Abstract
The 5′ and 3′ splice sites within an intron can, in principle, be joined to those within any other intron during pre-mRNA splicing. However, exons are joined in a strict 5′ to 3′ linear order in constitutively spliced pre-mRNAs. Thus, specific mechanisms must exist to prevent the random joining of exons. Here we report that insertion of exon sequences into an intron can inhibit splicing to the downstream 3′ splice site and that this inhibition is independent of intron size. The exon sequences required for splicing inhibition were found to be exonic enhancer elements, and their inhibitory activity requires the binding of serine/arginine-rich splicing factors. We conclude that exonic enhancers can act as barriers to prevent exon skipping and thereby may play a key role in ensuring the correct 5′ to 3′ linear order of exons in spliced mRNA.
Keywords: intronic splicing silencers, pre-mRNA splicing, accurate splice-site recognition and pairing
Most genes in higher eukaryotes contain multiple introns and exons that are transcribed to generate pre-mRNA. The production of mature mRNA requires the precise removal of introns and joining of exons by splicing. In constitutively spliced pre-mRNAs all of the introns are excised, and the exons are joined in the correct 5′ to 3′ order. This is a consequence of the fact that 5′ splice sites are ligated to a 3′ splice site exclusively within the same intron, thus avoiding the exclusion of exons (“exon skipping”). Two key problems in understanding the mechanisms of accurate pre-mRNA splicing are how the splicing machinery recognizes exons and how exons are joined in the correct 5′ to 3′ order (i.e., how exon skipping is avoided).
Significant advances have been made in understanding the mechanisms involved in accurate exon recognition (1–3). This process requires weakly conserved sequence elements located at the 5′ and 3′ splice sites and at the branchpoint sequence (BPS) within introns. In addition, exons contain sequence elements known as exonic splicing enhancers (ESEs), which function as binding sites for members of the serine/arginine-rich (SR) family of splicing factors. The bound SR proteins promote the use of flanking 5′ and 3′ splice sites through both protein–protein and protein–RNA interactions (4–6). Although splicing enhancer/SR protein complexes provide a mechanism for exon recognition, the mechanisms by which exon skipping is avoided are not understood.
An important insight into this problem was provided by the observation that many human genetic diseases are the consequence of single base mutations in ESEs and that these mutations cause exon skipping (7). Further insights were provided by studies of regulated alternative splicing, where it was shown that the inclusion of an alternate exon requires an ESE and the presence of specific SR proteins (8). Thus, exonic enhancer/SR protein complexes are required to avoid exon skipping in both constitutive and alternative splicing. Although these observations indicate that exon recognition is a necessary step in avoiding exon skipping, it is clearly not sufficient.
Here we use pre-mRNAs containing a single 5′ splice site and tandemly duplicated 3′ splice sites as a model system to study the mechanisms by which exon skipping is prevented. Our data reveal that splicing to the distal 3′ splice site is suppressed by proximal exonic sequences and that SR proteins are required for this suppression. Thus, SR protein/exonic enhancer complexes not only function in exon and splice-site recognition but also play a role in ensuring that 5′ and 3′ splice sites within the same intron are used, thus suppressing exon skipping.
Materials and Methods
Construction of Plasmids. The wild-type 3′ duplication constructs 3′D-14, 3′D-55, 3′D-115, and 3′D-205 were previously described (9) and were renamed WT14, WT55, WT115, and WT205, respectively, after subcloning in pcDNA3.1 vector. A 1,480-bp fragment from human thymidine kinase intron 2 (hTKi2) was cloned into the proximal intron of WT14, WT55, WT115, and WT205, generating WT14-long1, WT55-long1, WT115-long1, and WT205-long1, respectively. To create the mutated BPS (mBPS) constructs, the wild-type BPS sequence CACUGAC was changed to CAGAGUG by site-directed mutagenesis (10). The Δ3′ss series has a naturally occurring thalassemia deletion, βΔ3′ (11), of the pyrimidine tract, 3′ AG (italics), and the first two nucleotides of exon 2 (lowercase): Δ(UUGGUCUAUUUUCCCACCCUUAGgc). A 315-bp fragment from hTKi2 was cloned in the distal intron of WT55, WT205, Δ3′ss55, and Δ3′ss205, generating WT55-long2, WT205-long2, Δ3′ss55-long2, and Δ3′ss205-long2, respectively. A 212-bp fragment from hTKi3 was cloned into 3WT205 and Δ3′ss205 to produce WT205r-long2 and Δ3′ss205r-long2, respectively. SP64Hβ-ΔBam-X is a modification of the SP64Hβ-ΔBam vector containing human β-globin exon 1, IVS 1 with an XhoI linker, and exon 2. The pcD vector is a modification of the pcDNA3.1/myc-HisA vector with the myc-His portion replaced by an SP6 promoter site. The double duplication clone DD155-205 was described previously (9) and was subcloned in pcD vector. DD155-205Δi was constructed by inserting the HindIII–BamHI fragment of human β-globin cDNA in the XhoI linker of SP64Hβ-ΔBam-X before subcloning into pcD vector. A 358-bp fragment from the human β-globin IVS 2 (position 427–785) was cloned in an XhoI-digested SP64Hβ-ΔBam-X. A HindIII–EcoRI fragment was then subcloned in a pcD vector to produce the “control” construct.
The cis-competition constructs containing synthetic SR protein binding sites were constructed by inserting two copies of a strong 9G8 splicing enhancer (12) downstream of the proximal β-globin 3′ splice site and three multimerized copies of a strong SC35 splicing enhancer (12) downstream of the distal β-globin 3′ splice site. The appropriate oligonucleotides were subcloned into identical sites in each of the WT14 and Δ3′ss14 constructs to generate WT14 (distal SC35) and Δ3′ss14 (distal SC35). The 9G8 splicing enhancers were multimerized by annealing and ligation, and two copies were ligated into the PstI sites of WT14 (distal SC35) and Δ3′ss14 (distal SC35) to generate WT14 (proximal 9G8 and distal SC35) and Δ3′ss14 (proximal 9G8 and distal SC35).
Synthesis of Pre-mRNAs and in Vitro Splicing. All of the duplication constructs were linearized with BamHI except DD155-205Δi and the “control” construct, which were linearized with EcoRI. Pre-mRNAs were synthesized by in vitro transcription with T7 RNA polymerase in the presence of [α-32P]UTP (NEN). HeLa-S3 (National Cell Culture Center, Minneapolis) nuclear extracts were prepared as described (13). Splicing assays were performed in 25-μl reaction mixtures containing 50% nuclear extract, 0.5 mM ATP, 20 mM creatine phosphate, 3.2 mM MgCl2, 60 mM KCl, 12 mM Tris·HCl (pH 7.6), and 10–20 ng of 32P-labeled pre-mRNA. Reactions were incubated at 30°C for 90 min. The extracted RNAs were resolved on 6.5% (19:1) 7 M urea polyacrylamide gel (or as indicated in the figure legends) and visualized by Molecular Imager (Bio-Rad). Complementation assays using HeLa cell S100 extracts and recombinant SR proteins were performed as described (12, 14), with the following modification. Reactions containing 40% cytoplasmic S100 extract were supplemented with 200 nM of SC35 and varying amounts of recombinant 9G8 (0–300 nM) before incubation at 30°C.
Coupled RNA Polymerase II Transcription Splicing. An efficient system for coupled transcription and splicing has recently been established (R. Das and R.R., unpublished results). Briefly, 200 ng of linearized DNA template was incubated in 25-μl reaction mixtures containing 50% HeLa nuclear extract, 0.5 mM ATP, 20 mM creatine phosphate, 3.2 mM MgCl2, 60 mM KCl, 12 mM Tris·HCl (pH 7.6), and 6 μCi of [α-32P]UTP (800 Ci/mmol; 1 Ci = 37 GBq) at 30°C for 90 min.
Xenopus Oocyte Microinjection. Microinjection of 32P-labeled RNAs into Xenopus laevis oocytes was performed as described (15). Oocytes were incubated for 40 min at 18°C, the nuclei were dissected, and nuclear RNAs were extracted and resolved by electrophoresis on 8% denaturing polyacrylamide gels.
SR Proteins. Total SR proteins from HeLa-S3 cells were prepared according to Zahler (16). Recombinant SC35 and 9G8 were expressed and purified from baculovirus-infected cell lysates as described (17).
Calculations for the Rate of Splicing. The spliced products were quantified by using quantity one software (Bio-Rad), and the values obtained were normalized according to the number of uridines present in the RNAs. The splicing efficiency was calculated by using the following formula: [(normalized value of the proximal spliced product) + (normalized value of the distal spliced product)]/[(normalized value of the pre-mRNA) + (normalized value of the proximal spliced product) + (normalized value of the distal spliced product)]. The ratio of proximal to distal 3′ splice-site selection was calculated by using the following formula: (normalized value of the proximal spliced product)/(normalized value of the distal spliced product).
Results
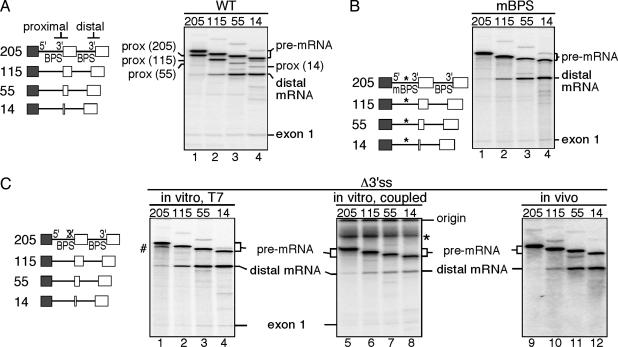
Suppression of Exon Skipping Requires Proximal Exon Sequences. Previous studies using model human β-globin pre-mRNAs containing a single 5′ splice site and tandemly duplicated 3′ splice sites showed that the proximal 3′ splice site is selected when both the proximal and distal 3′ splice sites are adjacent to a full-length exon (9). As the proximal exon is progressively truncated, use of the proximal 3′ splice site decreases with a concomitant increase in use of the distal 3′ splice site (Fig. 1A). This pattern of 3′ splice-site selection results, in part, from the loss of exonic enhancers in the proximal exon, which can no longer bind to SR proteins and promote the use of the proximal 3′ splice site (14). However, it is not known why only the proximal 3′ splice site is used when both the proximal and distal 3′ splice sites are adjacent to the full-length exon 2 (Fig. 1 A, lane 1).
Fig. 1.
Proximal exon sequences suppress use of the distal 3′ splice site. (Left) The structures of the pre-mRNAs derived from the wild-type (A), mBPS (B), and Δ3′ss (C) constructs are shown. Exonic sequences are represented by boxes; β-globin intronic sequences are represented by a straight line. The numbers refer to the length of the duplicated portion of exon 2. (A) For the proximal spliced products (prox), the lengths of the internal exon sequences are indicated in parentheses. (B) *, The mBPS. (C) The Δ3′ss pre-mRNAs were presynthesized by using a T7 promoter and then spliced in vitro in HeLa nuclear extract (Left), transcribed by using a cytomegalovirus (CMV) promoter and concomitantly spliced in HeLa nuclear extract (Center), or presynthesized by using a T7 promoter and then microinjected in Xenopus oocyte nuclei to be spliced in vivo (Right). The deletion of the pyrimidine tract at the proximal 3′ splice site is indicated by an X-marked 3′. #, Pre-mRNA breakdown products; *, nonspecific signal.
We investigated the mechanism by which the full-length proximal exon inhibits the use of the distal 3′ splice site. To eliminate the effects of competition between the proximal and distal 3′ splice sites, we inactivated the proximal 3′ splice site. We analyzed two mutants, a substitution mutant (mBPS) that inactivates the BPS (10) and a deletion mutant (Δ3′ss) lacking the 3′ splice site (18). As expected, no splicing to the proximal 3′ splice site was observed with the mBPS or Δ3′ss mutant pre-mRNAs (Fig. 1 B and C, lanes 1–4). Strikingly, however, when full-length exon 2 is adjacent to the inactivated proximal 3′ splice site, the pre-mRNAs largely remain unspliced (Fig. 1 B and C, lane 1). Thus, the full-length exon 2 alone inhibits use of the distal 3′ splice site. In support of this conclusion, the distal 3′ splice site is increasingly used in the pre-mRNAs containing progressively larger deletions of proximal exon 2 (Fig. 1 B and C, lanes 2–4). To confirm that the validity of this result is not restricted to in vitro conditions under which presynthesized T7 pre-mRNAs are used, we repeated the same experiments in a system where transcription and splicing are coupled. Significantly, we observed exactly the same pattern of 3′ splice-site use (compare Fig. 1C Left with Center). The same pattern of 3′ splice-site use was observed when the pre-mRNAs were microinjected into Xenopus oocyte nuclei (Fig. 1C, lanes 9–12). Taken together, these results show that exon sequences function to suppress exon skipping in vitro and in vivo.
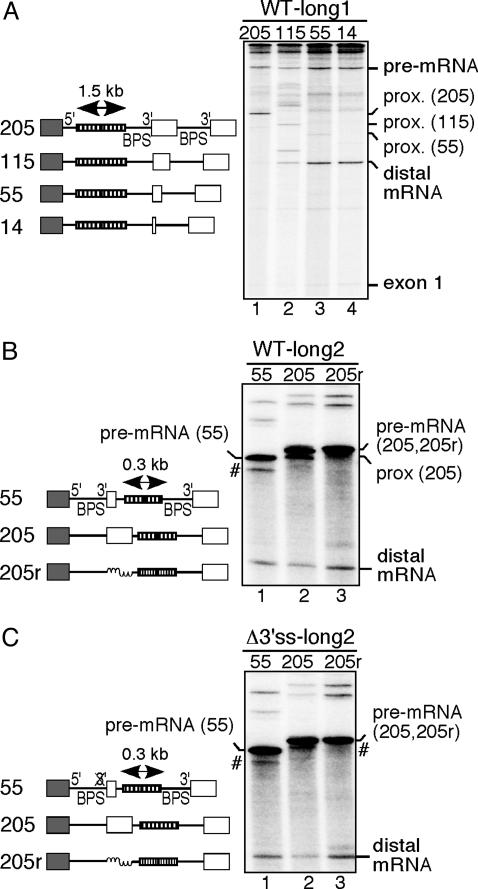
Intron Size Does Not Affect Exon Skipping. The average size of introns in the human genome is ≈1,000 nt, whereas the first intron of the β-globin gene is only 130 nt. To determine whether the failure to use the distal 3′ splice site results from the small size of the proximal intron, we inserted a 1.5-kb intronic spacer into the proximal intron of the wild-type constructs (Fig. 2A Left). Splicing of these constructs showed the same pattern of 3′ splice-site use as that observed with the 130-nt intron (Fig. 2 A Right). We conclude that the length of the proximal intron is not a critical determinant in the suppression of exon skipping in the model β-globin pre-mRNAs. We also considered the possibility that the small size of the distal intron affects the accessibility of the distal 3′ splice site to the splicing machinery. We therefore inserted a 0.3-kb intronic spacer just upstream of the distal BPS in the wild-type constructs (Fig. 2B Left). As a control, we also replaced the internal full-length exonic sequences with size-matched intronic sequences. As expected, when the internal exonic sequences were full-length, the proximal 3′ splice site was preferentially selected (Fig. 2B, lane 2). In contrast, when the internal exonic sequences were shortened to 55 nt, splicing to the distal 3′ splice site increased, whereas a proximal spliced product was almost undetectable (compare lanes 2 and 1 in Fig. 2B). Significantly, the replacement of internal exonic sequences with intronic sequences promoted the exclusive use of the distal 3′ splice site (compare lanes 3 and 2 in Fig. 2B). This result indicates that even a 55-nt internal exon sequence is sufficient to partially suppress the use of the distal 3′ splice site. We further confirmed, with constructs lacking a proximal 3′ splice site and containing a 0.3-kb intronic insertion in the distal intron, that a full-size internal exon suppresses the use of the distal 3′ splice site. A truncated exon had a weaker suppressor activity, and intronic sequences replacing the internal exon increased the use of the distal 3′ splice site (Fig. 2C).
Fig. 2.
The repression of the use of the distal 3′ splice site does not depend on the size of the proximal intron. (Left) The structures of the pre-mRNAs derived from the WT-long1 (A), WT-long2 (B), and Δ3′ss-long2 (C) constructs are shown. A double-headed arrow indicates the insertion of a spacer within the proximal β-globin intron. Thymidine kinase intronic sequences are represented by a coiled line. Pre-mRNA breakdown products are indicated by #. The transcripts were resolved on 4.5% denaturing polyacrylamide gels. For the proximal spliced products (prox) and some pre-mRNAs, the lengths of the internal exon sequences are indicated in parentheses.
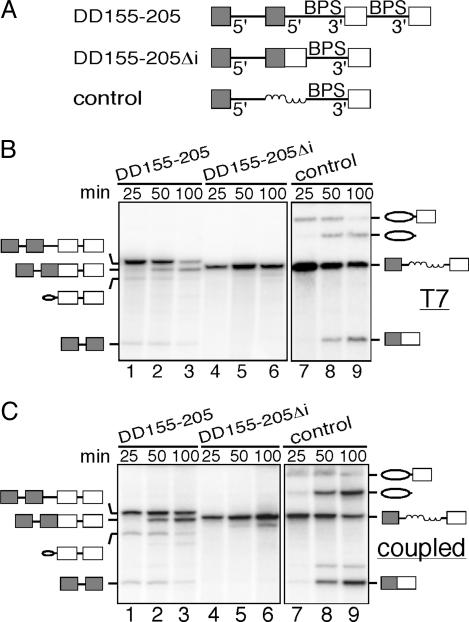
Spliced Exons Within an Intron Suppress Splicing. The observation that exon sequences can suppress exon skipping in pre-mRNAs containing duplicated 3′splice sites raises the possibility that, in general, splicing cannot occur over exon sequences. To further investigate this possibility, we examined splicing of a construct, DD155-205, containing duplicated 5′ splice sites flanked by full-length exon 1 and duplicated 3′ splice sites flanked by full-length exon 2 (Fig. 3A Top). In this context, only the internal pair of 5′and 3′ splice sites is used (Fig. 3B, lanes 1–3). The resulting spliced product contains ligated exons 1 and 2 within the intron and is itself a potential splicing substrate (Fig. 3A Middle). Significantly, however, this substrate (DD155-205Δi) is not spliced, not even when it is synthesized directly as a T7 transcript (Fig. 3B, lanes 4–6). To determine whether this splicing inhibition is due to the fused exons within the intron or to the presence of additional sequences, we examined the splicing of a control construct in which the internal spliced exons were replaced by a size-matched random sequence (Fig. 3A Bottom). In contrast to DD155-205Δi, the control construct is spliced efficiently (Fig. 3B, lanes 7–9). The same results were obtained in a coupled transcription/splicing system (Fig. 3C). Together, these data indicate that splicing cannot occur over exon sequences.
Fig. 3.
Spliced exons within an intron suppress splicing. (A) Schematic of the double duplication construct and its derivatives. (B) Spliced pre-mRNAs synthesized with T7 RNA polymerase. (C) Coupled transcription splicing system. Shaded boxes indicate full-length exon 1, and white boxes indicate full-length exon 2. Ovals indicate the position of the lariat exon intermediate or the excised intron. Coiled lines indicate a size-matched random sequence.
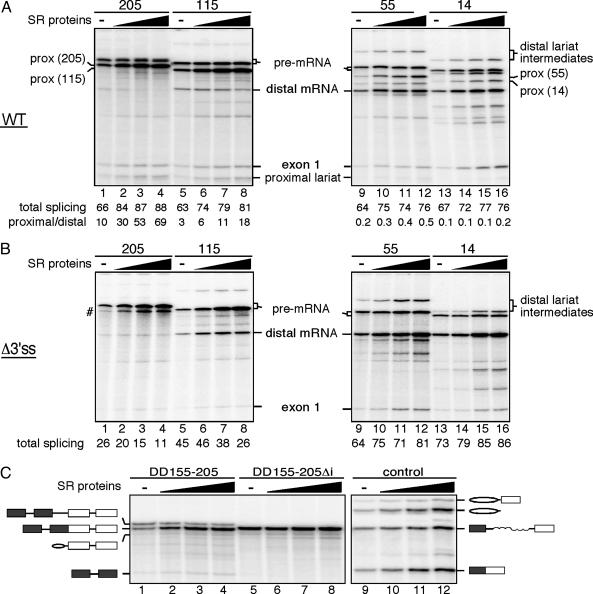
SR Proteins Are Required to Suppress Exon Skipping. β-Globin exon 2 contains several functional SR protein binding sites (14). Because SR proteins are usually limiting for splicing in nuclear extracts, we added increasing amounts of purified SR proteins to determine whether they affect the choice of proximal versus distal splice sites (Fig. 4). Consistent with previous studies, addition of SR proteins results in a dose-dependent stabilization of the pre-mRNAs. For the pre-mRNAs containing wild-type duplicated 3′ splice sites, an overall increase in splicing efficiency was also observed (Fig. 4A). Moreover, a dose-dependent increase in use of the proximal, but not the distal, 3′ splice site was observed for the pre-mRNAs containing full-length proximal exons (Fig. 4A, lanes 1–4). This SR protein-dependent effect is progressively decreased as the proximal exon is shortened (115, 55, and 14 nt in lanes 8–11, 9–12, and 13–16, respectively, of Fig. 4A). Thus, suppression of the distal 3′ splice site is both exon length-dependent and SR protein-dependent. This conclusion is further strengthened by the observation that SR proteins repress the distal 3′ splice site in the Δ3′ss pre-mRNA when the proximal exon 2 is either full-length or 115 nt long (Fig. 4B, lanes 1–4 and 5–8), whereas SR proteins activate the distal 3′ splice site in the Δ3′ss pre-mRNA when the proximal exon 2 is either 55 or 14 nt long (Fig. 4B, lanes 9–12 and 13–16). Finally, increasing SR protein levels enhances splicing of the internal intron in DD155-205 (Fig. 4C, lanes 1–4) but cannot overcome the block in splicing of DD155-205Δi (Fig. 4C, lanes 5–8). Together, these data indicate that SR proteins bound to exon sequences can prevent exon skipping.
Fig. 4.
SR proteins function in suppressing exon skipping. T7 precursors obtained from the wild-type series (A), the Δ3′ss series (B), or the double duplication construct and its derivative (C) were spliced in the absence (–) or presence (filled triangle) of increasing amounts of purified HeLa SR proteins. (A and B) The rate of splicing efficiency (A and B) and the ratio of proximal to distal 3′ splice-site selection mediated by the addition of SR proteins (A) are indicated below each lane. Pre-mRNA breakdown products are indicated by #. For the proximal spiced products (prox), the lengths of the internal exon sequences are indicated in parentheses.
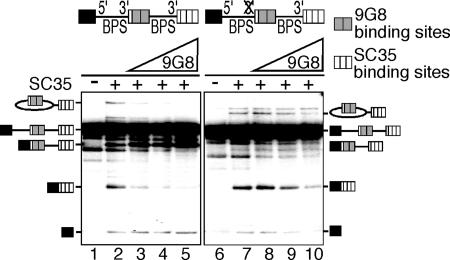
SR proteins function by binding to loosely conserved exonic enhancer elements. To directly test the possibility that SR protein/enhancer complexes play a role in preventing exon skipping, we examined pre-mRNAs containing multimerized SR protein binding sites replacing both the proximal and distal exon 2. Two copies of a 9G8 binding site (12, 19) were substituted for proximal exon 2, and three copies of an SC35 binding site (12) were substituted for distal exon 2. These pre-mRNAs were tested for splicing by using S100 extracts, which lack SR proteins. S100 extracts were complemented with a constant amount of recombinant SC35 and increasing amounts of recombinant 9G8 (Fig. 5). The addition of SC35 alone resulted in use of both the distal and proximal 3′ splice sites (compare lanes 1 and 2 in Fig. 5). In contrast, addition of increasing amounts of 9G8 in the presence of a constant amount of SC35 resulted in a dramatic increase in the use of the proximal 3′ splice site and a concomitant decrease in the use of the distal 3′ splice site (compare lanes 3–5 and 2 in Fig. 5). When the same assay was performed with the Δ3′ss substrate, the addition of recombinant SC35 alone resulted in exclusive use of the distal 3′ splice site as expected (compare lanes 6 and 7 in Fig. 5). Significantly, however, increasing amounts of 9G8 resulted in a dose-dependent decrease in use of the distal 3′ splice site and in overall splicing efficiency (compare lanes 8–10 and 7 in Fig. 5). We conclude that SR protein-enhancer complexes are required not only to activate the proximal 3′ splice site, but also to suppress exon skipping.
Fig. 5.
SR protein/enhancer complexes suppress the distal 3′ splice site. Splicing of wild-type (lanes 1–5) and mutant (lanes 6–10) pre-mRNAs in S100 extract alone (lanes 1 and 6), S100 extract complemented with 200 nM SC35 (lanes 2 and 7), or S100 extract complemented with SC35 and increasing amounts (0–300 nM) of 9G8 (lanes 3–5 and lanes 8–10).
Discussion
We have used model pre-mRNAs to investigate the mechanisms involved in preventing exon skipping during pre-mRNA splicing. These substrates contain a single 5′ splice site and two identical 3′ splice sites, each of which are followed by exon sequences. By inactivating the proximal 3′ splice site we have been able to uncouple the processes of exon recognition and prevention of exon skipping. We find that exon sequences downstream from the proximal 3′ splice site can suppress the use of the distal 3′ splice site and that SR proteins are required for this suppression. We also show that exon sequences within an intron can block splicing. We conclude that SR proteins bound to exonic enhancers can function to suppress exon skipping.
In previous studies (20, 21) and in the results reported here, the activities of ESEs were shown to be context-specific. ESEs promote early steps of spliceosome assembly when located upstream or downstream of 5′ or 3′ splice sites, respectively, but they suppress splicing when located within the intron (4). When promoting splicing, SR proteins interact with spliceosomal proteins that bind to the 5′ and 3′ splice sites (22, 23). Recent studies have shown that the RS domain of SR proteins bound to the exon can also interact directly with the BPS (6). Finally, SR proteins not only function by binding to ESEs, but they also play a critical role in the basic splicing mechanism within the intron, either by facilitating interactions between proteins bound to the 5′ and 3′ splice sites within the intron (24–26) and/or by contacting the 5′ splice site in the mature spliceosome (27).
The mechanism by which SR proteins that are bound to intron sequences suppress splicing is not understood. Computational analyses indicate that sequence motifs for ESEs are more frequent in exons than in introns (28). However, there are a number of examples in which SR protein binding sites are located within the intron and can function to promote splicing of upstream 5′ splice sites (8, 29–32). At the same time, negative intronic regulatory elements with which SR proteins interact have also been found (20, 29, 33, 34). For example, a splicing suppressor element that binds to SR proteins, 3RE, has been identified in the adenovirus major late IIIa pre-mRNA (20, 21). 3RE is located immediately adjacent to the BPS. The binding of SR proteins to 3RE seems to prevent the binding of U2 small nuclear ribonucleoprotein to the BPS. However, 3RE can function as a splicing silencer when moved 53 nt from the BPS and can also suppress splicing in a heterologous β-globin intron (21). Thus, splicing enhancers can suppress splicing when located within the introns of different splicing substrates, and we have shown that this suppression can be observed both in vitro and in microinjected Xenopus oocytes. Thus, there is compelling evidence that SR proteins can suppress splicing when bound to sequences located within introns.
How intronic SR protein binding sites can, on the one hand, promote and, on the other hand, inhibit splicing is not understood. It is important to note that the cases where intronic SR protein binding sites have been shown to promote splicing involve regulated alternative splicing and may therefore require the presence of specific regulatory proteins (in addition to SR proteins) that counteract the normally negative activity of the intronic SR protein binding site. There is evidence that the activities of ESEs may be determined by the ratio of heterogeneous nuclear ribonucleoprotein (hnRNP) proteins and SR proteins (35). When ESEs and hnRNP binding sites (exonic splicing silencers) are located adjacent to each other in exons, they can influence each other's activities. Thus, the overall activity of the exon sequence could be determined by the relative amounts of hnRNP proteins and SR proteins present. hnRNP proteins appear to spread from their initial binding sites in both directions, thus interfering with the binding of SR proteins to nearby sequences (36). Similarly, SR proteins are known to bind enhancer sequences cooperatively, to form highly stable complexes (37). These complexes could, in principle, interfere with the spread of hnRNP proteins from nearby binding sites. In the case of exons, positive splicing activity is determined by SR proteins, which promote spliceosome assembly (4), whereas hnRNP proteins block the activity of SR proteins through steric interference. The opposite may be true of constitutively spliced introns, where SR protein binding sites are rare and hnRNP proteins preferentially associate.
Recent studies of the SR protein ASF/SF2 provide evidence that the SR domain is required to promote splicing when bound to sites downstream of a 3′ splice site, whereas one of the two RNA binding domains, RBD2, is required to repress splicing when bound within the intron (21). These conclusions, based on studies of MS2/ASF/SF2 fusion proteins, do not apply to all SR proteins. We have shown that 9G8, which lacks an RBD2 domain, is also capable of repressing splicing when bound within an intron (Fig. 5). However, the fact that at least two distinct SR protein domains can promote or repress splicing suggests the possibility that the activity of the domain involves interactions with other proteins. These interacting proteins could either promote spliceosome recruitment or interfere with spliceosome assembly.
Regardless of the mechanisms involved, the data presented here strongly suggest that SR proteins bound to exons could play an important role in the prevention of exon skipping in constitutively spliced pre-mRNAs. According to this view, SR proteins bound to unspliced exons would not only direct the splicing machinery to the nearest 5′ or 3′ splice sites, they would also suppress splicing between upstream 5′ splice sites and downstream 3′ splice sites. Similarly, once two internal exons are joined, SR proteins bound to the spliced exons would provide an even stronger barrier to exon skipping.
Considering the importance of joining exons in the correct 5′ to 3′ order, it is likely that additional mechanisms are used to avoid exon skipping, especially for pre-mRNAs containing very large introns. For example, the extensive coupling between the transcription and splicing machineries (38) has suggested models in which exon skipping would be avoided by immobilizing the 5′ splice site on RNA polymerase until the 3′ splice from the same intron is synthesized.
Acknowledgments
We thank members of the R.R. and T.M. laboratories for helpful discussions; André Verdel, Claude Gazin, Jeanne Hsu, and Patricia Valencia for critical reading of the manuscript; and the National Cell Culture Center for providing the HeLa cells. This work was supported by National Institutes of Health Grants GM42231 (to T.M.), GM43375 (to R.R.), and GM62287 (to K.J.H.).
Author contributions: E.C.I., T.D.S., K.J.H., R.R., and T.M. designed research; E.C.I., T.D.S., and K.J.H. performed research; E.C.I., T.D.S., and K.J.H. contributed new reagents/analytic tools; E.C.I., T.D.S., K.J.H., R.R., and T.M. analyzed data; and E.C.I., T.D.S., K.J.H., R.R., and T.M. wrote the paper.
Abbreviations: BPS, branchpoint sequence; ESE, exonic splicing enhancer; SR, serine/arginine-rich; mBPS, mutated BPS; hnRNP, heterogeneous nuclear ribonucleoprotein.
See Commentary on page 4927.
References
- 1.Berget, S. M. (1995) J. Biol. Chem. 270, 2411–2414. [DOI] [PubMed] [Google Scholar]
- 2.Reed, R. (2000) Curr. Opin. Cell Biol. 12, 340–345. [DOI] [PubMed] [Google Scholar]
- 3.Hastings, M. L. & Krainer, A. R. (2001) Curr. Opin. Cell Biol. 13, 302–309. [DOI] [PubMed] [Google Scholar]
- 4.Graveley, B. R. (2000) RNA 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blencowe, B. J. (2000) Trends Biochem. Sci. 25, 106–110. [DOI] [PubMed] [Google Scholar]
- 6.Shen, H., Kan, J. L. & Green, M. R. (2004) Mol. Cell 13, 367–376. [DOI] [PubMed] [Google Scholar]
- 7.Cartegni, L., Chew, S. L. & Krainer, A. R. (2002) Nat. Rev. Genet. 3, 285–298. [DOI] [PubMed] [Google Scholar]
- 8.Black, D. L. (2003) Annu. Rev. Biochem. 72, 291–336. [DOI] [PubMed] [Google Scholar]
- 9.Reed, R. & Maniatis, T. (1986) Cell 46, 681–690. [DOI] [PubMed] [Google Scholar]
- 10.Reed, R. & Maniatis, T. (1988) Genes Dev. 2, 1268–1276. [DOI] [PubMed] [Google Scholar]
- 11.Reed, R. & Maniatis, T. (1985) Cell 41, 95–105. [DOI] [PubMed] [Google Scholar]
- 12.Schaal, T. D. & Maniatis, T. (1999) Mol. Cell. Biol. 19, 1705–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krainer, A. R., Maniatis, T., Ruskin, B. & Green, M. R. (1984) Cell 36, 993–1005. [DOI] [PubMed] [Google Scholar]
- 14.Schaal, T. D. & Maniatis, T. (1999) Mol. Cell. Biol. 19, 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo, M. J. & Reed, R. (1999) Proc. Natl. Acad. Sci. USA 96, 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zahler, A. M. (1999) Methods Mol. Biol. 118, 419–432. [DOI] [PubMed] [Google Scholar]
- 17.Tian, M. & Maniatis, T. (1993) Cell 74, 105–114. [DOI] [PubMed] [Google Scholar]
- 18.Treisman, R., Orkin, S. H. & Maniatis, T. (1983) Prog. Clin. Biol. Res. 134, 99–121. [PubMed] [Google Scholar]
- 19.Cavaloc, Y., Bourgeois, C. F., Kister, L. & Stevenin, J. (1999) RNA 5, 468–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanopka, A., Muhlemann, O. & Akusjarvi, G. (1996) Nature 381, 535–538. [DOI] [PubMed] [Google Scholar]
- 21.Dauksaite, V. & Akusjarvi, G. (2002) J. Biol. Chem. 277, 12579–12586. [DOI] [PubMed] [Google Scholar]
- 22.Wu, J. Y. & Maniatis, T. (1993) Cell 75, 1061–1070. [DOI] [PubMed] [Google Scholar]
- 23.Kohtz, J. D., Jamison, S. F., Will, C. L., Zuo, P., Luhrmann, R., Garcia-Blanco, M. A. & Manley, J. L. (1994) Nature 368, 119–124. [DOI] [PubMed] [Google Scholar]
- 24.Fu, X. D. & Maniatis, T. (1992) Proc. Natl. Acad. Sci. USA 89, 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hertel, K. J. & Maniatis, T. (1999) Proc. Natl. Acad. Sci. USA 96, 2651–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boukis, L. A., Liu, N., Furuyama, S. & Bruzik, J. P. (2004) J. Biol. Chem. 279, 29647–29653. [DOI] [PubMed] [Google Scholar]
- 27.Shen, H. & Green, M. R. (2004) Mol. Cell 16, 363–373. [DOI] [PubMed] [Google Scholar]
- 28.Liu, H. X., Zhang, M. & Krainer, A. R. (1998) Genes Dev. 12, 1998–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallego, M. E., Gattoni, R., Stevenin, J., Marie, J. & Expert-Bezancon, A. (1997) EMBO J. 16, 1772–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lou, H., Neugebauer, K. M., Gagel, R. F. & Berget, S. M. (1998) Mol. Cell. Biol. 18, 4977–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hastings, M. L., Wilson, C. M. & Munroe, S. H. (2001) RNA 7, 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Expert-Bezancon, A., Sureau, A., Durosay, P., Salesse, R., Groeneveld, H., Lecaer, J. P. & Marie, J. (2004) J. Biol. Chem. 279, 38249–38259. [DOI] [PubMed] [Google Scholar]
- 33.Pagani, F., Buratti, E., Stuani, C., Romano, M., Zuccato, E., Niksic, M., Giglio, L., Faraguna, D. & Baralle, F. E. (2000) J. Biol. Chem. 275, 21041–21047. [DOI] [PubMed] [Google Scholar]
- 34.Simard, M. J. & Chabot, B. (2002) Mol. Cell. Biol. 22, 4001–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Black, D. L. & Grabowski, P. J. (2003) Prog. Mol. Subcell. Biol. 31, 187–216. [DOI] [PubMed] [Google Scholar]
- 36.Zhu, J., Mayeda, A. & Krainer, A. R. (2001) Mol. Cell 8, 1351–1361. [DOI] [PubMed] [Google Scholar]
- 37.Lynch, K. W. & Maniatis, T. (1995) Genes Dev. 9, 284–293. [DOI] [PubMed] [Google Scholar]
- 38.Kornblihtt, A. R., De La Mata, M., Fededa, J. P., Munoz, M. J. & Nogues, G. (2004) RNA 10, 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]