Abstract
Topoisomerases manage the torsional stress associated with the separation of DNA strands during transcription and DNA replication. Eukaryotic Topoisomerase I (Top1) is a Type IB enzyme that nicks and rejoins only one strand of duplex DNA, and it is especially important during transcription. By resolving transcription-associated torsional stress, Top1 reduces the accumulation of genome-destabilizing R-loops and non-B DNA structures. The DNA nicking activity of Top1, however, can also initiate genome instability in the form of illegitimate recombination, homologous recombination and mutagenesis. In this review, we focus on the diverse, and often opposing, roles of Top1 in regulating eukaryotic genome stability.
Keywords: Top1, Supercoiling, Transcription, G4 DNA, R-loops, Illegitimate recombination, Deletions, Genome stability
1. Introduction
Topoisomerases transform the topological state of helical DNA by first creating a break in the DNA backbone and, following the swiveling of DNA strands, catalyzing religation of the broken strand(s). From bacteria to humans, these enzymes are critical for maintaining topological homeostasis and ensuring proper function and stability of a dynamic genome. Topoisomerases are classified as either Type I or Type II enzymes depending on whether they cleave one or both strands of DNA, respectively (reviewed in [1]). The Type I enzymes are further subdivided into Type IA and IB enzymes, which differ in their preference for double- versus single-stranded DNA as substrate and in the type of covalent linkage made with a nicked DNA strand. Type IA enzymes prefer single-stranded DNA and hence are specialized to deal with an underwound duplex, which has single-strand character and accumulates negative supercoils. Type IB enzymes only cleave double-stranded DNA [2] and can resolve both positive (overwound DNA) and negative supercoils. Like Type II enzymes, Type IA enzymes form a covalent link between an active-site tyrosine and the 5′-phosphate of a nicked DNA backbone, leaving a 3′-OH on the other side of nick. By contrast, Type IB enzymes form a 3′-phosphotyrosyl link when they nick DNA and leave a 5′-OH on the other side of the nick. In this perspective, we focus on the role of the highly conserved eukaryotic Topoisomerase I (Top1), which is a Type 1B enzyme. The myriad functions of Top1 related to genome stability can be divided into two opposing categories. Top1 is critically important for maintaining genome integrity, especially in areas facing the unique topological challenges associated with transcription. Even very transient breaking of the DNA backbone can be hazardous, however, turning Top1 from a helpful friend into a destabilizing foe that can initiate both small- and large-scale genetic changes. Here, we discuss these opposing roles of eukaryotic Top1.
2. Top1 as a regulator of genome stability
2.1. Top1 and transcription
The movement of the transcription machinery and the obligatory separation of DNA strands create twin domains of positive and negative supercoils ahead of and behind the transcription complex, respectively (Fig. 1; [3]). This necessitates topoisomerase action in order to avoid levels of helical tension that interfere with DNA metabolic processes. In bacteria, for example, activation of a single strong promoter in a plasmid results in negative supercoiling detectable by cruciform-structure formation at AT repeats embedded upstream of the transcribed gene [4]. In yeast, TOP1 deletion results in excessively negative-supercoiled plasmid DNA [5,6], which highlights the key role of Top1 in managing transcription-induced negative supercoiling.
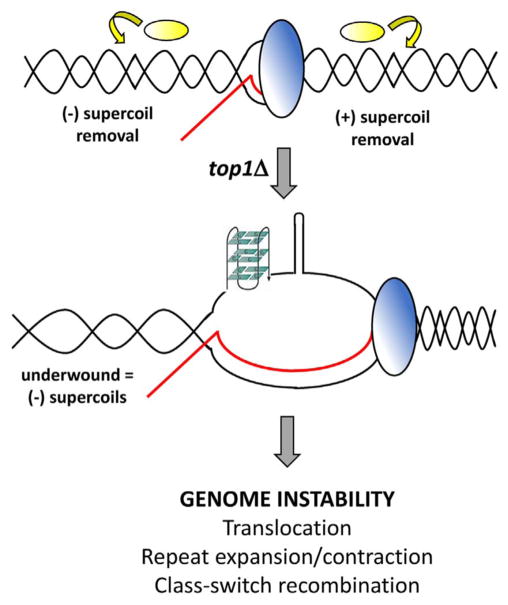
Fig. 1.
Genome stabilization by Top1 during transcription. During normal transcription by RNAP (blue oval), topological homeostasis is maintained by the activity of Top1 (yellow oval). In the absence of Top1, underwound and negatively supercoiled DNA that accumulates behind RNAP supports the formation of co-transcriptional R-loops in which the RNA transcript (red) pairs extensively with the DNA (black) template strand, and the non-template DNA strand is single-stranded. Single-stranded DNA folds into non-B secondary structures such as G4 DNA and hairpins. R-loops and non-B structures initiate genome instability.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A recent yeast study used two closely-spaced promoters to examine how eukaryotic topoisomerases deal with transcription-driven topological changes that have the potential to affect genome stability [7]. Activation of promoters arranged in a divergent configuration led to loss of a terminal segment of the corresponding chromosome arm, which reflects double-strand break (DSB) formation. Activation of two convergently arranged promoters, however, did not have any appreciable effect on such gross chromosomal rearrangements (GCRs). The DSBs initiating the GRC events associated with divergent promoters were attributed to excessive negative supercoils produced when two RNA polymerase (RNAP) complexes move away from each other, supporting the argument that negative torsional stress is the main transcription-associated source of genome instability. Neither loss of Top1 nor reduction of Top2 activity (Top2 is the sole Type II enzyme in yeast and is essential for chromosome segregation; [8]) affected the GCR rate when the promoters diverged. Reduced Top2 activity, however, sharply elevated GCRs when the promoters converged; loss of Top1 had no effect. These results suggest that Top2 can largely complement Top1 function in removing negative supercoils, but that Top1 cannot complement Top2 removal of positive supercoils that are potentially pathological. Top3, a Type IA topoisomerase that mainly functions during the resolution of Holliday junctions formed during homologous recombination [9], does not appear to be involved in regulating transcription-associated topological dynamics.
For transcribed genic regions, the regulation of transcription-associated topological stress by Top1 involves its physical association with the RNAPII complex. In yeast, the Top1 occupancy of a gene correlates with its level of transcription [10], and Top1 specifically interacts with the phosphorylated C-terminal repeat domain of the RNAPII catalytic subunit [11]. In human cell lines, Top1 occupancy also is enriched at highly transcribed genes and the N-terminal domain of Top1 mediates its physical interaction with the C-terminal domain of RNAPII [12]. Importantly, the DNA relaxation activity of Top1 in human cell lines is stimulated by interaction with the phosphorylated C-terminal domain of RNAPII and that this association facilitates promoter escape as well as elongation past natural pause sites. In addition to a direct interaction with RNAPII, human Top1 is recruited to transcriptionally active chromatin via interaction with chromatin remodeling factors [13].
In human cells, Top1 (and Top2) are required for the transcription of extremely long genes that are several hundred kb in length [14], and this could reflect the accumulation of inhibitory positive or negative supercoils. Negative supercoiling promotes the formation of R-loops (see below) whereas excessive positive supercoiling prevents the continued unwinding of DNA. Intriguingly, transcription-associated Top1 effects have been linked to several human neurological diseases, suggesting that the use of topoisomerase inhibitors could have therapeutic value [15]. In neurons, for example, genes linked to autism spectrum disorder are extremely long, and their expression is reduced by Top1 inhibitors [14]. Furthermore, a reduction in Top1 activity reduces expression of an anti-sense transcript that silences the paternal Ube3a allele in Angelman syndrome [16].
2.2. Top1 and RNA-DNA hybrids
The transcription bubble within elongating RNAP is ~15 nt, with pairing between the nascent transcript and template DNA strand extending 9 bp [17]. More extensive and stable hybridization between the template DNA strand and the nascent RNA can occur through a threadback mechanism after the transcript and duplex DNA exit the RNAP through separate channels [18]. The structure thus generated (a stable RNA:DNA hybrid and the displaced, non-template single strand) is referred to as an R-loop and occurs co-transcriptionally when RNA processing is disrupted. Reduction of the THO/TREX complex involved in mRNA packaging/export or the ASF1/SF2 splicing factor, for example, leads to transcription-dependent accumulation of extensive R-loops [19,20]. R-loops can be removed by the RNA:DNA hybrid-specific ribo-endonucleases RNase H1 and H2, or by the RNA:DNA helicases senataxin (SETX) and aquarius (AQR); disruption of any of these factors leads to accumulation of RNA:DNA hybrids [21–23]. Although R-loops have physiological functions (e.g., in transcription termination), they have emerged as an important pathological structure that instigates genome instability through disruption of transcription and replication (reviewed in [22,24,25]).
Negative torsional stress favors R-loop formation, and preventing the accumulation of R-loops is an important role of Type I enzymes that is conserved from prokaryotes to mammalian systems (Fig. 1). In E. coli, disruption of the Type IA topoisomerase topA leads to growth defects that are rescued by overexpression of RNase H, indicating RNA:DNA hybrid accumulation as the major consequence of excessive negative supercoils [26–28]. In yeast, loss of Top1 is associated with transient melting of the DNA duplex and R-loop accumulation in the very highly transcribed ribosomal RNA genes [29,30]. The accumulation of R-loops and the disruption of transcription worsens in top1Δ cells when the degradation of RNA:DNA hybrids is blocked by additional depletion of RNase H enzymes. In yeast cells lacking both RNase H1 and H2, the loss of Top1 is lethal [29]. Genome-wide mapping of R-loops using the RNA:DNA hybrid-specific monoclonal antibody S9.6 showed that conditional depletion of Top1 in yeast cells deficient in both RNase H1 and H2 leads to further accumulation of R-loops throughout the genome, especially at ribosomal RNA and transfer RNA genes [31]. A more recent study identified two additional features that predispose genomic regions to R-loop formation in the absence of RNase H1/H2: either high expression or presence of polyA tracts [32]. Whereas yeast cells tolerate the loss of Top1, RNase H1 or RNase H2 individually and in some combinations, each is an essential enzyme in mammals.
The function of Top1 in preventing R-loop accumulation is conserved in mammalian systems. Top1-deficient mouse lymphocytes accumulate stalled replication forks and DNA breaks at actively transcribed regions [33]. Both replication-fork stalling and DNA breaks are reduced by overexpression of RNase H1, implicating transcription-associated R-loops as the source of replication conflicts in these cells. In addition, treatment of post-mitotic neurons that are no longer replicating DNA with the Top1-inhibitor camptothecin (CPT) leads to activation of the ATM-dependent DNA damage response and formation of γH2AX foci, which are indicative of DNA DSBs [34]. In CPT-treated cells, either inhibition of transcription by α-amanitin or overexpression of RNase H1 prevents the damage response activation and γH2AX foci formation. R-loop accumulation upon Top1-inhibition can thus induce DNA breaks independent of replication. In mammalian cells, Top1 has kinase activity that regulates members of the SR family of splicing factors, including ASF1/SF2, through phosphorylation [33,35]. Depletion of ASF1/SF2 results in R-loop accumulation and an increase in transcription-associated genome instability [20], and inhibition of Top1 kinase activity by diospyrin treatment elevates DNA breaks in Top1-proficient cells. In Top1-deficient human cells, DNA breaks are suppressed by RNase H1 overexpression or by transcription-terminating nucleoside analogs, but are not fully suppressed by the expression of yeast Top1, which lacks the kinase domain. The kinase activity of mammalian Top1, although not involved in directly managing the topological state of transcribed regions, is nevertheless important for suppressing R-loop mediated, transcription-associated genome instability.
2.3. Top1 and non-B DNA
Despite the clear evidence that Top1 is important for regulating transcription-associated torsional stress, several studies measuring loss of heterozygosity (LOH) or gross chromosomal rearrangements have failed to detect a significant increase in yeast cells lacking Top1 [36–38]. However, other reports summarized below indicate that Top1 loss acutely affects the stability of genomic loci prone to forming DNA secondary structures. Double-stranded, B-form DNA is the low-energy, preferred conformation, even for repetitive sequences. Accumulation of torsional stress associated with transcription can shift the equilibrium toward alternative, non-B, structures such as hairpins, cruciform DNA, triplex DNA (H-DNA), and G-quadruplex (G4) DNA (reviewed in [39]). R-loop accumulation in the absence of Top1 and alternative DNA-structure formation are inter-related. During transcription, the existence of the non-transcribed strand in a single-stranded state is minimal and transient, precluding intra-strand interactions. Extensive and stable RNA-DNA hybrid (i.e. R-loop) formation, however, affords an opportunity for the non-transcribed strand to assume non-B secondary structures. Sequestration of the non-transcribed strand in a stable, non-B DNA conformation makes it inaccessible for pairing with the complementary, transcribed strand and leads to R-loop stabilization. The negative helical tension in DNA produced by transcription and maintained in the absence of functional Top1 favors the formation of both R-loops and non-B DNA, which then mutually promote stabilization of each other. Accordingly, Top1-regulated maintenance of topological homeostasis in transcribed regions is most consequential when there is the potential to form non-B structures (Fig. 1).
In the neurological disorders Huntington’s disease, Friedreich’s ataxia and fragile X mental retardation syndrome, disease pathology reflects the expansion of tri-nucleotide repeats (TNRs) CAG•CTG, GAA•TTC and CGG•CCG, respectively, in the corresponding disease gene (reviewed in [40]). These TNRs potentially assume non-B secondary structures such as slipped hairpins (CAG•CTG, CGG•CCG) and H-DNAs (GAA•TTC), and the expansion of TNRs increases the stability of these non-B conformations. In the context of plasmid DNA in Escherichia coli, GAA•TTC and CGG•CCG repeats become unstable when negative supercoils are induced by either transcription or by mutation in topA [41]. In human fibrosarcoma cell lines, TNR instability in the form of repeat contraction is significantly enhanced when Top1 function is reduced either by chemical inhibition or by siRNA-mediated knock-down [41,42]. As discussed for R-loop accumulation, the effect of Top1 depletion is made more severe by transcription through the TNRs, leading to a model in which excess negative torsional stress promotes non-B structure assembly that results in TNR instability. An alternative mechanism was suggested by the finding that disruption of Tdp1, a tyrosyl-DNA phosphodiesterase that removes the Top1-DNA adduct, leads to similar TNR instability. These mechanisms are not necessarily mutually exclusive, and further investigation will be needed to determine whether the effect of Top1-depletion on TNR stability is directly linked to accumulation of transcription-driven negative supercoils and/or to Top1-DNA adduct formation.
A strong correlation between non-B, extrahelical structures and Top1 has been demonstrated for sequences capable of adopting a G4-DNA structure. G4 DNA is a stable, four-stranded, ring-like structure containing multiple guanines that interact via Hoogsteen bonds. With regard to genome stability, G4 DNA can block critical cellular processes such as replication and transcription [43–46]. When a sequence with multiple runs of guanines is transcribed, the failure to remove transcription-associated helical stress is expected to shift the equilibrium toward G4 DNA formation and genome instability ensues. Studies in budding yeast have provided support for this model of topology-driven genome instability [47–49]. When G4-forming guanine runs were on the non-transcribed DNA strand, activation of transcription stimulated recombination, which was further increased upon deletion of TOP1. The effect of Top1 loss was not evident, however, when the direction of transcription was reversed so that the guanine runs were on the transcribed strand, likely in stable base-pairing with the nascent transcript. The over expression of E. coli topA reduced recombination associated with the actively transcribed G4 sequence, indicating that it is the Top1 removal of negative supercoils that prevents formation of G4 DNA [49]. In regions with highly negative topological tension, G4 DNA is very stable and apparently does not disassemble when the complementary DNA strand becomes available for pairing through RNase H1 destruction of the RNA:DNA hybrid.
G4 DNA is unique among non-B secondary DNA structures because Top1 specifically binds to G4 with high affinity [50–52]. This property of Top1 explains why mutation at the catalytic tyrosine (Y727F of yeast Top1) elevates G4-associated genome instability more than does complete deletion of the TOP1 gene [49]. Because the Top1-Y727F protein retains its DNA binding function, its unexpected deleterious effect with regard to G4 stability may reflect high-affinity interaction between Top1 and G4 DNA. Alternatively, the observed effect could be due to the sequestering and reduced function of Top1-interacting proteins. As further confirmation that Top1 reduces the adverse effect of G4 DNA, loss of Top1 significantly increases sensitivity of yeast cells to the G4-stabilizing ligand TmPyP4 [53]. Defining how the G4 binding properties of Top1 affect genome instability is an important area of future study. In addition, the specific interaction of Top1 with G4 DNA suggests it may be feasible to use G4-forming oligonucleotides as Top1 inhibitors.
2.4. Top1 and antibody diversification
Top1 is important in regulating acquired immunity in vertebrates and this appears to be closely related to its role in transcription-associated genome instability. During B lymphocyte development, immunoglobulin (Ig) genes encoding antigen receptor proteins or antibodies undergo several critical molecular changes. V(D)J recombination and somatic hypermutation occur for both heavy- and light-chain Ig genes, while class switch recombination (CSR) changes the Ig isotype and is unique to the heavy-chain locus [54]. CSR, similar to somatic hypermutation, is activated by antibody encounter with a cognate antigen and is initiated by AID (Activation-Induced cytosine Deaminase)-catalyzed conversion of cytosine to uracil in switch-region sequences (e.g., Sμ, Sγ, Sα). Following uracil excision and cleavage of the DNA backbone to create DSBs, the joining of two switch sequences deletes the sequence in between. The functional consequence of CSR is to change the antibody constant region that interacts with downstream effectors, while the antigen-interacting variable region remains unaltered. Mutation of the AID-encoding gene and the resulting defect in CSR manifest as a hyper-IgM syndrome characterized by acute susceptibility to opportunistic infections [55,56].
The potential role of Top1 in managing CSR emerged from the finding that expression of AID in B lymphocytes undergoing CSR correlates with the down-regulation of Top1 [57]. Further depletion of Top1 using siRNA resulted in significantly elevated CSR [58]. How can the down-regulation of Top1 effectively increase recombination between two switch-region sequences? One possible mechanism is that negative torsional stress accumulates at the switch regions when Top1 activity is reduced, thereby increasing deamination by AID. Although AID preferably acts on cytosines in single-strand DNA, it can deaminate cytosines in double-strand DNA that is supercoiled [59]. Alternatively, the correlation between the reduced level of Top1 and CSR efficiency could be related to another required element of the CSR process. There is a germline promoter upstream of each switch-region sequence [60], and its activation produces sterile (non-protein coding) transcripts required for CSR. The outcome of switch-region transcription is elevated topological strain that can have two consequences: first, to more efficiently target AID to cytosines on both strands of the switch region and second, to increase R-loop formation and stabilize the single-stranded character of the non-transcribed strand.
Extensive R-loop formation at activated switch regions in B lymphocytes as well as during in vitro transcription has been reported [18]. In addition, switch-region sequences are guanine-rich and have the potential to form G4 DNA, with the G-rich sequences located asymmetrically on the non-transcribed strand [43,61]. When these sequences are transcribed either in vitro or in E. coli cells, complex structures consisting of non-transcribed strand G4 DNA and a hybrid between the transcribed strand and the nascent RNA are visible by electron microscopy [61]. The requisite transcription of the switch regions in antigen-activated B cells will promote both R-loop and G4-DNA formation, and reduced Top1 activity is expected to increase both. In budding yeast, transcription of a fragment of the murine Sμ sequence integrated into the genome in its physiological orientation stimulates recombination [62]. Recombination is further elevated in a transcription- and orientation-specific manner in the absence of Top1. Whether the connection between CSR and Top1 is related to its role in preventing the formation of R-loops or of G4 DNA is an issue that needs further work to resolve. Another open question is whether G4-DNA formation affects CSR by modulating the efficiency of AID activity or by another yet-to-be identified pathway.
2.5. Top1 and oncogenic chromosomal translocations
Altered transcription patterns of oncogenes are often associated with the early stages of cancer development. The intimate connection between Top1 function and transcription broadly suggests a putative role for Top1 in the molecular changes associated with the early steps of cell transformation. A recent survey of ~20,000 cancer-associated translocations revealed a highly significant enrichment of G4-DNA forming sequences within 500 bp of the translocation breakpoints [63]. For blood cancers, nearly 70% of the genes involved in chromosomal rearrangements contain potential G4-DNA sequences. This correlation was earlier suggested by a smaller scale analysis that identified G4 motifs near many of the recurrent chromosomal translocation breakpoints in hematopoetic cancers, including those involving the BCL2, c-MYC, TCF3 (E2A) and k-RAS genes [64]. G4-forming IgH switch regions are also frequently involved in recurrent chromosomal translocations observed in multiple myeloma and Burkitt’s lymphoma, which are presumably an aberrant consequence of the CSR process [65,66]. Recent findings that ascribe a critical function for Top1 in maintaining stability of G4-forming sequences implicate Top1 in suppressing the translocations observed in multiple cancers, particularly various types of leukemia and lymphoma. The significance of Top1 in suppressing the accumulation of R-loop and/or G4-DNA structures at the switch regions is expected to be relevant to the effectiveness of acquired immunity as well as the development of B cell-derived malignancies. Indeed, the IgH-cMYC translocation frequency, along with CSR efficiency, was recently shown to be significantly elevated by knock-down of Top1 or SMARCA4, a chromatin remodeler responsible for recruiting Top1 to IgH switch regions in an AID-expressing B-cell line [13]. Making an indisputable connection between Top1 regulation in cancer development and its role as a regulator of genome instability at G4 sequences will require further research.
3. Top1 as an instigator of genome instability
As elaborated above, Top1 is critically important for maintaining genome integrity, especially in regions that face the unique topological challenges associated with transcription. The transient Top1-mediated cleavage of the DNA backbone, however, can also initiate mutagenic processes. Top1 is often overexpressed in human tumors [67,68] and an elevated Top1 level is presumably needed to sustain rapid growth. This not only enhances the sensitivity of tumor cells to Top1 inhibitors, it also may drive Top1-mediated instability that contributes to tumor evolution. It is thus important to understand how Top1 contributes to genetic instability. The covalent Top1-DNA cleavage intermediate is referred to as the Top1 cleavage complex (Top1cc). The equilibrium between Top1-mediated cleavage and religation strongly favors ligation, which historically has made it difficult to map Top1 cleavage sites in vitro. This equilibrium can be shifted, however, by the camptothecin (CPT) class of chemotherapeutic drugs or by DNA distortion near the cleavage site. CPT and its derivatives are interfacial, planar molecules that reversibly insert between the enzyme-DNA phosphotyrosyl bond and the 5′-OH, thereby blocking the nucleophilic attack required for religation (reviewed in [69]). By contrast, nearby helical distortions associated with DNA damage or mismatched bases result in misalignment of the 5′-OH relative to the phosphotyrosyl bond [70][118]. This misalignment not only reduces the probability of correct religation, it has the potential to promote ligation to a completely different 5′-OH. Stabilization/trapping of the Top1cc additionally may interfere with or preclude the repair of DNA lesions [71]. Top1 overexpression sensitizes yeast cells to a variety of DNA-damaging agents [72,73] and Top1 is partially responsible for hydrogen peroxide sensitivity in mammalian cells [73].
The encounter of a stabilized Top1cc-associated nick with a replication fork leads to potentially lethal DSBs, which arise either via replication run-off or as a result of torsion-driven fork reversal and processing [74–77]. In addition, an RNA polymerase encounter with a Top1cc on the transcribed strand creates a permanent single-strand break in vitro [78], which in vivo would subsequently be converted to a DSB by replication. Finally, Top1 can directly generate DSBs if it cleaves opposing DNA strands in close proximity, or if it cleaves opposite a nick. A Top1-generated DSB can be repaired via high-fidelity homologous recombination or error-prone non-homologous end joining (NHEJ), each of which requires processing of the Top1-generated breaks into clean DNA ends that can be extended and/or ligated (reviewed in [79]). Finally, Top1-generated single-strand nicks are the direct initiator of small deletions in budding yeast, and these nicks most often occur at sites of ribonucleotide incorporation into DNA (reviewed in [80]). Below, we review the types of genetic alterations that result from Top1-generated single-strand nicks and double-strand breaks.
3.1. Illegitimate recombination
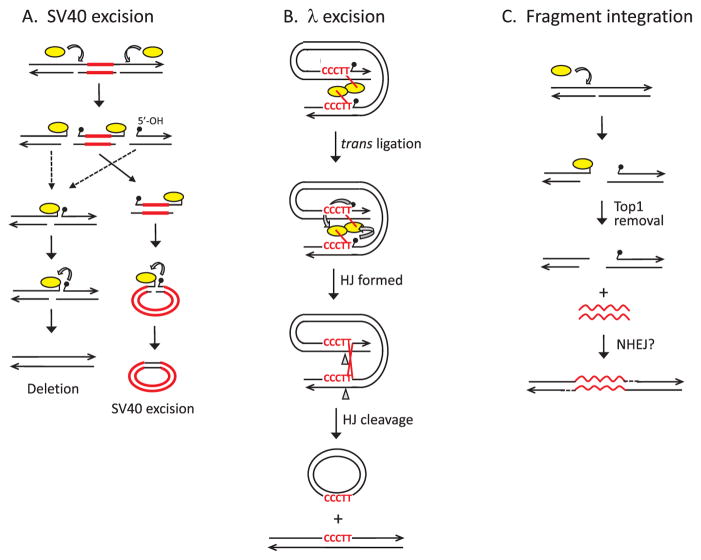
Illegitimate recombination (IR) broadly refers to recombination that is not driven by extended sequence homology and does not require a RecA-type strand-exchange protein. Examples include transposition and site-specific recombination, as well as NHEJ. Top1-mediated IR can be intra- or inter-molecular, and occurs when a donor duplex with trapped Top1cc at its end is ligated to an acceptor molecule containing a 5′-OH (reviewed in [81]). The first indication that Top1 activity promotes IR came through analysis of crossover junctions associated with the excision of virus SV40 in a rat cell line [82]. The crossover junctions contained several base pairs of homology, and when examined in vitro, coincided with Top1 cleavage sites. The model that emerged is shown in Fig. 2A and proposes that Top1 cleavage generates DSBs that flank the SV40 genome. Both cleavage sites are on the same DNA strand, and Top1 cleavage opposite a nick or gap in the non-scissile strand creates the initiating DSBs. The excised portion contains a Top1cc at one end and a 5′-OH at the other end, and can be circularized by Top1-mediated ligation. Although the focus was on the SV40 circularization junctions, the broken chromosome could similarly be “healed” by Top1-mediated ligation or by NHEJ. While this example specifically involves SV40 excision, a similar mechanism could be a potent source of spontaneous chromosomal deletions. Support for this comes from CPT-induced mutations in mammalian cells, which are often deletions or rearrangements [83,84]. In addition, NHEJ-dependent deletions in yeast are associated with the expression of a mutant form of Top1 that has reduced religation activity (J.E. Cho and S. Jinks-Robertson, unpublished).
Fig. 2.
Top1-mediated genome instability. (A) Top1 (yellow oval) catalyzes excision of SV40 (red lines) from the genome. Cleavage of Top1 opposite a nick generates a DSB with the enzyme trapped at the ends. The relevant cleavages occur on the same DNA strand so that the excised fragment contains a Top1cc at one end and a 5′-OH (small black popsicle) at the other. Attack of the Top1-DNA linkage by the 5′-OH produces a nicked circular molecule, and the similar reaction can seal the broken chromosome. (B) Excision of λ from the E. coli genome by vcTop1. vcTop1 cleaves at 5′-CCCTT consensus sites that flank the λ genome. The vcTop1 complexes interact to facilitate trans ligation in which the 5′-OH generated by one enzyme attacks the phosphotyrosyl bond of the other. This creates a Holliday junction (HJ) that is resolved into crossover products by nicking and ligating the complementary, 5′-AAGGG-containing strands. (C) Top1 cleaves opposite a nick, generating a genomic DSB. Exogenous DNA (wavy red lines) is then joined to the broken ends by the NHEJ pathway or by Top1-mediated ligation (not shown). Black lines correspond to single DNA strands, with arrowheads indicating 3′ ends.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A second example of Top1-mediated IR was observed following expression of vaccinia virus Top1 (vcTop1) in E. coli. A unique feature of vcTop1 is that it has a very well-defined and invariant cleavage site, 5′-(C/T)CCTT-3′ [85], and expression of vcTop1 was found to stimulate integrase-independent excision of a lambda (λ) prophage from the bacterial genome [86]. Significantly, the sequence of the phage at the site of excision matched the vcTop1 consensus site; a consensus site was also present on each side of the original integrated prophage. It was proposed that each flanking chromosomal site is cleaved by synapsed Top1 monomers, and that religation involves attack by the 5′-OH generated by the other enzyme (Fig. 2B). This reaction is analogous to the first of the two cleavages and trans ligations carried by site-specific recombinases, which create and then resolve a Holliday junction as a crossover event [87]. In the case of vcTop1-catalyzed λ excision, Holliday junction resolution by vcTop1, which has been observed in vitro [88], would simply reverse the initial reaction. Cleavage by an endogenous Holliday junction resolvase (e.g., RuvC), however, could generate the complete excision/crossover product.
A third example of Top1-mediated IR was from transformation studies in yeast that examined the integration of DNA fragments that contain no homology to the genome (Fig. 3C). Some of the integration events were enzyme-mediated, with a duplicated restriction site flanking the insertion, while others occurred where there is no homology [89]. The latter class occurred mostly at CTT or GTT sites that match the weak consensus cleavage site of Top1, which is 5′-(A/T) (G/C) (A/T) T-3′ for mammalian Top1 [90]. Most importantly, these IR events were reduced in a top1Δ background and were elevated upon overexpression of yeast or human Top1 [91,92], suggesting integration at sites of Top1-generated DSBs. Although a requirement for de-phosphorylation of the transforming DNA ends supports a Top1-mediated ligation mechanism, de-phosphorylation also would reduce circularization of the transforming fragment and might thereby promote its integration [92]. Because IR events in this system are strongly dependent on the Rad50 protein [93], which is required for the NHEJ pathway in yeast [94], fragment integration likely occurs via NHEJ. Interestingly, the yeast IR events are sometimes associated with the “capture” of mitochondrial DNA fragments, as well as the transforming DNA, at the integration site [89]. When considered in total, the available data suggest that Top1 initiates insertions and deletions, some of which depend on Top1-mediated ligation and others of which likely involve NHEJ. Though not yet reported, it is expected that additional rearrangements such as inversions and translocations will also reflect Top1 activity. With an appropriate selective system, it should be possible to observe these events as well.
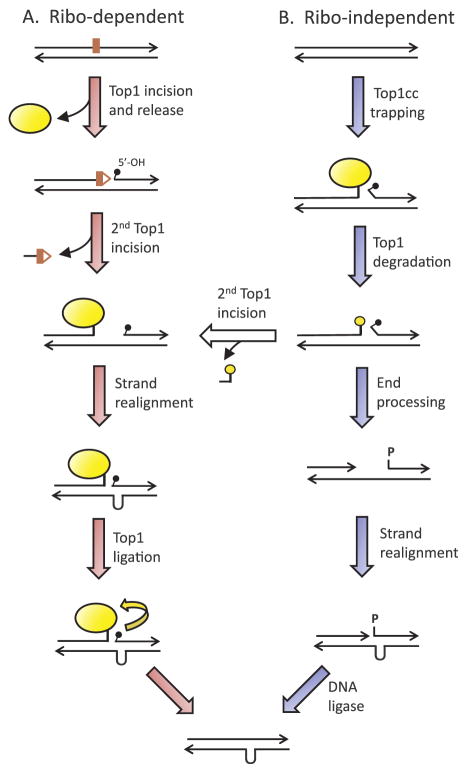
Fig. 3.
Mechanisms of Top1-dependent short deletions. (A) Ribo-dependent deletions are initiated when Top1 (yellow oval) cleaves at a ribo and is released from the DNA, leaving 2′,3′-cyclic phosphate (red triangle) and 5′-OH (small black popsicle) ends. A second Top1 cleavage upstream releases a small oligonucleotide and traps the enzyme. Repeat-mediated realignment of complementary strands brings the 5′-OH made by the first cleavage into the correct alignment for efficient enzyme-mediated ligation. (B) Ribo-independent deletions initiate with stabilization of a Top1cc. Following degradation of Top1, a second enzyme cleaves upstream and deletion formation proceeds as for ribo-dependent deletions. Alternatively, the ends can be processed into a gap flanked by a 3′-OH and 5′-phosphate. Realignment between complementary strands converts the gap to a nick that is sealed by ligase. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Homologous recombination
Ribonucleotides (ribos) are an abundant component of newly-synthesized eukaryotic DNA [95,96], and either are due to the sporadic incorporation of rNMPs or represent remnants of the RNA primers used to initiate DNA synthesis. Both RNase H1 and H2 degrade the RNA component of R-loops, but only RNase H2 can incise at the position of one or a few ribos embedded within duplex DNA [21]. RNase H2 incises 5′ of a ribo and initiates a ribonucleotide excision repair pathway in which the ribo is displaced and removed as part of a 5′-flap generated by nick translation [97]. The genetic consequences of RNase H2 loss have been studied in detail in budding yeast, where the enzyme is not required for viability (reviewed in [98]). Ribos that persist in yeast genomic DNA are associated with replication stress [99], elevated homologous recombination [100,101] and an increase in deletions within tandem repeats [102]. All of these ribo-associated effects are partially or completely dependent on Top1 activity [36,99,103].
Prior to the yeast studies, the biochemical consequence of Top1 incision at a single ribo had been determined using vcTop1 [104]. Substitution of a ribo for the terminal, scissile thymidine of the 5′-CC-CTT consensus sequence resulted in cleavage associated with release of the enzyme rather than religation. Release was the result of attack of the Top1-DNA phosphotyrosyl bond by the vicinal 2′-OH of ribose, leaving a nick flanked by a 2′,3′-cyclic phosphate and a 5′-OH. A similar reaction is catalyzed by yeast or human Top1, and is reversible [105–107]. In terms of ribo-stimulated homologous recombination, it recently was demonstrated that Top1 can directly generate DSBs by cleaving on opposing DNA strands in vitro. In this case, Top1 generates a nick at the site of a ribo on one strand, and then nicks the opposing strand to generate a DSB [108].
3.3. Short deletions in tandem repeats
As noted previously, Top1 is recruited to transcriptionally active DNA to relieve the associated torsional stress, and this occurs through direct interaction with both elongating RNAPII and chromatin modifications. In budding yeast, high levels of transcription are associated with elevated mutagenesis, and ~50% of transcription-associated mutations require the activity of Top1 [10,109]. Top1 generates a distinctive mutation signature comprised of short deletions within a tandem repeat containing only 2–4 repeat units. Not all such repeats accumulate Top1-dependent deletions, however, with events occurring at defined hotspots. The lack of an effect of either the homologous recombination or NHEJ machinery on Top1-dependent deletions suggested that the events reflect a single-strand nick rather than a DSB intermediate. Furthermore, the fact that the size of the deletion always corresponds precisely to the size of the repeat unit suggested a model in which Top1 (or the processing of the “dirty” ends it creates) produces a short gap that is converted into a ligatable nick by repeat-mediated realignment of complementary strands. Although Top1-dependent deletions were initially discovered as a signature of high transcription, this is not a prerequisite; with a more sensitive, selective assay, they are readily detected under low-transcription conditions as well [109].
RNase H2 loss is associated with a 2–5 bp mutation signature identical to that associated with high transcription [102], and ribo-initiated deletions are completely dependent on Top1 activity [103]. The activity of discrete Top1-dependent deletion hotspots in the presence of a mutant form of Top1 (Top1-T722A, which has reduced religation activity; [110]) or upon genetic manipulation of ribo levels in DNA suggests there are two types of hotspots [111]. Most Top1-dependent deletions initiate at ribos and are referred to as ribo-dependent; the remainding events are ribo-independent and presumably reflect the processing of a trapped Top1cc intermediate.
In vivo analysis of a ribo-dependent hotspot in a top1-T722A background revealed that the religation activity of Top1 is important for deletion formation [111], providing support for a sequential-cleavage model of deletion formation (Fig. 3A). In this model, Top1 incision at and release from a ribo is followed by a second Top1 incision within a few nucleotides 5′ of the ribo-associated nick. The short, intervening oligoncleotide is spontaneously released, trapping the Top1cc on the 5′ side of a small gap that is also flanked by a 5′-OH. The significance of the tandem repeat is that it allows realignment of complementary strands, which moves the 5′-OH next to the Top1cc and into the correct alignment for efficient, Top1-mediated ligation. Ligation can be prevented by the combined action of the Srs2 3′ > 5′ helicase and Exo1 5′ > 3′ exonuclease, which together unwind and remove the 5′-OH [101].
The sequential cleavage model has been confirmed in vitro using purified yeast or human Top1 and DNA fragments containing 2-bp deletion hotspots identified in vivo [106,107]. Furthermore, these studies support the importance of base-pairing of the re-located 5′-OH with the complementary strand for efficient ligation. Whereas base pairing of the 5′-OH acceptor end next to the Top1cc donor end is important for intra-molecular ligation, inter-molecular ligation requires no homology when the acceptor end is double-stranded [112]. In vitro, Top1 can generate insertions as well as deletions if a 5′-OH is available [113], but only deletions have been reported in vivo.
A duplication of the 5-bp vcTop1 cleavage site [(CCCTT)2] in a selective yeast system demonstrated that yeast Top1 generates ribo-dependent 5-bp deletions, with cleavage at each terminal T detected in parallel in vitro analyses [114]. Varying the distance between the cleavage sites changes the position between Top1-generated nicks in vitro and similarly alters the size of the deletion in vivo, confirming that the distance between the nicks determines deletion size. This approach was used to extend the deletion size to 7-bp in vivo, but whether larger deletions can be generated via the sequential cleavage mechanism has not been examined. Finally, a duplication of the CCCTT sequence with the ribo placed at the downstream cleavage site did not produce the full deletion reaction in vitro, while a triplication of the site with the ribo in the middle repeat supported robust 5-bp deletion formation. The difference between the duplication and the triplication is that the Top1-linked end must re-align to base pair with the gap when there is a duplication, while either end can realign when there is a triplication. This behavior is consistent with the tight clamping of Top1 around the duplex seen in crystal structures [115], which likely constrains strand realignment.
Although the molecular mechanism of ribo-dependent deletions has been well established, an interesting question is whether the same enzyme is responsible for both cleavages. In the case of ribo-independent deletions, the molecular mechanism remains unknown. As originally envisioned, it is possible that only a single Top1 cleavage is involved in these events, with subsequent end-processing producing a gap and ligase performing the final ligation (Fig. 3B). Alternatively, ribo-independent events could also involve a sequential Top1 cleavage mechanism, with the enzyme being trapped by the first cleavage. A second Top1 can interact with a Top1cc in vitro and cleaves 10–15 nt upstream, and it has been suggested that this could initiated removal of the Top1cc [116]. To access a second cleavage site only a few base pairs away, however, the Top1cc would have to be mostly or completely removed, and there are multiple activities that could potentially do this [79,117]. At this point, the mechanism of ribo-independent deletions converges with that of ribo-dependent deletions. A challenge of future studies will be to understand the molecular mechanism of ribo-independent deletions initiated by Top1, and whether Top1 initiates similar ribo-dependent and ribo-independent events in mammalian cells.
4. Summary
Top1 relieves transcription-associated torsional stress that can both interfere with continued gene expression and trigger genome instability. Genetic instability associated with reduced Top1 activity largely reflects the accumulation of negative supercoils, which promote the formation of co-transcriptional R-loops and stable non-B DNA structures. Both R-loops and non-B structures interfere with DNA replication and are associated with the formation of potentially toxic DSBs. Although reduced Top1 activity generally has pathological consequences, down-regulation of Top1 appears to be physiologically relevant during development of the immune system. Recent studies also suggest that a targeted reduction in Top1 activity, and the associated modulation of transcription, may have therapeutic applications in some neurological diseases. The management of torsional stress by Top1 requires that it nick and reseal duplex DNA, and this catalytic cycle is typically of no genetic consequence. The robust ability of Top1 to join non-cognate ends, however, has been implicated in various types of illegitimate recombination events. In addition, the single-strand nicks created by Top1 initiate a novel form of mutagenesis that is characterized by short deletions in low copy-number tandem repeats. It is likely that additional examples of the opposing roles of Top1 in shaping genome structure will emerge over the coming years.
Acknowledgments
Work in the SJR lab was supported by a grant from the National Institutes of Health (1R35-GM119077). NK was supported by grants from the Welch Foundation (AU1875) and the National Institutes of Health (1R01-GM116007).
Abbreviations
- Top1
topoisomerase I
- DSB
double-strand break
- RNAP
RNA polymerase
- GCR
gross chromosomal rearrangement
- CPT
camptothecin
- G4
guanine quadruplex
- TNR
trinucleotide repeat
- CSR
class-switch recombination
- AID
activation-induced cytosine deaminase
- NHEJ
non homologous end joining
- IR
illegitimate recombination
- ribo
ribonucleotide
- Top1cc
Top1 cleavage complex
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Been MD, Champoux JJ. Breakage of single-stranded DNA by eukaryotic type 1 topoisomerase occurs only at regions with the potential for base-pairing. J Mol Biol. 1984;180:515–531. doi: 10.1016/0022-2836(84)90025-1. [DOI] [PubMed] [Google Scholar]
- 3.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 4.Krasilnikov AS, Podtelezhnikov A, Vologodskii A, Mirkin SM. Large-scale effects of transcriptional DNA supercoiling in vivo. J Mol Biol. 1999;292:1149–1160. doi: 10.1006/jmbi.1999.3117. [DOI] [PubMed] [Google Scholar]
- 5.Brill SJ, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54:403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez X, Diaz-Ingelmo O, Martinez-Garcia B, Roca J. Chromatin regulates DNA torsional energy via topoisomerase II-mediated relaxation of positive supercoils. EMBO J. 2014;33:1492–1501. doi: 10.15252/embj.201488091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pannunzio NR, Lieber MR. Dissecting the roles of divergent and convergent transcription in chromosome Instability. Cell Rep. 2016;14:1025–1031. doi: 10.1016/j.celrep.2015.12.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 9.Bizard AH, Hickson ID. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol. 2014;6:a016477. doi: 10.1101/cshperspect.a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2011;108:692–697. doi: 10.1073/pnas.1012582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry. 2004;43:15702–15719. doi: 10.1021/bi048364h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baranello L, Wojtowicz D, Cui K, Devaiah BN, Chung HJ, Chan-Salis KY, Guha R, Wilson K, Zhang X, Zhang H, Piotrowski J, Thomas CJ, Singer DS, Pugh BF, Pommier Y, Przytycka TM, Kouzine F, Lewis BA, Zhao K, Levens D. RNA polymerase II regulates ropoisomerase 1 activity to favor efficient transcription. Cell. 2016;165:357–371. doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Husain A, Begum NA, Taniguchi T, Taniguchi H, Kobayashi M, Honjo T. Chromatin remodeller SMARCA4 recruits topoisomerase 1 and suppresses transcription-associated genomic instability. Nat Commun. 2016;7:10549. doi: 10.1038/ncomms10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King IF, Yandava CN, Mabb AM, Hsiao JS, Huang HS, Pearson BL, Calabrese JM, Starmer J, Parker JS, Magnuson T, Chamberlain SJ, Philpot BD, Zylka MJ. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501:58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKinnon PJ. Topoisomerases and the regulation of neural function. Nat Rev Neurosci. 2016;17:673–679. doi: 10.1038/nrn.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, Sciaky N, Dutton JW, Jr, Lee HM, Chen X, Jin J, Bridges AS, Zylka MJ, Roth BL, Philpot BD. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2011;481:185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Silva DA, Pardo-Avila F, Wang D, Huang X. Structural model of RNA polymerase II elongation complex with complete transcription bubble reveals NTP entry routes. PLoS Comput Biol. 2015;11:e1004354. doi: 10.1371/journal.pcbi.1004354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol. 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rondon AG, Jimeno S, Aguilera A. The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim Biophys Acta. 2010;1799:533–538. doi: 10.1016/j.bbagrm.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamperl S, Cimprich KA. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Rep. 2014;19:84–94. doi: 10.1016/j.dnarep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baaklini I, Hraiky C, Rallu F, Tse-Dinh YC, Drolet M. RNase HI overproduction is required for efficient full-length RNA synthesis in the absence of topoisomerase I in Escherichia coli. Mol Microbiol. 2004;54:198–211. doi: 10.1111/j.1365-2958.2004.04258.x. [DOI] [PubMed] [Google Scholar]
- 27.Masse E, Phoenix P, Drolet M. DNA topoisomerases regulate R-loop formation during transcription of the rrnB operon in Escherichia coli. J Biol Chem. 1997;272:12816–12823. doi: 10.1074/jbc.272.19.12816. [DOI] [PubMed] [Google Scholar]
- 28.Phoenix P, Raymond MA, Masse E, Drolet M. Roles of DNA topoisomerases in the regulation of R-loop formation in vitro. J Biol Chem. 1997;272:1473–1479. doi: 10.1074/jbc.272.3.1473. [DOI] [PubMed] [Google Scholar]
- 29.El Hage A, French SL, Beyer AL, Tollervey D. Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.French SL, Sikes ML, Hontz RD, Osheim YN, Lambert TE, El Hage A, Smith MM, Tollervey D, Smith JS, Beyer AL. Distinguishing the roles of topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol Cell Biol. 2011;31:482–494. doi: 10.1128/MCB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Hage A, Webb S, Kerr A, Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mi-tochondria. PLoS Genet. 2014;10:e1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016;30:1327–1338. doi: 10.1101/gad.280834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, Pommier Y, Tazi J, Coquelle A, Pasero P. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sordet O, Redon CE, Guirouilh-Barbat J, Smith S, Solier S, Douarre C, Conti C, Nakamura AJ, Das BB, Nicolas E, Kohn KW, Bonner WM, Pommier Y. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 2009;10:887–893. doi: 10.1038/embor.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuduri S, Crabbe L, Tourriere H, Coquelle A, Pasero P. Does interference between replication and transcription contribute to genomic instability in cancer cells? Cell Cycle. 2010;9:1886–1892. doi: 10.4161/cc.9.10.11539. [DOI] [PubMed] [Google Scholar]
- 36.Conover HN, Lujan SA, Chapman MJ, Cornelio DA, Sharif R, Williams JS, Clark AB, Camilo F, Kunkel TA, Argueso JL. Stimulation of chromosomal rearrangements by ribonucleotides. Genetics. 2015;201:951–961. doi: 10.1534/genetics.115.181149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersen SL, Sloan RS, Petes TD, Jinks-Robertson S. Genome-destabilizing effects associated with Top1 loss or accumulation of Top1 cleavage complexes in yeast. PLoS Genet. 2015;11:e1005098. doi: 10.1371/journal.pgen.1005098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen-Soltero S, Martinez SL, Putnam CD, Kolodner RD. A Saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability. Mol Cell Biol. 2014;34:1521–1534. doi: 10.1128/MCB.00960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirkin SM. Discovery of alternative DNA structures: a heroic decade (1979–1989) Front Biosci. 2008;13:1064–1071. doi: 10.2741/2744. [DOI] [PubMed] [Google Scholar]
- 40.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Napierala M, Bacolla A, Wells RD. Increased negative superhelical density in vivo enhances the genetic instability of triplet repeat sequences. J Biol Chem. 2005;280:37366–37376. doi: 10.1074/jbc.M508065200. [DOI] [PubMed] [Google Scholar]
- 42.Nakatani R, Nakamori M, Fujimura H, Mochizuki H, Takahashi MP. Large expansion of CTG-CAG repeats is exacerbated by MutSβ in human cells. Sci Rep. 2015;5:11020. doi: 10.1038/srep11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weitzmann MN, Woodford KJ, Usdin K. The development and use of a DNA polymerase arrest assay for the evaluation of parameters affecting intrastrand tetraplex formation. J Biol Chem. 1996;271:20958–20964. doi: 10.1074/jbc.271.34.20958. [DOI] [PubMed] [Google Scholar]
- 45.Belotserkovskii BP, Liu R, Tornaletti S, Krasilnikova MM, Mirkin SM, Hanawalt PC. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci U S A. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belotserkovskii BP, Neil AJ, Saleh SS, Shin JH, Mirkin SM, Hanawalt PC. Transcription blockage by homopurine DNA sequences: role of sequence composition and single-strand breaks. Nucleic Acids Res. 2013;41:1817–1828. doi: 10.1093/nar/gks1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams JD, Fleetwood S, Berroyer A, Kim N, Larson ED. Sites of instability in the human TCF3 (E2A) gene adopt G-quadruplex DNA structures in vitro. Front Genet. 2015;6:177. doi: 10.3389/fgene.2015.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yadav P, Harcy V, Argueso JL, Dominska M, Jinks-Robertson S, Kim N. Topoisomerase I plays a critical role in suppressing genome instability at a highly transcribed G-quadruplex-forming sequence. PLoS Genet. 2014;10:e1004839. doi: 10.1371/journal.pgen.1004839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yadav P, Owiti N, Kim N. The role of topoisomerase I in suppressing genome instability associated with a highly transcribed guanine-rich sequence is not restricted to preventing RNA:DNA hybrid accumulation. Nucleic Acids Res. 2016;44:718–729. doi: 10.1093/nar/gkv1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shuai L, Deng M, Zhang D, Zhou Y, Zhou X. Quadruplex-duplex motifs as new topoisomerase I inhibitors. Nucleosides Nucleotides Nucleic Acids. 2010;29:841–853. doi: 10.1080/15257770.2010.530635. [DOI] [PubMed] [Google Scholar]
- 51.Arimondo PB, Riou JF, Mergny JL, Tazi J, Sun JS, Garestier T, Helene C. Interaction of human DNA topoisomerase I with G-quartet structures. Nucleic Acids Res. 2000;28:4832–4838. doi: 10.1093/nar/28.24.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchand C, Pourquier P, Laco GS, Jing N, Pommier Y. Interaction of human nuclear topoisomerase I with guanosine quartet-forming and guanosine-rich single-stranded DNA and RNA oligonucleotides. J Biol Chem. 2002;277:8906–8911. doi: 10.1074/jbc.M106372200. [DOI] [PubMed] [Google Scholar]
- 53.Lopez CR, Singh S, Hambarde S, Griffin WC, Gao J, Chib S, Yu Y, Ira G, Raney KD, Kim N. Yeast Sub1 and human PC4 are G-quadruplex binding proteins that suppress genome instability at co-transcriptionally formed G4 DNA. Nucleic Acids Res. 2017;45:5850–5862. doi: 10.1093/nar/gkx201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kato L, Stanlie A, Begum NA, Kobayashi M, Aida M, Honjo T. An evolutionary view of the mechanism for immune and genome diversity. J Immunol. 2012;188:3559–3566. doi: 10.4049/jimmunol.1102397. [DOI] [PubMed] [Google Scholar]
- 55.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 56.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 57.Kobayashi M, Aida M, Nagaoka H, Begum NA, Kitawaki Y, Nakata M, Stanlie A, Doi T, Kato L, Okazaki IM, Shinkura R, Muramatsu M, Kinoshita K, Honjo T. AID-induced decrease in topoisomerase 1 induces DNA structural alteration and DNA cleavage for class switch recombination. Proc Natl Acad Sci U S A. 2009;106:22375–22380. doi: 10.1073/pnas.0911879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobayashi M, Sabouri Z, Sabouri S, Kitawaki Y, Pommier Y, Abe T, Kiyonari H, Honjo T. Decrease in topoisomerase I is responsible for activation-induced cytidine deaminase (AID)-dependent somatic hypermutation. Proc Natl Acad Sci U S A. 2011;108:19305–19310. doi: 10.1073/pnas.1114522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci U S A. 2004;101:12997–123002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin Immunol. 2004;16:257–275. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 61.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim N, Jinks-Robertson S. Guanine repeat-containing sequences confer transcription-dependent instability in an orientation-specific manner in yeast. DNA Rep. 2011:953–960. doi: 10.1016/j.dnarep.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bacolla A, Tainer JA, Vasquez KM, Cooper DN. Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Res. 2016:5673–5688. doi: 10.1093/nar/gkw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katapadi VK, Nambiar M, Raghavan SC. Potential G-quadruplex formation at breakpoint regions of chromosomal translocations in cancer may explain their fragility. Genomics. 2012;100:72–80. doi: 10.1016/j.ygeno.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 65.Bergsagel PL, Chesi M, Nardini E, Brents LA, Kirby SL, Kuehl WM. Promiscuous translocations into immunoglobulin heavy chain switch regions in multiple myeloma. Proc Natl Acad Sci U S A. 1996;93:13931–13936. doi: 10.1073/pnas.93.24.13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- 67.Grunnet M, Calatayud D, Schultz NA, Hasselby JP, Mau-Sorensen M, Brunner N, Stenvang J. TOP1 gene copy numbers are increased in cancers of the bile duct and pancreas. Scand J Gastroenterol. 2015;50:485–494. doi: 10.3109/00365521.2014.980318. [DOI] [PubMed] [Google Scholar]
- 68.Kumler I, Balslev E, Poulsen TS, Nielsen SL, Nygard SB, Romer MU, Christensen IJ, Hogdall E, Moreira J, Nielsen DL, Brunner N, Stenvang J. Topoisomerase-1 gene copy aberrations are frequent in patients with breast cancer. Int J Cancer. 2015;137:2000–2006. doi: 10.1002/ijc.29556. [DOI] [PubMed] [Google Scholar]
- 69.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pourquier P, Ueng LM, Kohlhagen G, Mazumder A, Gupta M, Kohn KW, Pommier Y. Effects of uracil incorporation DNA mismatches, and abasic sites on cleavage and religation activities of mammalian topoisomerase I. J Biol Chem. 1997;272:7792–7796. doi: 10.1074/jbc.272.12.7792. [DOI] [PubMed] [Google Scholar]
- 71.Lebedeva N, Auffret Vander Kemp P, Bjornsti MA, Lavrik O, Boiteux S. Trapping of DNA topoisomerase I on nick-containing DNA in cell free extracts of Saccharomyces cerevisiae. DNA Rep. 2006;5:799–809. doi: 10.1016/j.dnarep.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 72.Nitiss JL, Nitiss KC, Rose A, Waltman JL. Overexpression of type I topoisomerases sensitizes yeast cells to DNA damage. J Biol Chem. 2001;276:26708–26714. doi: 10.1074/jbc.M102674200. [DOI] [PubMed] [Google Scholar]
- 73.Daroui P, Desai SD, Li TK, Liu AA, Liu LF. Hydrogen peroxide induces topoisomerase I-mediated DNA damage and cell death. J Biol Chem. 2004;279:14587–14594. doi: 10.1074/jbc.M311370200. [DOI] [PubMed] [Google Scholar]
- 74.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 75.Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol. 2012;19:417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 76.Hsiang YH, Lihou MG, Liu LF. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 77.Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol Cell Biol. 2000;20:3977–3987. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu J, Liu LF. Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 1997;25:4181–4186. doi: 10.1093/nar/25.21.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao ZH, Seiler JA, Zhang H, Marchand C, Agama K, Nitiss JL, Redon C. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cho JE, Jinks-Robertson S. Ribonucleotides and tanscription-sssociated mutagenesis in yeast. J Mol Biol. 2016 doi: 10.1016/j.jmb.2016.08.005. http://dx.doi.org/10.1016/j.jmb.2016.08.005. [DOI] [PMC free article] [PubMed]
- 81.Auzanneau C, Pourquier P. DNA topoisomerase I and illegitimate recombination. In: Pommier Y, editor. DNA Topoisomerases and Cancer. Springer Science +Business Media; 2012. pp. 119–143. [Google Scholar]
- 82.Bullock P, Champoux JJ, Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985;230:954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- 83.Hashimoto H, Chatterjee S, Berger NA. Mutagenic activity of topoisomerase I inhibitors. Clin Cancer Res. 1995;1:369–376. [PubMed] [Google Scholar]
- 84.Balestrieri E, Zanier R, Degrassi F. Molecular characterisation of camptothecin-induced mutations at the hprt locus in Chinese hamster cells. Mutat Res. 2001;476:63–69. doi: 10.1016/s0027-5107(01)00083-5. [DOI] [PubMed] [Google Scholar]
- 85.Shuman S, Prescott J. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J Biol Chem. 1990;265:17826–17836. [PubMed] [Google Scholar]
- 86.Shuman S. Recombination mediated by vaccinia virus DNA topoisomerase I in Escherichia coli is sequence specific. Proc Natl Acad Sci U S A. 1991;88:10104–10108. doi: 10.1073/pnas.88.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jayaram M, Ma CH, Kachroo AH, Rowley PA, Guga P, Fan HF, Voziyanov Y. An overview of tyrosine site-specific recombination: from an Flp perspective. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0021-2014. [DOI] [PubMed] [Google Scholar]
- 88.Sekiguchi J, Seeman NC, Shuman S. Resolution of Holliday junctions by eukaryotic DNA topoisomerase I. Proc Natl Acad Sci U S A. 1996;93:785–789. doi: 10.1073/pnas.93.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schiestl RH, Dominska M, Petes TD. Transformation of Saccharomyces cerevisiae with nonhomologous DNA: illegitimate integration of transforming DNA into yeast chromosomes and in vivo ligation of transformating DNA to mitochondrial DNA sequences. Mol Cell Biol. 1993;13:2697–2705. doi: 10.1128/mcb.13.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jaxel C, Capranico G, Kerrigan D, Kohn KW, Pommier Y. Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin. J Biol Chem. 1991;266:20418–20423. [PubMed] [Google Scholar]
- 91.Zhu J, Schiestl RH. Topoisomerase I involvement in illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1805–1812. doi: 10.1128/mcb.16.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhu J, Schiestl RH. Human topoisomerase I mediates illegitimate recombination leading to DNA insertion into the ribosomal DNA locus in Saccharomyces cerevisiae. Mol Genet Genom. 2004;271:347–358. doi: 10.1007/s00438-004-0987-7. [DOI] [PubMed] [Google Scholar]
- 93.Schiestl RH, Zhu J, Petes TD. Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:4493–4500. doi: 10.1128/mcb.14.7.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 95.Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, Grimes G, Holt IJ, Hill RE, Taylor MS, Lawson KA, Dorin JR, Jackson AP. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM. RNase H2-initiated ribonucleotide excision repair. Mol Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Williams JS, Lujan SA, Kunkel TA. Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat Rev Mol Cell Biol. 2016;17:350–363. doi: 10.1038/nrm.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Williams JS, Smith DJ, Marjavaara L, Lujan SA, Chabes A, Kunkel TA. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell. 2013;49:1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ii M, Ii T, Mironova LI, Brill SJ. Epistasis analysis between homologous recombination genes in Saccharomyces cerevisiae identifies multiple repair pathways for Sgs1, Mus81-Mms4 and RNase H2. Mutat Res. 2011;714:33–43. doi: 10.1016/j.mrfmmm.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Potenski CJ, Niu H, Sung P, Klein HL. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature. 2014;511:251–254. doi: 10.1038/nature13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim N, Huang SY, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sekiguchi J, Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 105.Shuman S. Polynucleotide ligase activity of eukaryotic topoisomerase I. Mol Cell. 1998;1:741–748. doi: 10.1016/s1097-2765(00)80073-8. [DOI] [PubMed] [Google Scholar]
- 106.Huang SY, Ghosh S, Pommier Y. Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites. J Biol Chem. 2015;290:14068–14076. doi: 10.1074/jbc.M115.653345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sparks JL, Burgers PM. Error-free and mutagenic processing of topoisomerase 1-provoked damage at genomic ribonucleotides. EMBO J. 2015;34:1259–1269. doi: 10.15252/embj.201490868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang SN, Williams JS, Arana ME, Kunkel TA, Pommier Y. Topoisomerase I-mediated cleavage at unrepaired ribonucleotides generates DNA double-strand breaks. EMBO J. 2016;36:361–373. doi: 10.15252/embj.201592426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lippert MJ, Kim N, Cho JE, Larson RP, Schoenly NE, O’Shea SH, Jinks-Robertson S. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc Natl Acad Sci U S A. 2011;108:698–703. doi: 10.1073/pnas.1012363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Megonigal MD, Fertala J, Bjornsti MA. Alterations in the catalytic activity of yeast DNA topoisomerase I result in cell cycle arrest and cell death. J Biol Chem. 1997;272:12801–12808. doi: 10.1074/jbc.272.19.12801. [DOI] [PubMed] [Google Scholar]
- 111.Cho JE, Kim N, Li YC, Jinks-Robertson S. Two distinct mechanisms of topoisomerase 1-dependent mutagenesis in yeast. DNA Rep. 2013;12:205–211. doi: 10.1016/j.dnarep.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Christiansen K, Westergaard O. Characterization of intra- and intermolecular DNA ligation mediated by eukaryotic topoisomerase I. Role of bipartite DNA interaction in the ligation process. J Biol Chem. 1994;269:721–729. [PubMed] [Google Scholar]
- 113.Henningfeld KA, Hecht SM. A model for topoisomerase I-mediated insertions and deletions with duplex DNA substrates containing branches, nicks, and gaps. Biochemistry. 1995;34:6120–6129. doi: 10.1021/bi00018a015. [DOI] [PubMed] [Google Scholar]
- 114.Cho JE, Huang SY, Burgers PM, Shuman S, Pommier Y, Jinks-Robertson S. Parallel analysis of ribonucleotide-dependent deletions produced by yeast Top1 in vitro and in vivo. Nucleic Acids Res. 2016;44:7714–7721. doi: 10.1093/nar/gkw495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 116.Soe K, Dianov G, Nasheuer HP, Bohr VA, Grosse F, Stevnsner T. A human topoisomerase I cleavage complex is recognized by an additional human topisomerase I molecule in vitro. Nucleic Acids Res. 2001;29:3195–3203. doi: 10.1093/nar/29.15.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell. 2014;158:327–338. doi: 10.1016/j.cell.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 118.Pourquier P, Ueng LM, Fertala J, Wang D, Park HJ, Essigmann JM, Bjornsti MA, Pommier Y. Induction of reversible complexes between eukaryotic DNA topoisomerase I and DNA-containing oxidative base damages. 7, 8-dihydro-8-oxoguanine and 5-hydroxycytosine. J Biol Chem. 1999;274:8516–8523. doi: 10.1074/jbc.274.13.8516. [DOI] [PubMed] [Google Scholar]