein that was released from the PM was not dependent on the proteolytic cleavage of the N-terminal A subdomain under dehydration stress.
MfNACsa Plays a Positive Role in Drought Stress
To further elucidate the biological functions of MfNACsa under drought stress, MfNACsa was ectopically expressed in M. truncatula cv R108 plants. Given the difficulty of regenerating M. falcata cv PI502449, ‘R108’ was efficiently used for routine transformation (Cosson et al., 2006). In this study, two stable transgenic lines (MfNACsa-OE23 and MfNACsa-OE33) were selected by RT-qPCR and immunoblot analysis (Supplemental Figures 8A and 8B), and 10-d-old seedlings were submitted to three cycles of drought stress by withholding the water supply for 12, 14, and 18 d, respectively, until the leaves of wild-type plants exhibited primary wilted (Figure 3A). The wild-type plants displayed a survival rate of ∼20%, while lines in which MfNACsa was ectopically expressed had survival rates of over 80% (Figure 3B). Additionally, lines in which MfNACsa was ectopically expressed had remarkably lower levels of electrolyte leakage, which indicates injury of the plasma membrane, than did the wild-type plants (Kruskal-Wallis nonparametric test, P < 0.01) (Figure 3C). These results indicate that ectopic expression of MfNACsa enhanced drought stress tolerance in M. truncatula.
Figure 3.
MfNACsa Plays a Positive Role in Drought Stress Tolerance.
(A) Ten-day-old seedlings of the wild-type (R108) and lines ectopically expressing MfNACsa (OE23 and OE33) were subjected to three cycles of drought stress by withholding the water supply for 12, 14, and 18 d, respectively, until the leaves of wild-type plants exhibited primary wilting (ii). The mock plants were watered once a week, according to standard procedures (i).
(B) The survival rate of plants in (A), ii. The mean values and se were calculated from three independent experiments. The asterisk indicates a significant difference between ectopic expression of MfNACsa lines and wild-type plants (Kruskal-Wallis nonparametric test, **P < 0.01).
(C) The leaf relative electrical conductivity of the plants in (A), ii. The mean values and se were calculated from three independent experiments. The asterisk indicates a significant difference between ectopic expression of MfNACsa lines and wild-type plants (Kruskal-Wallis nonparametric test, **P < 0.01).
(D) The phenotypes of the wild-type (R108) and nacsa mutants (nacsa-1 and nacsa-2) subjected to three cycles of drought stress by withholding the water supply for 12, 14, and 14 d, respectively. Seedlings were photographed when the leaves of mutants exhibited primary wilting (ii). The mock plants were watered once a week, according to standard procedure (i).
(E) The survival rate of plants in (D), ii. The mean values and se were calculated from three independent experiments. The asterisk indicates a significant difference between nacsa mutants and wild-type plants (Kruskal-Wallis nonparametric test, **P < 0.01).
(F) The leaf relative electrical conductivity of plants in (D), ii. The mean values and se were calculated from three independent experiments. The asterisk indicates a significant difference between nacsa mutants and wild-type plants (Kruskal-Wallis nonparametric test, **P < 0.01).
MtNACsa, a homolog of MfNACsa in M. truncatula, shares 98.29% identity with MfNACsa. To further determine the function of MfNACsa in drought stress, we evaluated two homozygous Tnt1 insertion mutants of NACsa identified from a Tnt1 retrotransposon-tagged mutant population of M. truncatula (Cheng et al., 2014), NF5250 (designated nacsa-1) and NF9803 (designated nacsa-2), with insertions located in the first intron and third exon, respectively, via PCR (Supplemental Figures 8C and 8D, i). RT-PCR verified that the expression of MtNACsa was impaired (Supplemental Figure 8D, ii). The nacsa mutants and wild-type plants were subjected to three cycles of drought stress by withholding the water supply for 12, 14, and 14 d, respectively, until the leaves of the mutants exhibited primary wilting (Figure 3D). After drought stress, statistical analysis of survival rates revealed that ∼80% of the wild-type plants were still alive, whereas only 20 to ∼30% of the nacsa mutants survived (Figure 3E). The nacsa mutants displayed significantly higher electrolyte leakage than the wild-type plants (Kruskal-Wallis nonparametric test, P < 0.01) (Figure 3F), suggesting that the nacsa seedlings were sensitive to drought stress.
To further identify the function of NACsa, germinated seeds of nacsa-1, nacsa-2, MfNACsa-OE23, MfNACsa-OE33, and wild-type plants were dehydrated with 35% PEG-8000 for 14 d and were subsequently rehydrated for 7 d and the phenotypic characteristics were surveyed (Supplemental Figure 10A). The PEG concentrations used were selected based on a gradient sensitivity assessment in wild-type plants (Supplemental Figure 9). After rehydration, the lines in which MfNACsa was ectopically expressed exhibited remarkably more green leaf blades and longer primary roots than wild-type plants (Kruskal-Wallis nonparametric test, P < 0.01) (Supplemental Figures 10B and 10C), indicating that ectopic expression of MfNACsa enhanced dehydration stress tolerance. By contrast, both the nacsa-1 and nacsa-2 mutants exhibited significantly fewer green leaf blades than wild-type plants (Kruskal-Wallis nonparametric test, P < 0.05) (Supplemental Figure 10B), and the primary root length of nacsa-2 plants was markedly shorter than that of wild-type plants exposed to 35% PEG-8000 (Kruskal-Wallis nonparametric test, P < 0.01) (Supplemental Figure 10C), suggesting that the seedlings were sensitive to dehydration stress when the MtNACsa gene was knocked out. Based on these phenotypic and physiological observations, we deduced that MfNACsa plays an essential role in drought stress tolerance.
Transcriptional Control by MfNACsa under Dehydration Stress
To determine whether the nuclear translocation of MfNACsa modulates plant responses to drought stress, we profiled the transcriptome to identify the target genes regulated by MfNACsa under dehydration stress. The total number of clean reads was 7,121,390 for the wild-type plants and 7,248,791 for ectopic expression of MfNACsa lines at 4 h after 50% PEG-8000 treatment. Accordingly, a total of 21,418 and 21,317 transcripts were found to be expressed in these two samples (expected number of fragments per kilobase of transcript sequence per millions base pairs sequenced > 1), respectively, with 20,551 genes being present in both sets. Among the significantly differentially expressed transcripts, 40 were upregulated and 49 transcripts were downregulated (|log2foldchange| > 0.6, P value<0.05) in lines ectopically expressing MfNACsa compared with the wild type (Supplemental Data Sets 3 and 4). Gene Ontology (GO) term enrichment at the biological process level revealed that the differentially expressed genes were involved in oxidation-reduction processes, lipid transport, and lipid localization (Supplemental Figure 11A). The analysis of enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways showed that the upregulated genes were involved in fatty acid degradation (acyl-CoA synthetase and aldehyde dehydrogenase), carbon fixation in photosynthetic organisms (fructose-bisphosphate aldolase, ribulose bisphosphate carboxylase small chain), and pyruvate metabolism (aldehyde dehydrogenase, lactoylglutathione lyase) (Supplemental Figure 11B). The downregulated genes were enriched in pathways related to starch/sucrose metabolism (cell wall isozyme and pectin lyase-like superfamily protein), which catalyze the degradation of sucrose; biosynthesis of secondary metabolism (phenylpropanoid biosynthesis and tropane/piperidine/pyridine alkaloid biosynthesis), which affected the biosynthesis of coumarin and lignin; glutathione metabolism (glutathione S-transferase zeta class), which catalyzes the conversion of GSH into R-S-glutathione, thereby inhibiting the accumulation of GSH; and ubiquitin-mediated proteolysis (E3 ubiquitin-protein ligase SINA-like 10) (Supplemental Figure 11C).
Because MfNACsa possesses transactivation activity in vitro, we focused on the upregulated genes. These genes included MtRD22 (dehydration-responsive proteins) and MtBURP3 (BURP domain-containing protein), which play a vital role in drought stress resistance (Harshavardhan et al., 2014); MtCOL16 (zinc finger protein CONSTANS-LIKE16), which has an important role in the regulation of flowering by photoperiod (An et al., 2004); MtLTP (nonspecific lipid-transfer protein), MtKCS6 (fatty acid metabolism-associated 3-ketoacyl-CoA synthase 6) and MtLACS (long chain acyl-CoA synthetase), which show overlapping functions in plant wax and cutin synthesis, reducing the water loss in response to abiotic stress (Ni and Guo, 2008; Lü et al., 2009; Tapia et al., 2013); and MtGlyI (lactoylglutathione lyase, also known as glyoxalase I), which converts methyglyoxal to d-lactate, playing an important role in methyglyoxal detoxification and maintaining higher reduced glutathione levels under abiotic stress (Yadav et al., 2005). To determine whether these genes were transcriptionally regulated by dehydration stress, we analyzed their temporal expression patterns. The RT-qPCR results showed that stress- and lipid metabolism-related genes were induced by dehydration, while the expression of MtCO16 was inhibited under dehydration stress (Supplemental Figure 12).
To analyze whether MfNACsa TF regulates the above genes under dehydration stress, RT-qPCR analysis was performed and confirmed that the tested transcripts were increased in lines ectopically expressing MfNACsa compared with the wild-type plants under dehydration stress; MtGlyI was particularly sharply induced (Supplemental Figure 13A). By contrast, there was no significant difference in gene expression between lines ectopically expressing MfNACsa and wild-type plants under normal conditions (Supplemental Figure 13B). These results indicated that ectopic expression of MfNACsa increased the transcription of candidate genes in response to dehydration stress.
MfNACsa Transactivates the Expression of MtGlyI under Dehydration Stress
To examine whether MfNACsa could bind to the promoter region of MtGlyI, the 3-kb promoter region of MtGlyI was cloned for analysis. It has been reported that the CAAATNNNATTTG or [GC]AAA sequence was the specific NAC binding site of ATAF subfamily (Huh et al., 2012; Wang and Culver, 2012). An electrophoretic mobility shift assay (EMSA) confirmed the binding of MfNACsa to the MtGlyI promoter that contained the cis-element (CAAATNNNCTTTG) in vitro. In all cases, binding was abolished using an unlabeled competitor probe or was not detected when the predicted NAC binding site was mutated (Figure 4A).
Figure 4.
MfNACsa Directly Binds to the MtGlyI Gene Promoter to Activate Expression under Drought Stress.
(A) In vitro protein-DNA binding assay showing that His-MfNACsa protein interacted with a cis-element (CAAATNNNCTTTG) in the promoter region of MtGlyI. A purified His-MfNACsa protein (200 ng) was incubated with a biotin-labeled probe (20 fM) and a mutant probe (20 fM). For the competition assay, a nonlabeled probe (1000 times) was added. The cis-element (CAAATNNNCTTTG) of GlyI-P Wt and mutant element (CCCCCNNNCCCCC) of GlyI-P mut were showed in red font.
(B) In vivo ChIP assay showing binding of MfNACsa to the promoter of MtGlyI under PEG-imposed dehydration stress. The ChIP experiment was performed using 2-week-old Pro35S:MfNACsa-3xFLAG plants treated with 50% PEG-8000 for 4 h. Plants ectopically expressing MfNACsa without stress treatment were used as the negative control. The MtGlyI promoter fragment B containing the cis-element (CAAATNNNCTTTG) (–346 to ∼–175 bp), fragment A located upstream of cis-element (–1897 to ∼–1714bp), and fragment C located downstream of cis-element (+550 to ∼+718 bp). Because of the different Ct values of the A, B, and C fragments, mock samples (mock-actin, mock-A, mock-B, and mock-C) individually set to 1 and the PEG-8000 samples normalized relative to them, respectively, and actin used as a negative control. The mean values and se were calculated from the two independent experiments (Kruskal-Wallis nonparametric test, **P < 0.01).
(C) The Pro35S:MfNACsa-GFP effector plasmid and ProGlyI+Min35S:GUS reporter plasmid harboring the promoter region (–488bp to ∼–38bp) of MtGlyI, a 35S minimal promoter and GUS reporter gene were transiently expressed in N. benthamiana leaf cells, together with a negative control vector, respectively. Before starting with sampling, the plants were submitted to 10% PEG treatment for 2 h (dehydration) or not (mock). The GUS staining and GUS activity were assayed according to a previously described method with some modifications (Topping and Lindsey, 1997; Blazquez, 2007). The mean values and se were calculated from the two independent experiments (Kruskal-Wallis nonparametric test, **P < 0.01).
Furthermore, a chromatin immunoprecipitation (ChIP) experiment was performed using 2-week-old seedlings ectopically expressing 3x FLAG tagged MfNACsa upon exposure to the PEG-imposed dehydration stress or mock treatment. Following ChIP analysis with an anti-FLAG antibody, the MtGlyI promoter fragment B containing the cis-element (CAAATNNNCTTTG) was found to be enriched in MfNACsa-3xFLAG immunoprecipitation from stressed seedlings compared with nonstressed seedlings (Figure 4B), whereas the fragments lacking the cis-element (fragments A and C) were not enriched. These results suggested that MfNACsa directly binds to the promoter of MtGlyI under drought stress.
Using a transcriptional activity assay to identify whether the MtGlyI promoter region (–488 to –38 bp) harboring the MfNACsa binding site (CAAATNNNCTTTG) was induced by MfNACsa, the Pro35S:MfNACsa-GFP effector plasmid and ProGlyI:GUS reporter plasmid were transiently expressed in N. benthamiana leaf cells together with the negative control vector. A clear enhancement of GUS activity driven by cotransformation of the MfNACsa protein with the MtGlyI promoter region was detected under 10% PEG treatment for 2 h, whereas there was no remarkable difference in GUS activity under normal conditions (Figure 4C), suggesting that MfNACsa directly binds the Glyl promoter to activate its transcription under dehydration stress.
MtGlyI Is Necessary for Maintaining Glutathione Homeostasis in Response to Drought Stress
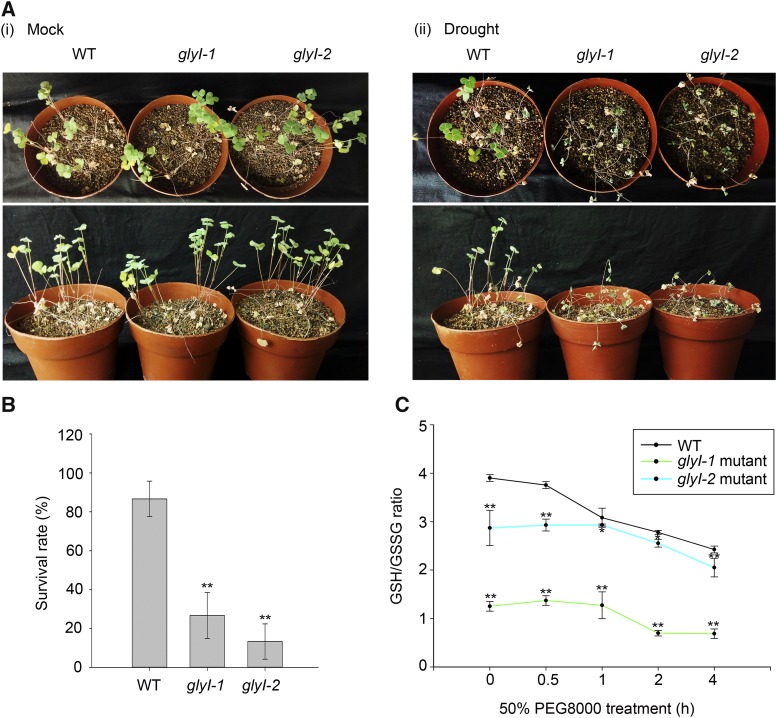
To investigate the role of MtGlyI in drought stress, two Tnt1 insertion mutants of the MtGlyI gene, NF20885 (designated glyI-1) and NF10049 (designated glyI-2), were identified from the Tnt1 retrotransposon-tagged mutant population of M. truncatula. These mutants exhibited insertions in the second exon and the sixth exon of MtGlyI, respectively (Supplemental Figure 14A), which were confirmed via PCR (Supplemental Figures 14B, i, and 14C, i), and RT-PCR analysis revealed that the expression of MtGlyI was impaired (Supplemental Figures 14B, ii, and 14C, ii). We evaluated the effect of three cycles of drought stress by withholding the water supply for 12, 14, and 14 d between glyI mutants and wild-type plants, until the leaves of mutants exhibited primary wilting (Figure 5A). Notably, after drought stress, wild-type plants exhibited a survival rate of ∼86.67%, while the glyI mutants showed survival rates of 26.67 and 13.33%, respectively (Figure 5B), suggesting that the glyI mutants were sensitive to drought stress.
Figure 5.
MtGlyI Is Necessary for Altering the GSH/GSSG Ratio in Response to Drought Tolerance.
(A) The wild-type (R108) plants and glyI mutants (glyI-1 and glyI-2) were subjected to three cycles of drought stress by withholding the water supply for 12, 14, and 14 d, respectively. Seedlings were photographed when the leaves of mutants exhibited primary wilting (ii). The mock plants were watered once a week, according to standard procedure (i).
(B) The survival rate of the plants in (A), ii. The mean values and se were calculated from three independent experiments. The asterisk indicates a significant difference between glyI mutants and wild-type plants (Kruskal-Wallis nonparametric test, **P < 0.01).
(C) The GSH/GSSG ratio of glyI mutants and wild-type plants under PEG-imposed dehydration stress. Total glutathione (GSH+GSSG) and GSSG were measured in glyI mutants and wild-type plants under 50% PEG-8000 treatment for 0, 0.5, 1, 2, and 4 h. The error bars represent the se of two samples. The asterisk indicates a significant difference between glyI mutants and wild-type plants under PEG-imposed dehydration stress (Kruskal-Wallis nonparametric test, **P < 0.01).
Elevated GlyI activity increases the GSH/GSSG ratio (Jain et al., 2002). To identify the redox state of glutathione associated with MtGlyI under drought stress, we measured the changes in the GSH/GSSG ratio in glyI mutants and wild-type plants under PEG-imposed dehydration stress. In the wild-type seedlings, the GSH/GSSG ratio continuously decreased during 4 h of 50% PEG-8000 treatment, but the glyI mutants showed a significantly lower GSH/GSSG ratio than wild-type plants under normal conditions or PEG-imposed dehydration stress (Figure 5C), indicating that MtGlyI is involved in the regulation of the GSH/GSSG redox potential under drought stress.
S-Palmitoylation at Cys26 Is Critical for Targeting MfNACsa to the PM
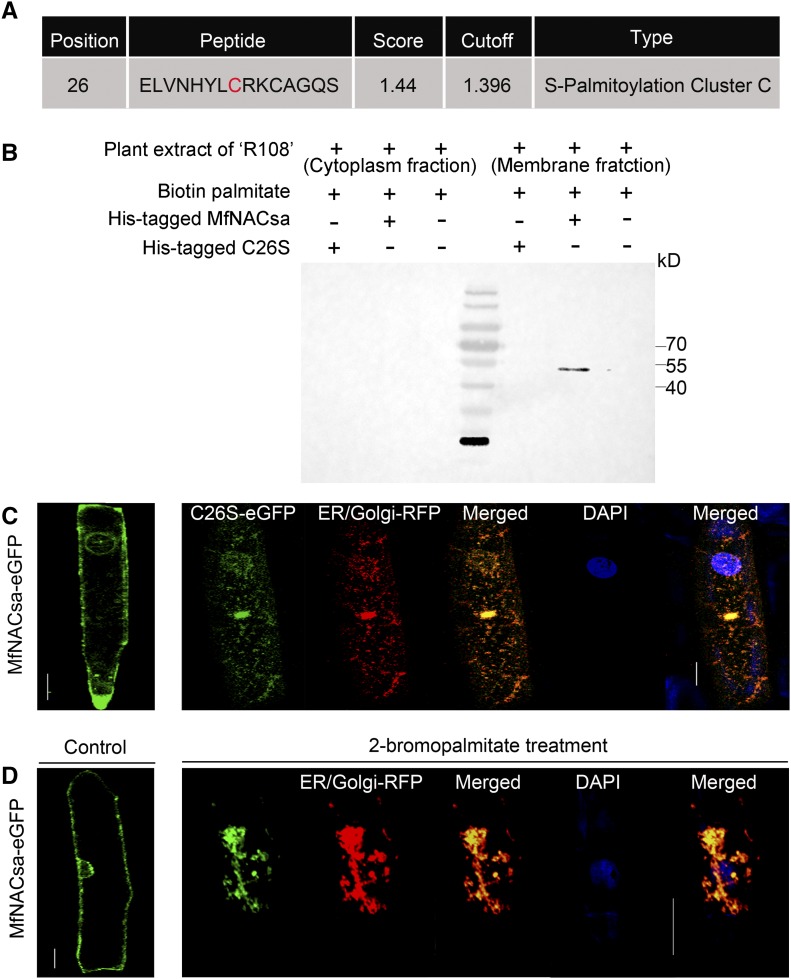
In this study, we were surprised to find that the anchoring of the MfNACsa TF to membranes does not depend on transmembrane domains, while the A subdomain of MfNACsa was sufficient for membrane targeting; the nuclear translocation of the MfNACsa protein was not dependent on the proteolytic cleavage. Moreover, MfNACsa can translocate to the nucleus to activate MtGlyI transcription and positively regulate drought stress tolerance. Therefore, we initially investigated the interaction manner of membrane and MfNACsa protein. First, an online tool (http://lipid.biocuckoo.org/webserver.php) was used to predict the potential for lipidation (palmitoylation, N-myristoylation, farnesylation, and geranylgeranylation) of MfNACsa, which is a posttranslational modification for targeting proteins to membranes. In this study, a threshold with high stringency was chosen for the prediction of lipid modification. MfNACsa was predicted to undergo S-palmitoylation at Cys26 of the N-terminal A subdomain (Figure 6A). S-palmitoylation is crucial for proteins from the ER/Golgi apparatus to undergo PM trafficking via a novel cellular targeting pathway (Batistic et al., 2008). We validated the S-palmitoylation of MfNACsa in the plant extract of M. truncatula cv R108. In the reaction system, the purified His-tagged wild-type MfNACsa protein or the mutated version of Cys-to-Ser (C26S) protein was incubated with biotin-modified palmitic acid for 4 h in the R108 extract of the cytoplasm or membrane fraction, respectively. Native protein gel electrophoresis was used to separate MfNACsa proteins, and labeled signals were detected using the streptavidin-HRP system. The immunoblotting results showed that a main band of approximately twice the molecular mass of the His-tagged MfNACsa monomer (∼35 kD) was biotin-palmitate labeled in the R108 extract of the membrane fraction, whereas this band was not labeled when His-tagged C26S mutant protein was added or in the R108 extract of cytoplasm extraction (Figure 6B). We hypothesize that some specific palmityl transferases from the M. truncatula R108 membrane extraction catalyze the S-palmitoylation of MfNACsa.
Figure 6.
S-Palmitoylation at Cys26 Determines PM Targeting of MfNACsa Protein.
(A) Cys26 in the N terminus of MfNACsa is predicted to be S-palmitoylated based on the GPS-lipid predictor analysis. The S-palmitoylation amino acid residue was shown in red font.
(B) Palmitoylation of MfNACsa in vitro. pET-30a(+)-His-MfNACsa and pET-30a(+)-His-C26S (200 ng) were incubated at 25°C for 4 h with pal-Lys (biotin) (0.1 μg) and the plant extracts of M. truncatula cv R108 (1 mg) from the cytoplasmic or membrane fraction, respectively. Protein expression was subsequently confirmed through native protein gel electrophoresis and immunoblot analysis. Labeled signals were detected through the streptavidin-HRP system.
(C) Subcellular localization of MfNACsa protein harboring the C26S substitution in the N terminus. C26S mutant protein was colocalized with the RFP-labeled Golgi and ER marker protein MAN1 from soybean, with the ICQ of 0.173, and not associated with the nucleus, as the negative (–0.021) ICQ between C26S-GFP and DAPI. The left panel is the subcellular localization of wild-type MfNACsa protein. The signal patterns were observed by fluorescence microscopy. Bars = 50 μm.
(D) Onion epidermal cells expressing MfNACsa-eGFP fusion proteins were treated with 2-bromopalmitate, an inhibitor of S-acyl transferase. After treatment with ethanol (Control, left panel) or 100 μM 2-bromopalmitate for 20 h, the individual cells were imaged. Upon 2-bromopalmitate treatment, MfNACsa was colocalized with the RFP-labeled Golgi and ER marker protein MAN1, with the ICQ of 0.18, and not associated with nucleus as the negative (–0.029) ICQ between MfNACsa-eGFP and DAPI. The signal patterns were observed by fluorescence microscopy (Olympus FluoView FV1000). Bars = 50 μm.
To determine whether the N-terminal Cys26 plays a vital role in the PM localization of MfNACsa, the C26S-eGFP fusion protein was transiently expressed in onion cells. Confocal imaging revealed spots distributed around the nucleus, and these spots merged with the Golgi and ER marker protein MAN1 from soybean (Glycine max) (Traverso et al., 2013). By contrast, the wild-type MfNACsa protein localized to the membrane (Figure 6C; Supplemental Figure 15A), showing that the C26S mutation caused MfNACsa ER/Golgi apparatus retention. Additionally, in the presence of a palmitoyl inhibitor (2-bromopalmitate dissolved in ethanol) (Lavy et al., 2002), the MfNACsa-eGFP proteins were colocalized with the ER/Golgi marker protein and absent at the PM. Whereas the control treatment (ethanol alone) did not affect the PM localization of MfNACsa (Figure 6D; Supplemental Figure 15B), indicating that inhibition of MfNACsa palmitoylation blocked its PM targeting.
MfNACsa Is Translocated to the Nucleus by De-S-Palmitoylation
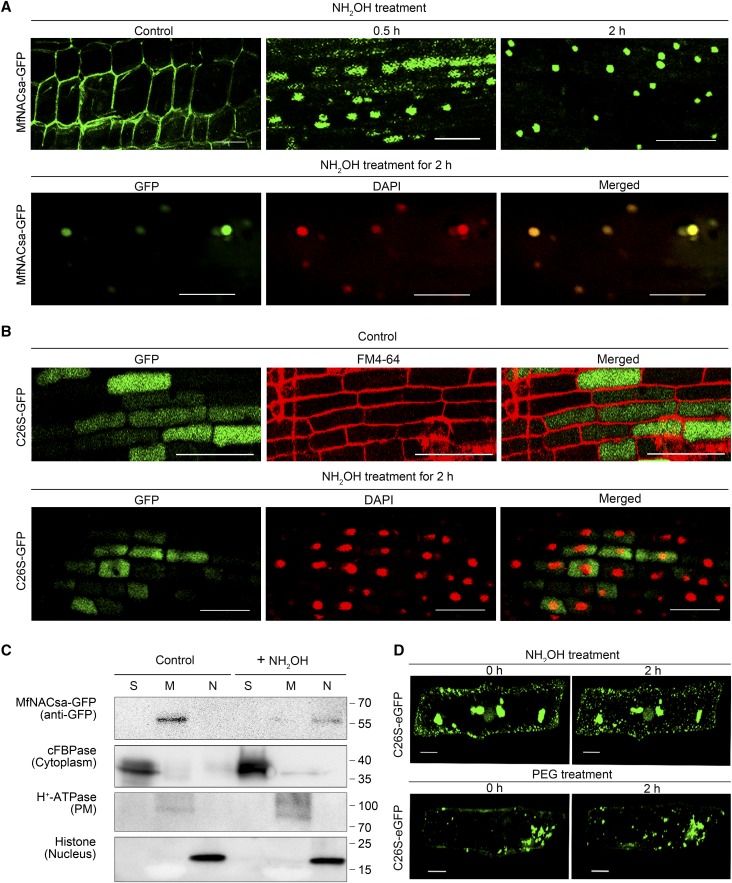
Unlike other lipid modifications that mediate membrane associations, S-palmitoylation is reversible (Conibear and Davis, 2010; Konrad et al., 2014). We speculated that the nuclear translocation of MfNACsa was based on de-S-palmitoylation. To verify this hypothesis, we detected the subcellular localization of MfNACsa upon hydroxylamine (NH2OH) treatment, which revealed protein de-S-palmitoylation through the cleavage of thioester linkages between cysteine residues and palmitate chains (Morrison et al., 1991). Transgenic M. truncatula hairy roots expressing MfNACsa-GFP fusion protein were treated with 0.05 M NH2OH. Membrane-localized MfNACsa-GFP fusion proteins were observed in the control. A small amount of the proteins were detected in the nucleus after a 0.5-h NH2OH treatment, and the nuclear-localized MfNACsa-GFP remarkably increased after treatment for 2 h, which was confirmed by DAPI staining (Figure 7A). Additionally, cellular fractionation and immunoblotting analysis showed that MfNACsa proteins were mainly localized in the nuclear component after 2 h of 0.05 M NH2OH treatment (Figure 7C). These results suggest that the mobilization mechanism of MfNACsa is dependent on de-S-palmitoylation.
Figure 7.
MfNACsa Is Translocated to the Nucleus through De-S-Palmitoylation.
(A) Transgenic M. truncatula hairy roots expressing the Pro35S:MfNACsa-GFP fusion proteins were treated with 0.05 M hydroxylamine (NH2OH). The GFP signal pattern was detected by confocal imaging at 0 (control), 0.5, or 2 h after NH2OH treatment. After 0.05 M NH2OH treatment for 2 h, the nucleus was stained with DAPI (pseudocolor, red), with the ICQ of 0.436. Bars = 50 μm.
(B) Transgenic M. truncatula hairy roots expressing the Pro35S:C26S -GFP mutant proteins were treated with 0.05 M NH2OH. The GFP signal pattern was detected by confocal imaging (Olympus FluoView FV1000) under control conditions, and the negative (–0.037) ICQ of C26S-GFP against FM4-64. After 0.05 M NH2OH treatment for 2 h, the nucleus was stained with DAPI (pseudocolor, red) and the negative (–0.006) ICQ of C26S-GFP against DAPI dye. Bars = 50 μm.
(C) Cellular fraction analysis of transient transgenic N. benthamiana leaves expressing MfNACsa-GFP proteins at 0 and 2 h after NH2OH treatment. Presence of MfNACsa-GFP was shown. cFBPase, H+-ATPase, and Histone H3 are used as cytoplasm, PM, and nuclear fraction markers, respectively.
(D) Onion epidermal cells expressing C26S-eGFP mutant proteins were treated with 0.05 M NH2OH or 10% PEG-8000 for 2 h, respectively. The signal patterns were observed by fluorescence microscopy (Olympus FluoView FV1000). Bars = 50 μm.
In mammalian cells, only the PM-localized fraction of the NFAT5α TF that was previously palmitoylated is imported to the nucleus (Eisenhaber et al., 2011). To further determine whether this is also the case in plants, the C26S mutant protein fused with GFP was transiently expressed in M. truncatula hairy roots, which were treated with NH2OH. As expected, when C26S-GFP fusion proteins expressed in M. truncatula hairy root cells were subjected to NH2OH treatment for 2 h, we did not observe the nuclear translocation of C26S mutant protein as the negative (–0.006) ICQ of C26S-GFP against DAPI dye (Figure 7B). Moreover, the expressed mutant C26S-eGFP protein in onion cells was retained in the ER/Golgi and not translocated to the nucleus under NH2OH or PEG treatment (Figure 7D). On the basis of the significant decrease in PM-localized wild-type MfNACsa protein (Figures 2B, 2C, and 7A), we deduced that de-S-palmitoylation is actively used to regulate the nuclear translocation of PM-localized MfNACsa.
MtAPT1 Mediates the Nuclear Translocation of MfNACsa
The reversal of S-palmitoylation in vivo is catalyzed by thioesterases that cleave the thioester bond between the protein and the acyl group (Camp and Hofmann, 1993). In mammalian cells, increasing evidence suggests that acyl-protein thioesterases APT1 and APT2 are the major APTs that mediate the depalmitoylation of diverse cellular substrates (Lin and Conibear, 2015). In this study, we generated a phylogenetic tree of potential thioesterase-like proteins from the Medicago JCVI database and identified a cluster consisting of MtAPT1, Medtr5g056230.1, Medtr5g056300.1, and Medtr5g056390.2, that shares a relatively high degree of amino acid sequence identity with AtALT family members, which belong to a group of single hotdog fold fatty acyl-ACP thioesterase that generate fatty acids and β-ketofatty acids (Pulsifer et al., 2014) (Supplemental Figure 16) and possess conserved active site residues (aspartate, glycine, and valine in the DXXGXV motif) (Supplemental Figure 17). Furthermore, the relative expression of the three indicated genes under 50% PEG-8000 treatment was monitored via RT-qPCR, which demonstrated that MtAPT1 was rapidly and strongly induced in response to dehydration stress (Figure 8A).
Figure 8.
MtAPT1 Interacts with MfNACsa and Mediates the Nuclear Translocation of MfNACsa.
(A) The relative expression of MtAPT1, Medtr5g056230, and Medtr5g056390 belong to a hotdog fold fatty acyl-ACP thioesterase in M. truncatula and were monitored by RT-qPCR under 50% PEG-8000 treatment for 8 h. Gene expression was calculated using the 2−ΔΔCT method and the MtActin as an endogenous control. The mean values and se were calculated from three independent experiments.
(B) The interaction of MfNACsa with MtAPT1 was tested by yeast two-hybrid analysis. The transformed yeast cultures were spotted onto SD plates without tryptophan, histidine, leucine, and adenine, but supplemented with 9 mM 3-aminotriazole. The plates were incubated for 3 d, and an α-Gal assay was performed. Positive control, DBD-p53 + AD-RecT; negative control, DBD- Lam + AD-RecT.
(C) Cotransformation of MfNACsa-eGFP with MtAPT1-RFP (i) or MtAPT1m-RFP (DXXGXV-AXXAXA) mutant protein (ii) in onion epidermal cells. The signal patterns were observed at 488 nm (eGFP), 546 nm (RFP), and 358 nm (DAPI) fluorescence excitation wavelength by confocal imaging (Olympus FluoView FV1000). The ICQ value of MfNACsa-eGFP against DAPI is 0.210 (i) and against MtAPT1m-RFP is 0.174 (ii). Bars = 50 μm.
(D) The BiFC assay of SPYNE173-MfNACsa with SPYCE(MR)-MtAPT1m (i), SPYNE173-MfNACsa with SPYCE(MR) (ii), and SPYNE173 with SPYCE(MR)-MtAPT1m (iii), which were coexpressed transiently in onion epidermal cells, and the YFP protein was visualized by confocal imaging (Olympus FluoView FV1000). Bars = 50 μm.
(E) Immunoblot assay of total, nuclear, and membrane components separated from N. benthamiana leaf cells that were cotransformed with Pro35S:MfNACsa-GFP and Pro35S:MtAPT1-HA (i) or Pro35S:MtAPT1m-HA (ii). H+-ATPase and Histone H3 were used as PM and nuclear fraction markers, respectively.
To investigate whether MtAPT1 interacts with MfNACsa, a yeast two-hybrid assay was performed. We cotransformed the GAL4 DBD-MtAPT1 construct, as bait, and the full-length MfNACsa protein fused with the GAL4 activation domain (GAL4 AD-MfNACsa) into the AH109 yeast strain. The coexistence of the GAL4AD-MfNACsa and GAL4DBD-MtAPT1 fusion proteins induced growth on SD minimal base medium lacking tryptophan, histidine, leucine, and adenine in the presence of 9 mM 3-aminotriazole and showed α-Gal activity. By contrast, none of the empty vectors grew in the same medium (Figure 8B). Thus, MtAPT1 interacted with MfNACsa in vitro.
To identify whether MtAPT1 was colocalized with MfNACsa in vivo, we cotransformed MfNACsa-eGFP and MtAPT1-RFP in onion cells. We found that GFP and RFP fluorescence signals were detected at the membranes and GFP fluorescence was observed in the nucleus (Figure 8C, i). When the active site residues of MtAPT1 were mutated (DXXGXV-AXXAXA), MtAPT1m was colocalized with MfNACsa only at membranes (Figure 8C, ii), suggesting that MtAPT1 induced the nuclear accumulation of MfNACsa, whereas mutation of the MtAPT1 catalytic site resulted in no cleavage activity, despite colocalization. Taken together, we speculated that GFP fluorescence observed in the nucleus was probably related to the activity of wild-type MtAPT1.
Considering the localization of MfNACsa was affected by active MtAPT1, the mutation of MtAPT1 at the active site residues (DXXGXV-AXXAXA) was used for a bimolecular fluorescence complementation (BiFC) assay to determine the interaction locations of MfNACsa and MtAPT1. YFP fluorescence was only detected at the membranes of cells cotransformed with MfNACsa-nYFP and MtAPT1m-cYFP (Figure 8D). Combining the results of colocalization, we deduced that wild-type MtAPT1 physically interacted with MfNACsa and possibly accelerated the release of MfNACsa from the PM.
To further investigate the role of MtAPT1 in the activation of MfNACsa nuclear translocation, the cells of N. benthamiana leaves cotransformed with MfNACsa-GFP and wild-type MtAPT1 or MtAPT1m mutant protein were used to generate extracts after separating the total, nuclear, and membrane components. Immunoblot analysis revealed that GFP-tagged MfNACsa was detectable in both the nuclear and membrane components following cotransformation with MfNACsa and wild-type MtAPT1 but was only detectable in the membrane component following cotransformation with MfNACsa and the mutated form of MtAPT1m (Figure 8E). Based on these results, we deduced that the acyl-protein thioesterase MtAPT1 is required to regulate MfNACsa nuclear translocation.
APT1 and NACsa Are Required for GlyI Expression and Glutathione Homeostasis in Response to Drought Stress
As APT1 mediates NACsa nuclear translocation, we were prompted to examine whether the activation of GlyI expression by NACsa was dependent on APT1. To this end, Tnt1 insertion mutants of APT1 were identified from the Tnt1 retrotransposon-tagged mutant population of M. truncatula. Only NF15130 (designated apt1) was screened. The Tnt1 retrotransposon was inserted in the second exon of APT1 (Supplemental Figure 19A), which was confirmed by PCR (Supplemental Figure 19B). The expression of APT1 transcript was significantly impaired, as confirmed by RT-PCR (Supplemental Figure 19C). We further examined the temporal expression pattern of GlyI in the apt1 mutant and wild-type plants. PEG-imposed dehydration stress enhanced GlyI expression to peak levels after 2 h in wild-type plants, whereas the GlyI transcript was not induced in the apt1 mutant under stress (Figure 9A), suggesting that APT1 is also essential for the activation of GlyI under dehydration stress.
Figure 9.
APT1 and NACsa Are Required for GlyI Expression and GSH/GSSG Ratio Regulation.
(A) Transcript levels of MtGlyI in 2-week-old wild-type (R108) plants or the apt1 mutant subjected to 50% PEG-8000 treatment for 0, 0.5, 1, 2, and 4 h. The error bars represent the se of two samples. The asterisk indicates a significant difference between apt1 mutants and wild-type plants under PEG-imposed dehydration stress (Kruskal-Wallis nonparametric test, **P < 0.01).
(B) The apt1 mutant and wild-type plant tissues (0.1 g) were used to measure the total glutathione (GSH+GSSG) and GSSG, and the GSH/GSSG ratio was calculated under 50% PEG-8000 treatment for 0, 0.5, 1, 2, and 4 h. The error bars represent se of two samples. The asterisk indicates a significant difference between apt1 mutants and wild-type plants under PEG-imposed dehydration stress (Kruskal-Wallis nonparametric test, **P < 0.01).
(C) Transcript levels of MtGlyI in 2-week-old of wild-type (R108) plants or nacsa-1 mutant with treated with 50% PEG-8000 for 0, 0.5, 1, 2, and 4 h. The error bars represent the se of two samples. The asterisk indicates a significant difference between nacsa mutants and wild-type plants under PEG-imposed dehydration stress (Kruskal-Wallis nonparametric test, **P < 0.01).
(D) The GSH/GSSG ratio of ectopic expression of MfNACsa, nacsa-1 mutant, and wild-type R108 plants under PEG-imposed dehydration stress. Total glutathione (GSH+GSSG) and GSSG were measured in leaf tissues of tested plants (0.1 g) subjected to 50% PEG-8000 treatment for 0, 0.5, 1, 2, and 4 h. The error bars represent the se of two samples. The asterisk indicates a significant difference between lines ectopically expressing MfNACsa or nacsa-1 mutant compared with wild-type plants under PEG-imposed dehydration stress (Kruskal-Wallis nonparametric test, **P < 0.01).
To identify the redox state of glutathione associated with the APT1 gene, we measured the ratio of GSH/GSSG in the apt1 mutant under dehydration stress. The GSH/GSSG ratio continuously decreased over 4 h upon 50% PEG-8000 treatment in wild-type plants, while the GSH/GSSG ratio markedly decreased in the apt1 mutant compared with that in wild-type plants (Figure 9B), suggesting that APT1 is necessary for the activation of GlyI expression and the regulation of the GSH/GSSG ratio mediated by MtGlyI in M. truncatula.
We speculated that the potential mechanism by which NACsa enhances drought tolerance involves alteration of the GSH/GSSG ratio. First, we determined the temporal expression pattern of GlyI in the nacsa mutant and wild-type plants under PEG-imposed dehydration stress. GlyI was significantly induced in wild-type plants, but was not induced in the nacsa mutant under 50% PEG-8000 treatment (Figure 9C). Furthermore, we determined the changes in the GSH/GSSG ratio in ectopic expression of NACsa lines and the nacsa mutant under dehydration stress. The result showed that the GSH/GSSG ratio decreased continuously over 4 h under 50% PEG-8000 treatment in wild-type plants. This decrease was more apparent in the nacsa mutant, suggesting that knockout of NACsa decreased the GSH/GSSG ratio under dehydration stress. By contrast, the GSH/GSSG ratio continuously increased, peaking after 2 h of stress in the ectopic expression of NACsa lines (Figure 9D), further confirming that NACsa affects the changes in the GSH/GSSG ratio under dehydration stress. We speculated that the decreased GSH/GSSG ratio observed in the ectopic expression of MfNACsa lines after 4 h compared with 2 h of drought stress may have been due to the serious oxidative damage caused by prolonged stress. These results further confirmed that the effect of GlyI on the GSH/GSSG redox potential was dependent on the nuclear translocation of NACsa.
In conclusion, S-palmitoylation-modified MfNACsa was translocated into the nucleus upon dehydration stress, which was mediated by the thioesterase APT1, and this regulation mode played a vital role in GlyI expression and glutathione homeostasis.
DISCUSSION
Drought-Induced Changes in the Cellular Glutathione Pool Are Correlated with the Regulatory Module of NACsa/APT1/GlyI
In this study, we characterized a lipid-anchored NAC domain-containing protein, NACsa, as a positive regulator of the glutathione redox balance mediated by activation of GlyI. The thioesterase APT1 is involved in this process. This conclusion is supported by the following observations. First, APT1 was rapidly and strongly induced in response to dehydration stress (Figure 8A). Second, coexpression of APT1 accelerated the nuclear translocation of NACsa, suggesting that de-S-palmitoylation mediated by APT1 is required for the nuclear import of NACsa (Figure 8D). Third, the coexistence of NACsa and APT1 is necessary for the activation of GlyI under drought stress. This notion was confirmed in nacsa and apt1 mutants (Figures 9A and 9C). Fourth, glutathione homeostasis under drought stress requires the activation of GlyI expression, which was confirmed in nacsa, apt1 and glyI mutants (Figures 5C, 9B, and 9D).
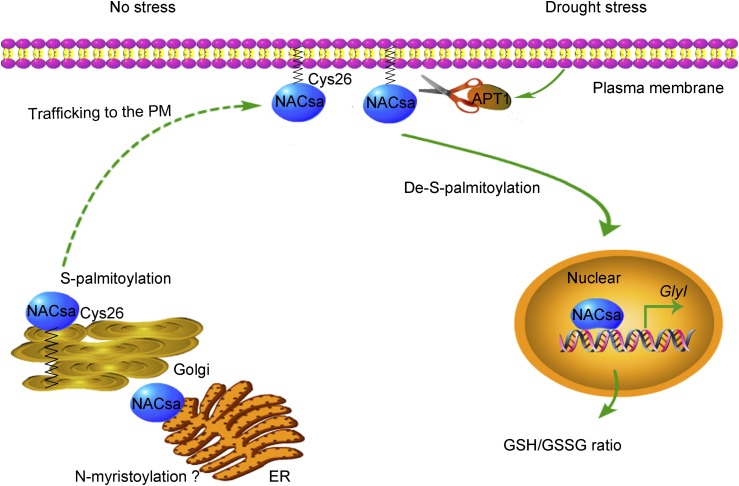
Based on the findings of this study, we propose a hypothetical working model for the nuclear translocation of lipid-anchored NACsa in response to drought tolerance (Figure 10). S-palmitoylated MfNACsa is transported to the PM via the ER and Golgi in an uninduced state. Under drought stress, MfNACsa is translocated to the nucleus after de-S-palmitoylation by the acyl-protein thioesterase APT1, where it activates the transcription of a glyoxylase GlyI, which maintains GSH homeostasis under drought stress.
Figure 10.
A Hypothetical Working Model for the Regulation of NACsa Translocation under Drought Stress.
NACsa is S-palmitoylated at Cys26 and anchored to the PM in an uninduced state. Under drought stress, NACsa is translocated to the nucleus after de-S-palmitoylation by the acyl-protein thioesterase APT1, where NACsa directly activates GlyI transcription and modulates the GSH/GSSG ratio.
It is worth noting that the model we proposed is focused on the regulation mechanism of de-S-palmitoylation for nuclear import of MfNACsa TF. However, little is known about the defined distribution between the ER/Golgi and the PM of MfNACsa, which are continuous with endosomes, although it is known that S-palmitoylation is crucial for ER-to-PM trafficking (Batistic et al., 2008), and it was reported that S-palmitoylated substrates were randomly and rapidly distributed over all membranes in mammalian cells (Rocks et al., 2010). We next examined why MfNACsa is retained in the ER/Golgi when protein palmitoylation is blocked. There are two hypotheses to address this issue: (1) MfNACsa initially undergoes an irreversible hydrophobic modification in the cytosol and then reversibly associates with the ER and Golgi membranes, where S-palmitoylation is processed for MfNACsa PM trafficking as GAD65 (GABA-synthesizing enzyme) that is palmitoylated in Golgi membranes and targeted to a post-Golgi vesicular pathway (Kanaani et al., 2008). (2) Other lipidations existed at the N terminus of MfNACsa. It was reported that an early cotranslational modification of N-terminal Met excision and myristoylation occur in the ribosomal tunnel to target calcineurin B-like protein 1 to the ER, and the consensus sequence of myristoylation is MGXXX(S/T) (Batistic et al., 2008). In our study, we analyzed the amino acid residues of MfNACsa and focused on the MQGXXX motif. The mutant form of G3A-eGFP was transiently expressed in onion cells to observe its subcellular localization. The confocal images revealed that G3A-eGFP was localized in the nucleus (Supplemental Figure 20). MfNACsaC26S was retained in the ER/Golgi when S-palmitoylation was prevented (Figures 6C and 6D). Taken together, we speculated that Gly-3 is a probable key residue for N-myristoylation of MfNACsa, which is a possible prerequisite for ER membrane tethering and the subsequent S-palmitoylation of MfNACsa. However, these possibilities need to be further validated.
We next wanted to know whether N-myristoylation at the Gly-3 residue of MfNACsa was affected by dehydration stress treatment. In our study, we found that PM-localized wild-type MfNACsa disappeared with prolonged PEG/NH2OH treatment (Figures 2B, 2C, and 7A), but the endomembrane-localized MfNACsaC26S mutant protein was not translocated into the nucleus upon stress signals (Figures 7B and 7D), suggesting that N-myristoylation of MfNACsa was not affected by the above stress treatment. These results indicated that de-S-palmitoylation played a vital role in regulating the mobilization of MfNACsa for nuclear import under dehydration stress. We speculate that, under dehydration stress, MfNACsa is released from the PM and the NLS is exposed, probably because of the protein conformational change, which makes MfNACsa translocate into the nucleus.
Therefore, the hypothesis model uncovered the regulatory module of NACsa/APT1/GlyI upon drought stress, whereas the possible influence on intracellular lipidation modification (N-myristoylation or S-palmitoylation) of MfNACsa under prolonged and extensive drought stress merits further study.
A General Mechanism for the Nuclear Translocation of S-Palmitoylated NAC TFs
In this study, we found that the N-terminal Cys26 of MfNACsa was important for the translocalization and function of the protein. However, it remains to be determined whether S-palmitoylation of NAC TFs that are imported into the nucleus is a more general phenomenon. To further investigate this possibility, we compared the phylogenetic trees with Arabidopsis, soybean, and M. truncatula to identify the homologous NACs of MfNACsa. Four Arabidopsis NACs cluster with MfNACsa and were most closely related to the ATAF group: ATAF1, ATAF2, ANAC102, and ANAC032 (Supplemental Figure 21). The online GPS-lipid predictor was used to predict potential lipidation, and a threshold with high stringency was chosen. Only ATAF1 and ATAF2 were predicted to undergo S-palmitoylation (Supplemental Figure 23). In soybean, GmNAC18, Glyma.04G208300.1, Glyma.05G195000.1, Glyma.04G249000.1, GmNAC2, Glyma.13G030900.1, and Glyma.14G152700.1 belong to the ATAF subfamily (Supplemental Figure 22), GmNAC18, Glyma.04G249000.1, GmNAC2, Glyma.13G030900.1, and Glyma.14G152700.1 were predicted to be S-palmitoylated, while Glyma.05G195000.1 was predicted to undergo both N-myristoylation and S-palmitoylation (Supplemental Figure 23). In M. truncatula, MtNACsa, Medtr3g096920.1, Medtr8g094580.1, and Medtr3g088110.1 belong to the ATAF subfamily (Supplemental Figure 2). MtNACsa, Medtr3g096920.1, and Medtr3g088110.1 were predicted to be S-palmitoylated, whereas there was no lipidation modification predicted for the Medtr8g094580.1 protein (Supplemental Figure 23).
Among these proteins, MtNACsa, ATAF2, and GmNAC2 were selected to determine the effect of S-palmitoylation/de-S-palmitoylation on their subcellular partitioning according to the GPS-lipid prediction (Supplemental Figure 24A). Confocal imaging showed that all three proteins were colocalized with the lipophilic dye FM4-64 (Supplemental Figure 24B, i). After 2 h of NH2OH treatment, the proteins translocated to the nucleus (Supplemental Figure 24B, ii). These results suggested that S-palmitoylation is vital for membrane association and de-S-palmitoylation is required for the nuclear translocation of ATAF subfamily members, which provided a general mechanism for the nuclear translocation of S-palmitoylated ATAF TFs and supplemented our current knowledge of the protein modifications of NAC TFs.
The Role of the Lipid-Anchored NACsa Transcription Factor
Whether the S-palmitoylated membrane-associated MfNACsa TF is only dormant or performs certain functions in the membrane system is an interesting question. In this study, we wanted to determine the role of PM-targeted MfNACsa. Palmitoylation of the soluble proteins is critical for lipid raft associations of the PM (Li et al., 2002; Fragoso et al., 2003), and the segregation of PM proteins into the lipid rafts is important for regulating many cellular signaling events and often requires palmitoylation (Levental et al., 2010). Such localization promotes interaction with functional partners and protein activity. Additionally, wild-type MfNACsa was significantly decreased at the PM and translocated into nucleus after PEG-simulated drought stress or NH2OH treatment, whereas the C26S mutant protein was retained at the endomembrane during PEG or NH2OH treatment, suggesting that only PM-localized/S-palmitoylated MfNACsa is translocated into the nucleus, which is consistent with the reported findings regarding the NFAT5α TF from H. sapiens (Eisenhaber et al., 2011). Therefore, we speculated that PM-localized MfNACsa specifically functions to rapidly respond and transduce the drought stress signal.
Interestingly, based on the transcriptome data, we found that some lipid transport/localization-related genes were differentially expressed in lines ectopically expressing MfNACsa (Supplemental Figure 13). Hence, we sought to decipher the role of the palmitic group released from the membrane: Does it return to the ER/Golgi for repalmitoylation, or is it involved in fatty acid metabolism under abiotic stress? These points merit further research.
The Role of Acyl-Protein Thioesterase in the Regulation of Drought Signal Transduction
Protein-bound palmitate is added posttranslationally and turns over more rapidly than the protein itself (Loisel et al., 1999; Conibear and Davis, 2010), suggesting that the regulation of protein de-S-palmitoylation rapidly modulates stress responses and cellular signal transduction. The signaling regulators in the animal system, such as H/N-Ras, heterotrimeric G-protein subunit Gi, and the nonreceptor tyrosine kinase Fyn, are de-S-palmitoylated to be released and returned to the Golgi for another round of repalmitoylation. The Golgi and PM pools of these proteins activate different downstream signaling cascades (Fehrenbacher et al., 2009; Conibear and Davis, 2010). Additionally, S-palmitoylated proteins that eventually undergo degradation and disposal in the lysosome are de-S-palmitoylated through the palmitoyl-protein thioesterase (PPT1) to cleave fatty acids from cysteine residues in proteins (Lu and Hofmann, 2006). Until recently, little was known about the role of de-S-palmitoylation in TFs during signal transduction in plant.
In animals, acyl-protein thioesterase 1 (APT1) was initially identified as a lysophospholipase that catalyzes fatty acid release (Sugimoto et al., 1998; Satou et al., 2010). Subsequently, APT1 was the first authentic participant in the regulated depalmitoylation of intracellular signaling proteins to be identified (Duncan and Gilman, 1998, 2002), and increasing evidence suggests that APT1 and APT2 are the major APTs that mediate the depalmitoylation of diverse cellular substrates (Lin and Conibear, 2015). In plants, acyl-protein thioesterase ALT-like genes, which are present in diverse plant taxa, are important for regulating lipid metabolism and lipid trafficking (Pulsifer et al., 2014). However, little is known about the role of acyl-protein thioesterases in signal transduction in plants. In this study, we demonstrate that a newly characterized acyl-protein thioesterase 1 (MtAPT1) regulates MfNACsa translocation, suggesting a role for MtAPT1 in the regulation of drought signal transduction. However, whether MtAPT1 is the only enzyme responsible for regulating the state of S-palmitoylation in Medicago and which factors regulate the access of APT1 to NACsa merit further investigation.
METHODS
Plant Materials
Medicago falcata ecotype (accession number PI502449) is a diploid cultivar of M. falcata, and the seeds originated from Russia; Medicago truncatula cv R108 seeds were provided by BRC (UMR 1097, INRA, Montpellier, France).
The Tnt1 insertion mutant lines, NF5250 (nacsa-1 mutant), NF9803 (nacsa-2 mutant), NF15130 (apt1 mutant), NF20885 (glyI-1 mutant), and NF10049 (glyI-2 mutant) were screened from a Tnt1 retrotransposon-tagged mutant population of M. truncatula (Tadege et al., 2008; Cheng et al., 2014).
The Pro35S:MfNACsa-3×FLAG plasmid construct was generated using the full-length MfNACsa gene amplified from M. falcata cDNA. The plant expression construct was transformed into the Agrobacterium tumefaciens EHA105 strain and used for the transformation of M. truncatula cv R108 as previously described (Cosson et al., 2006).
Pro35S:MfNACsa-GFP and ProMfNACsa:MfNACsa-GFP carried by the Agrobacterium rhizogenes strain ARqua1 were transformed into M. truncatula hairy roots as described in the Medicago truncatula Handbook (http://www.noble.org/Global/medicagohandbook/pdf/AgrobacteriumRhizogenes.pdf). Approximately 4 weeks after A. rhizogenes inoculation, plants expressing the MfNACsa-GFP fusion protein were subjected to 50% PEG-8000 or 0.05 M NH2OH treatment. Under treatment, the hair roots were divided into different portions, one was imposed into deionized water (mock), and others were soaked in PEG-8000 or NH2OH solution for various hours, respectively. The numerous cells/tissues were observed. The GFP fluorescence was excited at 488 nm, DAPI was used to stain nuclei and excited at 385 nm, and FM4-64 staining was observed at a 546-nm fluorescence excitation wavelength under a confocal microscope (Olympus FluoView FV1000), and the single slice image mode was performed because of the thickness of the hairy root tissues.
Seeds were surfaced-sterilized with 1 mL of H2SO4 for 8 min, sterilized five times with water, and then sterilized with 1 mL of 5% sodium hypochlorite containing 0.05% (v:v) Tween 20 for 15 min and subsequently washed five times with sterile water (Clean Bench). The sterilized seeds were plated on 0.8% agarose medium and allowed to grow for ∼3 d at 4°C, after which they were placed in the dark at 24°C for 1 d. The germinated seeds were subsequently planted in a soil/vermiculite (1:3, v/v) mixture and grown in a controlled culture room under a 22°C/18°C day/night regime, with 70% relative humidity, dim light (0.7–0.8 μmol s−1 m−2) (Philips; TL5 Essential Super 80) and a 14-h-light/10-h-dark light cycle.
Dehydration Treatment of M. falcata and RT-qPCR Assay
M. falcata cv PI502449 seeds were germinated as described above, and 4-week-old seedlings were planted in the soil-oriented plates for dehydration treatment according to previous work (Seki et al., 2002; Lee et al., 2012; Miao et al., 2015). Briefly, the seedlings were air-dried on 3 MM Whatman filter paper for various durations (1, 2, 3, and 4 h) under dim light (0.7–0.8 μmol s−1 m−2) (Philips; TL5 Essential Super 80), a relative humidity of 70%, and a 22°C/18°C day/night regime during the stress period. Whole plants were employed for the extraction of total RNA using the TRIzol reagent (Invitrogen), and first-strand cDNA synthesis was performed using 2.0 μg of total RNA using oligo-(dT18) and M-MLV reverse transcriptase (Promega). RT-qPCR analysis was performed in a CFX-96 real-time system (Bio-Rad) using SYBR Premix Ex Taq (TaKaRa). To validate the presumed stable expression of reference genes under dehydration, the stability of four candidate reference genes (MfActin, MfGAPDH, MfEFla, and MfeIF4A) during various dehydration periods was ranked according to a comparison of Ct values using GeNorm software, which defined the internal control gene-stability measure (M) as the average pairwise variation of a particular gene with all other control genes (Vandesompele et al., 2002). The expression of MfEF1a was found to be stable at the lowest M value (M = 0.035 < 0.5) and was selected as the reference gene for PI502449 under dehydration treatment. For the temporal expression of the MfNACsa transcript, the relative expression data were normalized with MfEF1a. The mean values and se were calculated from the results of two independent experiments. The primers that were used in this study are listed in Supplemental Data Set 8.
For the spatial analysis of MfNACsa expression after dehydration for 2 h, the roots, stems, and leaves of three individual plants were sampled. The experiments were independently repeated twice. Total RNA extraction, reverse transcription, and RT-qPCR analysis were performed as previously described.
Phenotypic Assay, Electrolyte Leakage, and GSH/GSSG Ratio Determination under Soil Drought Stress Treatment
Seedlings of lines ectopically expressing MfNACsa, nacsa mutants, glyI mutants, and the wild type (R108) were grown under normal watering, in individual pots containing an equal weight of a dry soil/vermiculite (1:3, v/v) mixture in a controlled culture room. Ten-day-old seedlings were subjected to three cycles of water-deficit treatments by withholding water for 12, 14, or more than 14 d, respectively. After the first and second cycles of drought stress treatment, the seedlings were rewatered with 600 mL water/plot for the next drought stress. The plants were photographed and the survival rates and relative electrical conductivity in plants were investigated until the wild-type or mutant leaves showed primary wilting. The mock-treated plants were watered once a week, according to standard procedures.
The seedlings were evaluated to determine the survival percentage based on the observation of actively regrowing seedlings as survivors, and nongrowing or wilted seedlings, as nonsurvivors. The experiments were performed using 15 seedlings of each line, and the vertical bars represent the se of three individual experiments.
Electrolyte leakage was assayed as previously described (Zhang et al., 2011) with some modifications. Briefly, three leaf discs detached from the drought-stressed plants described above were immersed in 50-mL tubes containing 25 mL of deionized water. After 15 min of vacuum treatment and shaking at 230 rpm for 2 h at 25°C, the electrical conductivity of the samples (C1) was determined with an electrical conductivity meter (HI8733; Hanna Instruments). The samples were then boiled for 15 min. After cooling to room temperature, the electrical conductivity of the samples (C2) and deionized water (C0) were determined again. The relative ion leakage% = (C1-C0/C2-C0) × 100% was subsequently calculated. The experiments were performed using 15 seedlings from each line, and the vertical bars represent the se of three individual experiments.
Total glutathione (GSH+GSSG) and GSSG were measured according the GSH and GSSG Assay Kit (Beyotime) using kinetic determination methods under a 405-nm wavelength (Bio-Rad; model 680), and the GSH/GSSG ratio (reduced/oxidized glutathione) was calculated. The vertical bars represent the se of two individual samples.
PEG-Simulated Dehydration Stress Treatment
The wild-type (R108) plants grown under treatment with different concentrations of PEG-8000 were germinated as described above. The sterilized seeds of wild-type plants were plated on 0.8% agarose medium and grown for 3 d at 4°C, followed by growth in darkness for 1 d at 24°C. The germinated seedlings were subsequently transferred to the agar-solidified half-strength Murashige and Skoog (MS) basal salt medium or 10% (w/v), 35% (w/v), and 70% (w/v) PEG-infused agar plates for 14 d. The PEG-infused plate system was described previously (Verslues and Bray, 2004). Then, the plants were transferred to half-strength MS salts for recovery for 14 d, and the relative length of the primary root and numbers of lateral roots were quantified. For each repeated experiment, ∼15 wild-type seedlings were used. The mean values and se were calculated from the results of two independent biological replicate.
The MfNACsa ectopic expression lines, nacsa mutants, and wild-type (R108) plants were plated on half-strength MS salts solid medium that supplemented with 35% (w:v) PEG-8000 as described above. The germinated seedlings were subsequently plated on half-strength MS salts solid medium that supplemented with 35% (w:v) PEG-8000 for 14 d. The seedlings were then transferred to half-strength MS salts solid medium for recovery for 7 d. In the mock control, the plants were cultured in normal half-strength MS salt solid medium for 21 d. The number of green leaf blades and the relative length of the primary roots were quantified. For each repeated experiment, ∼10 seedlings from the wild type, the MfNACsa ectopic expression lines, and the nacsa mutants were used. The mean values and se were calculated from the results of two independent biological replicates.
Transcriptome Analysis
Seedlings of lines ectopically expressing MfNACsa (OE23 and OE33) and the wild type were grown in half-strength MS liquid medium under a 16-h-light/8-h-dark cycle (Philips; TL5 Essential Super 80). Total RNA was extracted from 2-week-old seedlings treated with 50% PEG-8000 for 4 h. Total RNA was isolated from whole plants using TRIzol reagent (Invitrogen). RNA quality was monitored based on the absorbance at 260 and 280 nm (NanoVue plus), and the integrity of all RNA samples was examined via 1.2% agarose gel electrophoresis. The total RNA samples were pooled into four samples for RNA-seq analysis (Novogene), designated PWT-1, PWT-2, PMfNACsa (PMfNMASA)-1, and PMfNACsa (PMfNMASA)-2 (two genotypes and two biological replicates each). Then, 3 μg of total RNA from each sample was used to isolate poly(A) mRNA and prepare a nondirectional Illumina RNA-seq library. For each library, the SE50 single-read sequencing method was applied using the Illumina HiSeq 2500 platform. Based on the RNA-seq data, 30 of the most significantly enriched GO terms, analyzed with Goseq software, are presented in Supplemental Figure 11A. GO terms with a corrected P value of <0.05 were considered significantly enriched (Young et al., 2010). We used KOBAS software to test for statistical enrichment of differentially expressed genes in KEGG pathways (Mao et al., 2005), and 20 of the most significantly enriched pathway terms were assessed.
To determine the relative expression of upregulated genes based on RNA-seq analysis, dehydration stress treatment of wild-type plants and of plants ectopically expressing MfNACsa was performed as described previously in the RNA-seq assay. The MtActin gene (GenBank: JQ028731.1) was used as the reference gene, and all primers employed in this study are listed in Supplemental Data Set 8.
To determine the temporal expression patterns of MtNACsa, MtAPT1, and MtGlyI under 50% PEG-8000 treatment in M. truncatula cvR108, total RNA was extracted from leaf tissues of 2-week-old seedlings treated with or without 50% PEG-8000 for 0.5, 1, 2, and 4 h. Real-time RT-PCR analysis was performed with the CFX-96 real-time system (Bio-Rad) using SYBR Premix Ex Taq (TaKaRa). MtActin was used as the reference gene, and the primers employed in this assay are listed in Supplemental Data Set 84.
hiTAIL-PCR
Genomic DNAs were prepared from M. falcata cv PI502449 leaf tissues. The LAD primers, universal primer AC1, and known sequence-specific primers are listed in Supplemental Data Set 8. The preamplification, primary TAIL-PCR, and secondary TAIL-PCR reactions and the amplification procedure were performed as previously described (Liu and Chen, 2007).
Phylogenetic Analysis
NAC domains in the N-terminal region were classified into two large groups: Groups I and II, and each group could be divided into several subgroups on the basis of similarities in NAC domain structures (Ooka et al., 2003). Group I was divided into 14 subgroups (TERN, ONAC022, SENU5, NAP, AtNAC3, ATAF, OsNAC3, NAC2, ANAC011, TIP, OsNAC8, OsNAC7, NAC1, and NAM), and Group II was divided into four subgroups (ANAC001, ONAC003, ONAC001, and ANAC063). The N-terminal consensus NAC domains (including subdomains A–E) for each subfamily from Ooka et al. (2003), Figure 3, were used as an input for the phylogenetic tree analysis (Supplemental Data Set 1). According to the classification of NACs in Medicago (Supplemental Figure 1C), MfNACsa belonged to the ATAF subgroup of proteins that the N-terminal consensus NAC domain was subdivided into the five subdomains on N-terminal NAC domain (A–E). The NLS (IKKALVFYAGKAPKGVKTN) was located in the D motif; and the conserved TAR (EVQSEPKW) that is only found in ATAF group was previously reported in GmNAC30 from soybean (Glycine max; Mendes et al., 2013). A phylogenetic tree was constructed using MEGA 5.0 software through the neighbor-joining method with 1000 bootstrap replicates.
For the phylogenetic analysis of thioesterase-like proteins, sequences were obtained from the JCVI database (Mt genome v 4.0) (Supplemental Table 1 and Supplemental Data Set 5). The phylogenetic tree was constructed using MEGA 5.0 software through the neighbor-joining method with 1000 bootstrap replicates.
For the phylogenetic analysis of MfNACsa and homologous NACs in M. truncatula, Arabidopsis thaliana, and soybean, sequences were obtained from Phytozome v11.0 (http://www.phytozome.net) (see Supplemental Data Sets 2, 6, and 7). The multiple sequence alignment of NAC amino acids was analyzed using ClustalW2, and the phylogenetic tree was constructed by the neighbor-joining method. The numbers beside the branches represent bootstrap values based on 1000 replicates.
Transactivation Assays in Yeast Cells and Nicotiana benthamiana Leaves
The functional transactivation domain of MfNACsa was determined using a yeast transactivation assay as previously described (Shan et al., 2014). The coding region of MfNACsa was fused to the GAL4 binding domain (DBD). According to the manufacturer’s instructions, DBD-MfNACsa, DBD-MfNACsa-N (harboring N-terminal region without TAR), the positive control DBD-p53 + AD-RecT, and the negative control DBD-Lam + AD-RecT were employed to transform the AH109 yeast strain using the lithium acetate method. The transformed strains were then spotted onto SD medium lacking leucine/histidine/adenine and cultured for 3 d at 28°C. α-Gal activity was colorimetrically analyzed based on x-α-Gal staining.
The reporter construct ProGlyI:GUS was generated using the sequence of the promoter region harboring the MfNACsa binding site (CAAATNNNCTTTG) of MtGlyI (–488 to –38 bp) fused with the GUS reporter gene through restriction digestion, using the EcoRI and HindIII sites of the pCAMBIA1381 vector harboring the minimal 35S promoter. The Pro35S:MfNACsa-GFP effector construct and ProGlyI:GUS reporter construct were coinfiltrated into N. benthamiana leaves using the Agrobacterium EHA105 strain. GUS staining and GUS activity were assayed according to previously described methods with some modifications (Topping and Lindsey, 1997; Blazquez, 2007). The plants were incubated at 22°C for 3 d and were subjected to treatment with 10% PEG-8000 for 2 h or not prior to staining with X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) for 12 h. To determine the GUS activity, the crude protein was extracted from the leaves of infiltrated plants that were subjected to PEG treatment or not, using GUS extraction buffer (50 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 0.1% sodium lauryl/sarcosine, and β-mercaptoethanol). Enzymatic reactions were performed using 0.8 mg of total protein extract with 4-methylumbelliferyl-β-d-glucuronide (M-5664; Sigma Chemical) as substrate at 37°C for 30 min using an F4500 microtiter fluorometer.
Biolistic Bombardment in Onion Epidermal Cells
To examine subcellular localization, MfNACsa fused with eGFP at its N or C terminus, respectively, MfNACsa fused with mCherry at its N terminus and eGFP at its C terminus, the associated variations (C26S, G3A, and G10A), MtNACsa, the truncated forms of MfNACsa, ATAF2, and GmNAC2 fused with eGFP at their C terminus, and MtAPT1/MtAPT1m, AtPIP2A (PM marker), and GlyMan1 (ER/Golgi marker) fused with RFP at their C terminus were transiently expressed or coexpressed in onion cells. The transformed onion cells were cultured on MS medium for at least 2 h and bombarded with DNA-coated gold particles using a PDS-1000/He system (Bio-Rad) at 1100 p.s.i. The transformed cells were cultured on MS medium at 25°C for 18 to 24 h and subsequently observed under an Olympus FluoView FV1000 confocal fluorescence microscope. Confocal Z-stack, 1- to 3-μm/slice images were captured. The eGFP fluorescence was excited at 488 nm, and red RFP and mCherry were excited at 546 nm. DAPI was used to stain nuclei and excited at 385 nm. For staining with the cellular membrane-specific lipophilic dye FM4-64 (Lam et al., 2007; Zhou et al., 2013), transformed cells were stained with 30 μg/mL FM4-64 for more than 5 min and were observed at a 546-nm fluorescence excitation wavelength. For various treatments, such as PEG-8000, 2-bromopalmitate, and homologous control, the solution was added to the onion cells, and the same cells were imaged before and after PEG treatment. Concentration gradient experiments were repeated at least twice using samples derived from different specimens.
Intensity Correlation Analysis
Colocalization analysis of the fluorescence signals or dyes in the transformed onion cells or Medicago hairy roots cells was conducted using ImageJ. The intensity correlation analysis plug-in was employed as previously described (Li et al., 2004). When the intensities of two images varied synchronously, they varied around their respective mean image intensities together. The ICQ is based on nonparametric sign-test analysis of the product of the differences from the mean (PDM) values: dependent staining 0<ICQ≤+0.5; random staining: ICQ∼0; segregated staining: 0>ICQ≥−0.5.
Cellular Fractionation and Immunoblot Assays
Agrobacterium EHA105 cells carrying the Pro35S:MfNACsa-GFP or ProMfNACsa:MfNACsa-GFP plasmid were infiltrated into N. benthamiana leaves. The leaves of transformed N. benthamiana plants were harvested at 3 d after infiltration, and the fusion proteins were extracted as previously described (Lei et al., 2015) with some modifications. To separate the soluble cytoplasm and insoluble membrane components, ProMfNACsa:MfNACsa-GFP and Pro35S:MfNACsa-GFP fusion proteins under treatment with 50% PEG-8000 or 0.05 M hydroxylamine or normal conditions were extracted from N. benthamiana by homogenization in buffer I (50 mM HEPES-KOH, pH 7.5, 10% sucrose, 50 mM NaCl, 5 mM MgCl2, 1 mM 2-mercaptoethanol, protease inhibitor cocktail, and 2 mM PMSF). The homogenates were centrifuged for 10 min at 4°C at 25,000g, and the supernatants and the pellets were collected, respectively. The supernatants were centrifuged again at 100,000g for 1 h at 4°C, and the supernatant samples were collected as the soluble fraction. The collected pellets were incubated at 4°C for 30 min in buffer II (50 mM HEPES-KOH, pH 7.5, 10% sucrose, 50 mM NaCl, 5 mM MgCl2, 1 mM 2-mercaptoethanol, 0.2% Triton X-100, plant protease inhibitor cocktail, and 2 mM PMSF) and were centrifuged for 10 min at 4°C at 25,000g, The supernatants were collected as the insoluble membrane fraction. Protein concentrations were measured using a Coomassie (Bradford) protein assay kit (Thermo). The soluble fractions were 1× concentrated compared with the membrane component. The soluble fractions and insoluble membrane fraction were brought up to equal concentrations, and the fractions (1 mg) were analyzed by 10% SDS-PAGE (Liu et al., 2017), and immunoblotted using a 1:2000 dilution of GFP monoclonal antibody (M20004M; Abmart; ∼27 kD). A 1:5000 dilution of H+-ATPase (Agrisera; ∼95 kD) and a 1:5000 dilution of cFBPase (Agrisera; ∼37 kD) were used as plasma membrane and cytosolic fraction markers, respectively.
To isolate the cytoplasm and nuclei, the above fusion proteins were extracted from N. benthamiana using the nuclear/cytoplasmic method (Du et al., 2015). The 1.5 g powder was homogenized with 3 mL lysis buffer (20 mM Tris-HCl, pH 7.4, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 25% glycerol, 5 mM DTT, and 1× protease inhibitor cocktail), the supernatant was recentrifuged and collected as the cytoplasmic fraction, and the pellet was washed gently five times with 5 mL precooled NRBT buffer (20 mM Tris-HCl, pH 7.4, 2.5 mM MgCl2, 25% glycerol, and 0.2% Triton X-100). Then, the pellet was resuspended in 300 μL extraction buffer II (10 mM Tris-HCl, pH 8.0, 250 mM sucrose, 10 mM MgCl2, 5 mM β-mercaptoethanol, 1% Triton X-100, and protease inhibitor cocktail) and gently placed on a suspension of 300 μL extraction buffer III (10 mM Tris-HCl, pH 8.0, 1.7 M sucrose, 2 mM MgCl2, 5 mM β-mercaptoethanol, 0.15% Triton X-100, and 1× protease inhibitor cocktail). The pellet was resuspended in 100 μL lysis buffer and used as the nuclear fraction. Protein concentrations were measured with a Coomassie (Bradford) protein assay kit (Thermo). The nuclear fractions were 30-fold more concentrated than the soluble fractions. The samples were brought up to equal concentrations and the fractions (1 mg) were analyzed by 10% SDS-PAGE (Liu et al., 2017), and immunoblotted using a 1:2000 of dilution of GFP monoclonal antibody (M20004M; Abmart), 1:5000 dilution of Histone H3 (Agrisera; ∼17 kD), and 1:5000 dilution of cFBPase as nuclear and cytoplasmic fraction markers, respectively.
Palmitoylation of the MfNACsa Protein in Vitro
The pET-30a(+)-His-MfNACsa and pET-30a(+)-His-C26S (Cys26-Ser26) constructs were transformed into Escherichia coli strain BL21 (DE3), and the proteins were purified via Ni affinity chromatography according to the manufacturer’s instructions. The synthesis of biotin-modified palmitate (pal-Lys [biotin]) was performed at Synpeptide with 98% purity. For dissolution, 10% HAC + 90% CAN was employed, and the concentration was 200 ng/mL. M. truncatula cv R108 plant extracts were obtained from the insoluble membrane fraction and soluble cytoplasmic fraction as described previously (Lei et al., 2015).
The pET-30a(+)-His-MfNACsa and pET-30a(+)-His-C26S proteins (200 ng) were incubated at 25°C for 4 h with pal-Lys (biotin) (0.1 μg) and M. truncatula cv R108 plant extracts, respectively. Protein expression was subsequently confirmed through native protein gel electrophoresis and immunoblot analysis. Labeled signals were detected using the streptavidin-HRP system.
Yeast Two-Hybrid Assay
pGBKT7-MtAPT1 was generated to be used as a bait vector, and MfNACsa was cloned into the pGADT7 prey vector. The positive control pGBKT7-p53 + pGADT7-RecT and the negative control pGBKT7-Lam + pGADT7-RecT were used to transform the AH109 yeast strain using the lithium acetate method according to the manufacturer’s protocol (Clontech). The colonies were assayed on SD medium lacking leucine /histidine/adenine supplemented with 9 mM 3-aminotriazole and cultured for 3 d at 28°C. The α-galactosidase activity was colorimetrically analyzed based on α-Gal staining.
BiFC Assay
For BiFC visualization of the interaction between MfNACsa and MtAPT1/MtAPT1m when transiently coexpressed in onion epidermal cells, one group of vectors harbored the eYFP N terminus encompassing amino acids 1 to 173, and the target genes were cloned into the N terminus of eYFP (pSPYNE173) or the C terminus of eYFP [pSPYNE(R)173]. The other group of vectors harbored the eYFP C terminus encompassing amino acids 156 to 239 and the point mutation A206K in the eYFP C terminus, and the target genes were cloned into the N terminus of eYFP [pSPYCE(M)] or C terminus of eYFP [pSPYCE(MR)] (Waadt et al., 2008). To evaluate the interaction between MfNACsa and MtAPT1, the pSPYNE(R)173MfNACsa and pSPYCE(M)-MtAPT1/pSPYCE(MR)-MtAPT1; pSPYNE173-MfNACsa and pSPYCE (M)-MtAPT1/pSPYCE (MR)-MtAPT1; pSPYNE(R)173-MtAPT1 and pSPYCE(M)-MfNACsa/pSPYCE(MR)-MfNACsa; and pSPYNE173-MtAPT1 and pSPYCE(M)-MfNACsa/pSPYCE(MR)-MfNACsa of combined constructs were used to identify the physical interaction. The MfNACsa and MtAPT1m constructs were similarly combined and analyzed. All of the constructs and the respective negative controls were coexpressed in onion cells via gene-gun bombardment transient transformation, and YFP fluorescence signals were analyzed via confocal fluorescence microscopy with excitation wavelength at 513 or 488 nm, and the emission wavelength at 527 or 507 nm.
EMSA
MfNACsa was fused with a His tag and expressed in the E. coli BL21 strain using the pET-30a (+) vector. The proteins were purified through Ni affinity chromatography according to the manufacturer’s instructions. The EMSA experiments were performed using a LightShift chemiluminescent EMSA kit (Thermo). The reaction mixture was loaded into an 8% nondenatured polyacrylamide gel and blotted onto a nylon membrane under UV cross-linking. The labeled signals were detected using the streptavidin-HRP system. The DNA probes employed in the EMSA are listed in Supplemental Table 2.
ChIP Assay
The ChIP experiments were performed as previously described (Saleh et al., 2008) with some modifications. Two-week-old seedlings ectopically expressing MfNACsa were treated with 50% PEG-8000 for 4 h or not. The DNA-bound proteins from leaves were cross-linked with cross-linking buffer I (0.4 M sucrose,10 mM Tris-HCl, pH 8.0, and 1 mM EDTA) and resuspended in nuclei isolation buffer (0.25 M sucrose, 15 mM PIPES, pH 6.8, 5 mM MgCl2, 60 mM KCl, 15 mM NaCl, 1 mM CaCl2, and 0.9% Triton X-100). After centrifugation, the pellets (nuclei component) were resuspended in cold nuclei lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, pH 8.0, 0.1% SDS, 0.1% deoxycholate, and 1% Triton X-100), followed by sonication for 7 m at 0°C (Bioruptor Next Gen; Diagenode; high power, 15 s on, 15 s off, 14 cycles). An anti-FLAG antibody (F3165; Sigma-Aldrich) was used to immunoprecipitate the target MfNACsa-3×FLAG fusion protein. RT-qPCR was performed to identify enriched DNA fragments in the immunoprecipitations compared with inputs. The degenerated primers are listed in Supplemental Data Set 8. To analyze the relative immunoprecipitations of samples, mock samples (mock-actin, mock-A, mock-B, and mock-C) were individually set to 1 and the PEG-8000-treated samples were normalized relative to the mock samples, respectively (Wang et al., 2011). Actin was used as a negative control. The vertical bars represent the sd of two individual experiments.
Statistical Analysis
The values are presented as the mean ± se. Tests for significance were conducted using the nonparametric tests of Kruskal-Wallis H-test or the nonparametric tests of one-way ANOVA and Duncan multiple range test with SPSS statistical software. The P values are reported for each statistical test.
Accession Numbers
Sequence data from this article can be found in the GenBank data library under accession numbers KY673691 (MfEF1α), KY673692 (MfNACsa), KY673693 (MtNACsa), KY673694 (MtGlyI), KY673695 (MtAPT1), At1g01720.1 (ATAF1), At5g08790.1 (ATAF2), At5g63790.1 (ANAC102), At1g77450.1 (ANAC032), Glyma.06G114000.1 (GmNAC2), and Glyma.06G157400.1 (GmNAC18). The raw data for RNA-seq analysis have been submitted to the NCBI database under accession number PRJNA383288.
Supplemental Data
Supplemental Figure 1. MfNACsa is induced by dehydration stress and encodes an ATAF NAC-domain transcriptional activator.
Supplemental Figure 2. The phylogenetic tree of the homologous NACs of MfNACsa in M. truncatula.
Supplemental Figure 3. The amino acid sequence and domain structure of ATAF members in M. truncatula.
Supplemental Figure 4. The intensity correlation analysis plots of colocalization of MfNACsa with membrane markers and membrane/nuclear dyes.
Supplemental Figure 5. Subcellular localization of N-terminal GFP fusion MfNACsa protein.
Supplemental Figure 6. A subdomain at the N terminus of MfNACsa was sufficient to mediate plasma membrane targeting.
Supplemental Figure 7. The intensity correlation analysis of MfNACsa translocation under PEG-imposed dehydration stress.
Supplemental Figure 8. Molecular identification of the lines ectopically expressing MfNACsa or Tnt1 insertion.
Supplemental Figure 9. The phenotype of wild-type R108 under PEG-8000 treatment.
Supplemental Figure 10. NACsa positively regulates plant dehydration tolerance.
Supplemental Figure 11. Gene Ontology and pathway enrichment analysis for differentially expressed genes regulated by MfNACsa.
Supplemental Figure 12. The temporal expression pattern of stress-, lipid transport-, and localization-related genes under dehydration stress.
Supplemental Figure 13. MfNACsa positively regulates the stress-, lipid transport-, and localization-related genes expression under PEG-imposed drought stress.
Supplemental Figure 14. Molecular identification of glyI mutants.
Supplemental Figure 15. The intensity correlation analysis of mutant C26S-eGFP or MfNACsa-eGFP subjected to 2-BP treatment.
Supplemental Figure 16. MtAPT1 belongs to a single hotdog fold fatty acyl-ACP thioesterase.
Supplemental Figure 17. MtAPT1 possesses conserved active site residues (aspartate, glycine, and valine) in the DXXGXV motif of acyl-ACP thioesterase members.
Supplemental Figure 18. The intensity correlation analysis of MfNACsa colocalized with wild-type MtAPT1 and mutant MtAPT1m.
Supplemental Figure 19. Molecular identification of the apt1 mutant.
Supplemental Figure 20. Subcellular localization of MfNACsaG3A-eGFP fusion protein.
Supplemental Figure 21. The phylogenetic tree of the homologous NACs of MfNACsa in Arabidopsis.
Supplemental Figure 22. The phylogenetic tree of the homologous NACs of MfNACsa in soybean.
Supplemental Figure 23. The lipidation prediction of ATAF family members in Arabidopsis, Glycine max, and M. truncatula.
Supplemental Figure 24. A general mechanism for the nuclear relocation of S-palmitoylated ATAF transcription factors.
Supplemental Table 1. List of potential thioesterase-like proteins (JCVI database, Mt genome v 4.0).
Supplemental Table 2. Oligonucleotide probes used in EMSA.
Supplemental Data Set 1. Text file of the amino acid sequence of MfNACsa and the N-terminal consensus NAC domains from Ooka et al. (2003) that were used as an input for the phylogenetic tree analysis.
Supplemental Data Set 2. Text file of the homologous NACs of MfNACsa in M. truncatula.
Supplemental Data Set 3. Upregulated genes in lines ectopically expressing MfNACsa under PEG-simulated drought stress.
Supplemental Data Set 4. Downregulated genes in lines ectopically expressing MfNACsa under PEG-simulated drought stress.
Supplemental Data Set 5. Text file of the amino acid sequences of potential thioesterase-like proteins in M. truncatula.
Supplemental Data Set 6. Text file of the homologous NACs of MfNACsa in Arabidopsis.
Supplemental Data Set 7. Text file of the homologous NACs of MfNACsa in Glycine max.
Supplemental Data Set 8. Primers used in this study.
Acknowledgments
We thank Jean Marie Prosperi and Magalie Delalande (BRC for Medicago truncatula, UMR 1097, INRA, Montpellier, France) for providing seeds of M. truncatula cv R108. We thank the U.S. National Plant Germplasm System of the USDA for providing M. falcata cv PI502449 seeds. This work was supported by grants from the National Natural Science Foundation of China (31571587 and 31371689).
AUTHOR CONTRIBUTIONS
T.W. and J.D. led the study and helped revise the manuscript. M.D. performed the main experiments and wrote the manuscript. R.Z. participated in the plant materials preparation, stress treatment, and data analysis. F.Z. participated in the modifications of some constructs and genetic transformation. Z.Z. participated in the ChIP assay. L.G. participated in genetic transformation and figures arrange. J.W. provided the Tnt1 mutants. All authors read and approved the final manuscript.
Glossary
- TF
transcription factor
- PM
plasma membrane
- ER
endoplasmic reticulum
- TAR
transcriptional activation region
- NLS
nuclear localization signal
- SD
synthetic dropout
- ICQ
intensity correlation quotient
- DAPI
4′,6-diamidino-2-phenylindole
- GO
Gene Ontology
- EMSA
electrophoretic mobility shift assay
- ChIP
chromatin immunoprecipitation
- BiFC
bimolecular fluorescence complementation
- MS
Murashige and Skoog
Footnotes
Articles can be viewed without a subscription.
References
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudo-Ibáñez L., Herrero A., Barbacid M., Crespo P. (2015). H-ras distribution and signaling in plasma membrane microdomains are regulated by acylation and deacylation events. Mol. Cell. Biol. 35: 1898–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambawat S., Sharma P., Yadav N.R., Yadav R.C. (2013). MYB transcription factor genes as regulators for plant responses: an overview. Physiol. Mol. Biol. Plants 19: 307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626. [DOI] [PubMed] [Google Scholar]
- Baldoni E., Genga A., Cominelli E. (2015). Plant MYB transcription factors: their role in drought response mechanisms. Int. J. Mol. Sci. 16: 15811–15851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O., Sorek N., Schültke S., Yalovsky S., Kudla J. (2008). Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20: 1346–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez M. (2007). Quantitative GUS Activity Assay of Plant Extracts. Cold Spring Harb. Protoc. 2007: doi/10.1101/pdb.prot4609. [DOI] [PubMed]
- Camp L.A., Hofmann S.L. (1993). Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J. Biol. Chem. 268: 22566–22574. [PubMed] [Google Scholar]
- Chen M., Zhao Y., Zhuo C., Lu S., Guo Z. (2015). Overexpression of a NF-YC transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol. J. 13: 482–491. [DOI] [PubMed] [Google Scholar]
- Cheng X., Wang M., Lee H.K., Tadege M., Ratet P., Udvardi M., Mysore K.S., Wen J. (2014). An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol. 201: 1065–1076. [DOI] [PubMed] [Google Scholar]
- Christianson J.A., Dennis E.S., Llewellyn D.J., Wilson I.W. (2010). ATAF NAC transcription factors: regulators of plant stress signaling. Plant Signal. Behav. 5: 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E., Davis N.G. (2010). Palmitoylation and depalmitoylation dynamics at a glance. J. Cell Sci. 123: 4007–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson V., Durand P., d’Erfurth I., Kondorosi A., Ratet P. (2006). Medicago truncatula transformation using leaf explants. Methods Mol. Biol. 343: 115–127. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K.J., Vogel M.O., Viehhauser A. (2010). AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245: 3–14. [DOI] [PubMed] [Google Scholar]
- Du J.L., Zhang S.W., Huang H.W., Cai T., Li L., Chen S., He X.J. (2015). The splicing factor PRP31 is involved in transcriptional gene silencing and stress response in Arabidopsis. Mol. Plant 8: 1053–1068. [DOI] [PubMed] [Google Scholar]
- Duncan J.A., Gilman A.G. (1998). A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J. Biol. Chem. 273: 15830–15837. [DOI] [PubMed] [Google Scholar]
- Duncan J.A., Gilman A.G. (2002). Characterization of Saccharomyces cerevisiae acyl-protein thioesterase 1, the enzyme responsible for G protein alpha subunit deacylation in vivo. J. Biol. Chem. 277: 31740–31752. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Sammer M., Lua W.H., Benetka W., Liew L.L., Yu W., Lee H.K., Koranda M., Eisenhaber F., Adhikari S. (2011). Nuclear import of a lipid-modified transcription factor: mobilization of NFAT5 isoform a by osmotic stress. Cell Cycle 10: 3897–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Liao K., Du H., Xu Y., Song H., Li X., Xiong L. (2015). A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 66: 6803–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher N., Bar-Sagi D., Philips M. (2009). Ras/MAPK signaling from endomembranes. Mol. Oncol. 3: 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso R., Ren D., Zhang X., Su M.W., Burakoff S.J., Jin Y.J. (2003). Lipid raft distribution of CD4 depends on its palmitoylation and association with Lck, and evidence for CD4-induced lipid raft aggregation as an additional mechanism to enhance CD3 signaling. J. Immunol. 170: 913–921. [DOI] [PubMed] [Google Scholar]
- Golldack D., Li C., Mohan H., Probst N. (2014). Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 5: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomord V., Denmat L.A., Fitchette-Lainé A.C., Satiat-Jeunemaitre B., Hawes C., Faye L. (1997). The C-terminal HDEL sequence is sufficient for retention of secretory proteins in the endoplasmic reticulum (ER) but promotes vacuolar targeting of proteins that escape the ER. Plant J. 11: 313–325. [DOI] [PubMed] [Google Scholar]
- Harshavardhan V.T., Van Son L., Seiler C., Junker A., Weigelt-Fischer K., Klukas C., Altmann T., Sreenivasulu N., Bäumlein H., Kuhlmann M. (2014). AtRD22 and AtUSPL1, members of the plant-specific BURP domain family involved in Arabidopsis thaliana drought tolerance. PLoS One 9: e110065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley P.A., Grierson C.S. (2008). Multiple roles for protein palmitoylation in plants. Trends Plant Sci. 13: 295–302. [DOI] [PubMed] [Google Scholar]
- Hemsley P.A., Taylor L., Grierson C.S. (2008). Assaying protein palmitoylation in plants. Plant Methods 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley P.A., Weimar T., Lilley K.S., Dupree P., Grierson C.S. (2013). A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol. 197: 805–814. [DOI] [PubMed] [Google Scholar]
- Hu H., Dai M., Yao J., Xiao B., Li X., Zhang Q., Xiong L. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 103: 12987–12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Wang Y., Li B., Chang J., Chen M., Li K., Yang G., He G. (2015). TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 15: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh S.U., Lee S.B., Kim H.H., Paek K.H. (2012). ATAF2, a NAC transcription factor, binds to the promoter and regulates NIT2 gene expression involved in auxin biosynthesis. Mol. Cells 34: 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Choudhary D., Kale R.K., Bhalla-Sarin N. (2002). Salt- and glyphosate-induced increase in glyoxalase I activity in cell lines of groundnut (Arachis hypogaea). Physiol. Plant. 114: 499–505. [DOI] [PubMed] [Google Scholar]
- Jennings P., Bertocchi C., Frick M., Haller T., Pfaller W., Dietl P. (2005). Ca2+ induced surfactant secretion in alveolar type II cultures isolated from the H-2Kb-tsA58 transgenic mouse. Cell. Physiol. Biochem. 15: 159–166. [DOI] [PubMed] [Google Scholar]
- Jeong J.S., Kim Y.S., Baek K.H., Jung H., Ha S.H., Do Choi Y., Kim M., Reuzeau C., Kim J.K. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaani J., Patterson G., Schaufele F., Lippincott-Schwartz J., Baekkeskov S. (2008). A palmitoylation cycle dynamically regulates partitioning of the GABA-synthesizing enzyme GAD65 between ER-Golgi and post-Golgi membranes. J. Cell Sci. 121: 437–449. [DOI] [PubMed] [Google Scholar]
- Kim M.J., Park M.J., Seo P.J., Song J.S., Kim H.J., Park C.M. (2012). Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem. J. 448: 353–363. [DOI] [PubMed] [Google Scholar]
- Kim S.G., Lee A.K., Yoon H.K., Park C.M. (2008). A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. 55: 77–88. [DOI] [PubMed] [Google Scholar]
- Kim S.G., Lee S., Seo P.J., Kim S.K., Kim J.K., Park C.M. (2010). Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95: 56–65. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Kim S.G., Park J.E., Park H.Y., Lim M.H., Chua N.H., Park C.M. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18: 3132–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad S.S., Popp C., Stratil T.F., Jarsch I.K., Thallmair V., Folgmann J., Marin M., Ott T. (2014). S-acylation anchors remorin proteins to the plasma membrane but does not primarily determine their localization in membrane microdomains. New Phytol. 203: 758–769. [DOI] [PubMed] [Google Scholar]
- Krasensky J., Jonak C. (2012). Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63: 1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.K., Siu C.L., Hillmer S., Jang S., An G., Robinson D.G., Jiang L. (2007). Rice SCAMP1 defines clathrin-coated, trans-golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19: 296–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M., Yalovsky S. (2006). Association of Arabidopsis type-II ROPs with the plasma membrane requires a conserved C-terminal sequence motif and a proximal polybasic domain. Plant J. 46: 934–947. [DOI] [PubMed] [Google Scholar]
- Lavy M., Bracha-Drori K., Sternberg H., Yalovsky S. (2002). A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14: 2431–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Seo P.J., Lee H.J., Park C.M. (2012). A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 70: 831–844. [DOI] [PubMed] [Google Scholar]
- Lei M.J., et al. (2015). The small GTPase ROP10 of Medicago truncatula is required for both tip growth of root hairs and nod factor-induced root hair deformation. Plant Cell 27: 806–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I., Lingwood D., Grzybek M., Coskun U., Simons K. (2010). Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl. Acad. Sci. USA 107: 22050–22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Yang C., Tong S., Weidmann A., Compans R.W. (2002). Palmitoylation of the murine leukemia virus envelope protein is critical for lipid raft association and surface expression. J. Virol. 76: 11845–11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Lau A., Morris T.J., Guo L., Fordyce C.B., Stanley E.F. (2004). A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci. 24: 4070–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Li H., Zhou F., Li H., Liu J., Hao Y., Wang Y., Zhao H., Han S. (2015). Subcellular distribution of NTL transcription factors in Arabidopsis thaliana. Traffic 16: 1062–1074. [DOI] [PubMed] [Google Scholar]
- Lin D.T., Conibear E. (2015). Enzymatic protein depalmitoylation by acyl protein thioesterases. Biochem. Soc. Trans. 43: 193–198. [DOI] [PubMed] [Google Scholar]
- Liu Y.G., Chen Y. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43: 649–650, 652, 654. [DOI] [PubMed] [Google Scholar]
- Liu Z., Jia Y., Ding Y., Shi Y., Li Z., Guo Y., Gong Z., Yang S. (2017). Plasma membrane CRPK1-Mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 66: 117–128. [DOI] [PubMed] [Google Scholar]
- Loisel T.P., Ansanay H., Adam L., Marullo S., Seifert R., Lagacé M., Bouvier M. (1999). Activation of the β(2)-adrenergic receptor-Galpha(s) complex leads to rapid depalmitoylation and inhibition of repalmitoylation of both the receptor and Galpha(s). J. Biol. Chem. 274: 31014–31019. [DOI] [PubMed] [Google Scholar]
- Lu J.Y., Hofmann S.L. (2006). Thematic review series: lipid posttranslational modifications. Lysosomal metabolism of lipid-modified proteins. J. Lipid Res. 47: 1352–1357. [DOI] [PubMed] [Google Scholar]
- Lü S., Song T., Kosma D.K., Parsons E.P., Rowland O., Jenks M.A. (2009). Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 59: 553–564. [DOI] [PubMed] [Google Scholar]
- Mao X., Cai T., Olyarchuk J.G., Wei L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21: 3787–3793. [DOI] [PubMed] [Google Scholar]
- Mendes G.C., Reis P.A., Calil I.P., Carvalho H.H., Aragão F.J., Fontes E.P. (2013). GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc. Natl. Acad. Sci. USA 110: 19627–19632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z., et al. (2015). De novo transcriptome analysis of Medicago falcata reveals novel insights about the mechanisms underlying abiotic stress-responsive pathway. BMC Genomics 16: 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D.F., O’Brien P.J., Pepperberg D.R. (1991). Depalmitylation with hydroxylamine alters the functional properties of rhodopsin. J. Biol. Chem. 266: 20118–20123. [PubMed] [Google Scholar]
- Ng S., et al. (2013). A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25: 3450–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Y., Guo Y.J. (2008). Progress in the study on genes encoding enzymes involved in biosynthesis of very long chain fatty acids and cuticular wax in plants. Yi Chuan 30: 561–567. [DOI] [PubMed] [Google Scholar]
- Ooka H., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10: 239–247. [DOI] [PubMed] [Google Scholar]
- Pulsifer I.P., Lowe C., Narayaran S.A., Busuttil A.S., Vishwanath S.J., Domergue F., Rowland O. (2014). Acyl-lipid thioesterase1-4 from Arabidopsis thaliana form a novel family of fatty acyl-acyl carrier protein thioesterases with divergent expression patterns and substrate specificities. Plant Mol. Biol. 84: 549–563. [DOI] [PubMed] [Google Scholar]
- Rigal A., Doyle S.M., Robert S. (2015). Live cell imaging of FM4-64, a tool for tracing the endocytic pathways in Arabidopsis root cells. Methods Mol. Biol. 1242: 93–103. [DOI] [PubMed] [Google Scholar]
- Rocks O., Gerauer M., Vartak N., Koch S., Huang Z.P., Pechlivanis M., Kuhlmann J., Brunsveld L., Chandra A., Ellinger B., Waldmann H., Bastiaens P.I. (2010). The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141: 458–471. [DOI] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Satou M., Nishi Y., Yoh J., Hattori Y., Sugimoto H. (2010). Identification and characterization of acyl-protein thioesterase 1/lysophospholipase I as a ghrelin deacylation/lysophospholipid hydrolyzing enzyme in fetal bovine serum and conditioned medium. Endocrinology 151: 4765–4775. [DOI] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31: 279–292. [DOI] [PubMed] [Google Scholar]
- Shan W., Kuang J.F., Lu W.J., Chen J.Y. (2014). Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ. 37: 2116–2127. [DOI] [PubMed] [Google Scholar]
- Sharma C., Yang X.H., Hemler M.E. (2008). DHHC2 affects palmitoylation, stability, and functions of tetraspanins CD9 and CD151. Mol. Biol. Cell 19: 3415–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipston M.J. (2011). Ion channel regulation by protein palmitoylation. J. Biol. Chem. 286: 8709–8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaraja R.R., Kang M.H., Vaid K., Sanders S.S., Vilas G.L., Arstikaitis P., Coutinho J., Drisdel R.C., El-Husseini Ael.D., Green W.N., Berthiaume L., Hayden M.R. (2009). Palmitoylation of ATP-binding cassette transporter A1 is essential for its trafficking and function. Circ. Res. 105: 138–147. [DOI] [PubMed] [Google Scholar]
- Singh D., Laxmi A. (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 6: 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H., Odani S., Yamashita S. (1998). Cloning and expression of cDNA encoding rat liver 60-kDa lysophospholipase containing an asparaginase-like region and ankyrin repeat. J. Biol. Chem. 273: 12536–12542. [DOI] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., Cayrel A., Endre G., Zhao P.X., Chabaud M., Ratet P., Mysore K.S. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54: 335–347. [DOI] [PubMed] [Google Scholar]
- Tapia G., Morales-Quintana L., Parra C., Berbel A., Alcorta M. (2013). Study of nsLTPs in Lotus japonicus genome reveal a specific epidermal cell member (LjLTP10) regulated by drought stress in aerial organs with a putative role in cutin formation. Plant Mol. Biol. 82: 485–501. [DOI] [PubMed] [Google Scholar]
- Tarnowski B.I., Spinale F.G., Nicholson J.H. (1991). DAPI as a useful stain for nuclear quantitation. Biotech. Histochem. 66: 297–302. [PubMed] [Google Scholar]
- Topping J.F., Lindsey K. (1997). Promoter trap markers differentiate structural and positional components of polar development in Arabidopsis. Plant Cell 9: 1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso J.A., Micalella C., Martinez A., Brown S.C., Satiat-Jeunemaître B., Meinnel T., Giglione C. (2013). Roles of N-terminal fatty acid acylations in membrane compartment partitioning: Arabidopsis h-type thioredoxins as a case study. Plant Cell 25: 1056–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed]
- Verslues P.E., Bray E.A. (2004). LWR1 and LWR2 are required for osmoregulation and osmotic adjustment in Arabidopsis. Plant Physiol. 136: 2831–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Wang L., Hua D., He J., Duan Y., Chen Z., Hong X., Gong Z. (2011). Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 7: e1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Culver J.N. (2012). DNA binding specificity of ATAF2, a NAC domain transcription factor targeted for degradation by Tobacco mosaic virus. BMC Plant Biol. 12: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24: 482–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S.K., Singla-Pareek S.L., Ray M., Reddy M.K., Sopory S.K. (2005). Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem. Biophys. Res. Commun. 337: 61–67. [DOI] [PubMed] [Google Scholar]
- Yang Z.T., Wang M.J., Sun L., Lu S.J., Bi D.L., Sun L., Song Z.T., Zhang S.S., Zhou S.F., Liu J.X. (2014). The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 10: e1004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H.K., Kim S.G., Kim S.Y., Park C.M. (2008). Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol. Cells 25: 438–445. [PubMed] [Google Scholar]
- You J., Zong W., Hu H., Li X., Xiao J., Xiong L. (2014). A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiol. 166: 2100–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.L., Zhao M.G., Tian Q.Y., Zhang W.H. (2011). Comparative studies on tolerance of Medicago truncatula and Medicago falcata to freezing. Planta 234: 445–457. [DOI] [PubMed] [Google Scholar]
- Zhou L.Z., Li S., Feng Q.N., Zhang Y.L., Zhao X., Zeng Y.L., Wang H., Jiang L., Zhang Y. (2013). Protein S-ACYL Transferase10 is critical for development and salt tolerance in Arabidopsis. Plant Cell 25: 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]