Abstract
Human psychiatric disorders such as schizophrenia, bipolar disorder, and attention-deficit/hyperactivity disorder often include adverse behaviors including increased aggressiveness. Individuals with psychiatric disorders often exhibit social withdrawal, which can further increase the probability of conducting a violent act. Here, we used the inbred, sequenced lines of the Drosophila Genetic Reference Panel (DGRP) to investigate the genetic basis of variation in male aggressive behavior for flies reared in a socialized and socially isolated environment. We identified genetic variation for aggressive behavior, as well as significant genotype-by-social environmental interaction (GSEI); i.e., variation among DGRP genotypes in the degree to which social isolation affected aggression. We performed genome-wide association (GWA) analyses to identify genetic variants associated with aggression within each environment. We used genomic prediction to partition genetic variants into gene ontology (GO) terms and constituent genes, and identified GO terms and genes with high prediction accuracies in both social environments and for GSEI. The top predictive GO terms significantly increased the proportion of variance explained, compared to prediction models based on all segregating variants. We performed genomic prediction across environments, and identified genes in common between the social environments that turned out to be enriched for genome-wide associated variants. A large proportion of the associated genes have previously been associated with aggressive behavior in Drosophila and mice. Further, many of these genes have human orthologs that have been associated with neurological disorders, indicating partially shared genetic mechanisms underlying aggression in animal models and human psychiatric disorders.
Keywords: Drosophila melanogaster, social isolation, social experience, GBLUP, GFBLUP, GenPred, Shared data resource, Genomic Selection
VIOLENT behavior is a common comorbidity among humanpsychiatric disorders such as schizophrenia, bipolar disorder and attention-deficit/hyperactivity disorder (Volavka and Citrome 2008; Retz and Rösler 2010; Hodgins et al. 2014; Volavka 2014; Hoptman 2015). Not all patients with a diagnosis of a psychiatric disorder will show elevated aggressiveness; however, such individuals have an increased risk of performing an aggressive act (Volavka and Citrome 2008; Retz and Rösler 2010; Hodgins et al. 2014). Social withdrawal is frequently observed for patients with a psychiatric disorder, which further contributes to an increased risk of performing a violent act (Retz and Rösler 2010; Hansen et al. 2013) and could point to a potential genotype-by-social environmental interaction (GSEI). Some data indicate that GSEI for aggressiveness in human populations does exist. Caspi et al. (2002) provided the first evidence for an interaction between genotype and early life maltreatment affecting the probability of developing antisocial behavior later in life. These findings were later replicated in a laboratory study (Gallardo-Pujol et al. 2013).
Studying the genetic basis of aggressive behavior in human populations is challenging due to the difficulty in quantifying the phenotype, genetic heterogeneity, and uncontrolled environmental conditions. Animal models not only provide a valuable tool for understanding the genetic basis of aggressiveness, which is evolutionarily conserved (Loren et al. 2003; Mosca et al. 2012; Jones and Norton 2015), but also provide a way to investigate how the social environment affects the degree and direction of individual levels of aggressive behavior. Aggressive behavior has been studied in several model organisms, including mice (Miczek et al. 2001), voles (Gobrogge and Wang 2011), zebrafish (Jones and Norton 2015), and fruit flies (Chen et al. 2002; Edwards et al. 2006; Wang et al. 2008; Kravitz and Fernandez 2015; Shorter et al. 2015). In the fly model, studies have demonstrated repeatedly that socially isolated individuals exhibit increased levels of aggressive behavior compared to socially experienced individuals (Wang et al. 2008; Zhou et al. 2008; Dankert et al. 2009). However, it is unknown whether the effect of social isolation is genetically variable (i.e., exhibits GSEI) or what the underlying genetic basis of GSEI for aggressive behavior is.
The Drosophila Genetic Reference Panel (DGRP) consists of 205 largely unrelated inbred lines derived from a natural population with full publicly available genome sequence data (http://dgrp2.gnets.ncsu.edu) (Mackay et al. 2012; Huang et al. 2014). Since the DGRP lines are inbred, it is possible to assess many individuals of the same genotype in multiple environments, an ideal design for detecting and quantifying the magnitude of genotype-by-environment interactions. To investigate the underlying genetic mechanisms of aggressive behavior, and potentially the genetic mechanisms driving GSEI, we quantified the aggressive behavior of 87 DGRP lines for multiple individuals reared under socialized or socially isolated conditions.
We performed a quantitative genetic analysis and showed there was significant genetic variation for aggression in both social environments, as well as significant GSEI. We then performed several genomic analyses to identify genes and gene ontology (GO) terms associated with male aggression, and genomic features determining the interaction term. The genomic analyses included a genome-wide association (GWA) using single marker regression to assess the contribution of individual polymorphic markers on aggressive behavior in the two environments, and for the difference in aggressiveness between socialized and socially isolated individuals (GSEI). The small sample size of the DGRP, expected small effects of individual genetic markers, and the large number of tests to be performed with sequence data results in limited statistical power to detect true associated genetic variants (Hirschhorn and Daly 2005; McCarthy et al. 2008; Wang et al. 2010a, 2011; Fridley and Biernacka 2011; Rohde et al. 2016a). Methods that combine the signals from multiple genetic markers, including set-test approaches [i.e., the sequence kernel association test (Wu et al. 2011) and covariance association test (Rohde et al. 2016a)] and genomic prediction models [genomic best linear unbiased prediction (GBLUP) (Meuwissen et al. 2001)], may better capture the signal from numerous genetic markers with small effect sizes. Extending GBLUP by fitting multiple genetic components has been shown to increase predictive ability (PA) (Speed and Balding 2014; Tucker et al. 2015). In particular, fitting multiple genetic components can be used to partition genetic variance by pathway (Edwards et al. 2015), or to test the PA of predefined sets of genes using genomic feature BLUP (GFBLUP) (Edwards et al. 2016; Sarup et al. 2016; Fang et al. 2017). The second set of genomic analyses therefore included genomic prediction models where the aims were to identify sets of genes predictive of the trait variation, but also to investigate means to predict aggression across social rearing conditions.
Materials and Methods
Drosophila and establishment of social rearing conditions
87 DGRP lines (Mackay et al. 2012) and an isogenic control strain, Canton-SB (CSB) (Norga et al. 2003), were maintained in the Mackay laboratory at North Carolina State University (Raleigh, NC) under controlled conditions (cornmeal-molasses-agar medium, 25°, 70% humidity, and a 12 hr light-dark cycle). Two social rearing environments were established by initially setting up the DGRP lines and an equal number of CSB flies in two replicate bottles per social group, each consisting of 20 males and 20 females, and allowing them to mate and lay eggs for four consecutive days.
Socially experienced flies:
Twelve days after the initial cross, the bottles were cleared for emerged flies. The following morning, flies that had emerged within the last 16 hr were sorted into two vials, with 24 mixed-sex flies per vial. Three days later, the flies were lightly anesthetized and sorted by sex. Up to 24 males/DGRP line were kept, and 16 hr prior to the aggression assay the socially experienced DGRP and CSB males (see below) were transferred to the Flydiator arena (Figure 1), where they had access to food.
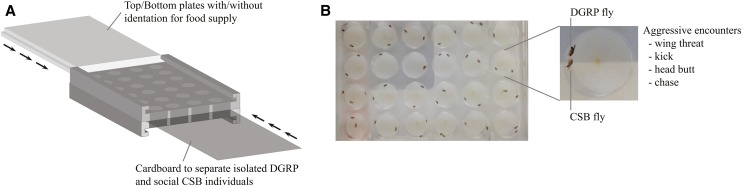
Figure 1.
Assay used to quantify aggressive behavior. (A) Illustration of the Flydiator arena. (B) Top view of the Flydiator arena loaded with pairs of CSB and DGRP flies. CSB, Canton-SB; DGRP, Drosophila Genetic Reference Panel.
Socially isolated flies:
Eleven days after the initial cross, the bottles were cleared. The next morning, newly-eclosed (determined by meconium) DGRP males were separated into collection vials, 24–32 per DGRP line. Individual males were then placed into cells of a 96-well plate (2.2 ml deep) containing fly food. 16 hr prior to the assay, the isolated DGRP and the social CSB males (see below) were added to the Flydiator arena with access to food (Figure 1), but separated by cardboard.
Control flies:
The same day the socially experienced DGRP flies were established, two vials of 24 mixed-sex CSB flies were prepared. Three days later, CSB males were lightly anesthetized and sorted by sex. Twenty-four males were placed in vials corresponding to the number of DGRP lines to be assayed.
Note that our definition of social isolation includes physical isolation from both males and females and thus includes a mating component (socially isolated males are virgins while the normally socialized males are most likely mated) and a male–male interaction component (socially isolated flies have no experience with other males while normally socialized males will likely have participated in, and learned from, previous aggressive encounters).
Drosophila aggression assay: the Flydiator arena
A behavioral arena, the Flydiator arena, was developed to facilitate high-throughput acquisition of aggression data on pairs of individuals; one DGRP and one control fly (CSB). The arena was constructed in transparent polycarbonate (3.5 in. × 5.0 in. × 1 in.) with 4 × 6 wells (diameter of 0.625 in.). The top and bottom were removable, which allowed exchange between smooth plates and plates with indentations (diameter of 0.25 in.) for food supply. Each well could be divided into separate containers (diameter of 0.625 in., height 0.5 in.) by an interchangeable cardboard barrier to keep the pair of flies separated until the start of the assay (Figure 1).
All assays were performed between 8–11 am, and flies were starved for 90 min prior to the aggression assay. New food was introduced 2 min before the initiation of the assay, and the barrier separating the isolated DGRP flies and social CSB flies was removed. Flies were filmed for 2 min using an iPad Mini (Apple, Cupertino, CA). Subsequently, the total number of aggressive encounters for the DGRP and the CSB individuals were manually scored using Jwatcher software (Blumstein et al. 2012). These behaviors included wing threat (the fly raises both wings to a 45 angle toward the opponent or flicks wings at a 45 angle while facing away from the opponent), head butt (strikes the opponent with the head), kicking (extends leg and makes contact with the opponent), and chasing (runs after the opponent with close proximity) (Zwarts et al. 2012). The manual scoring was accomplished by the same person to minimize variation caused by different persons scoring and to avoid potential bias.
The aggression assay was conducted in five blocks with 21, 17, 15, 23, and 11 DGRP lines represented in each block (Supplemental Material, Figure S1 in File S1), and phenotypes were obtained one block at a time.
Genomic data
SNP genotypes were obtained from whole-genome sequence data (available at http://dgrp2.gnets.ncsu.edu/). A total of 1,725,755 SNPs with a minor allele frequency (MAF) of ≥ 0.05 were present in the 87 DGRP lines, distributed on the six chromosome arms; 2L, 2R, 3L, 3R, 4, and X. SNPs were annotated to genes (defined as SNPs within the transcribed region of the gene) using FlyBase annotation v5.49 (flybase.org, Figure S2 in File S1), and mapped to a total of 10,517 genes. Genes were linked to GO terms using the org.Dm.eg.db package (Carlson 2015) for Bioconductor (Gentleman et al. 2004). We only considered GO terms with ≥ 10 directly evidenced genes and a minimum of 199 SNPs. This cutoff resulted in a total of 1134 GO terms and a total of 963,207 unique SNPs.
Quantitative genomic parameters
GSEI:
An individual’s phenotype can be described as the sum of its genotypic and environmental effects. Genotype-by-environmental interaction occurs when different genotypes respond differentially to environmental changes (Falconer and Mackay 1996; Lynch and Walsh 1998). A two-way factorial mixed model ANOVA was fitted to test for GSEI; where y is a vector of phenotypic values of flies from both rearing environments (square root transformed to fulfill assumptions of normality), B is a fixed experimental block effect, E is a fixed environmental rearing effect (i.e., social or socially isolated environment), L is a random line effect, is a random interaction term between line and rearing environment, and e is the residual. The significance of was obtained using a likelihood ratio test comparing the model to a reduced model neglecting the interaction term.
GSEI can be viewed as a trait itself. Here, GSEI was quantified as the difference in adjusted phenotypic values () for socially experienced and socially isolated flies within each DGRP line, i.e., (see Adjusted phenotypes used in genomic association and prediction models).
Estimating variance components:
Variance components were estimated using the R package lme4 (Bates et al. 2015) for flies reared in a socialized environment, flies reared in social isolation, and for the two-way interaction model (see above). The models within social environment were: where y is a vector of phenotypic values of flies from one of the rearing environments (social or isolated, square root transformed to fulfill assumptions of normality), B is a fixed experimental block effect, L is a random line effect, and ϵ is residual effect. Segregating polymorphic chromosomal inversions (Huang et al. 2014) and Wolbachia infection status were tested for association with aggressive behavior; none had major effects (results not shown), thus, these were not accounted for.
The proportion of phenotypic variance explained by genetic variance within social environment was estimated as and across social environments as is the genetic variance among lines, is the genotype-by-environment interaction variance, and is the pooled within-line variance.
Cross-environment phenotypic and genetic correlations:
Phenotypic and genetic correlations were computed between DGRP lines from the two social rearing conditions. The phenotypic correlation () was computed as Pearson’s correlation using line means, and the cross-environment genetic correlation was computed as using the estimated variance components from the two-way ANOVA.
Adjusted phenotypes used in genomic association and prediction models
The phenotypes used in the single marker regressions and the prediction models were the observed number of aggressive encounters, transformed by the square root to achieve an approximation of a Gaussian distribution, or GSEI, adjusted for experimental block effect by fitting the linear mixed model
| (1) |
where y is a vector of repeated phenotypic observations, X and Z are design matrices linking fixed and random effects to the phenotypic values, b is a vector of fixed effects, g is a vector of random genetic effects defined as and ϵ is a vector of residual effects defined as G is the genomic relationship matrix computed using all segregating markers, where m is the total number of genetic markers, and W is a centered and scaled genotype matrix. Each column vector of W was computed as where is the MAF of the i-th marker, and is the i-th column vector of the allele count matrix, A, containing the genotypes encoded as 0, 1, or 2, counting the number of minor alleles (VanRaden 2008).
Model (1) is an animal model with repeated phenotypes per DGRP line, resulting in the number of estimated genetic effects () corresponding to the number of DGRP lines. The residual effects () have the same dimension as the vector of phenotypes, thus, to retain the replicated structure in the data for the following analyses the adjusted phenotypic values () were computed as
| (2) |
where is a vector of adjusted phenotypic values for the i-th DGRP line, is the estimated genetic effect for DGRP line i, and is a vector of residual effects for line i. The estimated genetic and residual effects were derived using average information restricted maximum likelihood, as implemented in DMU (Madsen et al. 1994).
Single marker regression
Single marker regression, also known as GWA, evaluates the contribution of each segregating genetic marker one at a time, allowing detection of markers with large effects. The test for association of each genetic marker was a t-test on the regression coefficient from the regression of estimated genetic effects ( equation 1) on each polymorphic marker (i.e., 1,725,755 SNPs at MAF > 0.05). A nominal P-value was used as the significance threshold. The genetic effects were used as the response variable because these comprise the DGRP line means adjusted for fixed effects and the genetic structure among DGRP lines.
Identification of predictive SNP sets
The set of causal genetic variants for a given trait should be predictive of the trait value. Therefore, statistical models that allow genomic predictions based on multiple groups of genetic variants could provide a valuable tool for identification of SNP sets containing causal variants. Genetic markers associated with trait variation may not be uniformly distributed across the genome, but may be enriched for genes connected in pathways (Allen et al. 2010; Lage et al. 2012; O’Roak et al. 2012). GO terms are a useful resource of groups of genes with shared biological, molecular, or cellular functions (The Gene Ontology Consortium 2000). An extension of the standard GBLUP (Meuwissen et al. 2001), which allows the addition of an extra genetic component for a set of genetic markers, e.g., all those within a certain GO term, is GFBLUP (Edwards et al. 2015, 2016; Ehsani et al. 2015; Sarup et al. 2016; Fang et al. 2017). The genomic feature model is described below, along with a method to decompose the genetic variance within a GO term to the variance explained per gene within a GO term.
Genomic feature models:
The following methodology was used to identify groups of genetic markers predictive of the phenotype. First, a NULL model (GBLUP) based on all genetic markers was obtained
| (3) |
Then, for all sets of genetic markers (i.e., GO terms), a separate model was fitted (GFBLUP),
| (4) |
where f denotes the genetic effects captured by the genetic markers within a subset of markers, and r was the genetic effects captured by all genetic markers not included in the first set. In particular, the random genetic effects were defined as and where and
Using a standard 10-fold cross validation scheme adapted from animal breeding, the PA was measured as the correlation between the observed genetic values (corresponds to the adjusted line means), and the predicted genetic effects in the validation set. To ensure accurate estimation of PAs, a total of 100 training (t) and validation (v) sets were generated (Figure S3 in File S1). For each pair of training and validation sets the GBLUP (equation 5) and GFBLUP (equation 6) models were fitted. The genetic effects in the validation set () for the GBLUP model were obtained as
| (5) |
and for the GFBLUP model,
| (6) |
Predictive GO terms were defined as those that significantly increased PA compared to the NULL model. To assess whether a GO term increased PA compared to the NULL model, Welch’s t-test was applied (Welch 1947). Subsequently, all t-test P-values were adjusted for multiple testing (Bonferroni correction) and the significance threshold was set at P < 0.05.
In addition, 10 random sets of SNPs were generated for each predictive GO term. The properties of these random sets corresponded to that of the true GO term. That is, the architecture of the set was retained, such that the number of SNPs was the same, and the SNPs were clustered in groups corresponding to the number of genes in the true GO term. The predictive abilities of these random sets were compared to the PA of the true GO term using a t-test.
Genetic decomposition of predictive SNP sets:
GO terms are composed of several genes, and it is unlikely that all genes within a GO term contribute equally to the PA. Therefore, to dissect the genetic contribution of the genes within the predictive GO terms, the genetic variation within the set of predictive GO terms was decomposed to gene level (Rohde et al. 2016b).
For each predictive GO term (i.e., those with an adjusted P-value < 0.05), the marker effects for those genetic markers belonging to a particular GO term () were obtained by multiplication of the centered and scaled genotypes () and the estimated genetic effects corresponding to that particular GO term (f):
| (7) |
Using the genetic effects for the genes constituting the GO term () were computed as:
| (8) |
where is the marker effect of the i-th marker computed for feature f, and is the number of markers within gene Thus, if a GO term has the genetic effect and consists of x genes, then A measure of the genetic variation for each feature per gene adjusted for the number of SNPs within gene (), was approximated as:
| (9) |
Across-environment prediction
Two prediction approaches were used to determine the degree of shared genetic signal for aggressive behavior for flies reared in the socialized or socially isolated environment, as well as for GSEI.
Genomic prediction using sets of candidate genes:
The GFBLUP models (equation 4) were used to test if the combined set of predictive GO terms and genes within one environment was predictive of the genetic effects within another environment. In this scenario, the feature group contained all the GO terms or genes predictive within a given environment. A total of 18 predictions were performed: nine for the set of predictive GO terms (e.g., social-to-social, social-to-isolated, and social-to-GSEI), and nine for the set of genes capturing 10% of the genetic variance within the set of predictive GO terms.
Genomic prediction based on single marker regression:
Single marker regressions were performed for each training set (and environment), and genomic predictions (equation 4) were performed within the validation set for each environment, e.g., social-to-social, social-to-isolated, and social-to-GSEI. Here, the feature group contained sets of SNPs that, from the single marker regression, had a nominal P-value or The second random genetic component was the genetic effects captured by all genetic variants.
For each within-environment prediction (i.e., social-to-social, isolated-to-isolated, or GSEI-to-GSEI) we applied a resampling scheme to obtain the PA of randomly sampled SNPs. A total of 100 randomly sampled SNP sets were generated for each P-value threshold. The number of sampled SNPs within each set corresponded to the average of the observed number of SNPs below that threshold (Figure S4 in File S1).
Enrichment of associated variants within the predictive SNP sets
To link the results from GFBLUP with the single marker regressions, the set of predictive GO terms and the set of genes explaining 10% of the genetic variance were tested for enrichment of individual genetic markers with a nominal P-value < 0.001. The enrichment test was performed using a standard procedure by applying a hypergeometric distribution (Rivals et al. 2007; de Leeuw et al. 2016). The enrichment test was performed both within and across environments, i.e., the predictive set of GO terms and genes explaining 10% of genetic variance was tested for enrichment of associated genetic markers from both environments and from GSEI (18 comparisons).
Software
All statistical analyses were performed within R version 3.1.0 (R Core Team 2016). The mixed models were fitted using DMU software (Madsen and Jensen 2013) using the DMU interface within the R package “qgg” (http://psoerensen.github.io/qgg/). In particular, GBLUP and GFBLUP models were fitted with the “reml” function, the genetic decomposition was done with the function “covSets,” and the enrichment test was performed with the function “setTest.”
Data availability
The DGRP genotypes can be accessed via the website http://dgrp2.gnets.ncsu.edu, and the phenotypic data are available in Table S1.
Results
Quantitative genetics of aggressive behavior
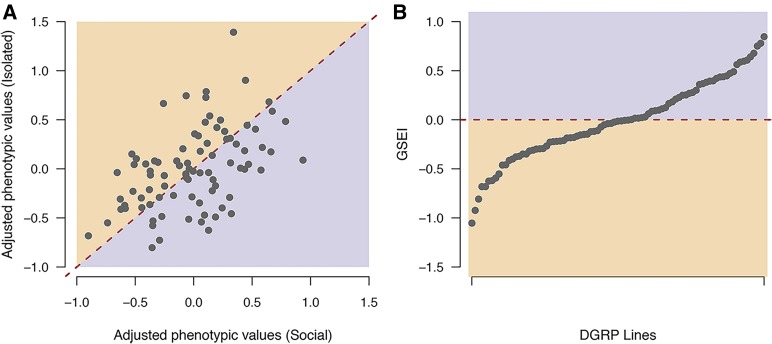
We quantified the total number of aggressive encounters (i.e., the sum of wing threats, head butting, kicking, and chasing) for 87 DGRP lines reared in a socialized or socially isolated environment (Figure 2A and Table S1). We used linear mixed models to infer the genetic variation among lines as well as genetic variation in the social environment interaction term (GSEI). We observed significant genetic variation for aggressive behavior in each social environment and averaged across the two environments, as well as significant GSEI (Table 1). GSEI can be viewed as a trait itself, here quantified as the difference in aggressive behavior between the social and social isolation environments (Figure 2B). The nature of the GSEI was that some DGRP lines showed elevated aggressive behavior when reared in the socially isolated environment, some DGRP lines became less aggressive when reared in social isolation, and some lines were not affected by the rearing regime (Figure 2). We estimated the broad sense heritability averaged across the two social environments ( = 0.14) as well as for socially reared ( = 0.14) and socially isolated ( = 0.12) flies (Table 1). An increase in residual variance ( Table 1) within the social isolation environment resulted in the decreased estimate for this environment (Table 1).
Figure 2.
Variation in aggression among DGRP lines in two social environments. (A) Mean adjusted phenotypic values of aggressive behavior for flies reared in an isolated environment as a function of the mean adjusted phenotypic values for socially experienced flies. The red dashed line illustrates the expected point with no difference in phenotype from the two social environments. (B) GSEI () ordered by increasing trait value. The horizontal dashed line indicates the expected value in absence of GSEI. Shaded areas illustrate in which social environment the DGRP lines became more aggressive (orange, flies from the socially isolated environment; purple, flies from the socialized environment). DGRP, Drosophila Genetic Reference Panel; GSEI, genotype-by-social environmental interaction.
Table 1. Estimated quantitative genetic parameters for socialized individuals, socially isolated individuals, and for a joint analysis with the social rearing regime as a factor to determine potential genotype-by-social environment interaction.
| Parameters | SOC:ISO | SOC | ISO |
|---|---|---|---|
| 0.89 | 0.81 | 0.97 | |
| 0.072*** | 0.11*** | 0.12*** | |
| 0.049*** | — | — | |
| 0.77 | 0.70 | 0.84 | |
| 0.14 | 0.14 | 0.12 | |
| 0.59 | — | — | |
| 0.52 | — | — |
The estimated parameters were the variance components ( total phenotypic variance, genetic variance, genotype-by-social environment variance, and residual variance), the broad sense heritability (), the cross-environment genetic correlation (), and the phenotypic correlation estimated by Pearson’s correlation (). *** indicates statistical significance (P-value ) of variance components. SOC:ISO, for a joint analysis with the social rearing regime as a factor to determine potential genotype-by-social environment interaction; SOC, estimated quantitative genetic parameters for socialized individuals; ISO, estimated quantitative genetic parameters for socially isolated individuals.
The estimate of the phenotypic correlation () between the number of aggressive encounters for flies reared in the social and socially isolated environment was = 0.52, and the cross-environment genetic correlation () was = 0.59 (Table 1). Thus, from our quantitative genetic analysis, we infer that the genetic basis of aggression is in part shared between social environments and in part genetically distinct within each environment.
An aggressive encounter requires interaction between two (or more) individuals, and successive encounters may require the individuals to move. Thus, a correlation between measures of locomotor activity and aggression might be expected. Previously, it has been shown that eliminating social interactions affects locomotor activity (McCarthy et al. 2015). To assess whether the differences in aggressive encounters were mainly driven by phenotypic differences in locomotor activity, we computed Pearson’s correlations between aggressive behavior in the social rearing conditions and previously published measures of locomotor activity obtained for the DGRP (Mackay et al. 2012; Harbison et al. 2013). None of the available measures of locomotor activity were significantly correlated with aggressive behavior (Figure S5 in File S1).
Genetic markers and genomic features associated with aggressive behavior
An advantage of inbred genetic resource populations, such as the DGRP, is the ability to phenotype multiple animals of the same genotype because it increases the precision of the estimate of the genotypic value of the trait for each genetic background. The heritability of aggressive behavior was low in both the social and socially isolated environment, as expected for behavioral traits. However, the heritability of line means is where and are, respectively, the among-line and within-line variance of the individual data, and n is the number of individuals scored per line. The average number of flies per DGRP line assessed for aggressive behavior was 20 for the social environment and 19 for the socially isolated environment. Therefore, the broad sense heritabilities of line means for the socialized and socially isolated flies are, respectively, = 0.76 and = 0.73. This increases the power of association mapping as well as PA (Edwards et al. 2016).
We first performed GWA analyses using single marker regressions to identify variants associated with aggressive behavior for flies reared in a socialized environment and flies reared in a socially isolated environment, as well as GSEI. At a nominal P-value 25 SNPs in 15 unique genes were associated with variation in aggressiveness for socially reared flies, eight SNPs in six unique genes were associated with variation in aggressive behavior for flies reared in a socially isolated environment, and 24 SNPs in 17 genes were associated with GSEI (Table S2). None of the variants were significant following correction for multiple tests (Table S2). A quantile–quantile plot of did not show any evidence of confounding factors inflating the overall signal; however, deviations from the expected probabilities below toward more extreme values were observed, suggesting enrichment of true positive associations (Figure S6 in File S1).
Because methods that consider all variants jointly can capture small polygenic effects, which is not possible in underpowered GWA analyses, we asked to what extent a GBLUP NULL model using all variants could predict aggressive behavior using cross validation. The PA is then the correlation (r) between the predicted and observed phenotypes in the validation set. We found low PAs (SE) for normally socialized flies (PA = 0.18 0.03), socially isolated flies (PA = 0.25 0.04), and GSEI (PA = 0.24 0.03) (Table S3). Similar low predictive abilities for the GBLUP NULL model were observed previously for other quantitative traits measured in the DGRP (Ober et al. 2012, 2015; Edwards et al. 2016). To put this in perspective, the maximum PA is (Mrode 2005; Goddard 2009). Thus, for normally socialized flies, the GBLUP NULL model only explains 0.03/0.76 = 3.9% of the observed heritability of line means, while for socially isolated flies the NULL model explains 0.06/0.73 = 8.2% of the observed heritability of line means.
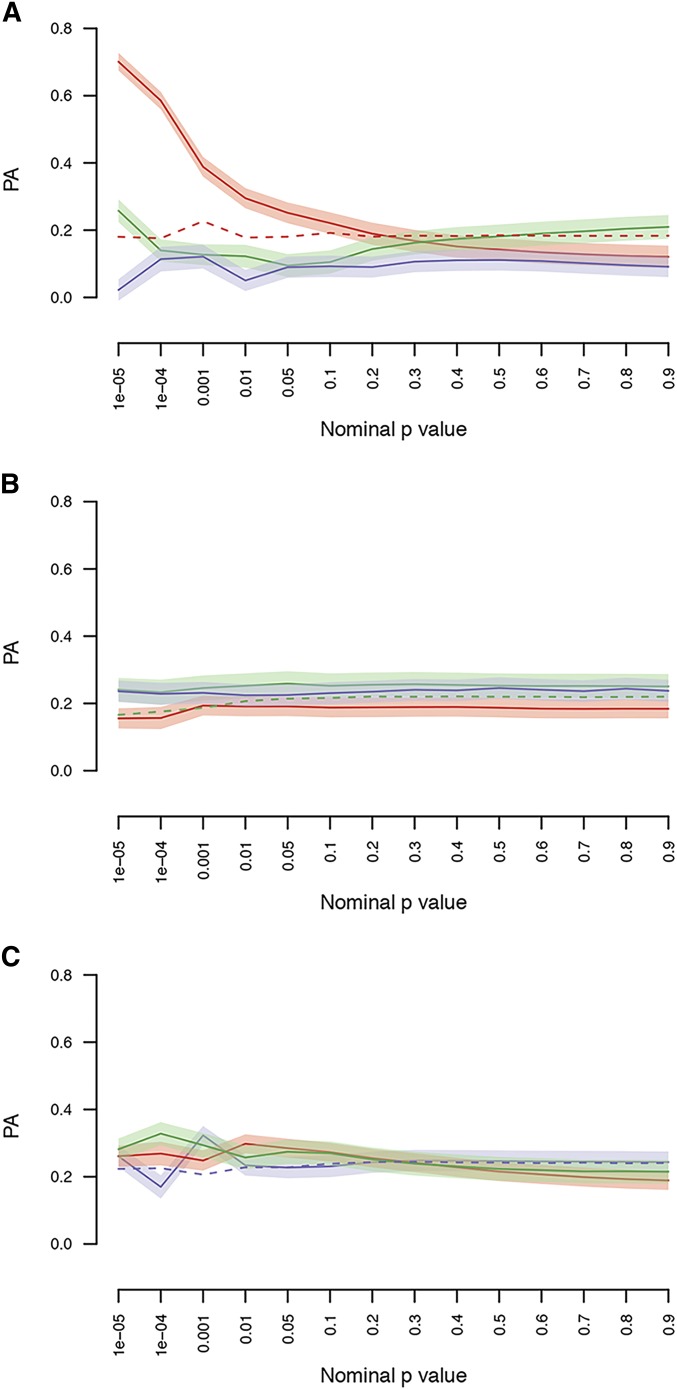
However, the NULL model assumes, unrealistically, that all markers contribute equally to variation in the trait. It is more likely that only a subset of markers affects the trait variation, while the majority have no effect. In this case, strategies to weight markers by the strength of their association in the training set (Ober et al. 2015) or to group markers according to prior biological information and utilize these feature groupings in prediction models may offer significant improvements over the NULL model (Speed and Balding 2014; Edwards et al. 2016; Sarup et al. 2016). Therefore, within each environment we performed GWA analyses in the training set and developed prediction models using SNPs at various P-value thresholds to assess PA in the validation set [Figure 3A (red line), Figure 3B (green line), and Figure 3C (blue line)]. This strategy resulted in a significant increase in PA for the normally socialized flies for markers with (PA = 0.70 0.03), but not for socially isolated flies (PA = 0.24 0.04) nor for GSEI (PA = 0.26 0.03) (Figure 3). The PA of the randomly selected SNP sets was stable across the different P-value thresholds and corresponded to the GBLUP PA (dashed lines Figure 3). We then used GO terms to generate sets of genetic markers, used these sets as priors in genomic prediction models, and assessed whether these marker sets increased the model PA compared to the NULL model that used all segregating genetic markers. We found that 50 GO terms significantly increased PA for flies reared in the social environment, 14 GO terms increased PA for socially isolated flies, and 11 GO terms increased PA for GSEI (Table S3). All predictive GO terms also performed better than sets of random SNPs defined as having the same structural properties as the true predictive set (Figure S7 in File S1). The lack of strong correlation between the GO term PA for flies reared in the socialized environment, the socially isolated environment, and for GSEI (Figure S8 in File S1), and having few GO terms in common across environments (Figure S3 in File S1 and Table S3), further supports the inferences of both distinct and shared genetic control of aggressive behavior under the different social rearing conditions. Two GO terms (phosphotransferase activity, alcohol group as acceptor, and epithelial cell migration) were predictive for aggressive behavior in both social environments, and one of the predictive GO terms (regulation of neurotransmitter secretion) was in common for flies reared in the socialized environment and for GSEI (Table S3). In contrast to the NULL model, the PA of the top GO term explains a substantial fraction of the heritability of lines means: 53.6% for normally socialized flies and 46.7% for flies reared in social isolation.
Figure 3.
Genomic predictions within and across environments using GWA results with bins defined by P-value cutoffs. The data were divided into training and validation sets (10-fold), and GWA analyses were performed on each training set. Each training GWA analysis was then used to predict the genetic values in the validation set [socially experienced flies (red), socially isolated flies (green), or GSEI (blue)] using GFBLUP with feature sets defined by P-value bins. Each panel shows the predictive ability using GWA results from the social environment (A), the socially isolated environment (B), or GSEI (C). Shaded areas indicate the SEM. As comparison, the dashed lines represent the PA (within environment) from sets of randomly selected SNPs of size corresponding to the number of SNPs within environment below the P-value cutoff. GFBLUP, genomic feature best linear unbiased prediction; GSEI, genotype-by-social environmental interaction; GWA, genome-wide association; PA, predictive ability; SNP, single nucleotide polymorphism.
When a set of SNPs increases PA, it is likely because the set is enriched for trait-specific causal variants. We assessed whether the predictive sets contained features discriminating them from nonpredictive sets, in particular with respect to MAF. The average MAF within the predictive sets did not deviate from the mean MAF across the genome, of randomly selected GO terms, or similar sized nongene regions (Figure S9 in File S1); however, we did observe a tendency for more genes within the predictive GO terms to be significantly enriched for low-frequency variants (Figure S10 in File S1). We asked whether low-frequency variants contributed more to increased PA than common variants by fitting the GFBLUP models (equation 4) and removing either low-frequency (MAF < 0.1 or MAF < 0.2) or common variants (MAF > 0.1 or MAF > 0.2) from the feature group of the predictive GO terms. These analyses did not reveal any specific pattern as the PA of the selected GO terms were increased, decreased, or not changed (Figure S11 in File S1), indicating little bias in allele frequency for variants affecting aggressive behavior (and GSEI).
GO terms are composed of several genes, and the same gene can be associated with multiple GO terms. Therefore, we computed and ranked the proportion of variance explained per gene within each predictive GO term to estimate the importance of individual genes. Of the 50 GO terms predictive of aggressive behavior for flies reared in the social environment, a total of 97 genes explained > 10% of the total genetic variance. Nine of these genes (AP-2σ, CG17821, CG31141, Egfr, Elo68β, Frq2, pnt, sgl, and wg) were represented by two or three GO terms (85 unique genes, Table S4). For socially isolated flies, 39 unique genes explained > 10% of the genetic variation within the set of predictive GO terms for aggressive behavior; one gene (chb) appeared in two GO terms (Table S4). A total of 16 genes explained > 10% of the genetic variation within the predictive GO terms for GSEI; one gene (cv-c) was associated with two GO terms and one other (Dys) was associated with three GO terms (13 unique genes, Table S4). Of the genes capturing 10% of the genetic variance within the predictive GO terms, four genes (Ady43A, CG2794, Egfr, and rhea) were in common between the socialized and social isolation environment, three (CASK, Dys, Frq2) between the socialized environment and GSEI, and one (cv-c) between socially isolated flies and GSEI (Table S4).
Predictions within environment based on predictive feature sets
We determined the PA of a GFBLUP model where the feature group contained all predictive GO terms, or genes explaining 10% of the genetic variance within the set of predictive GO terms, within each environment, and for GSEI. In all cases, these models performed significantly better than the NULL model (Table 2). For flies reared in the social environment, the PA from GO terms (PA = 0.56 0.02) was not significantly different from the PA from top genes (PA = 0.60 0.02). However, the PA from GO terms (PA = 0.60 0.02) gave improved performance over top genes (PA = 0.47 0.03) for socially isolated flies, while for GSEI, the PA from top genes (PA = 0.71 0.02) was better than that from GO terms (PA = 0.52 0.03) (Table 2). Next, we asked to what extent these GO categories or genes were enriched for GWA analysis variants () from the analysis of the whole data set, thus, integrating association analyses and prediction. We found considerable enrichment for socially reared flies and for GSEI, but not for socially isolated flies (Table 2).
Table 2. Within- and across-environment genomic predictions using the set of predictive gene ontology terms, or the set of genes explaining 10% of genetic variance within predictive gene ontology terms.
| Train | Validate | PA[GO] | p(t) | Enrichment | PA[Fb] | p(t) | Enrichment | p(GO < Fb) |
|---|---|---|---|---|---|---|---|---|
| Social | Social | 0.56 (0.02) | *** | *** | 0.60 (0.02) | *** | *** | Ns |
| Social | Isolated | 0.29 (0.04) | Ns | Ns | 0.24 (0.04) | Ns | Ns | Ns |
| Social | GSEI | 0.44 (0.03) | *** | * | 0.40 (0.03) | *** | Ns | Ns |
| Isolated | Isolated | 0.60 (0.02) | *** | Ns | 0.47 (0.03) | *** | Ns | Ns |
| Isolated | Social | 0.22 (0.03) | Ns | Ns | 0.12 (0.03) | Ns | Ns | Ns |
| Isolated | GSEI | 0.28 (0.03) | * | Ns | 0.31 (0.03) | * | Ns | Ns |
| GSEI | GSEI | 0.52 (0.03) | *** | *** | 0.71 (0.02) | *** | *** | *** |
| GSEI | Social | 0.34 (0.03) | *** | *** | 0.41 (0.03) | ** | ** | * |
| GSEI | Isolated | 0.35 (0.03) | Ns | Ns | 0.34 (0.03) | Ns | Ns | Ns |
The predictive abilities were compared to the corresponding NULL model [p(t)], and for each set an enrichment test was performed to test whether the set was enriched for genetic variants with low nominal P-values (i.e., P-value < 0.001). p(GO < Fb) is a P-value from Welch’s t-test on whether the predictive ability based on target genes had higher predictive ability than prediction based on selected GO terms. Input GO terms for PA[GO] are found in Table S3. Input genes for PA[Fb] are found in Table S4. * P-value < 0.05, ** P-value < 0.01, and *** P-value < 0.00001. PA[GO], predictive gene ontology terms; PA[Fb], genes explaining > 10% of genetic variance between predictive gene ontology terms; Ns, not significant; GSEI, genotype-by-social environmental interaction.
Predictions across environments
The genetic correlation of aggressive behavior between the two social groups was significantly different from both zero and one, indicating both a shared and distinct genetic basis of aggression when flies are reared separately or together. To the extent that the same genes affect variation in aggression in the two environments, we expect that GO categories and genes that are predictive in one environment should also be predictive across environments. We used two methods for predicting across environments; the first was based on single marker association results and the second on results from the GFBLUP and the genetic decomposition of GO terms to genes.
Predictions based on bins of single marker regression P-values showed that when predictions were performed using the social environment as the training environment, the PA increased within the social isolation environment (Figure 3A) at a P-value threshold of However, when the prediction models were trained on the socially isolated environment or GSEI, no improvement of PA was observed (Figure 3, B and C). The lack of improvement is not easily explained, other than the constraint of small sample size.
When we used the set of predictive GO terms or the subset of genes capturing 10% of the genetic variance from the social environment to predict aggression of flies reared in isolation or vice versa, we found no significant improvement over the NULL model (Table 2). However, we did find a significant increase in PA over the NULL model when we used the predictive GO terms or genes from the social environment to predict GSEI, and vice versa, and in both cases we found that the predictive GO terms or genes (Table S3 and Table S4) were also enriched for associated single genetic variants (Table 2).
By selecting the genes capturing 10% of the genetic variance within the set of predictive GO terms, we expected an increased PA compared to the prediction models based on the complete set of predictive GO terms. However, this was only observed when training on GSEI and predicting within the social environment (Table 2), which indicates that more genes with even smaller effects (10%) are contributing to the variation in aggressive behavior.
Discussion
This study was designed to determine the effect of social isolation on aggressive behavior; to investigate whether the effect of removing prior social experiences was genetically variable; and to gain insight to the genetic basis of this phenomenon by mapping the phenotypic variation of aggressive behavior in both social environments and the response to social isolation to genetic markers, genes, and GO terms.
Aggressive behavior and the effect of social isolation
Aggressive behavior has been studied in several model systems, such as mice (Miczek et al. 2001), voles (Gobrogge and Wang 2011), zebrafish (Jones and Norton 2015), and fruit flies (Chen et al. 2002; Edwards et al. 2006; Wang et al. 2008; Kravitz and Fernandez 2015; Shorter et al. 2015), as well as in humans (Caspi et al. 2002; Gallardo-Pujol et al. 2013; Fernàndez-Castillo and Cormand 2016). In this study, we developed and used the Flydiator arena (Figure 1) to achieve high-throughput acquisition of aggressive behavior of multiple animals of the same genome-wide genotype. The design of the Flydiator arena enabled us to keep two flies within the same arena but separated until the start of the assay (Figure 1A), allowing us to test the immediate effect of social isolation without the confounding effects of recent exposure to CO2 or excess aggravation from vigorous handling of the flies.
Detection of genotype-by-environmental interaction in general, and GSEI interaction in particular can be difficult because of the need to control environmental factors and to test the same genotype in multiple environments. The DGRP is a resource that can mitigate both of these challenges. Thus, it was possible to investigate the effect of social isolation on naturally occurring variation for aggressive behavior by combining the DGRP with the Flydiator arena.
Previous studies investigating the effect of social isolation on aggressive behavior in fruit flies have reported increased levels of aggressiveness when flies were reared in a socially isolated environment (Wang et al. 2008; Zhou et al. 2008; Dankert et al. 2009). The results of this study partially support these prior findings (Figure 2), and more importantly demonstrate the existence of genetic variation in the effect of social isolation, GSEI (Table 1). This finding is consistent with human studies indicating a genotype-by-environment interaction between prior social experience and certain genetic polymorphisms on aggressive behavior (Caspi et al. 2002; Gallardo-Pujol et al. 2013).
Determining the genotype–phenotype map
With advances in sequencing technologies and reduced cost of genome sequence data acquisition, it has become increasingly important to develop new methods to identify genetic markers contributing to the variation in phenotypes, particularly when the majority of markers are expected to have individually small phenotypic effects. Single marker regression methods are extensively used to associate genetic markers with trait variation (Hirschhorn and Daly 2005; McCarthy et al. 2008). However, as a consequence of multiple testing penalties, most of the signal will disappear and leave a large fraction of the true causal variants undetected. In addition, quantitative traits are likely to be genetically complex; thus, statistical methods that exploit prior information to group genetic markers, such as set tests (Wang et al. 2007, 2010a, 2011; Mooney et al. 2014) or prediction models (Speed and Balding 2014; Edwards et al. 2016; Sarup et al. 2016), might be better suited for identifying genomic features containing SNPs with small effects, and have the potential to improve biological inference.
The different statistical models have different pros and cons; therefore, applying a range of models might provide new insight into the distinctive layers of the underlying biology. Moreover, the choice of statistical model might also dictate the genetic model. Single marker regression models detect genetic markers with large effect sizes (Hirschhorn and Daly 2005), whereas genomic prediction using GBLUP assumes an infinitesimal genetic model (Meuwissen et al. 2001). The GFBLUP model is an intermediate between single marker regression and GBLUP in that it assumes a quasi-infinitesimal genetic model (Edwards et al. 2016). Because the genetic model for a given complex trait is unknown, restricting the analysis to one type of statistical model will set a limit to what can be identified given the model. Here, we used both single marker regressions and GFBLUP to gain insight into the genetic basis of aggression in socialized and socially isolated individuals.
Previous work has demonstrated that prediction models incorporating prior biological information enriched for causal variants can increase the PA of complex traits (Edwards et al. 2016; Sarup et al. 2016; Fang et al. 2017). Recently it was shown in the DGRP that the level of PA of complex traits depends on the underlying genetic architecture (Edwards et al. 2016). An advantage of inbred reference populations is the ability to phenotype multiple animals of the same genotype, which increases the precision of the estimate of the genotypic value of each line and increases the broad sense heritability, which in turn can increase the PA (Edwards et al. 2016). The measure of GSEI used in this study was based on mean differences in aggressive behavior between the two social rearing environments, resulting in loss of the replicated design, thus potentially limiting the PA of GSEI. However, comparing the GBLUP predictive abilities for flies reared in a socialized and socially isolated environment with the PA for GSEI did not indicate any reduction in predictive power.
Using the GFBLUP models resulted in a substantial increase in predictive abilities (Figure 3 and Table S3). This was somewhat surprising given the small sample size. However, our findings were robust in the sense that predictions based on randomly selected SNPs did not increase the PA (Figure 3 and Figure S7 in File S1), and indicates that the predictive SNP sets do tag causal variants. However, estimation bias and nonindependence among genomic features could contribute to artificially high accuracies, similar to the overestimation of variance explained by the predictive genes (Table S4), which is known as the Beavis effect (Beavis 1998). In this case, the overestimation could also be due to some degree of nonindependence among the GO terms (i.e., genes and SNPs represented within less than one GO term).
Previously, it has been shown that partitioning of genetic variance into multiple components can increase the PA (Speed and Balding 2014; Tucker et al. 2015). The great increase in PA observed here could in part be due to the precise measurement of each genotype obtained by multiple phenotypic records per genotype, but it may also be the case that partitioning genetic variance into multiple components allows separation of the true genetic signal and noise if the partitioning is correct. Low predictive abilities are commonly seen for complex trait prediction when based on GBLUP, especially for unrelated populations, such as humans (de los Campos et al. 2013; Speed and Balding 2014; Tucker et al. 2015). Therefore, prediction models such as the GFBLUP model might provide similar improved results when applied to other data sets and species. Here, the genomic features were specified according to GO categories, but classifications based on expression profiles, protein–protein interactions, and other types of “omic” data might also perform well.
Insights about the molecular genetic basis of natural variation for aggressive behavior
The single marker GWA regressions and the GFBLUP analyses of GO terms and genes give different perspectives about the genetic basis of naturally occurring aggressive behavior in socialized and socially isolated flies. Only a few genetic variants were associated with aggressive behavior in the two social environments and GSEI from the single marker regression analyses, and none of the individual variants or genes in close physical proximity to them was in common. However, the function of the genes harboring the index SNPs had common functional themes. Both dally and spg were associated with aggressive behavior in the social environment. dally affects the development of sensory organs (Fujise et al. 2001), and spg is involved in the embryonic development of the central nervous system (Biersmith et al. 2011). Rbfox1, which affects the development of the nervous system, was associated with variation in aggression of flies from the social isolation environment. Two genes associated with aggressive behavior in the socially isolated environment, CG34371 and zormin, have been implicated in sensory perception of pain (Neely et al. 2010). Three of the genes associated with GSEI were: osa, which is believed to be involved in neurogenesis (Neumüller et al. 2011); Ten-a, which has shown to affect synaptic growth (Mosca et al. 2012); and trv, which has shown to regulate dendritic morphogenesis (Honjo et al. 2016). In addition, two of the associated genes have previously been associated with sleep [CG13868 and DNaseII (Thimgan et al. 2015)], and two have been found to influence memory [Rbfox1 (Guven-Ozkan et al. 2016) and Syn (Michels et al. 2011). tow was found to be associated with GSEI. tow interacts with fz, which has previously been associated with Drosophila aggressive behavior (Edwards et al. 2006; Shorter et al. 2015), and was functionally validated by RNA interference suppression of gene expression (Shorter et al. 2015).
As noted above, the statistical model used will impose a certain genetic model. Therefore, an overlap in the genes found with single marker regression and GFBLUP was not expected a priori, and only one gene, RecQ4, was found associated with GSEI using both methods (Table 3). GFBLUP, in combination with partitioning of predictive GO terms to the genes capturing a large fraction of the genetic variance, identified a large number of genes associated with GSEI and aggressive behavior in both social environments. Two additional sources of evidence can further strengthen the support for a subset of those genes. First, GO terms are constructed in a hierarchical structure (The Gene Ontology Consortium 2000), therefore a gene may be present in multiple GO terms. If the same gene is identified within multiple GO terms, this may indicate stronger support for the particular gene. Second, the positive genetic correlation between the two social conditions suggests some shared underlying genetic mechanism, thus, an overlap in the genes identified would be expected, which was also observed (Table 3).
Table 3. Overview of selected candidate genes for aggressive behavior of flies reared in a socialized or socially isolated environment, and GSEI; identified with single marker regression or with genomic prediction following genetic decomposition to genes 10% of the genetic variance.
| Drosophila Gene Symbol | Drosophila Gene Name | SMR | GFBLUP | Prior Associations with Drosophila Aggression | Human Ortholog | Selected Human Associations (Reference) |
|---|---|---|---|---|---|---|
| abd-A | abdominal A | S | 5 | HOXA6 | ||
| Ady43A | Ady43A | S, I | ADK | |||
| AP-2σ | Adaptor Protein complex 2, σ subunit | Sa | AP2S1 | |||
| Cad87A | Cadherin 87A | G | 2, 3, 5 | CDH23 | Bipolar disorder (Winham et al. 2013) | |
| CASK | CASK | S, G | CASK | |||
| CG11486 | CG11486 | S | 3 | PAN3 | ||
| CG13868 | CG13868 | I | 4 | |||
| CG17821 | CG17821 | Sa | ELOVL5 | |||
| CG2794 | CG2794 | S, I | ||||
| CG31141 | CG31141 | Sa | ELOVL1 | |||
| CG42458 | CG42458 | G | 3 | HNRNPC | Bipolar disorder (Smith et al. 2011) | |
| chb | chromosome bows | Ia | CLASP1 | |||
| CtBP | C-terminal Binding Protein | S | 2 | CTBP1, CTBP2 | ||
| cv-c | crossveinless c | I, Ga | 2, 3, 4 | DLC1 | ||
| dally | division abnormally delayed | S | 3 | GPC5 | ||
| DNaseII | Deoxyribonuclease II | G | 5 | DNASE2, DNASE2B | ||
| Dys | Dystrophin | S, Ga | 2, 5 | DMD | Anxiety in depression (Schosser et al. 2013), response to antidepressant treatment in depression (Clark et al. 2012), and bipolar disorder and schizophrenia (Wang et al. 2010b) | |
| eca | eclair | S | 6 | TMED4 | ||
| EcR | Ecdysone receptor | S | 3 | NR1H3 | ||
| egl | egalitarian | S | 5 | EXD1 | ||
| Egfr | Epidermal growth factor receptor | Sa, I | ERBB4/EGFR | |||
| Eip75B | Ecdysone-induced protein 75B | S | 2 | NR1D2 | ||
| Elo68β | Elongase 68β | Sa | ELOVL4 | |||
| Fas3 | Fasciclin 3 | G | CADM1, CADM4, NECTIN3 | |||
| Frq2 | Frequenin 2 | Sa, G | NCS1 | Alzheimer’s disease (Sherva et al. 2014) | ||
| ftz-f1 | ftz transcription factor 1 | S | 2 | NR5A1, NR5A2 | ||
| Gug | Grunge | S | 3, 5 | RERE | Schizophrenia (Sherva et al. 2014) | |
| kirre | kin of irre | S | 3 | KIRREL, KIRREL3 | ||
| lbl | ladybird late | S | 2 | LBX1 | ||
| Myo61F | Myosin 61F | I | 5 | MYO1C | ||
| nwk | nervous wreck | I | 1 | FCHSD2 | ||
| osa | osa | G | ARID1A, ARID1B | |||
| pnt | pointed | Sa | ETS1 | Depression and manic episodes in bipolar disorder (Fabbri and Serretti 2016) | ||
| Ptp99A | Protein tyrosine phosphatase 99A | S | 2, 5 | PTPRG | Alzheimer’s disease (Herold et al. 2016) | |
| pyd | polychaetoid | S | 2 | TJP1, TJP2 | Response to antipsychotic treatment (Clark et al. 2012) | |
| rb | ruby | S | 5 | AP3B1 | ||
| Rbfox1 | RNA-binding Fox protein 1 | I | 2, 3, 4 | RBFOX1, RBFOX2, RBFOX3 | Alzheimer’s disease (Herold et al. 2016) and schizophrenia (Goes et al. 2015) | |
| RecQ4 | RecQ4 helicase | G | G | RECQL4 | ||
| rhea | rhea | S, I | TLN2 | Sleep (Byrne et al. 2013) and brain structure (Stein et al. 2010) | ||
| salm | spalt major | S | 5 | SALL1 | ||
| sgl | sugarless | Sa | 5, 6 | UGDH | ||
| shg | shotgun | S | 3 | CDH20 | ||
| sina | seven in absentia | S | 3 | SIAH1 | ||
| slou | slouch | S | 2 | NKX1-1 | ||
| spg | sponge | S | DOCK3 | |||
| Syn | Synapsin | G | SYN3 | |||
| Ten-a | Tenascin accessory | G | 3, 5 | TENM3 | Schizophrenia (Goes et al. 2015) | |
| Timp | Tissue inhibitor of metalloproteases | G | 2 | TIMP1, TIMP2, TIMP3 | ||
| Tl | Toll | G | 5 | TLR3, TLR9 | Bipolar disorder (Psychiatric GWAS Consortium Bipolar Disorder Working Group 2011) and schizophrenia (Ripke et al. (2013) | |
| tow | target of wingless | G | C1orf21 | |||
| trv | trivet | G | 3 | TIA1 | ||
| wg | wingless | Sa | WNT1 | |||
| zormin | zormin | I | 5 | PALLD, MYPN |
Genes previously associated with Drosophila aggressive behavior are indicated by the numbers 1–6: 1, GWAS on the DGRP (Shorter et al. 2015); 2, GWAS on advanced intercross population created from the DGRP (Shorter et al. 2015); 3, epistatic interaction network derived from the DGRP (Shorter et al. 2015); 4, functional validation of candidate genes identified by Shorter et al. (2015); 5, differentially expressed genes between Drosophila lines selected for high and low aggressive behavior (Edwards et al. 2006); 6, mutational screen using P-elements (Edwards et al. 2009). Human orthologs and their selected associations with human quantitative traits and psychiatric disorders are listed. SMR, single marker regression; GFBLUP, genomic feature best linear unbiased prediction; S, socialized; I, socially isolated; GSEI, genotype-by-social environmental interaction; GO, gene ontology; DGRP, Drosophila Genetic Reference Panel; GWAS, genome-wide association study.
Indicates a gene (capturing 10% of genetic variance within the predictive GO term) that was identified in multiple GO terms.
Nine genes were present in two or three GO terms for flies reared in a socialized environment (Table 3). Among these were: AP-2σ, which is involved in neurogenesis (Neumüller et al. 2011); Egfr, which is believed to be related to differentiation of the nervous system (Kim et al. 2015); a transcription factor that promotes neural progenitors, pnt (Zhu et al. 2011); wg, which controls the growth of dendrites of periphery sensory neurons (Li et al. 2016); Frq2, which is believed to control neurotransmitter secretion (Attrill et al. 2016); and sgl, which has been previously associated with Drosophila aggressive behavior (Edwards et al. 2006). Three genes (CG17821, CG31141, and Elo68β), about which there is no prior knowledge, were also represented by two or more GO terms in the social environment. For flies reared in social isolation, only chb was present in multiple GO terms. chb has been shown to mediate axon guidance (Lee et al. 2004). Two genes were represented by two and three GO terms for GSEI, cv-c and Dys, both of which previously have been associated with Drosophila aggression (Edwards et al. 2006; Shorter et al. 2015).
Four of the genes capturing 10% of the genetic variance within the predictive GO terms were associated with aggression for both social environments (Table 3). These were: Egfr (which was also represented in multiple GO terms, see above); rhea, which regulates transcription (Bécam et al. 2005); and two genes about which there is no prior molecular or biological knowledge, Ady43A and CG2794. Three genes were in common between flies reared in the social environment and GSEI. These were CASK, Dys, and Frq2; Dys and Frq2 were also were represented by multiple GO terms (see above). CASK has been shown to regulate neurotransmitter release (Zordan et al. 2005) and affect locomotor activity (Sun et al. 2009; Slawson et al. 2011). cv-c (also represented by multiple GO terms, see above) was associated with both GSEI and aggression of flies from the socially isolated environment.
A total of 35 genes that were associated with Drosophila aggressive behavior in any of the analyses performed in this study (Table S3 and Table S4) have been associated with Drosophila aggressive behavior in other studies (Table 3), and eight of the genes identified in this study have orthologs that have been previously associated with aggressive behavior in mice (Table 4). Furthermore, several of the genes with mouse orthologs affecting aggression were also previously implicated in Drosophila aggressive behavior (kirre, ftz-f1, and Fas3) (Shorter et al. 2015), suggesting that they are true positive associations (Table 3 and Table 4). For all the genes discussed here (Table 3 and Table 4), except for CG13868 and CG2794, we identified human orthologs using the Drosophila RNAi Screening Center Integrative Ortholog Prediction Tool (Hu et al. 2011): 24% (14/59) of them have a human ortholog that has previously been associated with human neurological disorders, such as bipolar disorder or schizophrenia (Table 3 and Table 4).
Table 4. Summary of genes associated with aggressive behavior for flies reared in a socialized or socially isolated environment, or with GSEI using single marker regression or with genomic prediction, that have been associated with aggressive behavior in mouse models.
| Drosophila Gene Symbol | Drosophila Gene Name | SMR | GFBLUP | Mouse Ortholog | Reference |
|---|---|---|---|---|---|
| cac | cacophony | S | Cacna1b | Kim et al. (2015), Murakami et al. (2007) | |
| dysfa | dysfusion | I | Npas4 | Coutellier et al. (2012) | |
| Fas2b | Fasciclin 2 | I | Ncam1 | Stork et al. (1997, 2000) | |
| Fas3 | Fasciclin 3 | G | Cadm1 | Takayanagi et al. (2010) | |
| ftz-f1 | ftz transcription factor 1 | S | Nr5a1 | Grgurevic et al. (2008) | |
| kirre | kin of irre | S | Kirrel3 | Prince et al. (2013) | |
| loco | locomotion defects | S | Rgs2 | Oliveira-Dos-Santos et al. (2000) | |
| rst | roughest | S | Kirrel3 | Prince et al. (2013) |
SMR, single marker regression; GFBLUP, genomic feature best linear unbiased prediction; S, socialized; I, socially isolated; GSEI, genotype-by-social environmental interaction.
The human ortholog is NPAS4, which has been associated with bipolar disorder (Psychiatric GWAS Consortium Bipolar Disorder Working Group 2011).
The human ortholog is NCAM1, which has been associated with suicide attempts in depression and bipolar disorder (Mullins et al. 2014).
In summary, we established two social rearing conditions, a normally social and a socially isolated environment, and assayed aggressive behavior among a subset of DGRP lines for multiple individuals per line per environment in the Flydiator arena. We found that the genetic basis of aggression is in part shared between social environments, and in part genetically distinct within each environment. Furthermore, we provide evidence that the phenotypic response to social isolation is genetically variable, that is, evidence for GSEI. The phenotypic variation was mapped to individual genetic variants, genes, and GO terms. The large number of associated genes is consistent with a highly polygenic basis of aggressive behavior, with many genes with individually small effects. However, many of these genes have been previously associated with other behavioral traits and neurological processes, as well as with aggressive behavior in Drosophila. Candidate genes identified in this study have mouse orthologs that have been associated with aggressive behavior, and human orthologs with prior associations with neurological disorders. Thus, these findings suggest a partially conserved mechanism for aggressive behavior across animals, but also between animal aggression and human psychiatric disorders resulting in altered, undesirable behaviors.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.200642/-/DC1.
Acknowledgments
This study was in part funded by grants from the Lundbeck Foundation (R155-2014-1724) the Danish Strategic Research Council (GenSAP: Centre for Genomic Selection in Animals and Plants, contract no. 12-132452) to P.S. and T.F.C.M., and the National Institutes of Health (R01-AA016560 and R01-AG043490) to T.F.C.M.
Footnotes
Communicating editor: A. A. Palmer
Literature Cited
- Allen H. L., Estrada K., Lettre G., Berndt S. I., Weedon M. N., et al. , 2010. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467: 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attrill H., Falls K., Goodman J. L., Millburn G. H., Antonazzo G., et al. , 2016. FlyBase: establishing a gene group resource for Drosophila melanogaster. Nucleic Acids Res. 44: 786–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B. M., Walker S. C., 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67: 1–48. [Google Scholar]
- Beavis W. D., 1998. QTL analyses: power, precision, and accuracy, pp. 145–162 in Molecular Dissection of Complex Traits, edited by Paterson A. H. CRC Press, New York. [Google Scholar]
- Bécam I. E., Tanentzapf G., Lepesant J.-A., Brown N. H., Huynh J.-R., 2005. Integrin-independent repression of cadherin transcription by talin during axis formation in Drosophila. Nat. Cell Biol. 7: 510–516. [DOI] [PubMed] [Google Scholar]
- Biersmith B., Liu Z. C., Bauman K., Geisbrecht E. R., 2011. The DOCK protein sponge binds to ELMO and functions in Drosophila embryonic CNS development. PLoS One 6: e16120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumstein, D. T., J. C. Daniel, and C. S. Evans, 2012 Jwatcher TM (version 1.0). Available at: http://www.jwatcher.ucla.edu. Accessed: February 2014.
- Byrne E. M., Gehrman P. R., Medland S. E., Nyholt D. R., Heath A. C., et al. , 2013. A genome-wide association study of sleep habits and insomnia. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 162B: 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, M., 2015 org.Dm.eg.db: Genome wide annotation for Fly. (R package version 3.2.3).
- Caspi A., McClay J., Moffitt T. E., Mill J., Martin J., et al. , 2002. Role of genotype in the cycle of violence in maltreated children. Science 297: 851–854. [DOI] [PubMed] [Google Scholar]
- Chen S., Lee A. Y., Bowens N. M., Huber R., Kravitz E. A., 2002. Fighting fruit flies: a model system for the study of aggression. Proc. Natl. Acad. Sci. USA 99: 5664–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. L., Adkins D. E., Aberg K., Hettema J. M., McClay J. L., et al. , 2012. Pharmacogenomic study of side-effects for antidepressant treatment options in STAR*D. Psychol. Med. 42: 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutellier L., Beraki S., Ardestani P. M., Saw N. L., Shamloo M., 2012. Npas4: a neuronal transcription factor with a key role in social and cognitive functions relevant to developmental disorders. PLoS One 7: e46604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankert H., Wang L., Hoopfer E. D., Anderson D. J., Perona P., 2009. Automated monitoring and analysis of social behavior in Drosophila. Nat. Methods 6: 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw C. A., Neale B. M., Heskes T., Posthuma D., 2016. The statistical properties of gene-set analysis. Nat. Rev. Genet. 17: 353–364. [DOI] [PubMed] [Google Scholar]
- de los Campos G., Vazquez A. I., Fernando R., Klimentidis Y. C., Sorensen D., 2013. Prediction of complex human traits using the genomic best linear unbiased predictor. PLoS Genet. 9: e1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. C., Rollmann S. M., Morgan T. J., Mackay T. F. C., 2006. Quantitative genomics of aggressive behavior in Drosophila melanogaster. PLoS Genet. 2: 1386–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. C., Zwarts L., Yamamoto A., Callaerts P., Mackay T. F. C., 2009. Mutations in many genes affect aggressive behavior in Drosophila melanogaster. BMC Biol. 7: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. M., Thomsen B., Madsen P., Sørensen P., 2015. Partitioning of genomic variance reveals biological pathways associated with udder health and milk production traits in dairy cattle. Genet. Sel. Evol. 47: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. M., Sørensen I. F., Sarup P., Mackay T. F. C., Sørensen P., 2016. Genomic prediction for quantitative traits is improved by mapping variants to gene ontology categories in Drosophila melanogaster. Genetics 203: 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehsani A., Janss L., Pomp D., Sørensen P., 2015. Decomposing genomic variance using information from GWA, GWE and eQTL analysis. Anim. Genet. 47: 165–173. [DOI] [PubMed] [Google Scholar]
- Fabbri C., Serretti A., 2016. Genetics of long-term treatment outcome in bipolar disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 65: 17–24. [DOI] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F., 1996. Introduction to Quantitative Genetics, Ed. 4 Longman Group, Harlow, Essex. [Google Scholar]
- Fang L., Sahana G., Ma P., Su G., Yu Y., et al. , 2017. Exploring the genetic architecture and improving genomic prediction accuracy for mastitis and milk production traits in dairy cattle by mapping variants to hepatic transcriptomic regions responsive to intra-mammary infection. Genet. Sel. Evol. 49: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernàndez-Castillo N., Cormand B., 2016. Aggressive behavior in humans: genes and pathways identified through association studies. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 171: 676–696. [DOI] [PubMed] [Google Scholar]
- Fridley B. L., Biernacka J. M., 2011. Gene set analysis of SNP data: benefits, challenges, and future directions. Eur. J. Hum. Genet. 19: 837–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujise M., Izumi S., Selleck S. B., Nakato H., 2001. Regulation of dally, an integral membrane proteoglycan, and its function during adult sensory organ formation of Drosophila. Dev. Biol. 235: 433–448. [DOI] [PubMed] [Google Scholar]
- Gallardo-Pujol D., Andrés-Pueyo A., Maydeu-Olivares A., 2013. MAOA genotype, social exclusion and aggression: an experimental test of a gene-environment interaction. Genes Brain Behav. 12: 140–145. [DOI] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., et al. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobrogge K. L., Wang Z. W., 2011. Genetics of aggression in voles. Adv. Genet. 75: 121–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard M., 2009. Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136: 245–257. [DOI] [PubMed] [Google Scholar]
- Goes F. S., McGrath J., Avramopoulos D., Wolyniec P., Pirooznia M., et al. , 2015. Genome-wide association study of schizophrenia in Ashkenazi Jews. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 168B: 649–659. [DOI] [PubMed] [Google Scholar]
- Grgurevic N., Büdefeld T., Tobet S. A., Rissman E. F., Majdic G., 2008. Aggressive behaviors in adult SF-1 knockout mice that are not exposed to gonadal steroids during development. Behav. Neurosci. 122: 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T., Busto G. U., Schutte S. S., Cervantes-Sandoval I., O’Dowd D. K., et al. , 2016. MiR-980 is a memory suppressor microRNA that regulates the autism-susceptibility gene A2bp1. Cell Rep. 14: 1698–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C. F., Torgalsbøen A.-K., Røssberg J. I., Romm K. L., Andreassen O. A., et al. , 2013. Object relations, reality testing, and social withdrawal in schizophrenia and bipolar disorder. J. Nerv. Ment. Dis. 201: 222–225. [DOI] [PubMed] [Google Scholar]
- Harbison S. T., McCoy L. J., Mackay T. F. C., 2013. Genome-wide association study of sleep in Drosophila melanogaster. BMC Genomics 14: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold C., Hooli B. V., Mullin K., Liu T., Roehr J. T., et al. , 2016. Family-based association analyses of imputed genotypes reveal genome-wide significant association of Alzheimer’s disease. Mol. Psychiatry 21: 1608–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn J. N., Daly M. J., 2005. Genome-wide association studies for common diseases and complex traits. Natl. Rev. 6: 95–108. [DOI] [PubMed] [Google Scholar]
- Hodgins S., Piatosa M. J., Schiffer B., 2014. Violence among people with schizophrenia: phenotypes and neurobiology. Curr. Top. Behav. Neurosci. 17: 329–368. [DOI] [PubMed] [Google Scholar]
- Honjo K., Mauthner S. E., Wang Y., Skene J. P., Tracey W. D., 2016. Nociceptor-enriched genes required for normal thermal nociception. Cell Rep. 16: 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoptman M. J., 2015. Impulsivity and aggression in schizophrenia: a neural circuitry perspective with implications for treatment. CNS Spectr. 20: 280–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., et al. , 2011. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ràmia M., et al. , 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24: 1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L. J., Norton W. H. J., 2015. Using zebrafish to uncover the genetic and neural basis of aggression, a frequent comorbid symptom of psychiatric disorders. Behav. Brain Res. 276: 171–180. [DOI] [PubMed] [Google Scholar]
- Kim H. J., Ahn H. J., Lee S., Kim J. H., Park J., et al. , 2015. Intrinsic dorsoventral patterning and extrinsic EGFR signaling genes control glial cell development in the Drosophila nervous system. Neuroscience 307: 242–252. [DOI] [PubMed] [Google Scholar]
- Kravitz E. A., Fernandez M. P., 2015. Aggression in Drosophila. Behav. Neurosci. 129: 549–563. [DOI] [PubMed] [Google Scholar]
- Lage K., Greenway S. C., Rosenfeld J. A., Wakimoto H., Gorham J. M., 2012. Genetic and environmental risk factors in congenital heart disease functionally converge in protein networks driving heart development. Proc. Natl. Acad. Sci. USA 109: 14035–14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Engel U., Rusch J., Scherrer S., Sheard K., et al. , 2004. The microtubule plus end tracking protein orbit/MAST/CLASP acts downstream of the tyrosine kinase Abl in mediating axon guidance. Neuron 42: 913–926. [DOI] [PubMed] [Google Scholar]
- Li X., Wang Y., Wang H., Liu T., Guo J., et al. , 2016. Epithelia-derived wingless regulates dendrite directional growth of Drosophila ddaE neuron through the Fz-Fmi-Dsh-Rac1 pathway. Mol. Brain 9: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loren C. E., Englund C., Grabbe C., Hallberg B., Hunter T., et al. , 2003. A crucial role for the Anaplastic lymphoma kinase receptor tyrosine kinase in gut development in Drosophila melanogaster. EMBO Rep. 4: 781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen, P., and J. Jensen, 2013 A users guide to DMU, version 6, Release 5.2, Aarhus University. Available at: http://dmu.agrsci.dk. Accessed: June 2016.
- Madsen P., Jensen J., Thompson R., 1994. Estimation of (co)-variance components by REML in multivariate mixed linear models using average of observed and expected information, pp. 455–462 in Fifth World Congress of Genetics Applied to Livestock Production, Guelph, Ontario, Canada. [Google Scholar]
- McCarthy K., Kjærsgaard A., Bahrndorff S., Schou T. M., Manenti T., et al. , 2015. The effect of social isolation on locomotor activity in the houseflies (Musca Domestica). J. Insect Behav. 28: 288–296. [Google Scholar]
- McCarthy M. I., Abecasis G. R., Cardon L. R., Goldstein D. B., Little J., et al. , 2008. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9: 356–369. [DOI] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels B., Chen Y.-C., Saumweber T., Mishra D., Tanimoto H., et al. , 2011. Cellular site and molecular mode of synapsin action in associative learning. Learn. Mem. 18: 332–344. [DOI] [PubMed] [Google Scholar]
- Miczek K. A., Maxson S. C., Fish E. W., Faccidomo S., 2001. Aggressive behavioral phenotypes in mice. Behav. Brain Res. 125: 167–181. [DOI] [PubMed] [Google Scholar]
- Mooney M. A., Nigg J. T., McWeeney S. K., Wilmot B., 2014. Functional and genomic context in pathway analysis of GWAS data. Trends Genet. 30: 390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca T. J., Hong W., Dani V. S., Favaloro V., Luo L., 2012. Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature 484: 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrode R. A., 2005. Linear Models for the Prediction of Animal Breeding Values, Ed. 2 Centre for Agriculture and Biosciences International Publishing, Wallingford, Oxfordshire. [Google Scholar]
- Mullins N., Perroud N., Uher R., Butler A. W., Cohen-Woods S., et al. , 2014. Genetic relationships between suicide attempts, suicidal ideation and major psychiatric disorders: a genome-wide association and polygenic scoring study. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 165: 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Nakagawasai O., Yanai K., Nunoki K., Tan-No K., et al. , 2007. Modified behavioral characteristics following ablation of the voltage-dependent calcium channel beta-3 subunit. Brain Res. 1160: 102–112. [DOI] [PubMed] [Google Scholar]
- Neely G. G., Hess A., Costigan M., Keene A. C., Goulas S., et al. , 2010. A genome-wide Drosophila screen for heat nociception identifies α2δ3 as an evolutionarily conserved pain gene. Cell 143: 628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller R. A., Richter C., Fischer A., Novatchkova M., Neumüller K. G., et al. , 2011. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell 8: 580–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norga K. K., Gurganus M. C., Dilda C. L., Yamamoto A., Lyman R. F., et al. , 2003. Quantitative analysis of bristle number in Drosophila mutants identifies genes involved in neural development. Curr. Biol. 13: 568–574. [DOI] [PubMed] [Google Scholar]
- Ober U., Ayroles J. F., Stone E. a., Richards S., Zhu D., et al. , 2012. Using whole-genome sequence data to predict quantitative trait phenotypes in Drosophila melanogaster. PLoS Genet. 8: e1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober U., Huang W., Magwire M., Schlather M., Simianer H., et al. , 2015. Accounting for genetic architecture improves sequence based genomic prediction for a Drosophila fitness trait. PLoS One 10: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Dos-Santos A. J., Matsumoto G., Snow B. E., Bai D., Houston F. P., et al. , 2000. Regulation of T cell activation, anxiety, and male aggression by RGS2. Proc. Natl. Acad. Sci. USA 97: 12272–12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B. J., Vives L., Girirajan S., Karakoc E., Krumm N., et al. , 2012. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince J. E. a., Brignall A. C., Cutforth T., Shen K., Cloutier J.-F., 2013. Kirrel3 is required for the coalescence of vomeronasal sensory neuron axons into glomeruli and for male-male aggression. Development 140: 2398–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group , 2011. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2016. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria: URL http://www.R-project.org/. [Google Scholar]
- Retz W., Rösler M., 2010. Association of ADHD with reactive and proactive violent behavior in a forensic population. Atten. Defic. Hyperact. Disord. 2: 195–202. [DOI] [PubMed] [Google Scholar]
- Ripke S., O’Dushlaine C., Chambert K., Moran J. L., Kähler A. K., et al. , 2013. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 45: 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivals I., Personnaz L., Taing L., Potier M. C., 2007. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23: 401–407. [DOI] [PubMed] [Google Scholar]
- Rohde P. D., Demontis D., Cuyabano B. C. D., Genomic Medicine for Schizophrenia Group. Børglum A. D., et al. , 2016a Covariance association test (CVAT) identifies genetic markers associated with schizophrenia in functionally associated biological processes. Genetics 203: 1901–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde P. D., Krag K., Loeschcke V., Overgaard J., Sørensen P., et al. , 2016b A quantitative genomic approach for analysis of fitness and stress related traits in a Drosophila melanogaster model population. Int. J. Genomics 2016: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarup P., Jensen J., Ostersen T., Henryon M., Sørensen P., 2016. Increased prediction accuracy using a genomic feature model including prior information on quantitative trait locus regions in purebred Danish Duroc pigs. BMC Genet. 17: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schosser A., Butler A. W., Uher R., Ng M. Y., Cohen-Woods S., et al. , 2013. Genome-wide association study of co-occurring anxiety in major depression. World J. Biol. Psychiatry 14: 611–621. [DOI] [PubMed] [Google Scholar]
- Sherva R., Tripodis Y., Bennett D. A., Chibnik L. B., Crane P. K., et al. , 2014. Genome-wide association study of the rate of cognitive decline in Alzheimer’s disease. Alzheimers Dement. 10: 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Couch C., Huang W., Carbone M. A., Peiffer J., et al. , 2015. Genetic architecture of natural variation in Drosophila melanogaster aggressive behavior. Proc. Natl. Acad. Sci. USA 112: 3555–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson J. B., Kuklin E. A., Ejima A., Mukherjee K., Ostrovsky L., et al. , 2011. Central regulation of locomotor behavior of Drosophila melanogaster depends on a CASK isoform containing CaMK-Like and L27 domains. Genetics 187: 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. N., Koller D. L., Panganiban C., Szelinger S., Zhang P., et al. , 2011. Genome-wide association of bipolar disorder suggests an enrichment of replicable associations in regions near genes. PLoS Genet. 7: e1002134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D., Balding D. J., 2014. MultiBLUP: improved SNP-based prediction for complex traits. Genome Res. 24: 1550–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein J. L., Hua X., Morra J. H., Lee S., Hibar D. P., et al. , 2010. Genome-wide analysis reveals novel genes influencing temporal lobe structure with relevance to neurodegeneration in Alzheimer’s disease. Neuroimage 51: 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork O., Welzl H., Cremer H., Schachner M., 1997. Increased intermale aggression and neuroendocrine response in mice deficient for the neural cell adhesion molecule (NCAM). Eur. J. Neurosci. 9: 1117–1125. [DOI] [PubMed] [Google Scholar]
- Stork O., Welzl H., Wolfer D., Schuster T., Mantei N., et al. , 2000. Recovery of emotional behaviour in neural cell adhesion molecule (NCAM) null mutant mice through transgenic expression of NCAM180. Eur. J. Neurosci. 12: 3291–3306. [DOI] [PubMed] [Google Scholar]
- Sun M., Liu L., Zeng X., Xu M., Liu L., et al. , 2009. Genetic interaction between Neurexin and CAKI/CMG is important for synaptic function in Drosophila neuromuscular junction. Neurosci. Res. 64: 362–371. [DOI] [PubMed] [Google Scholar]
- Takayanagi Y., Fujita E., Yu Z., Yamagata T., Momoi M. Y., et al. , 2010. Impairment of social and emotional behaviors in Cadm1-knockout mice. Biochem. Biophys. Res. Commun. 396: 703–708. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium , 2000. Gene ontology: tool for the identification of biology. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimgan M. S., Seugnet L., Turk J., Shaw P. J., 2015. Identification of genes associated with resilience/vulnerability to sleep deprivation and starvation in Drosophila. Sleep 38: 801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker G., Loh P. R., MacLeod I. M., Hayes B. J., Goddard M. E., et al. , 2015. Two-variance-component model improves genetic prediction in family datasets. Am. J. Hum. Genet. 97: 677–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. [DOI] [PubMed] [Google Scholar]
- Volavka J., 2014. Comorbid personality disorders and violent behavior in psychotic patients. Psychiatr. Q. 85: 65–78. [DOI] [PubMed] [Google Scholar]
- Volavka J., Citrome L., 2008. Heterogeneity of violence in schizophrenia and implications for long-term treatment. Int. J. Clin. Pract. 62: 1237–1245. [DOI] [PubMed] [Google Scholar]
- Wang K., Li M., Bucan M., 2007. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 81: 1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H., 2010a Analysing biological pathways in genome-wide association studies. Natl. Rev. 11: 843–854. [DOI] [PubMed] [Google Scholar]
- Wang K. S., Liu X. F., Aragam N., 2010b A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr. Res. 124: 192–199. [DOI] [PubMed] [Google Scholar]
- Wang L., Dankert H., Perona P., Anderson D. J., 2008. A common genetic target for environmental and heritable influences on aggressiveness in Drosophila. Proc. Natl. Acad. Sci. USA 105: 5657–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Jia P., Wolfinger R. D., Chen X., Zhao Z., 2011. Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics 98: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch B. L., 1947. The generalization of “Student’s” problem when several different population variances are involved. Biometrika 34: 28–35. [DOI] [PubMed] [Google Scholar]
- Winham S. J., Cuellar-Barboza A. B., Oliveros A., McElroy S. L., Crow S., et al. , 2013. Genome-wide association study of bipolar disorder accounting for effect of body mass index identifies a new risk allele in TCF7L2. Mol. Psychiatry 19: 1010–1016. [DOI] [PubMed] [Google Scholar]
- Wu M. C., Lee S., Cai T., Li Y., Boehnke M., et al. , 2011. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Rao Y., Rao Y., 2008. A subset of octopaminergic neurons are important for Drosophila aggression. Nat. Neurosci. 11: 1059–1067. [DOI] [PubMed] [Google Scholar]
- Zhu S., Barshow S., Wildonger J., Jan L. Y., Jan Y.-N., 2011. Ets transcription factor Pointed promotes the generation of intermediate neural progenitors in Drosophila larval brains. Proc. Natl. Acad. Sci. USA 108: 20615–20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan M. A., Massironi M., Ducato M. G., te Kronnie G., Costa R., et al. , 2005. Drosophila CAKI/CMG protein, a homolog of human CASK, is essential for regulation of neurotransmitter vesicle release. J. Neurophysiol. 94: 1074–1083. [DOI] [PubMed] [Google Scholar]
- Zwarts L., Versteven M., Callaerts P., 2012. Genetics and neurobiology of aggression in Drosophila. Fly (Austin) 6: 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DGRP genotypes can be accessed via the website http://dgrp2.gnets.ncsu.edu, and the phenotypic data are available in Table S1.