Abstract
Increased incidence of multidrug resistant (MDR) Gram negative infection has resulted in high rates of morbidity and mortality. Klebsiella pneumoniae is one of the commonest MDR pathogens causing bacteraemia with limited therapeutic options such as colistin and tigecycline. Present study focused on molecular characterisation of MDR K. pneumoniae from bloodstream infection and their clinical outcome. A total of 115 K. pneumoniae from January 2015 to September 2016 were included in the study which comprised of phenotypically identified ESBL and carbapenem resistant (CR) isolates. Multiplex PCR was performed for detection of resistance genes encoding β-lactam resistance. This includes blaSHV, blaTEM, blaVEB, blaPER, blaCTX-M, blaDHA, blaCIT, blaFOX, blaACC, blaACT, blaNDM, blaOXA48-like, blaVIM and blaKPC. Co-expression of blaSHV, blaTEM and blaCTX-M was predominant with 64% (74/115) prevalence. CTX-M-1 was the variant produced by all the isolates producing CTX-M. AmpC was uncommon, seen in 5% of the isolates (6/115). Among the carbapenemases co-expression of blaNDM and blaOXA48-like was observed in 28% (32/115) and blaNDM in 19% (22/115) and blaOXA48-like in 13% (15/115). blaKPC was absent. Overall mortality was observed to be 57% (64/113) and mortality among CR K. pneumoniae (Kp) was 68% (50/73). The antibiotics that were administered for treatment of CRKp were colistin in 90% (66/73) and tigecycline in 7% (5/73) and in 99% combined with meropenem (72/73). Prevalence of community acquired and nosocomial infections were 5% (4/73) and 95% (69/73) respectively among CRKp. Minocycline and meropenem susceptibilities were comparable and hence minocycline can be a carbapenem sparing agent. The resistance to β-lactam antibiotics is steadily increasing and are plasmid mediated, their containment in healthcare setting is a challenge.
Keywords: K. pneumoniae, carbapenem resistance, NDM, OXA-48-like, mortality, bacteremia
Introduction
The incidence of infections with extended spectrum β-lactamase (ESBL) producing Gram negative Enterobacteriaceae has steadily increased in nosocomial setting and is associated with significant morbidity and mortality. This situation has been worsened further by the emergence of carbapenemase producing strains that render carbapenem therapy suboptimal. Klebsiella pneumoniae and Acinetobacter baumannii-calcoaceticus complex (Abcc) are the most common multidrug resistant pathogens, and are frequently associated with a bacteremic illness. Other organisms are not as frequently reported to be resistant to commonly used antimicrobials as K. pneumoniae and Acinetobacter spp.
According to the Centre for Disease Dynamics, Economics and Policy (CDDEP), 80% of the Indian K. pneumoniae isolates are resistant to cephalosporins and up to 60% resistant to carbapenems. A review of data from our centre reveals a carbapenem resistance of 32 and 73% amongst K. pneumoniae and Abcc isolates respectively. India has witnessed a rise in carbapenem resistance rates from 9% in 2008 to 44% in 2010 [1]. In Italy, the prevalence of carbapenem resistant (CR) K. pneumoniae isolates, non-existent in 2008, has risen to 60% in 2013 (CDDEP). This rapid rise in carbapenem resistance reflects a worrisome trend.
Currently the Klebsiella pneumoniae carbapenemase (KPC), an Ambler class-A beta-lactamase is the most globally prevalent carbapenem resistance enzyme and is particularly widespread in Europe and North America. The New Delhi metallo-β-lactamase (NDM), an Ambler class-B enzyme, is the main cause of carbapenem resistance in India and has now spread across the globe [2]. This is slowly being accompanied by an increased incidence of the OXA48- like group, Ambler class D enzyme, in India [3−5].
Carbapenem resistant (CR) K.pneumoniae infections are associated with mortality rates as high as 30 to 44% [6−9]. Ben-David et al., report a 22 and 48% infection related mortality rate amongst patients infected with ESBL and CR K. pneumoniae respectively [10]. In this context, a clear understanding of the prevalent mechanisms for carbapenem resistance is essential. Despite a high disease burden, reports from India, on the prevalence of resistance mechanisms in multi drug resistant K. pneumoniae isolates are limited. The objective of this study is molecular characterization of the enzymatic mechanisms of resistance to β-lactam antibiotics in K. pneumoniae blood stream isolates.
Materials and methods
Isolates and phenotypic characterisation
A total of 115 K. pneumoniae blood culture isolates from January 2015 to September 2016 at the department of Clinical Microbiology, Christian Medical College, Vellore, were included in the study. Semiautomated BacT/ALERT system (Biomerieux) was used for blood culture. The isolates were identified using standard culture and biochemical reactions [11]. Duplicate isolates from the same patient were excluded from the study.
Antimicrobial susceptibility testing for different classes of antimicrobials such as cephalosporins - cefotaxime (30 μg), ceftazidime(30 μg); β-lactam/β-lactamase inhibitors - piperacillin/tazobactam(100/10 μg), cefoperazone/sulbactam (75/30 μg); carbapenems- imipenem(10 μg), meropenem (10 μg); fluoroquinolones- ciprofloxacin (5 μg), levofloxacin (5 μg), aminoglycosides- amikacin (30 μg), gentamicin (10 μg), tetracycline- minocycline (30µg), glycylcycline- tigecycline (15µg), was performed for all the isolates by Kirby Bauer disk diffusion method as recommended by Clinical Laboratory Standards Institute (CLSI) and interpreted according to CLSI 2015 guidelines [12,13] except for cefoperazone/sulbactam interpreted according to manufacturer, Pfizer (http://www.thefilipinodoctor.com/brand_pdf/Sulperazone.pdf) and tigecycline interpreted as per FDA breakpoints (http://www.accessdata.fda.gov/drugsatfda_docs/label/2009/021821s016lbl.pdf). ATCC E. coli 25922 was used as the control strain for susceptibility testing. The isolates were classified as ESBL producers if the zone diameter for ceftazidime was ≤22 mm and ≤27 mm for cefotaxime. They were screened for carbapenem resistance using meropenem and were included in the study if the zone diameter for meropenem was ≤19 mm.
In-house CarbaNP test as recommended by CLSI for the detection of carbapenemase activity was performed for a subset of randomly selected 73 isolates that were phenotypically resistant to imipenem and meropenem.
Genotypic characterisation
Bacterial DNA was extracted from 18 h cultures by Qiasymphony as per manufacturer’s instructions. Multiplex PCR was performed for the detection of genes encoding beta-lactam resistance which includes EBSL, CTX-M, ampC and carbapenemases. Multiplex PCR for EBSL genes included blaSHV, blaTEM, blaPER, blaVEB, and blaGES and was performed as previously described [14]. Multiplex PCR for CTX-M genes such as blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, blaCTX-M-9 and blaCTX-M-25 was performed according to Woodford et al. [15]. Multiplex PCR for AmpCβ-lactamases including blaACT, blaACC, blaCIT, blaDHA and blaFOX was performed as previously described [16]. Carbapenemase genes detected by multiplex PCR were blaNDM, blaVIM, blaIMP, blaSPM, blaOXA 48-like, and blaKPC and was performed as previously described [14,17−19]. Known positive controls for each gene was used with every run (courtesy: IHMA, USA).
Whole genome sequencing for a subset of eleven isolates was performed using Ion Torrent PGM platform with 400 bp chemistry. The Raw reads were assembled de novo using Assembler SPAdes software v5.0 in Torrent suite server version 4.4.3. Annotation of the genomes was performed with RAST (Rapid Annotation using Subsystems Technology- http://rast.nmpdr.org/) and PATRIC (Pathosystems Resource Integration Centre - https://www.patricbrc.org/). The genomes were submitted to GenBank to obtain accession numbers. Using the annotated whole genome sequences (WGS), the presence of any antibiotic resistance genes was detected from the ResFinder version 2.1 (https://cge.cbs.dtu.dk/services/ResFinder/).
Clinical data collection
Data from eligible subjects were retrospectively collected using a systematic data abstraction form. Patient data included age, gender, admission date, discharge date or date of death and admission diagnosis. Data was entered in software EpiData Entry v 3.0 to rule out errors such as repeat entry. K. pneumoniae isolates were classified into community acquired vs. nosocomial based on Centers for Disease Control and Prevention (CDC) guidelines for the same. Antibiotic susceptibility profiles and dates for all positive blood cultures were recorded. For each antibiotic, the dose, frequency and duration were documented. In bacteremic patients repeat blood cultures following the index blood cultures were recorded to document microbiological clearance. Mortality data at hospitalization and 90 days post-discharge were also recorded. The data was analysed using EpiData Analysis software v 3.0.
Results
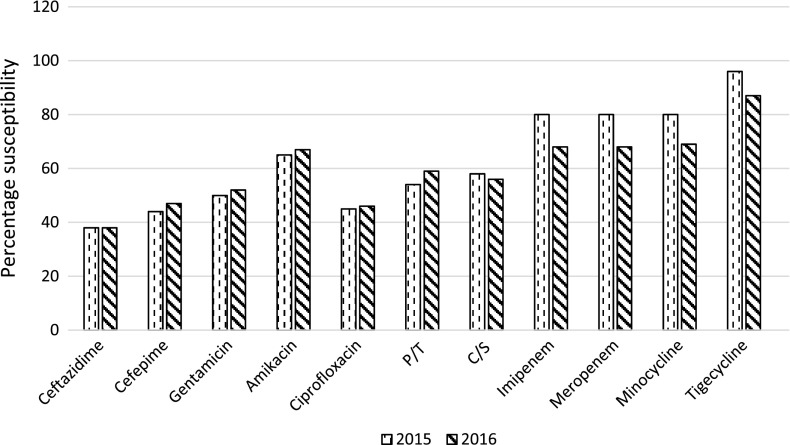
Susceptibility profile of K. pneumoniae from blood culture isolated during 2015 and 2016 is shown in Figure 1. During 2016, the susceptibility of most antimicrobials are comparable except for imipenem, meropenem, minocycline and tigecycline. Carbapenem susceptibility has reduced from 80 to 68% and minocycline to 69%. Susceptibilities of carbapenems and minocycline seem to be comparable. Susceptibility to tigecycline has reduced from 96 to 87% from 2015 to 2016.
Figure 1.
Antimicrobial susceptibility of K. pneumoniae from bloodstream infections at CMC.
P/T: Piperacillin/tazobactam; C/S: Cefoperazone/sulbactam.
CarbaNP test was found to be positive in 86% (n = 63/73) of K. pneumoniae tested. The results of multiplex PCR for β-lactamases are mentioned in Table 1. Among the ESBLs, 64% of the isolates showed co-expression of blaSHV, blaTEM and blaCTX-M genes. All the isolates tested for blaCTX-M showed the presence of blaCTX-M-1. AmpC was uncommon and seen in 5% of the isolates (6/115) which expressed blaDHA and blaCIT. Co-expression of blaNDM and blaOXA-48 like was seen in 28% of the isolates. The blaKPC gene was absent in the study isolates. Forty isolates that were phenotypically resistant to meropenem lacked carbapenemases.
Table 1.
β-lactamases seen in K. pneumoniae as reported by various studies from India including present study.
| Reference | ESBL | CTX-M | ESBL and CTX-M | AmpC | Class A carbapenemase | Class B carbapenemase (MBLs) | Class D carabpenemase | Class B and D carbapenemase | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaSHV | blaTEM | blaSHV, TEM | blaCTX-M | blaTEM, CTX-M | blaSHV, CTX-M | blaSHV, TEM, CTX-M | blaDHA | blaCIT | blaMOX | blaKPC | blaNDM | blaVIM | blaNDM, VIM | blaOXA-48-LIKE | blaNDM, OXA-48 LIKE | blaVIM, OXA-48 LIKE | blaNDM, OXA-48 LIKE, VIM | |
| Veeraraghavan et al. (n = 115)(this study) | 16% | 0 | 1.7% | 0 | 2.6% | 5.2% | 64.3% | 4.3% | 0.8% | 0 | 0 | 19% | 1.7% | 3.4% | 13% | 28% | 0 | 0 |
| (n = 18) | 0 | (n = 2) | (n = 3) | (n = 6) | (n = 74) | (n = 5) | (n = 1) | (n = 22) | (n = 2) | (n = 4) | (n = 15) | (n = 32) | ||||||
| Goyal et al., 2009 [23] | 14% | 19% | 0 | 10% | 29% | 10% | 19% | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| (n = 3) | (n = 4) | (n = 2) | (n = 6) | (n = 2) | (n = 4) | |||||||||||||
| (n = 21) | ||||||||||||||||||
| Sharma et al., 2010 [38] | 72% | 60% | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND | ND | ND | ND | ND | ND | ND |
| (n = 18) | (n = 15) | |||||||||||||||||
| (n = 25) | ||||||||||||||||||
| Manoharan et al., 2011 [22] | 7% | 0 | 5% | 8% | 15% | 8% | 43% | 0 | 0 | 0 | ND | ND | ND | ND | ND | ND | ND | ND |
| (n = 4) | (n = 3) | (n = 5) | (n = 9) | (n = 5) | (n = 26) | |||||||||||||
| (n = 61) | ||||||||||||||||||
| Nagaraj et al., 2012 [24] | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0 | 75% | 13.8% | 0 | ND | ND | 0 | 0 |
| (n = 25) | (n = 5) | |||||||||||||||||
| (n = 36) | ||||||||||||||||||
| Mohamudha et al., 2012 [39] | ND | ND | ND | ND | ND | ND | ND | 17.4% | 10% | 1.8% | ND | ND | ND | ND | ND | ND | ND | ND |
| (n = 19) | (n = 11) | (n = 2) | ||||||||||||||||
| (n = 109) | ||||||||||||||||||
| Krishnamurthy et al., 2013 [40] | 36% | 7% | 0 | 43% | 0 | 0 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| (n = 10) | (n = 2) | (n = 12) | ||||||||||||||||
| (n = 28) | ||||||||||||||||||
| Chaudhary et al., 2013 [41] (n = 150) |
45.33% | 47.33% | 0 | 37.33% | 0 | 0 | 0 | ND | ND | ND | 7.9% | 6.25% | 17.3% | 0 | ND | ND | 0 | 0 |
| (n = 68) | (n = 71) | (n = 56) | (n = 19) | (n = 24) | (n = 26) | |||||||||||||
| Roy et al., 2015 [42] | 0 | 0 | 0 | 0 | 33% | 0 | 67% | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| (n = 2) | (n = 4) | |||||||||||||||||
| (n = 6) | ||||||||||||||||||
| Anandan et al., 2015 [25] | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0 | 34% | 0 | 0 | 44% | 16% | 0 | 0 |
| (n = 30) | (n = 39) | (n = 14) | ||||||||||||||||
| (n = 88) | ||||||||||||||||||
| Pragasam et al., 2016 [4] | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0 | 24% | 0 | 2.3% | 55% | 16% | 0 | 0 |
| (n = 20) | (n = 2) | (n = 47) | (n = 14) | |||||||||||||||
| (n = 85) | ||||||||||||||||||
| Sharma et al., 2016 [3] | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | – | 27% | 1% | 1% | 36% | 15% | 10% | 4% |
| (n = 36) | (n = 2) | (n = 2) | (n = 48) | (n = 20) | (n = 14) | (n = 6) | ||||||||||||
| (n = 134) | ||||||||||||||||||
From the WGS analysis, apart from confirming the PCR results, the variants of β-lactamase genes such as blaSHV, blaTEM, blaCTX-M, blaNDM and blaOXA-48-like were identified. The variants of SHV obtained were SHV-1, SHV-2, SHV-11 and SHV-12. TEM1A and TEM-1B were the only two variants of TEM observed in this study. CTX-M-15 was the only member of CTX-M-1 group seen in this study. Among the carbapenemases, NDM-1 was common and in the group of OXA48-like enzymes, OXA181 and OXA232 were commonly seen. The whole genome sequences were deposited at GenBank with accession numbers as follows: MOXM00000000, MIEJ00000000, MPCT00000000, MDZG00000000, MOXL00000000, MEBR00000000, MOXN00000000, LZYN00000000, MCFO00000000, MCFP00000000 and MCFQ00000000. Plasmid mediated colistin resistance due to mcr-1 and mcr-2 was absent.
Clinical data from 113 patients were collected and analysed. The overall mortality rate was noted to be 57% (64/113) while the mortality rate in patients with CR K. pneumoniae infections was 68% (50/73). Antimicrobials used in patients with CR K.pneumoniae infection included colistin 90% (66/73) and tigecycline 7% (5/73). Colistin or tigecycline were not initiated in 2 patients since they succumbed to their illness within 3 h of presentation, before the availability of blood culture reports. Colistin or tigecycline were administered in combination with meropenem in 99% (72/73) patients. Amongst patients with CR K. pneumoniae infections, 5% (4/73) had a community acquired infection while 95% (69/73) had a nosocomial infection. Nosocomial CR K. pneumoniae infections presented either as infective complications following chemotherapy for hematologic or solid organ malignancies 20% (14/69), or occurred in ICU patients as ventilator or central venous catheter related complications 80% (55/69).
Discussion
Globally, the highest rate of carbapenem resistance has been reported in Greece with 68% resistance followed by India and eastern Mediterranean regions with 54% resistance [20] as mentioned in Table 2. USA, China and Africa have low resistance rates with 11, 11 and 4% respectively. The predominant enzymatic mechanism of resistance in Europe is KPC followed by OXA-48-like and NDM [21]. While in USA, it is KPC followed by NDM and minimal due to OXA-48-like [21].
Table 2.
Epidemiology of carbapenem resistance in K. pneumoniae [20,21].
| Region | Resistant rates (%) | Mechanism of resistance |
|---|---|---|
| Africa | 4 | OXA-48-like, NDM |
| America (USA) | 11 | KPC, NDM, OXA-48-like |
| Western pacific region (Mongolia, China) | 11 | KPC, NDM, OXA-48-like |
| Eastern Mediterranean | 54 | NDM, OXA-48-like |
| India | 54 | NDM, OXA-48-like, KPC |
| Europe (Greece, Georgia, Italy) | 68 | KPC, OXA-48-like, NDM |
While in India, amongst the ESBLs, SHV, TEM and CTX-M have been commonly reported (Table 1). Similar to this study observation, co-existence of SHV, TEM and CTX-M in Enterobacteriaceae have been reported by Manoharan et al. and Goyal et al. [22,23]. However, the number of isolates included in these studies are relatively less. In India, carbapenem resistance is predominantly due to NDM and OXA-48-like and KPC is hardly seen [4,24,25]. Though OXA-48 like group of carbapenemases are emerging in India, most Indian studies, have been unable to capture this trend adequately (Table 1). It is essential for all the hospitals in the county to monitor the carbapenemases prevalent in their regions so that the change in trend from NDM to OXA-48 like group of enzymes can be demonstrated. VIM metallo-β-lactamase producing isolates are also being increasingly reported from India, overall this study detected only two VIM producing isolates. While KPC is the predominant carbapenemase produced in Europe it has been rarely reported from India.
A recent study by Falagas et al. showed that mortality in critically ill patients with CR K. pneumoniae infections continued to exceed 60% irrespective of whether they were on a combination regimen or monotherapy [26]. The mortality rate in the study was also noted to be high (68%) despite patients being on a combination of colistin and meropenem. Due to this compelling need for newer antimicrobial therapy in CR K. pneumoniae infections, older drugs such as minocycline are also being evaluated. On analysing isolates obtained between August 2015 and July 2016, we found that amongst ESBL K. pneumoniae isolates the susceptibility to minocycline and tigecycline was 85 and 91% respectively. Amongst CR K. pneumoniae isolates the susceptibility to minocycline and tigecycline was 55 and 73% respectively [27]. Minocycline will be of great use for ESBL positive isolates and can be a carbapenem sparing agent while among CR isolates its usefulness is low. Since the susceptibilities of meropenem and minocycline were same for the K. pneumoniae isolated in the present study centre for the past two years, minocycline can be used as an alternate for carbapenems. There is a need for studies from other parts of the country for meropenem and minocycline susceptibilities which can enable a changes in national antibiotic policies.
Eight colistin resistant isolates were identified among the study isolates [28]. Three of these had significant mutations in the mgrB gene while one isolate had a mutation in pmrB and mgrB genes as previously reported. These genes are responsible for lipid A modifications leading to colistin resistance. One of the colistin resistant isolates with a high MIC did not have an identifiable mutation but arnT gene had novel mutations when compared to mutations seen in other seven. Two isolates though had mutations in many genes, none were previously reported and their significance is yet to be established [28]. Among the ESKAPE pathogens, colistin resistance is rising in Klebsiella spp. than the other organisms. Highest resistance to colistin among K. pneumoniae has been reported from Netherlands with 55% followed by Italy (43%) and Greece (20%) [29–31].
Earlier at the study centre, the novel β-lactam- β-lactam inhibitor combinations that are yet to be made available in the Indian market such as aztreonam-avibactam, ceftaroline-avibactam, ceftazidime-avibactam and ceftolozane-tazobactam were evaluated. Among these, susceptibility among the overall study isolates was found to be 84, 53, 58 and 53% for aztreonam-avibactam, ceftaroline-avibactam, ceftazidime-avibactam and ceftolozane-tazobactam respectively (unpublished data). Among the CR isolates, the susceptibility to aztreonam-avibactam, ceftaroline-avibactam, ceftazidime-avibactam and ceftolozane-tazobactam were 67, 17, 26 and 10% respectively (unpublished data). These combination drugs do not seem to be promising agents for the treatment of CRKp.
For CR isolates, tigecycline and colistin remain antimicrobials of choice. But with increased usage, resistance to tigecycline and colistin are rapidly emerging. From experience at the present study centre, the resistance to tigecycline has significantly increased from 2015 to 2016. However, in the present study, combination of meropenem and tigecycline was used for therapy only in 7% of the patients which is in contrast to meropenem-colistin combination being used in 90% of the patients. Though susceptibility to colistin remains about 98%, there has been emergence of plasmid mediated resistance [32,33] which has the potential to rapidly spread among many genera. With resistance to the last resort antibiotics, soon there will be a threat to management of infections especially in critically ill patients.
There is however, a paucity of data regarding the effectiveness of combination antimicrobial regimens when compared to monotherapy. This is primarily due to the fact that most of the existing evidence has been derived from retrospective data obtained from small patient numbers. In critically ill bacteremic patients with CR K. pneumoniae, evidence seems to suggest that a combination regimen may be associated with better survival than monotherapy alone. Three studies also show that meropenem addition to a colistin-tigecycline combination may result in a significant increase in survival, even in those patients who are infected with CR isolates, possibly due to potential colistin-carbapenem synergy [34–36]. The results of these studies however may be limited by small patient numbers.
Common combinations used in CR K. pneumoniae infections include tigecycline with colistin, tigecycline with gentamicin and carbapenem with colistin. Patients included in our study were predominantly treated with a combination of colistin with meropenem and with tigecycline-meropenem to a lesser extent. None were treated with monotherapy which limits the comparison of outcomes of monotherapy vs. combination therapy.
Among K. pneumoniae, there has been emergence of the hypervirulent strains which initially were prevalent among the isolates susceptible to antimicrobials. Presently, they have been reported among MDR isolates [37], as also seen in the study centre, which further poses a challenge for treatment since virulence combined with antimicrobial resistance will be very worrisome leading to very high mortality rates.
Conclusion
Understanding the prevalent resistance mechanisms for β-lactam antibiotics is essential for individual treatment and hospital infection control. Steady increase in carbapenem and colistin resistance is seen in K. pneumoniae among the ESKAPE pathogens which is a worrisome condition. In this study, we predominantly see SHV, TEM and CTX-M-1 contributing to ESBLs while NDM and OXA48-like group are responsible for carbapenem resistance. Though previously uncommon, now OXA48-like group of carbapenemase is becoming widespread especially in the Indian setting. The mortality rate among the CRKp infections of 68% was similar to other studies reported worldwide. Colistin with meropenem was the most frequently used combination for treatment while tigecycline with meropenem was less frequent. But there is also simultaneous increase in resistance to colistin and tigecycline. Mostly infections in our setting were nosocomial in nature and management of MDR nosocomial infection is challenging. There is a need for better therapeutic options for CRKp infections. Among the β-lactam- β-lactam inhibitor combinations, aztreonam-avibactam is promising and minocycline can also be a potential carbapenem sparing agent.
Conflicts of interest
The authors declare that they have no conflicts of interest.
References
- [1].Parveen RM, Harish BN, Parija SC. Emerging carbapenem resistance among nosocomial isolates of Klebsiella pneumoniae in South India. Int J Pharma Biosci. 2010;1(2):1–11. [Google Scholar]
- [2].Johnson AP, Woodford N. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol. 2013;62(4):499–513. 10.1099/jmm.0.052555-0 [DOI] [PubMed] [Google Scholar]
- [3].Sharma A, Bakthavatchalam YD, Gopi R, et al. Mechanisms of carbapenem resistance in K. pneumoniae and E. coli from bloodstream infections in India. J Infect Dis Ther. 2016;4:293. [Google Scholar]
- [4].Pragasam AK, Sahni RD, Anandan S, et al. A pilot study on carbapenemase detection: do we see the same level of agreement as with the CLSI Observations. J Clin Diagn Res. 2016;10(7):DC09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Singh SK, Gupta M. blaOXA-48 carrying clonal colistin resistant-carbapenem resistant Klebsiella pneumoniae in neonate intensive care unit India. Microb Pathog. 2016;30(100):75–77. 10.1016/j.micpath.2016.09.009 [DOI] [PubMed] [Google Scholar]
- [6].Patel G, Huprikar S, Factor SH, et al. Outcomes of carbapenem-resistant klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29(12):1099–1106. 10.1086/592412 [DOI] [PubMed] [Google Scholar]
- [7].Schwaber MJ, Klarfeld-Lidji S, Navon-Venezia S, et al. Predictors of carbapenem-resistant klebsiella pneumoniae acquisition among hospitalized adults and effect of acquisition on mortality. Antimicrob Agents Chemother. 2008;52(3):1028–1033. 10.1128/AAC.01020-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Falagas ME, Rafailidis PI, Kofteridis D, et al. Risk factors of carbapenem-resistant Klebsiella pneumoniae infections: a matched case–control study. J Antimicrob Chemo. 2007;60(5):1124–1130. 10.1093/jac/dkm356 [DOI] [PubMed] [Google Scholar]
- [9].Daikos GL, Markogiannakis A. Carbapenemase-producing Klebsiella pneumoniae: (when) might we still consider treating with carbapenems? Clin Microbiol Infect. 2011;17(8):1135–1141. 10.1111/j.1469-0691.2011.03553.x [DOI] [PubMed] [Google Scholar]
- [10].Ben-David D, Kordevani R, Keller N, et al. Outcome of carbapenem resistant Klebsiella pneumoniae bloodstream infections. Clin Microbiol Infect. 2012;18(1):54–60. 10.1111/j.1469-0691.2011.03478.x [DOI] [PubMed] [Google Scholar]
- [11].Koshi M. Myer’s and Koshi’s manual of diagnostic procedures in medical microbiology and immunology/serology. Vellore, India: Christian Medical College and Hospital; 2001. [Google Scholar]
- [12].Clinical and Laboratory Standards Institute Performance standards for antimicrobial disc susceptibility testing; approved standard. 12th ed. CLSI document M02-A12. 35(1). Wayne (PA); 2015. [Google Scholar]
- [13].Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing; twenty fifth informational supplement. Wayne (PA): Clinical and Laboratory Standards Institute. CLSI document; 2015. p. M100–S25. [Google Scholar]
- [14].Dallenne C, Da Costa A, Decré D, et al. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- [15].Woodford N, Ward ME, Kaufmann ME, et al. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK. J Antimicrob Chemother. 2004;54(4):735–743. 10.1093/jac/dkh424 [DOI] [PubMed] [Google Scholar]
- [16].Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162. 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Poirel L, Walsh TR, Cuvillier V, et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- [18].Ellington MJ, Kistler J, Livermore DM, et al. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob Chemother. 2007;59(2):321–322. [DOI] [PubMed] [Google Scholar]
- [19].Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151–1161. 10.1128/AAC.45.4.1151-1161.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].World Health Organization Antimicrobial resistance: 2014 Global report on surveillance. World Health Organization; 2014. [Google Scholar]
- [21].Lee CR, Lee JH, Park KS, et al. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Manoharan A, Premalatha K, Chatterjee S, et al. Correlation of TEM, SHV and CTX-M extended-spectrum beta lactamases among Enterobacteriaceae with their in vitro antimicrobial susceptibility. Indian J Med Microbiol. 2011;29(2):161. 10.4103/0255-0857.81799 [DOI] [PubMed] [Google Scholar]
- [23].Goyal A, Prasad KN, Prasad A, et al. Extended spectrum β-lactamases in Escherichia coli & Klebsiella pneumoniae & associated risk factors. Indian J Med Res. 129(6):695–700. [PubMed] [Google Scholar]
- [24].Nagaraj S, Chandran SP, Shamanna P, et al. Carbapenem resistance among Escherichia coli and Klebsiella pneumoniae in a tertiary care hospital in south India. Indian J Med Microbiol. 2012;30(1):93. [DOI] [PubMed] [Google Scholar]
- [25].Anandan S, Damodaran S, Gopi R, et al. Rapid screening for carbapenem resistant organisms: current results and future approaches. J Clin Diagn Res: JCDR. 2015;9(9):DM01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Falagas ME. Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerg Infect Dis J-CDC. 2014;20(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Veeraraghavan B, Shankar C, Vijayakumar S. Can minocycline be a carbapenem sparing antibiotic? current evidence. Indian J Med Microbiol. 2016;34(4):513. 10.4103/0255-0857.195380 [DOI] [PubMed] [Google Scholar]
- [28].Pragasam AK, Shankar C, Veeraraghavan B, et al. Molecular mechanisms of colistin resistance in Klebsiella pneumoniae causing bacteremia from India – a first report. Front Microbiol. 2016;7:2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Halaby T, Kucukkose E, Janssen AB, et al. Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob Agents Chemother. 2016;60(11):6837–6843. 10.1128/AAC.01344-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Neonakis IK, Samonis G, Messaritakis H, et al. Resistance status and evolution trends of Klebsiella pneumoniae isolates in a university hospital in Greece: ineffectiveness of carbapenems and increasing resistance to colistin. Chemotherapy. 2010;56:448–452. 10.1159/000320943 [DOI] [PubMed] [Google Scholar]
- [31].Kontopidou F, Plachouras D, Papadomichelakis E, et al. Colonization and infection by colistin-resistant Gram-negative bacteriain a cohort of critically ill patients. Clin Microbiol Infect. 2011;17:E9–E11. 10.1111/j.1469-0691.2011.03649.x [DOI] [PubMed] [Google Scholar]
- [32].Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- [33].Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium. Eurosurveillance. [cited July 7,2016];21(27). [DOI] [PubMed] [Google Scholar]
- [34].Qureshi ZA, Paterson DL, Potoski BA, et al. Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob Agents Chemother. 2012;56(4):2108–2113. 10.1128/AAC.06268-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tumbarello M, Viale P, Viscoli C, et al. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin Infect Dis. 2012;55(7):943–950. 10.1093/cid/cis588 [DOI] [PubMed] [Google Scholar]
- [36].Daikos GL, Tsaousi S, Tzouvelekis LS, et al. Carbapenemase-producing klebsiella pneumoniae bloodstream infections: lowering mortality by antibiotic combination schemes and the role of carbapenems. Antimicrob Agents Chemother. 2014;58(4):2322–2328. 10.1128/AAC.02166-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shankar C, Nabarro LE, Ragupathi NK, et al. Draft genome sequences of three hypervirulent carbapenem-resistant Klebsiella pneumoniae isolates from bacteremia. Genome Announcements. 2016;4(6):e01081–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sharma J, Sharma M, Ray P.. Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res. 2010;132: 332–336. [PubMed] [Google Scholar]
- [39].Mohamudha PR, Harish BN, Parija SC. Molecular description of plasmid-mediated AmpC β-lactamases among nosocomial isolates of Escherichia coli & Klebsiella pneumoniae from six different hospitals in India. Indian J Med Res. 2012;135(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Krishnamurthy V, Vijaykumar GS, Kumar S, et al. Phenotypic and genotypic methods for detection of extended spectrum β lactamase producing Escherichia coli and Klebsiella pneumoniae isolated from ventilator associated pneumonia. J Clin Diagn Res: JCDR. 2013;7(9):1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chaudhary M, Payasi A. Antimicrobial susceptibility patterns and molecular characterization of Klebsiella pneumoniae clinical isolates from north Indian patients. Int J Med Med Sci. 2013;46:1218–1224. [Google Scholar]
- [42].Roy S, Gaind R, Chellani H, et al. Neonatal septicaemia caused by diverse clones of Klebsiella pneumoniae & Escherichia coli harbouring blaCTX-M-15. Indian J Med Res. 2013;137(4):791. [PMC free article] [PubMed] [Google Scholar]