Abstract
Analysis of transcripts of 75 genes encoding putative basic leucine zipper (bZIP) transcription factors in the Arabidopsis genome identified AtbZIP60, which was induced by tunicamycin. AtbZIP60 encodes a predicted protein of 295 aa with a putative transmembrane domain near its C terminus after a bZIP domain. A truncated form of AtbZIP60 without a transmembrane domain (AtbZIP60ΔC) fused with GFP localized to the nucleus, suggesting translocation of native protein to the nucleus by release from the membrane. AtbZIP60 was also induced by DTT and azetidine-2-carboxylate, which induce the endoplasmic reticulum (ER) stress response (also called the unfolded protein response). Expression of AtbZIP60ΔC clearly activated any of three BiP and two calnexin promoters in a dual luciferase assay using protoplasts of cultured cells. The induction was considered to be through cis-elements plant-specific unfolded protein response element and ER stress-response element. Interestingly, AtbZIP60ΔC also appeared to induce the expression of AtbZIP60 through an ER stress-response element-like sequence in the promoter of AtbZIP60. These characteristics of AtbZIP60 imply a signal transduction pathway of the ER stress response unique to plants.
Keywords: Arabidopsis thaliana, BiP, tunicamycin, unfolded protein response
The endoplasmic reticulum (ER) consists of a three-dimensional structure in eukaryotic cells where proteins for the secretary pathway are synthesized. Proper folding and assembly of proteins synthesized in the ER are necessary for transport to their final destinations. When folding or assembly of proteins in the ER is disordered, unfolded proteins accumulate in the ER and expression of genes for ER-resident chaperones, such as BiP, and folding enzymes are induced. This phenomenon is conserved among eukaryotic cells and is referred to as the ER stress response or the unfolded protein response (UPR) (1–4). Recent studies conducted in yeast and mammalian cells have shown that the ER stress response plays essential roles not only under specific stresses but also under normal growth conditions (5–8). In plants, the ER stress response has been implicated in plant-specific processes, such as seed development and pathogen response (9).
The mechanism of signal transduction for the ER stress response has been extensively characterized in yeast and mammalian cells. In yeast cells, IRE1, an ER membrane-located protein kinase/ribonuclease, plays a pivotal role for the perception of ER stress (10, 11). Sensing ER stress, IRE1 dimerizes and transautophosphorylates, activating its ribonuclease activity (12, 13). Activated IRE1 catalyzes the spliceosome-independent splicing of Hac1 mRNA, encoding a basic leucine zipper (bZIP) transcription factor. Hac1 protein is efficiently synthesized from spliced Hac1 mRNA and binds to a cis-element, UPR element (UPRE; consensus sequence CAGCGTG), resulting in induction of downstream chaperone genes, such as BiP (14–16).
The ER stress response pathways of mammalian cells are multiple, in contrast to that of yeast, which is explained by a linear pathway consisting of IRE1, Hac1, UPRE, and the induction of chaperone genes. In mammals, at least two bZIP transcription factors, XBP1 and ATF6, have been identified that function in the ER stress response. The XBP1 mRNA is spliced by IRE1α through unconventional splicing, similarly to yeast Hac1 (17). This splicing removes 26 nucleotides from authentic XBP1 mRNA, resulting in a frame shift. XBP1 protein, with an activation domain at the C terminus, is synthesized after splicing and enhances target gene expression through the cis-elements ER stress-response element (ERSE; consensus sequence CCAAT-N9-CCACG), ERSE-II (consensus sequence ATTGG-N-CCACG), or XBP1-BS [consensus sequence GA-TGACGT-G(T/G)] (18–22). Another protein, ATF6, is a transmembrane protein located in the ER membrane with a bZIP domain on the cytoplasmic side. In response to ER stress, ATF6 protein is processed by S1P and S2P proteases in the transmembrane domain (TMD) (23, 24). The processing localizes the cytoplasmic bZIP domain to the nucleus and it activates downstream genes through ERSE or ERSE-II, cooperating with the NF-Y transcription factor complex (25, 26). The active form of ATF6 is produced before that of XBP1 in response to ER stress, because the former protein is derived from a preexisting precursor protein, whereas the latter must be newly translated from transcriptionally induced mRNA and then processed by IRE1-dependent splicing (17). Because XBP1 contains ERSE in its promoter, ER stress signaling can be amplified through the transcription of XBP1 as long as IRE1 is activated.
By using a model plant, Arabidopsis thaliana, we previously isolated two IRE1 homologs (27) and identified the cis-element plant-specific UPRE (P-UPRE) responsible for the ER stress response in the BiP2 (locus tag At5g42020) promoter (28). Interestingly, P-UPRE consisted of two cis-elements identified in the mammalian ER stress response, ERSE-II and XBP1-BS. In addition to the BiP2 promoter, P-UPRE was found in the promoters of other ER chaperone genes, including BiP1 (locus tag At5g28540). A transcriptomic approach using microarrays showed that ERSEs were also found in promoters of several genes induced by ER stress (29, 30). Furthermore, the third BiP, BiP-L (locus tag At1g09080) (referred to as BiP3 in the present study), also contains two functional ERSEs, because mutation of ERSE in the BiP3 promoter abolishes induction in response to ER stress (30). By analogy with yeast and mammals, bZIP transcription factors are predicted to function in the ER stress response of plants. An exhaustive search of the Arabidopsis genomic database, however, did not succeed in finding sequence homologs of XBP1 or ATF6. The present study was conducted to isolate a transcription factor involved in the ER stress response in plants, because plants also show a clear ER stress response (29, 31–35) although knowledge of the molecular mechanism for the response is limited.
Materials and Methods
Genome-Wide Analysis of bZIP Transcripts. Total RNA was extracted from Arabidopsis (Col-0 ecotype) leaves treated with or without 5 μg/ml tunicamycin for 12 h. From each RNA sample, cDNA synthesis and subsequent PCR was conducted by using the RNA PCR kit (avian myeloblastosis virus) version 2.1 (Takara, Otsu, Japan) according to the manufacturer's instructions. The size and signal intensity of PCR products using specific primers for 75 bZIP genes were examined by gel electrophoresis.
RNA Blot Analysis. Arabidopsis seedlings were grown in one-half-strength MS medium supplemented with 2% (wt/vol) sucrose in a 16-h light/8-h dark cycle. Total RNA was extracted by using the aurintricarboxylic acid method (36) from 2-week-old seedlings treated with 5 μg/ml tunicamycin, 2 mM DTT, or 5 mM azetidine-2-carboxylate. Five micrograms of RNA per lane was fractionated on a 1.2% agarose gel containing 2% formaldehyde, capillary-blotted onto a nylon membrane (Hybond-N, Amersham Biosciences) in 20× standard saline citrate (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7), and fixed by UV irradiation. Hybridization probes of BiP and AtbZIP60 cDNAs were labeled with [α-32P]dCTP by using a DNA labeling kit (BcaBEST labeling kit, Takara). The membrane was washed with 0.2× SSC/0.1% SDS at 65°C three times then exposed to x-ray film.
DNA Constructs for Protoplast Transformation. For observation of the subcellular localization of truncated AtbZIP60, a cDNA fragment corresponding to amino acids 1–216 in AtbZIP60 was PCR-amplified by using primers GTCGACATGGCGGAGGAATTTGGAAGCATAG and CCATGGTAGACTCCTGCTTCGACATCATGG. The PCR product was then fused to the N terminus of the sGFP in the cauliflower mosaic virus (CaMV) 35S-sGFP(S65T)-NOS3′ vector, a gift of Y. Niwa (University of Shizuoka, Shizuoka, Japan) (37).
For transient luciferase assays, the β-glucuronidase gene (GUS) in pBI221 (Clontech) was replaced with the firefly luciferase gene derived from pGL3-Basic (Promega), which produced the plasmid pBI221-Luc. We then amplified ≈1.2 kb of BiP, calnexin (CNX), and Hsp70 promoters by PCR with primers CTCGAGAGAGGAGGTTGAGAGAGAAGATAGAC and ACTAGTAGCCATATCGGAAACTTTTGCGTACG for BiP1, CTCGAGTGTATTGTAAAAGCCCTTAGCGTTACCGG and GGATCCAGCCATATCGGAAACTTTTGCGTACG for BiP2, CTCGAGCAAACATAGCACCGAACGACTTACTAC and CGCATGGATCCAATCATTTTTCGTTGTTGAGAACTCTTCTTCG for BiP3, CTCGAGGACGAGATGGTTGCTTTGGGTCTA and GGATCCTCTCATTCTCGGAATCTCTAAAAT for CNX1, CTCGAGCGTCGTTTCTCTATGATTCATTTG and GGATCCTCTCATTATCGCAATCTCAAGAGA for CNX2, and CTCGAGCGAACATTTTGCTGAACTGATTAG and GGATCCCGCCATTATTAGAGATCAGAATTG for Hsp70. PCR products were translationally fused to the firefly luciferase gene by replacing the CaMV 35S promoter of pBI221-Luc and were designated BiP1pro-Luc, BiP2pro-Luc, BiP3pro-Luc, CNX1proLuc, CNX2pro-Luc, and Hsp70pro-Luc, respectively. A P-UPRE hexamer fused with the CaMV 35S–46 minimal promoter (min) and firefly luciferase, designated P-UPREx6-min-Luc, was used as described in ref. 28. For ERSE, a TTACCAATCACTTCTTGACACGAGA hexamer was synthesized and used to replace that of P-UPREx6-min-Luc to generate ERSEx6-min-Luc. For overexpression of intact and truncated AtbZIP60, cDNA sequences encoding each polypeptide were substituted with the GUS gene of pBI221. Resulting constructs were designated 35S-AtbZIP60 and 35S-AtbZIP60ΔC. For overexpression of HY5, a cDNA fragment amplified with primers GGATCCATGCAGGAACAAGCGACTAGCTCT and GAGCTCTCAAAGGCTTGCATCAGCATTAGA was substituted with the GUS gene of pBI221. The resulting construct was designated 35S-HY5. For promoter analysis of AtbZIP60, an ≈1.2-kb region of promoter amplified by PCR with primers AAGCTTCGTAAAACAATTTAATAGATGTTAATG and GGATCCCATGGTCAAAAAAAAAAAAATATACAAAGAAGAAAAAAAAAAGC was translationally fused to the firefly luciferase gene by replacing the CaMV 35S promoter of pBI221-Luc (AtbZIP60pro-Luc). To obtain mutations in the promoter, two mutated PCR fragments were amplified by using a combination of AAGCTTCGTAAAACAATTTAATAGATGTTAATG and AGATGAGAGAAGGCTTAGTTCTGGAAGAATAGGATCACAG as well as GAACTAAGCCTTCTCTCATCTTGTGTGACGGCACATAAAA and GGATCCCATGGTCAAAAAAAAAAAAATATACAAAGAAGAAAAAAAAAAGC. Subsequent PCR was performed by using AAGCTTCGTAAAACAATTTAATAGATGTTAATG and GGATCCCATGGTCAAAAAAAAAAAAATATACAAAGAAGAAAAAAAAAAG to obtain a full-length mutated promoter, which was substituted for the CaMV 35S promoter of pBI221-Luc (AtbZIP60mpro-Luc).
Stable Transformation with a Chimeric Gene Consisting of the AtbZIP60 Promoter and the GUS Gene. The promoter region used to construct AtbZIP60pro-Luc was fused with the GUS gene by replacing the CaMV 35S promoter of pBI121 to generate AtbZIP60pro-GUS. Stable transformation of Arabidopsis was carried out according to Clough and Bent (38). The GUS activity of T1 plants was measured by using 4-methylumbelliferyl-β-d-glucuronide as described in ref. 28.
Transient Expression Analysis by Fluorescent GFP and Dual Luciferase Assays. Protoplasts were isolated from Arabidopsis suspension cells and transiently transformed by using polyethylene glycol according to Ueda et al. (39). Fluorescence of GFP was observed by an LSM510 confocal laser scanning microscope (Carl Zeiss) after incubation at 23°C for 16 h. For the dual luciferase assay, transformed protoplasts were incubated at 23°C for 16 h in the dark, and luciferase activities were measured by using the dual luciferase assay system (Promega) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity.
Results
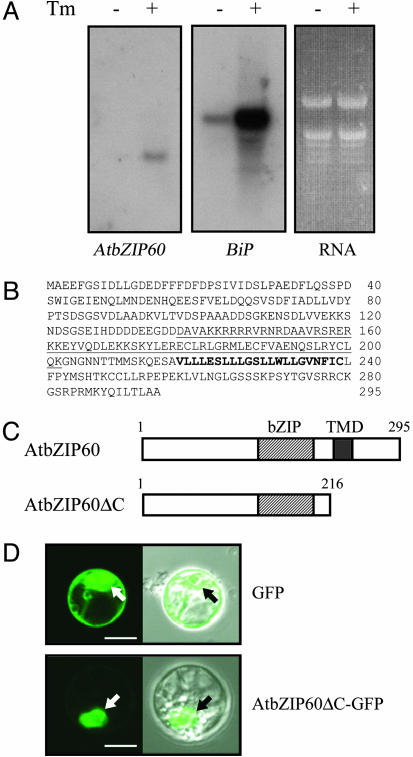
AtbZIP60 Induced by Tunicamycin Was Identified by Using Genomic Information. We assumed that bZIP transcription factors play roles in the ER stress response of plants, because bZIPs are involved in the response of yeast (Hac1) and mammals (XBP1 and ATF6). Thus, according to the prediction of 75 bZIP genes in the Arabidopsis genome (40), they were screened one by one by RT-PCR using RNA prepared from Arabidopsis leaves treated with and without tunicamycin, an inhibitor of asparagine-linked glycosylation that is generally used to induce the ER stress response. Among the transcripts detected, only transcripts of AtbZIP60 (locus tag At1g42990) were highly induced by tunicamycin, and the induction was confirmed by RNA gel blotting analysis (Fig. 1A). AtbZIP60 encoded an ORF consisting of 295 aa (Fig. 1B) having a bZIP DNA binding domain followed by a putative TMD (Fig. 1C). The putative TMD implies conversion of AtbZIP60 to a soluble protein by proteolysis in response to ER stress in analogy to ATF6 in mammals. Indeed, a truncated AtbZIP60 containing amino acids 1–216 (AtbZIP60ΔC), which are fused to GFP, localized to the nucleus when transiently expressed in Arabidopsis protoplasts (Fig. 1D).
Fig. 1.
Identification and characterization of AtbZIP60. (A) RNA blot analysis of AtbZIP60 and BiP. Total RNA was extracted from 2-week-old Arabidopsis seedlings that had been placed in water with DMSO (as a solvent control; –) or 5 μg/ml tunicamycin (+) for 12 h and used for RNA blot analysis. AtbZIP60 or BiP cDNA was used as a probe. (B) Deduced amino acid sequence of AtbZIP60. The bZIP domain is underlined, and a putative TMD is indicated in bold. (C) A schematic structure of AtbZIP60 protein. The locations of the bZIP domain and the TMD are indicated. AtbZIP60ΔC represents the truncated form used in later experiments. (D) Observation of fluorescence of GFP alone and of the AtbZIP60ΔC–GFP fusion protein expressed transiently in protoplasts. Confocal and brightfield images were captured from the same cells. Arrows indicate position of the nucleus. (Bar, 10 μm.)
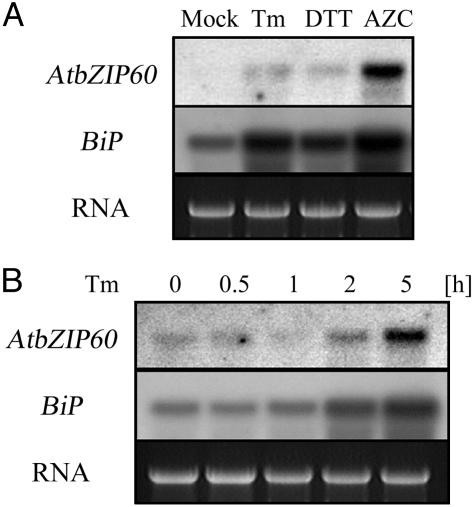
Expression of AtbZIP60 Was Regulated by Other ER Stresses. To examine whether other agents inducing ER stress affect the expression of AtbZIP60, Arabidopsis seedlings treated with tunicamycin, DTT (a reducing agent inhibiting disulfide bond formation), or azetidine-2-carboxylate (a proline analog that perturbs protein structure) were subjected to RNA blot analysis. As shown in Fig. 2A, these agents also induced AtbZIP60 as well as BiP. As shown in Fig. 2B, the time course of AtbZIP60 induction in response to tunicamycin treatment was quite similar to that of BiP transcript induction in response to ER stress.
Fig. 2.
Expression profiles of AtbZIP60 and BiP transcripts. (A) Effects of various reagents inducing the ER stress response. Total RNA was extracted from Arabidopsis seedlings treated with DMSO (Mock), 5 μg/ml tunicamycin (Tm), 2 mM DTT, or 5 mM azetidine-2-carboxylate (AZC) for 5 h and analyzed by RNA blotting. (B) Induction of time course after tunicamycin treatment. Arabidopsis seedlings were treated with 5 μg/ml tunicamycin, and RNA was extracted and analyzed at the indicated time periods. The exposure for AtbZIP60 in B was conducted for five times longer than that in A.
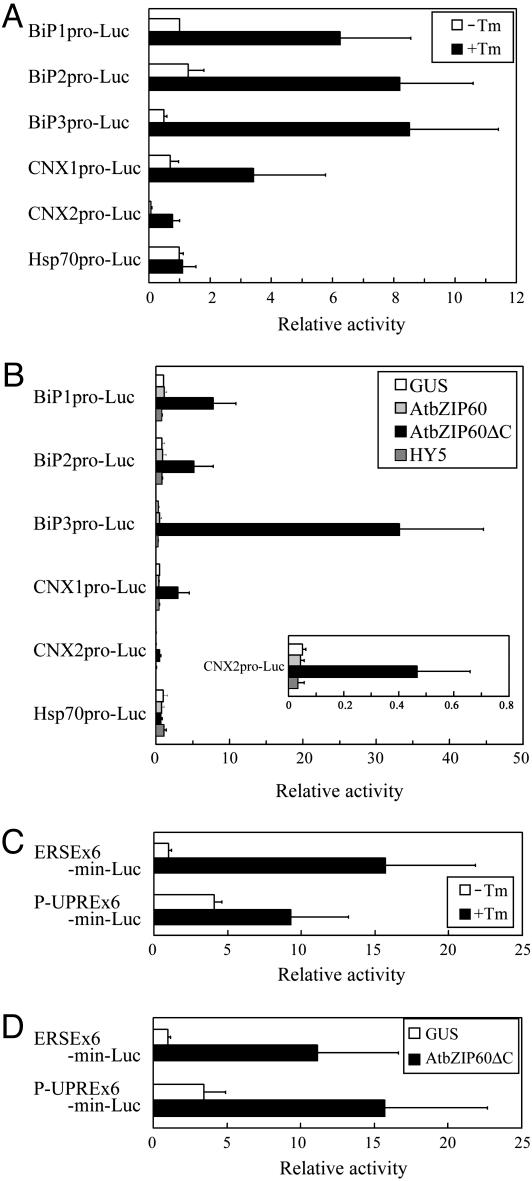
Promoters of BiP and CNX Were Activated by Truncated AtbZIP60. The Arabidopsis genome contains three BiP genes. Two of them, BiP1 and BiP2, including promoter and intron sequences, are closely related to each other (41), whereas BiP3 is different to some extent (30). The promoter of BiP3 lacks P-UPRE, a cis-element responsible for the ER stress response that is found in BiP1 and BiP2. Instead, the BiP3 promoter has two copies of ERSE, which is also assumed to be a cis-element responsible for the ER stress response. Because the induction of BiP represents the ER stress response, the effect of AtbZIP60 on induction of the three BiP genes was examined by a dual luciferase assay in protoplasts of Arabidopsis. To investigate the effect on other genes upregulated by ER stress, we tested the promoters of the CNX genes (CNX1 and CNX2), which encode lectin-like ER-resident chaperones and contain ERSE in the promoters (27).
First, each of the three BiP promoters and two CNX promoters (≈1.2 kb), all of which are fused to the firefly luciferase gene, was introduced into protoplasts prepared from Arabidopsis suspension cells. In transient assays, treatment with tunicamycin clearly enhanced luciferase activity driven by all BiP and CNX promoters, indicating they were responsible for responding to ER stress (Fig. 3A). High induction of endogenous BiP was also confirmed by RNA blots (data not shown). As a control promoter, the promoter of cytosolic heat shock-inducible Hsp70 (locus tag At3g12580) (42) was tested, which was unaffected by tunicamycin treatment (Fig. 3A).
Fig. 3.
Effect of AtbZIP60 on gene expression in the ER stress response. (A) Activation of BiP and CNX promoters with tunicamycin (Tm) treatment. Protoplasts were transiently transformed with plasmids carrying either the firefly luciferase gene under the control of each BiP and CNX promoter or Renilla luciferase driven by the CaMV 35S promoter. After transformation, protoplasts were incubated with or without 5 μg/ml tunicamycin for 16 h. Luciferase activities were normalized by the ratio of firefly and Renilla luciferase activities. Relative activity represents activities relative to basal activity obtained from the construct with the BiP1 promoter. (B) Effects of AtbZIP60 and AtbZIP60ΔC on BiP and CNX promoters. Transient assays were carried out as described above. Instead of tunicamycin treatment, effector plasmids carrying GUS, AtbZIP60, AtbZIP60ΔC, or HY5 driven by the CaMV 35S promoter were cotransformed. (Inset) An enlarged view of the activity of the CNX2 promoter. Relative activity represents activity relative to basal activity obtained from constructs with the BiP1 promoter and GUS. Expression of full-length or truncated AtbZIP60 protein was confirmed by using antibodies for AtbZIP60 (data not shown). (C) Activation of ERSE and P-UPRE by tunicamycin. Transient transformation, including tunicamycin treatment and dual luciferase assay, was carried out as described in A. Plasmids consisting of hexamers of ERSE or P-UPRE, the minimal promoter of CaMV 35S (min) and firefly luciferase (Luc), were introduced. Relative activity represents activities relative to basal activity obtained from the construct with the ERSE hexamer. (D) Effects of AtbZIP60ΔC on ERSE and P-UPRE. Transient assays were carried out as described in B. Although the data from intact AtbZIP60 is not shown, it had little effect on induction. Relative activity represents activity relative to that of constructs with the ERSE hexamer and GUS.
Using this assay system, either intact AtbZIP60 or the truncated form, AtbZIP60ΔC (amino acids 1–216), was coexpressed under the CaMV 35S promoter. As a control effector, we used GUS and HY5, a bZIP transcription factor involved in signal transduction of photomorphogenesis (43), because HY5 showed the highest similarity with XBP1 in a database search of the Arabidopsis genome. Although GUS, HY5, and the intact AtbZIP60 did not affect induction of luciferase activity, coexpression of AtbZIP60ΔC clearly enhanced luciferase activity driven by all BiP and CNX promoters (Fig. 3B). The level of induction was higher for the BiP3 promoter than for BiP1 and BiP2. The Hsp70 promoter was again not affected.
Activation of Promoters Was Through P-UPRE and ERSE. As described above, P-UPRE and ERSE have been considered responsible for the ER stress response. Thus, it was likely that activation of BiP and CNX promoters by AtbZIP60 depends on these ciselements. To examine whether this hypothesis were true, the effect of AtbZIP60ΔC on P-UPRE- and ERSE-dependent induction was analyzed. A hexamer of either P-UPRE or ERSE fused to the CaMV 35S–46 minimal promoter and the luciferase gene was subjected to a luciferase reporter assay using protoplasts in the same way as described above. As shown in Fig. 3C, P-UPRE responded to tunicamycin as reported in ref. 28. ERSE also responded to tunicamycin, indicating that ERSE was sufficient for the ER stress response in plants as predicted. When AtbZIP60ΔC was coexpressed under the CaMV 35S promoter, luciferase activities driven by P-UPRE and ERSE were clearly enhanced, in contrast to GUS's lack of effect on coexpression (Fig. 3D). These results suggested that AtbZIP60 activates the BiP and CNX promoter in an ERSE- and P-UPRE-dependent manner in response to ER stress.
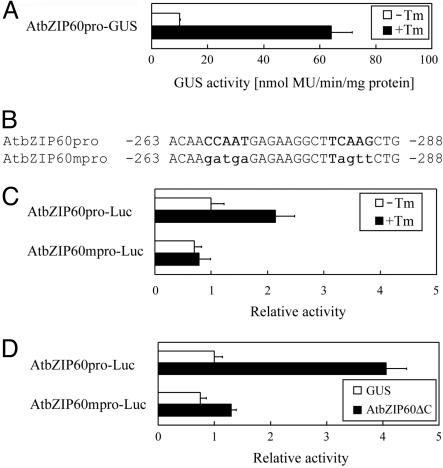
AtbZIP60 Induces Its Own Transcription Through an ERSE-Like Sequence. We fused ≈1.2 kb of the promoter region of AtbZIP60 to a GUS reporter gene and introduced the promoter into Arabidopsis. GUS activity was clearly induced in the transgenic plants by tunicamycin, suggesting that the promoter responded to ER stress (Fig. 4A). Because AtbZIP60 was induced during the ER stress response similarly to BiP, AtbZIP60 may induce its own transcription. Indeed, the promoter of AtbZIP60 contains a sequence, CCAAT-N9-TCAAG, similar to the general ERSE sequence CCAAT-N9-CCACG. As indicated in Fig. 4B, CCAAT is conserved exactly, and TCAAG has two base mismatches with the CCACG. We predicted that this ERSE-like sequence functions in the induction of AtbZIP60. To test this prediction, a mutation was introduced into the ERSE-like sequence (Fig. 4B) and subjected to transient luciferase assay. As shown in Fig. 4C, an authentic AtbZIP60 promoter responded to tunicamycin; however, the mutated promoter showed little response to tunicamycin. This result indicates that the induction of AtbZIP60 depends on the ERSE-like sequence. Comparison of the induction rate between Fig. 4 A and C suggests that clear induction is easily observed in stable transformants.
Fig. 4.
Regulation of AtbZIP60 promoter through an ERSE-like sequence. (A) GUS activity in transgenic Arabidopsis harboring a chimeric gene consisting of the AtbZIP60 promoter and GUS gene. Extracts from leaves treated with or without 5 μg/ml tunicamycin (Tm) for 12 h were subjected to quantitative GUS assay. (B) Nucleotide sequence from –288 to –263 of the AtbZIP60 promoter. The ERSE-like sequence and mutated sequence (AtbZIP60mpro) used in later experiments are indicated in bold. (C) Activation of the AtbZIP60 promoter by tunicamycin in a transient assay. Transient transformation of protoplasts was carried out as described in the legend for Fig. 3A. Reporter plasmids consisting of authentic or mutated promoter and the firefly luciferase gene were used for transfection. Tunicamycin (5 μg/ml) treatment was conducted for 16 h. Relative activity represents activity relative to basal activity obtained from constructs with the intact AtbZIP60 promoter. (D) Effect of AtbZIP60ΔConthe AtbZIP60 promoter. Protoplasts were transfected with reporter plasmids carrying authentic and mutated promoters fused to the firefly luciferase gene and effector plasmid carrying GUS and AtbZIP60ΔC genes driven by the CaMV 35S promoter. Relative activity represents activity relative to that obtained from constructs with intact AtbZIP60 promoter and GUS.
Subsequently, the effect of AtbZIP60ΔC on the AtbZIP60 promoter was examined. As shown in Fig. 4D, coexpression of AtbZIP60ΔC clearly activated the authentic AtbZIP60 promoter. However, this activation was almost completely abolished by mutation of the ERSE-like sequence, suggesting that AtbZIP60 activates its own transcription through the ERSE-like sequence.
Discussion
According to the prediction of the involvement of bZIP transcription factors in the ER stress response, AtbZIP60 was identified by genome-wide screening based on genomic information on Arabidopsis. Tunicamycin and other reagents activating the ER stress response induced transcripts of AtbZIP60. From these results, we predicted that AtbZIP60 plays a role in the ER stress response. Because the expression profile of AtbZIP60 was close to that of BiP, induction of AtbZIP60 transcript was not considered to be the first trigger of activation for BiP expression. Instead, it was assumed that a conformational change of AtbZIP60 activates the expression of chaperone genes, such as BiP. This prediction was based on the fact that AtbZIP60 contains a putative TMD like that of ATF6 in mammalian cells. Specifically, it was hypothesized that AtbZIP60 is converted to a soluble form by ER stress and becomes localized to the nucleus, resulting in the activation of chaperone genes. Indeed, a truncated form of AtbZIP60 fused with GFP localized to the nucleus, supporting this hypothesis.
To test the hypothesis, intact and truncated forms of AtbZIP60 were coexpressed with constructs consisting of BiP and CNX promoters and a luciferase gene. The truncated form clearly enhanced luciferase activity for all BiP and CNX promoters, but the intact form did not. This result strongly supports our hypothesis that cleaved AtbZIP60 enhances BiP promoter activity. Although HY5 is the Arabidopsis bZIP with the highest similarity to XBP1, HY5 did not affect these promoters, indicating that simple homology could not identify a functional homolog.
Subsequent experiments clearly indicated that activation of BiP and CNX promoters by AtbZIP60 depends on the cis-elements P-UPRE in BiP1 and BiP2 and ERSE in BiP3, CNX1, and CNX2. As described in the introduction, P-UPRE contains ERSE-II, and a previous study indicated that ERSE-II was sufficient for response to ER stress (28). Thus, our results indicated activation of ERSE and ERSE-II by AtbZIP60, even though conservation of the two sequences is low. The most probable interpretation is that the conformation of ERSE-II (consensus sequence ATTGG-N-CCACG) is similar to that of ERSE (consensus sequence CCAAT-N9-CCACG), as reported in mammalian cells, because ERSE-II also contains two motifs, CCAAT (complementary to ATTGG) and CCACG, although the orientation and the spacing are different (20, 22). It is likely that ERSE has a higher binding affinity for AtbZIP60, because higher induction was observed in assays with ERSE. This result was consistent with the observation that the induction rate of BiP3 is higher than that of BiP1 and BiP2. We assume that AtbZIP60 also regulates expression of other ER chaperones, such as calreticulin and protein disulfide isomerase, because they have ERSE in their promoters (28, 29).
The characteristics of AtbZIP60 are similar in part to those of ATF6. That is, conformational change of the protein is considered to be the first trigger for the response. However, it is not clear whether AtbZIP60 is cleaved by a protease, like ATF6, because no conserved sequence necessary for cleavage by S1P and S2P proteases was found around the putative TMD of AtbZIP60 (44). In addition, the C-terminal region of AtbZIP60 is much shorter than that of ATF6, which is considered to function as a sensor for ER stress that interacts with BiP (45). Thus, the mechanism of conformational change of AtbZIP60 to the active form is still unknown. Even the putative TMD may not be a TMD but a hydrophobic region masking the active domain of AtbZIP60. Further analysis to clarify the mechanism of signal perception and conversion to the active form is necessary. Another interesting characteristic of AtbZIP60 is autoregulation of its transcription through the ERSE-like element in its promoter. This amplification of its own transcript is similar to that of XBP1 in mammalian cells, whereas, in this case, activation of XBP1 is by IRE1-dependent mRNA splicing (17).
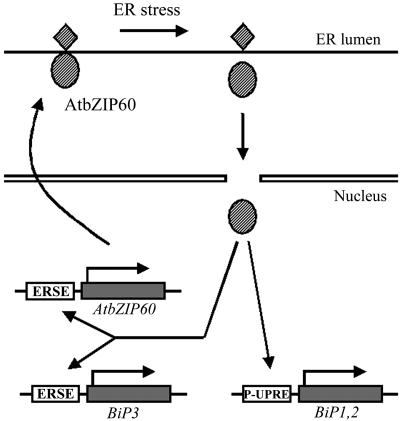
As summarized in Fig. 5, processed AtbZIP60 is considered to enhance BiP expression through P-UPRE or ERSE. The initial trigger of activation of AtbZIP60 seems to be conformational change of the protein, likely conversion to a soluble form that functions as a transcription factor in the nucleus. After activation, transcription of AtbZIP60 would also be enhanced through the ERSE-like element in its promoter. We would like to emphasize that the structure of AtbZIP60 and the current model for signaling in the ER stress response in plants proposed in the present study is different from those for yeast and mammals. The mechanism of the initial perception of the ER stress is still unclear and needs to be clarified. Although further studies will be needed, IRE1 homologs may play roles similar to those in other organisms. Because we have already isolated T-DNA mutants of AtbZIP60 and two IRE1 homologs, we look forward to further studies that provide more information about the signaling pathway for the ER stress response in plants.
Fig. 5.
A proposed model for the function of AtbZIP60 in the ER stress signaling pathway. AtbZIP60 is synthesized at a low level as a precursor protein that may be anchored in the ER membrane under unstressed conditions. Sensing ER stress by an unknown mechanism, the N-terminal domain of AtbZIP60, which is similar to AtbZIP60ΔC, is cleaved and translocated to the nucleus. Soluble AtbZIP60 or AtbZIP60ΔC activates transcription of target genes, such as BiP genes, through either P-UPRE or ERSE. Transcription of AtbZIP60 is also activated through an ERSE-like sequence to amplify the signal.
Acknowledgments
We thank Dr. Yasuo Niwa for kindly providing the CaMV 35S-sGFP(S65T)-NOS3′ plasmid and Ms. Naoko Yamaguchi in our laboratory for technical assistance. This work was supported in part by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grant-in-Aid for Scientific Research 1638023 (to N.K.). Y.I. was supported by a Grant-in-Aid for 21st Century Center of Excellence Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author contributions: N.K. designed research; Y.I. performed research; Y.I. analyzed data; and Y.I. and N.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: bZIP, basic leucine zipper; CNX, calnexin; ER, endoplasmic reticulum; GUS, β-glucuronidase; TMD, transmembrane domain; UPR, unfolded protein response; UPRE, UPR element; P-UPRE, plant-specific UPRE; ERSE, ER stress-response element; CaMV, cauliflower mosaic virus.
References
- 1.Kaufman, R. J., Scheuner, D., Schroder, M., Shen, X., Lee, K., Liu, C. Y. & Arnold, S. M. (2002) Nat. Rev. Mol. Cell Biol. 3, 411–421. [DOI] [PubMed] [Google Scholar]
- 2.Mori, K. (2000) Cell 101, 451–454. [DOI] [PubMed] [Google Scholar]
- 3.Patil, C. & Walter, P. (2001) Curr. Opin. Cell Biol. 13, 349–355. [DOI] [PubMed] [Google Scholar]
- 4.Rutkowski, D. T. & Kaufman, R. J. (2004) Trends Cell Biol. 14, 20–28. [DOI] [PubMed] [Google Scholar]
- 5.Reimold, A. M., Iwakoshi, N. N., Manis, J., Vallabhajosyula, P., Szomolanyi-Tsuda, E., Gravallese, E. M., Friend, D., Grusby, M. J., Alt, F. & Glimcher, L. H. (2001) Nature 412, 300–307. [DOI] [PubMed] [Google Scholar]
- 6.Harding, H. P., Zeng, H., Zhang, Y., Jungries, R., Chung, P., Plesken, H., Sabatini, D. D. & Ron, D. (2001) Mol. Cell 7, 1153–1163. [DOI] [PubMed] [Google Scholar]
- 7.Scheuner, D., Song, B., McEwen, E., Liu, C., Laybutt, R., Gillespie, P., Saunders, T., Bonner-Weir, S. & Kaufman, R. J. (2001) Mol. Cell 7, 1165–1176. [DOI] [PubMed] [Google Scholar]
- 8.Iwakoshi, N. N., Lee, A. H., Vallabhajosyula, P., Otipoby, K. L., Rajewsky, K. & Glimcher, L. H. (2003) Nat. Immunol. 4, 321–329. [DOI] [PubMed] [Google Scholar]
- 9.Vitale, A. & Ceriotti, A. (2004) Plant Physiol. 136, 3420–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, J. S., Shamu, C. E. & Walter, P. (1993) Cell 73, 1197–1206. [DOI] [PubMed] [Google Scholar]
- 11.Mori, K., Ma, W., Gething, M. J. & Sambrook, J. (1993) Cell 74, 743–756. [DOI] [PubMed] [Google Scholar]
- 12.Shamu, C. E. & Walter, P. (1996) EMBO J. 15, 3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 13.Bertolotti, A., Zhang, Y., Hendershot, L. M., Harding, H. P. & Ron, D. (2000) Nat. Cell Biol. 2, 326–332. [DOI] [PubMed] [Google Scholar]
- 14.Kohno, K., Normington, K., Sambrook, J., Gething, M. J. & Mori, K. (1993) Mol. Cell. Biol. 13, 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori, K., Sant, A., Kohno, K., Normington, K., Gething, M. J. & Sambrook, J. F. (1992) EMBO J. 11, 2583–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori, K., Kawahara, T., Yoshida, H., Yanagi, H. & Yura, T. (1996) Genes Cells 1, 803–817. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida, H., Matsui, T., Yamamoto, A., Okada, T. & Mori, K. (2001) Cell 107, 881–891. [DOI] [PubMed] [Google Scholar]
- 18.Wang, Y., Shen, J., Arenzana, N., Tirasophon, W., Kaufman, R. J. & Prywes, R. (2000) J. Biol. Chem. 275, 27013–27020. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida, H., Haze, K., Yanagi, H., Yura, T. & Mori, K. (1998) J. Biol. Chem. 273, 33741–33749. [DOI] [PubMed] [Google Scholar]
- 20.Kokame, K., Kato, H. & Miyata, T. (2001) J. Biol. Chem. 276, 9199–9205. [DOI] [PubMed] [Google Scholar]
- 21.Shen, X., Ellis, R. E., Lee, K., Liu, C. Y., Yang, K., Solomon, A., Yoshida, H., Morimoto, R., Kurnit, D. M., Mori, K. & Kaufman, R. J. (2001) Cell 107, 893–903. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto, K., Yoshida, H., Kokame, K., Kaufman, R. J. & Mori, K. (2004) J. Biochem. (Tokyo) 136, 343–350. [DOI] [PubMed] [Google Scholar]
- 23.Lee, K., Tirasophon, W., Shen, X., Michalak, M., Prywes, R., Okada, T., Yoshida, H., Mori, K. & Kaufman, R. J. (2002) Genes Dev. 16, 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haze, K., Yoshida, H., Yanagi, H., Yura, T. & Mori, K. (1999) Mol. Biol. Cell 10, 3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M. & Mori, K. (2000) Mol. Cell. Biol. 20, 6755–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M. & Mori, K. (2001) Mol. Cell. Biol. 21, 1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koizumi, N., Martinez, I. M., Kimata, Y., Kohno, K., Sano, H. & Chrispeels, M. J. (2001) Plant Physiol. 127, 949–962. [PMC free article] [PubMed] [Google Scholar]
- 28.Oh, D. H., Kwon, C. S., Sano, H., Chung, W. I. & Koizumi, N. (2003) Biochem. Biophys. Res. Commun. 301, 225–230. [DOI] [PubMed] [Google Scholar]
- 29.Martinez, I. M. & Chrispeels, M. J. (2003) Plant Cell 15, 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noh, S. J., Kwon, C. S., Oh, D. H., Moon, J. S. & Chung, W. I. (2003) Gene 311, 81–91. [DOI] [PubMed] [Google Scholar]
- 31.Leborgne-Castel, N., Jelitto-Van Dooren, E. P., Crofts, A. J. & Denecke, J. (1999) Plant Cell 11, 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koizumi, N. (1996) Plant Cell Physiol. 37, 862–865. [DOI] [PubMed] [Google Scholar]
- 33.Koizumi, N., Ujino, T., Sano, H. & Chrispeels, M. J. (1999) Plant Physiol. 121, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boston, R. S., Fontes, E. B., Shank, B. B. & Wrobel, R. L. (1991) Plant Cell 3, 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jelitto-Van Dooren, E. P., Vidal, S. & Denecke, J. (1999) Plant Cell 11, 1935–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez, R. G., Haxo, R. S. & Schleich, T. (1980) Biochemistry 19, 4299–4303. [DOI] [PubMed] [Google Scholar]
- 37.Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H. & Sheen, J. (1996) Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- 38.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- 39.Ueda, T., Yamaguchi, M., Uchimiya, H. & Nakano, A. (2001) EMBO J. 20, 4730–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T. & Parcy, F. (2002) Trends Plant Sci. 7, 106–111. [DOI] [PubMed] [Google Scholar]
- 41.Koizumi, N. & Sano, H. (1997) Plant Physiol. 113, 664. [Google Scholar]
- 42.Sung, D. Y., Vierling, E. & Guy, C. L. (2001) Plant Physiol. 126, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chattopadhyay, S., Ang, L. H., Puente, P., Deng, X. W. & Wei, N. (1998) Plant Cell 10, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye, J., Rawson, R. B., Komuro, R., Chen, X., Dave, U. P., Prywes, R., Brown, M. S. & Goldstein, J. L. (2000) Mol. Cell 6, 1355–1364. [DOI] [PubMed] [Google Scholar]
- 45.Shen, J., Chen, X., Hendershot, L. & Prywes, R. (2002) Dev. Cell 3, 99–111. [DOI] [PubMed] [Google Scholar]