Abstract
Many population genetic activities, ranging from evolutionary studies to association mapping, to forensic identification, rely on appropriate estimates of population structure or relatedness. All applications require recognition that quantities with an underlying meaning of allelic dependence are not defined in an absolute sense, but instead are made “relative to” some set of alleles other than the target set. The 1984 Weir and Cockerham estimate made explicit that the reference set of alleles was across populations, whereas standard kinship estimates do not make the reference explicit. Weir and Cockerham stated that their estimates were for independent populations, and standard kinship estimates have an implicit assumption that pairs of individuals in a study sample, other than the target pair, are unrelated or are not inbred. However, populations lose independence when there is migration between them, and dependencies between pairs of individuals in a population exist for more than one target pair. We have therefore recast our treatments of population structure, relatedness, and inbreeding to make explicit that the parameters of interest involve the differences in degrees of allelic dependence between the target and the reference sets of alleles, and so can be negative. We take the reference set to be the population from which study individuals have been sampled. We provide simple moment estimates of these parameters, phrased in terms of allelic matching within and between individuals for relatedness and inbreeding, or within and between populations for population structure. A multi-level hierarchy of alleles within individuals, alleles between individuals within populations, and alleles between populations, allows a unified treatment of relatedness and population structure. We expect our new measures to have a wide range of applications, but we note that their estimates are sensitive to rare or private variants: some population-characterization applications suggest exploiting those sensitivities, whereas estimation of relatedness may best use all genetic markers without filtering on minor allele frequency.
Keywords: allele matching, correlation of alleles, FST, identity by descent, rare variants
WE offer here a unified treatment of relatedness and population structure with an underlying framework of allelic dependence, where the degree of dependence can be quantified as the probability the alleles are identical by descent (ibd) or as the correlation of allelic-state indicators. We follow Thompson (2013) in regarding ibd for a set of alleles as being relative to some other, reference, set: “There is no absolute measure of ibd: ibd is always relative to some reference population.” In other words, ibd implies a reference point, and ibd status for different alleles at this point is often implicitly assumed to be zero. The need for a reference set for allelic-state correlations was made explicitly by Wright (1951): “the correlation between random gametes, drawn from the same subpopulation, relative to the total, is given by …” (emphasis added), and for inbreeding by Wright (1943): “The inbreeding coefficient is zero relative to the unit groups, relative to the intermediate groups and relative to the total.”
A function of allelic dependence of particular interest to us is which we will show below can be expressed as the probability of ibd of pairs of alleles within populations relative to that for pairs of alleles from different populations. The uses of estimates of this quantity are widespread, and here we note, for instance, a recent discussion by McTavish and Hillis (2015) who used “pairwise for all pairs of populations using Weir and Cockerham’s method.” We suggest that a more informative analysis may result from our population-specific estimates (Weir and Hill 2002; Weir et al. 2005; Browning and Weir 2010). Other authors (e.g., Balding and Nichols 1995; Beaumont and Balding 2004; Shriver et al. 2004; Gaggiotti and Foll 2010) have also discussed the advantages of working with population-specific values instead of single values for a set of populations, or of values for each pair of populations, and our recognition of allele frequency correlations among populations extends their work. Interpopulation correlations have also been considered by Fu et al. (2003), and in the Bayesian treatments of Fu et al. (2005), Song et al. (2006), Karhunen and Ovaskainen (2012) and Günther and Coop (2013). Here we allow for correlations in providing explicit moment estimates that apply to both populations and individuals.
The usual global measure can be regarded as an unweighted average of population-specific values, and, because it is an average, it masks the variation among populations that can indicate the effects of past selection (Beaumont and Balding 2004; Weir et al. 2005). The global measure can diminish signals of population history, and this diminution has become more pronounced as genetic marker data have become richer, and real differences among populations have become more evident.
As Astle and Balding (2009) noted “population structure and [cryptic] relatedness are different aspects of a single confounder: the unobserved pedigree defining the (often distant) relationships among the study subjects.” A similar point was made by Kang et al. (2010): “The presence of related individuals within a study sample results in sample structure, a term that encompasses population stratification and hidden relatedness.” Our goal is to provide a unified approach to characterizing population structure and individual relatedness and inbreeding, in terms of both the underlying parameters and the methods of estimation. By working with proportions of pairs of alleles that match, or are the same type, we can give a single estimator for where the pairs are from the same or different populations, and for inbreeding or coancestry, where the pairs are from the target individual(s) or from all pairs of individuals in a study. Measures of population structure are seen to be averages of coancestry measures for individuals within those populations as has been noted by Karhunen and Ovaskainen (2012).
Ibd refers to the history of pairs of alleles, and a consideration of historical “genetic sampling” (Weir 1996) shows that ibd measures allow quantification of the variance of allele frequencies among evolutionary replicates of these histories. Data from a single population or a single individual have no information about this variance, and so do not allow estimation of ibd probabilities. We might regard multiple loci as providing replication of the genetic sampling process, or we might collect data from multiple populations. An exception is when allele frequencies and ibd status in the reference population are assumed known, as is implied for standard methods for estimating relatedness and inbreeding (e.g., Ritland 1996; Purcell et al. 2007; Yang et al. 2011; Wang 2014) or in forensic science if the frequencies are taken from databases (e.g., Balding 2003). If, instead, estimation methods make use of frequencies from a sample of individuals, they are providing estimates of the inbreeding or relatedness ibd measures relative to those measures for all individuals in the sample. This point was also made by Yu et al. (2006), who spoke of “adjusting the probability of identity by state between two individuals with the average probability of identity by state between random individuals” in order to address ibd. Existing relatedness estimation methods that do not use allele frequencies (e.g., KING-robust, Manichaikul et al. 2010) estimate ibd between individuals (coancestry) relative to that within individuals (inbreeding).
For both population structure and relatedness, we propose the use of allelic matching proportions within and between individuals or populations in order to characterize ibd for an individual or a population relative to a reference set of ibd values. We use allele matching, equivalent to homozygosity and complementary to heterozygosity as used by Nei (1973), rather than components of variance (Weir and Cockerham 1984: hereafter WC84). Although our matching proportions can be translated into the sums of squares used by WC84 we believe they may have more intuitive appeal. Our present treatment also differs from that in WC84 by using unweighted averages of statistics over populations instead of the weighted averages that were more appropriate for the WC84 model of independent populations. We return to this aspect in the Discussion.
The size of current genetic studies requires computationally feasible methods for estimating relatedness between all pairs of individuals, potentially 5 billion pairs for the TOPMed project (http://www.nhlbiwgs.org). The scale of the task may well rule out maximum likelihood approaches (e.g., Thompson 1975; Ritland 1996; Milligan 2003) and Bayesian methods (e.g., Gaggiotti and Foll 2010), and Karhunen and Ovaskainen (2012) have reviewed the challenges of selecting the allele frequency distributions needed for likelihood- and Bayesian-based methods. Moment estimates seem still to be relevant, therefore, and will be presented here.
Materials and Methods
Allele-pair dependencies
Our discussion involves two dualities: the dependencies between pairs of alleles expressed either as correlations or as probabilities of ibd; and the identification of allele pairs either by the individuals or the populations from which they are drawn. Although we generally sample individuals and score genotypes, we begin with allelic descriptors: for a locus of interest, and allele A identified by individual and population (see Table 1), we assign the allelic indicator the value 1 if A is of type u, and the value 0 if it is of not of type u. We will assume alleles within diploids are defined unambiguously, although we have previously (Hill and Weir 2004) discussed the situation when they are not, as have others (e.g., Holsinger et al. 2002). We write the dosage of allele u for a diploid individual as the sum of the x’s for the two alleles carried by the individual, and for haploids the dosage is For SNPs, we write X as the dosage of the reference allele.
Table 1. Notation.
| Quantity | Notation |
|---|---|
| Allele | for allele individual population |
| Allelic indicator | for allele being of type u |
| Allele frequency | expected value of for all |
| actual frequency for allele type u in population i | |
| observed frequency for allele type u in sample from population i | |
| Theta | is probability of ibd between allele k in individual j from population i and allele in individual from population |
| Inbreeding coefficient | is the ibd probability for the two alleles for individual j in population i: |
| Coancestry coefficient | Coancestry is the ibd probability for a pair of alleles drawn from individuals in population i: is the average of for all pairs is the average over populations of For any two distinct alleles drawn from population i, the ibd probability is The average over populations of is is the average of ibd probabilities for alleles from different populations |
| Relative inbreeding | The relative inbreeding coefficient for individual j in population i is reference is or |
| Relative coancestry | The relative coancestry coefficient for individuals in population i is reference is or |
| Population-specific | is probability two alleles drawn from population i are ibd, relative to the probability an allele drawn from one population is ibd to an allele drawn from another population. is for alleles drawn from two individuals in population i |
We stipulate that the expected value of where expectation is over replicates of the evolutionary history of that allele, is the probability a random allele is of type u, regardless of which individual carries that allele or which population contains that individual. The essence of our treatment rests on the expectation of the products of two xu’s, or the probabilities that pairs of alleles are both of type u. For alleles A and with indicators and we stipulate the expectation to be
| (1) |
As is also we see that the variance of is for any allele at the locus of interest. We also see, from Equation 1, that the covariance of and is and it follows that the quantity θ is the correlation of the indicators for pairs of alleles as in the writing of Cockerham (e.g., Cockerham 1969). There is no requirement in Equation 1 for θ to be positive, and, for example, negative values are expected for the two alleles carried by one individual in populations for which there is avoidance of mating between relatives. We add individual and population identifiers to θ in Table 1.
Following the work of Malécot (see review by Epperson 1999), we can also interpret Equation 1 with θ defined as the probability that alleles are ibd. It is then the case that θ cannot be negative. Either of the two alleles has probability of being of type u. The other allele has probability θ of being ibd to the first, and so is also of type u, and it has probability of not being ibd to the first, and so is of type u with probability If we follow Thompson (2013), and regard ibd alleles as those that descend from a single allele in a reference population, the allele probability refers to the reference population. We distinguish the expected value from the actual allele frequency in a population, and from the frequency in a sample from the population, as listed in Table 1.
We will phrase much of our subsequent discussion in terms of ibd probabilities, but will return to the allelic indicator correlations on occasion. Our estimation procedures rest on Equation 1 and so will hold for both interpretations. We turn first, however, to some predictions of ibd probabilities.
Predicted ibd probabilities
Individuals:
For a single diploid individual j, the inbreeding coefficient is the probability its two alleles are ibd. The coancestry, or kinship, coefficient for individuals is defined here as the average of the four ibd probabilities for one allele from each individual. It follows that the coancestry of individual j with itself is Generally, however, we will follow WC84 and reserve the term coancestry for distinct individuals. For haploids, inbreeding coefficients are not needed, and kinship is the ibd probability of the allele in individual j with the allele in individual We will have occasion to use the average over pairs of individuals of the coancestries for (samples from) a population. In Table 1, we have added superscripts to indicate the populations from which the individuals are drawn.
If diploid individual J is ancestral to both j and and if there are n individuals in the pedigree path joining j to through J, including j and then where is the inbreeding coefficient of J, and the sum is over all ancestors J and all paths joining j to through J (Wright 1922). The coancestry is also the inbreeding coefficient for an individual with parents If ancestor J is further back in time than the reference time, then it does not contribute to the relatedness of individuals j and
Populations:
For a single population, the average coancestry coefficient will refer to pairs of distinct alleles, one in each of two distinct individuals. For populations in which there is random union of gametes for all j and and θ will refer to a random pair of distinct alleles in the population regardless of the individuals in which they are carried. If we wish to distinguish this allele-based quantity from genotype-based as we do below, then we write it as In Table 1, we show superscripts to denote population, and we adopt that convention now to describe the accrual of ibd in random-mating population i with constant population size Without mutation, values for t discrete generations after the time when the population had ibd probability satisfy
| (2) |
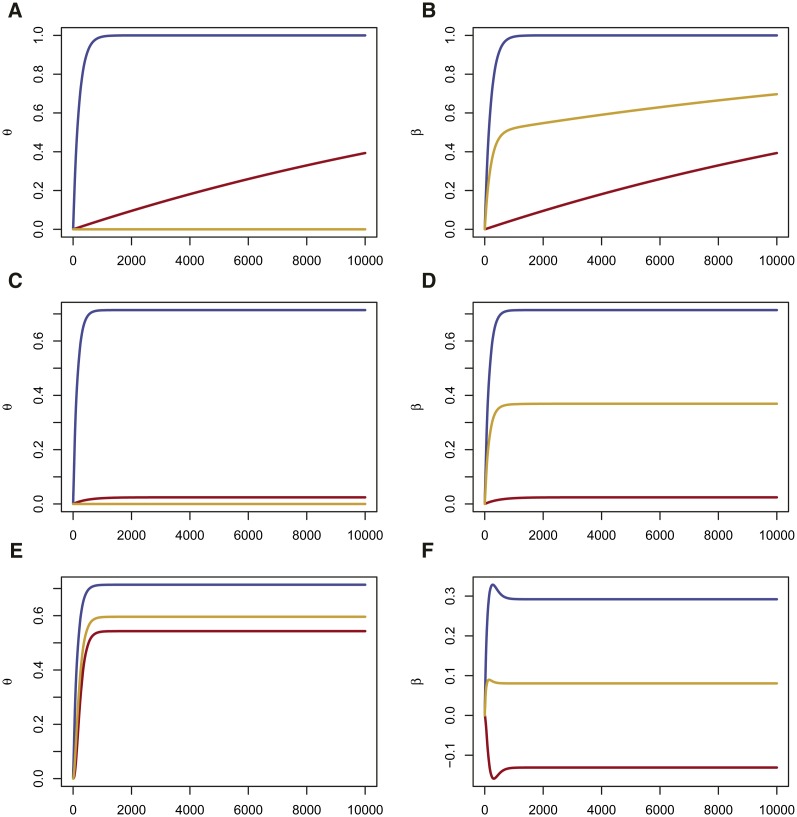
This result was discussed by Wright (1931), although not quite in this form, with shown explicitly. We plot θ from Equation 2 in the first row of Figure 1.
Figure 1.
Effects of Drift, Mutation and Migration on θ and β as a function of generation. For all panels, . Left column (A, C, E) in red, in blue, in orange. Right column (B, D, F) in red, in blue, in orange. (A, B) Drift only (no mutation nor migration). and β tend to 1, (C, D) Drift and Mutation θ and β have positive limits <1. At equilibrium, (E, F) Drift, Mutation and Migration. θ positive and <1, is positive but is negative. At equilibrium,
As for pairs of individuals, the coancestry for pairs of populations is defined here as the average ibd probability for pairs of alleles, one in each population. For populations the quantity is the average over all such pairs of alleles and it does not matter whether or not there is random mating within each population. If there is random mating within each of two populations with constant population sizes however, then genetic drift in the t distinct generations since they diverged from a common ancestral population where was the ibd probability provides
In the absence of mutation and migration, the between-population ibd probability at present time t is the same as it was, in the common ancestral population. To avoid having to specify the ancestral value we define the relative coancestries within populations as for It is pairs of alleles, one from each of populations 1 and 2, that serve as a reference for describing the ibd status for alleles within each of populations 1 and 2, and there is zero ibd between the two populations relative to this reference. For a study in which there are only these two populations, we write and We also write and we could write but this is zero for two populations.
For a set of r populations, we make use of the average over populations of the between-individual, within-population, coancestries, and the average over pairs of populations of the population-pair coancestries, We now have two possible reference sets for within-population coancestries. Relative to all pairs of individuals in population i, the coancestry for individuals is and this has an average value of zero. Relative to all pairs of alleles, one in each of two distinct populations, the coancestry is and we write the average of these quantities over all pairs of individuals as the “population-specific FST.” Averaging over populations gives the usual “population-average FST,” now written as
| (3) |
to stress it is the within-population coancestry relative to the between population-pair coancestry. Recall that our use of for within-population pairs of alleles indicates that we are referring to genotypes, whereas, if we work only with alleles, we write and allele-based is
| (4) |
This is the average over populations of the This expression has been given previously (e.g., Karhunen and Ovaskainen 2012). For random-mating populations, there will be no need for this distinction between and
We acknowledge a notational difficulty in our use of superscript B rather than T and the loss of an immediate similarity to the work of Sewall Wright (e.g., Wright 1951). We use B to stress that our reference set of alleles is between pairs of populations or individuals, whereas T would suggest a total of all pairs, including those within populations or individuals, and the subsequent need to specify population size for the proportion of pairs from the same allele in one individual. Our formulation is simpler by having a reference be “between” rather than “total.”
In WC84, we had set to zero but we do not need that restriction to extend the result of Reynolds et al. (1983) that for a set of populations provides a measure of the time since those populations separated from an ancestral population under a pure drift model. Population-specific and population-average FST values are defined for a set of populations, and are not defined when the set has a single population. For a single population i, we still have the ibd probability and we note that Balding (2003) refers to this as
This development with the θ values regarded as ibd probabilities can be replicated with θ regarded as a correlation of allelic state indicators. Transition equations can be established for the probability a random pair of alleles, one from population i and one from population are both of type u. Adding over allele types leads to the same transition equation for correlations as for ibd probabilities, so that Equation 4 applies to correlations, and brings us back to Wright’s original definition of (Wright 1951).
F-statistics:
The quantity is one of a set of three functions of allelic-state correlations introduced by Wright (1951) for alleles within individuals I within subpopulations S of a total population T. The three quantities are collectively referred to in population genetics as F-statistics. Reich et al. (2009) worked with functions of allele frequencies in two, three, or four populations. For a SNP reference allele, their two-population functions involved the squared difference of allele frequencies in the two populations, and were termed f-statistics. Subsequently, Peter (2016) defined “F-statistics” with, for example, where p is the actual allele frequency in population i. In our notation, omitting W subscripts,
Drift, mutation, and migration:
Nontrivial equilibria for populations drifting apart are obtained when there is mutation and migration, and we illustrate some aspects of our population-specific approach by considering the case of two randomly mating populations exchanging alleles each generation when there is infinite-alleles mutation. A similar treatment (Rousset 1996) allows for symmetric mutation rates among a fixed finite set of alleles. The ibd probability transition equations for an arbitrary number of populations, but with equal population sizes and equal migration rates between all pairs of populations, were given by Maruyama (1970). In our case of two unequal population sizes and unequal migration rates, they are, omitting W subscripts,
| (5) |
where the mutation rate is μ, and population receives a fraction of its alleles each generation from population A consequence of these equations is that or that and is positive. However, it is not necessary that each of exceeds In Figure 1, second row, we show that mutation leads to equilibrium values of different from 1, and, in the third row, that migration can lead to cases where In the absence of migration, mutation drives to zero, so that are both positive. For two populations, is always zero.
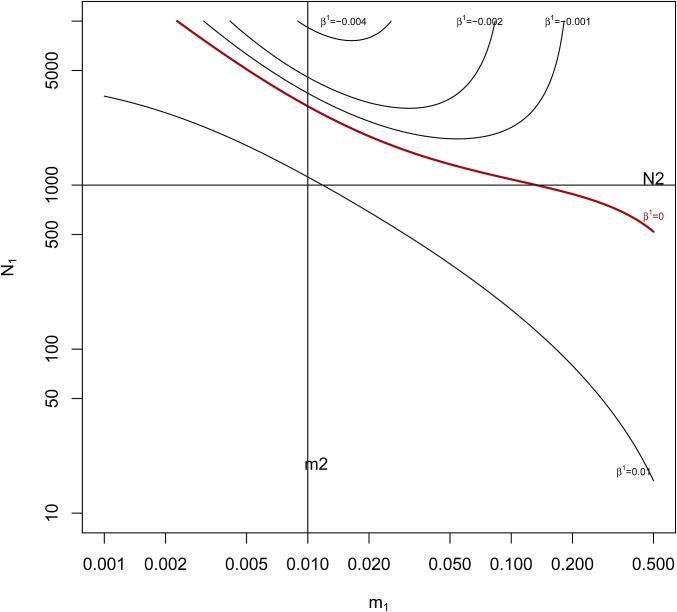
We used numerical methods to find the equilibria for Equation 5, and in Figure 2 we show the region in the space of values where for fixed Averaging over the two to work with hides any difference in the sign of . We note that, in this model, migrants do not come from a “unique and common migrant pool,” as is assumed in the F-model of Balding (2003), Beaumont (2005) and Gaggiotti and Foll (2010).
Figure 2.
Contour plots for at equilibrium obtained by solving the system of Equation 5. and fixed at 1000 and 0.01 respectively (solid horizontal and vertical black lines). The region above and to the right of the red line has equilibrium values of i.e., In that region, a pair of alleles within population 1 has a smaller probability of ibd than does an allele from population 1 with an allele from population 2.
Actual vs. predicted θ:
The probabilities of ibd calculated from path-counting methods for pedigrees of individuals, or from transition equations for populations, can be regarded as the expected values, over evolutionary replicates, of the actual identity status of a pair of alleles. We have previously discussed the variation of actual identity about the predicted value (Hill and Weir 2011, 2012), as did Speed and Balding (2015). The variance of an actual ibd measure for two alleles, whose predicted value is θ, is (Cockerham and Weir 1983), where is the joint probability of ibd for each of two pairs of alleles. The coefficient of variation of the actual coancestry for two individuals is >1 for individuals with predicted coancestry θ <0.125, and it increases as the degree of relationship decreases. The implication of this is that, for a particular pair of populations or individuals, estimated values may not match those expected from pedigrees or transition equations. Evaluation of estimation procedures should, therefore, be performed over many replicates.
Estimation
Allelic matching:
We find intuitive appeal in working with proportions of pairs of alleles that are identical by state (ibs). The matching (allele sharing) proportion for pairs of distinct alleles drawn from individual j in a sample from population i is using the notation in Table 1. From Equation 1 this matching proportion has expected value where Similarly, the matching proportion for pairs of alleles drawn from distinct individuals respectively in population i is and this has expectation In Table 2 we display all the matching proportions needed for data consisting of genotypes from individuals drawn from the ith of r populations, along with expected values of these proportions. Within populations, it is convenient to express matching proportions in terms of individual allelic dosages rather than allelic indicators. Between populations, it is convenient to use sample allele frequencies.
Table 2. Allele-pair matching proportions.
| Matching of two distinct alleles within individual j in population i |
| Average within-individual matching in population i |
| Average over populations of within-individual matching |
| Matching of one allele from each of individuals in population i |
| Average between-individual matching in population i |
| Average over populations of between-individual within-population matching |
| Matching of two distinct alleles, ignoring genotypes, within population i |
| Average over populations of within-population allele matching, ignoring genotypes |
| Matching of an allele from individual j in population i with an allele from individual in population |
| Matching of one allele from each of populations |
| Average over pairs of populations of between-population-pair matching |
Individuals:
If data are available only from a single population, it is possible to estimate only the probability of two alleles, within or between individuals, being ibd relative to the ibd probability of pairs of alleles in a reference set defined by these data. We take the reference set to be an allele from one individual in the sample, paired with an allele from another individual in the sample, averaged over all pairs of distinct individuals in the sample. The estimates are shown in Table 3, and for SNPs they are as shown in Equation 6 without designating the population:
| (6) |
where, for a sample of n individuals, Recall that is the reference-allele dosage for individual j. Averaging the inbreeding coefficient over individuals in the sample gives an estimate of the within-population inbreeding coefficient for the sampled population, whereas the average coancestry is zero by construction.
Table 3. Estimates of inbreeding, coancestry, and relatedness.
| Allele matching in individual j of population i, relative to individual-pair matching in population i. |
| Average within-individual matching in population i, relative to individual-pair matching in population i population-specific |
| Population average of within-individual matching, relative to individual-pair matching in each population. |
| Population average of within-individual matching, relative to allele matching between populations. |
| Allele matching between individuals in population i relative to between-individual matching in that population. with zero average over pairs of individuals. |
| Average individual matching within population i, relative to allele matching between populations. population-specific for genotypic data. |
| Population average of within-population individual-pair matching, relative to allele matching between populations. overall for genotypic data. |
| Distinct allele matching within population i, ignoring genotypes, relative to allele matching between populations. population-specific for allelic data. |
| Population average of within-population allele matching, relative to allele matching between populations. overall for allelic data. |
| Matching of one allele from each of populations relative to allele matching between all populations. with zero average over pairs of populations. |
Notice that we construct estimates as the ratio of expressions that each have expected values proportional to As we did in WC84, we assume the expected value of the ratio of two expressions is approximately the ratio of their expectations. The values cancel, and we are left with expected values that are our “relative to” functions of ibd probabilities. This first-order Taylor series approximation to the expectation of a ratio has proven robust for since 1984 (e.g., Goudet et al. 1996), and the results shown in Figure 7 below suggest it is also robust for relatedness estimation. Being able to cancel the M terms means we avoid having to know, or estimate, (squares of) the allele frequencies and so we avoid having to specify ancestral populations or individuals. Our work results in ranking populations or individuals by estimates of their ibd status.
Figure 7.
Comparison of β (A) and standard coancestry (B) estimates, when founders are drawn from a single population.
The new estimators we display in Equation 6 differ from the standard estimators (e.g., Ritland 1996; Yang et al. 2011; Wang et al. 2017). For biallelic loci these estimators are
| (7) |
for all These estimators use the sample allele frequencies for the sampled population, and are intended to estimate when and when There is no simple translation from these estimates to those we propose in Equation 6.
Ochoa and Storey (2016a,b) have estimates equivalent to those in Equation 6. Their expressions are a little different because their reference is for all pairs of alleles in a sample, including those within individuals, whereas ours are for pairs of alleles in different individuals. Astle and Balding (2009) (Equation 2.3) gave similar estimates although, in effect, they set the average coancestry of all pairs of individuals in a sample, to zero.
We estimate inbreeding and coancestry relative to the average coancestry of all pairs of individuals in a study. Yang et al. (2010) also discuss estimates relative to the study population, and say “Estimates of relationships are always relative to an arbitrary base population in which the average relationship is zero. We use the individuals in the sample as the base so that the average relationship between all pairs of individuals is 0 and the average relationship of an individual with him- or herself is 1.” Although our estimates of pairwise relationship sum to zero when we use data from a single population, we retain the unknown value in their expectations. We cannot estimate and we may prefer to report estimates relative to those for the least related pairs as described below in Equation 11.
Populations:
With data from a set of r populations, the matching proportions and estimates are also shown in Table 2 and Table 3. In each table these population-based entries reduce to individual-based entries if the sample sizes are one, Regardless of sample size, we can estimate inbreeding and coancestry relative to pairs of alleles, one from each of all pairs of populations in the study. In that case, we would replace a population-specific in Equation 6 by a population-pair average The average inbreeding coefficient estimate over individuals in a population i is now an estimate of the population-specific value, and averaging these over populations gives an estimate of Averaging the coancestries for pairs of individuals in population i gives an estimate of the population-specific and averaging those over populations gives an estimate of
With genotypic data, the estimates in Table 3 provide the usual relationship
| (8) |
although our use of the whole set of populations as a reference does not allow alleles to be drawn from the same population for the matching proportion This shows the composite nature of and we note that, if one is interested in an overall inbreeding coefficient, it might be better estimated by not accounting for the subpopulations. Note that Equation 8 holds for the overall quantities as well as the population-specific quantities.
If we ignore genotypes and use only allelic data, then we return to estimation of population-specific and population-average with and compared to
The population-average value has been given previously by Hudson et al. (1992) (Equation 3).
For SNPs, where the sample frequency of the reference allele for population i is the allele-based population-specific, and population-average estimates for large sample sizes can be written as
| (9) |
where and For a large number of sampled populations, and only then, is the common estimate (e.g., Hartl and Clark 1997, Equation 4.6). For all r it is an estimate of For the case the single-population and population-pair estimates are
| (10) |
Each of the estimates in Equation 10 reflects difference of the two sample allele frequencies. Either or can be negative as shown in Figure 2 for predicted values, but is positive.
Note that the pairwise coancestry estimates and population-pair estimates sum to zero by construction. Although it is not possible to find estimates for each θ when the sampled individuals within a population are related, or when sampled populations have correlated sample allele frequencies, or when there is just a single sampled population, it is possible to rank values, and, we expect these to have the same ranking as their expected values θ.
Combining over loci:
Single-locus analyses do not provide meaningful results, and combining estimates over loci l has often been considered in the literature. In a parallel discussion of weighting over alleles u at a single locus, Ritland (1996) considered weights chosen to minimize variance.
If the locus-l estimates for individuals (Equations 6 and 7) or populations (Equations 9 and 10), are written as then a weighted average over loci is Two extreme weights are and The first may be called “unweighted” and the second “weighted.” For population structure, Bhatia et al. (2013) refer to the first estimate as the “average of ratios” and the second as the “ratio of averages.” WC84 advocated the second, with justification given in the Appendix to that paper, as did Bhatia et al. (2013).
The unweighted estimate is unbiased for all allele frequencies, but is susceptible to the effects of rare variants, when the denominators of the single-locus estimates can be very small. Rare variants may have little effect on the weighted average, and the variance of the estimate is seen in simulations to be less than for the unweighted average, but it is unbiased only if every locus has the same β value. A more extensive discussion was given in the Appendix of WC84 for population structure, and by Ritland (1996) for inbreeding and relatedness. More recently, Ochoa and Storey (2016b) discussed weights for their estimates, and Wang et al. (2017) discuss weighting in the context of known allele frequencies.
Regardless of weighting scheme, the use of several loci allows us to use bootstrapping over loci (Weir 1996) to generate empirical sampling distributions for our estimates. We used bootstrapping for confidence intervals in the Results section. We discussed sampling properties previously (Weir and Hill 2002; Weir et al. 2005), and will give more details elsewhere. We note here that it is increasing the number of loci, rather than the number of individuals, that lead to the greatest reduction in variance—providing the parametric values do not vary too much over loci.
Private alleles:
Current sequence-based studies are revealing large numbers of low-frequency variants, including those found in only one population. These private alleles were identified by Slatkin (1985) and Mathieson and McVean (2012) as being of particular interest. They are very frequent in the 1000 genomes project data (1000 Genomes Project Consortium 2010). We show estimates in Table 4 for the case of an allele observed in only one of a set of r populations.
Table 4. Population-level estimates for private alleles.
| Quantity | Observation or estimate |
|---|---|
| Private allele frequency | in population 1, zero in populations |
| Sample matching proportions | |
| β estimates | |
The estimate of for a private allele is, approximately, its own-population sample frequency, but the population-specific value for its own population ranges from approximately when is very small to 1 when This amplifies the comment “populations can display spatial structure in rare variants, even when Wright’s fixation index is low” of Mathieson and McVean (2012). A population with many private alleles at low to intermediate frequencies will thus likely have a negative and how negative will depend on how many populations have been sampled. Note that this implies must be allowed to go negative, whereas Bayesian and maximum likelihood estimators of population specific are often forced to belong to although this assumption can be relaxed (Ritland 1996).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Population structure
We conducted a series of simulations to evaluate the performance of our estimates, and we looked at 1000 Genomes SNP data to explore the role of rare variants on the estimates. Some of the simulations were conducted with sim.genot.metapop.t available in the hierfstat package (Goudet 2005). The migration model we used allows for a matrix of migration rates between each pair of populations, and the mutation model allows for multiple alleles at a locus. The notation for a two-population model was given above. Our approach of estimating values that are population-specific, and of allowing allele frequencies to be correlated among populations, means that we are estimating different (combinations of) parameters than have other authors (e.g., Gaggiotti and Foll 2010).
Drift with mutation
We first simulated genotypic data under a scenario of pure genetic drift from a common ancestral population. Populations of different sizes ( and ) were investigated, and 50 diploid individuals, each genotyped at 1000 loci with up to 20 alleles were sampled from each population at three time points: generations. The results are reported in Table 5. In all situations, the estimates are close to their expectations, and the 95% confidence intervals obtained by bootstrapping over loci include the expected value The credible intervals for obtained from Bayescan 2.1 (Foll and Gaggiotti 2008) include the expected values for only three of the nine reported situations. The Bayescan estimate tends to overestimate when it is large, and to underestimate it when it is small. A possible reason for this discrepancy is that the Dirichlet distribution used in Bayescan is an approximation of allele frequency distribution under an equilibrium island model (Gaggiotti and Foll 2010). We note that an alternative to the Dirichlet distribution often used, the truncated normal distribution (Nicholson et al. 2002), might be more appropriate for the simulated data, but we are unaware of available implementations of such estimator of Moreover, both the Dirichlet and the truncated normal are just convenient approximations of the true distribution of allele frequencies [see figure S1 and file S2 in Karhunen and Ovaskainen (2012)].
Table 5. Predicted and estimated population-specific values for two populations without migration.
| t | N | |||
|---|---|---|---|---|
| 50 | 100 | 0.221 | 0.222 (0.215, 0.229) | 0.332 (0.325, 0.340) |
| 50 | 1,000 | 0.025 | 0.026 (0.024, 0.028) | 0.026 (0.025, 0.027) |
| 50 | 10,000 | 0.002 | 0.003 (0.001, 0.005) | 0.0003 (0.0001, 0.0005) |
| 500 | 100 | 0.891 | 0.887 (0.875, 0.899) | 0.918 (0.911, 0.925) |
| 500 | 1,000 | 0.211 | 0.211 (0.204, 0.219) | 0.289 (0.283, 0.296) |
| 500 | 10,000 | 0.023 | 0.025 (0.021, 0.029) | 0.002 (0.001, 0.002) |
| 5000 | 100 | 0.962 | 0.958 (0.950, 0.965) | 0.958 (0.953, 0.964) |
| 5000 | 1,000 | 0.693 | 0.698 (0.684, 0.713) | 0.683 (0.673, 0.694) |
| 5000 | 10,000 | 0.143 | 0.145 (0.138, 0.152) | 0.056 (0.053, 0.058) |
Mutation rate 1000 multi allelic loci. 95% confidence intervals for from bootstrapping over loci. 95% credible intervals obtained with Bayescan 2.1 for
Drift with mutation and migration
Model 1. Same migration rates, different population sizes:
We considered two populations under the model described by Equation 5, with sizes and migration rates The mutation rate was After 400 generations, β has expected values and We simulated 50 individuals from each population under this scenario, with 1000 loci and up to 20 alleles per locus. From the resulting allelic data we obtained estimates, and 95% confidence intervals by bootstrapping over loci. The results are shown in Table 6. The predicted values are contained in the confidence intervals, and there are negative values for both the parametric and the estimated value of Note that we cannot estimate with data from two populations.
Table 6. Predicted and estimated population-specific values for two populations with migration.
| Model | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 100 | 1000 | 0.01 | 0.01 | 0.156 | 0.159 (0.148, 0.169) | −0.037 | −0.031 (−0.038, −0.023) | 0.059 |
| 2 | 100 | 1000 | 0.01 | 0.01 | 0.198 | 0.203 (0.196, 0.211) | 0.024 | 0.025 (0.022, 0.027) | 0 |
| 3 | 100 | 1000 | 0.01 | 0.01 | 0.277 | 0.268 (0.254, 0.282) | −0.061 | −0.059 (−0.067, −0.050) | 0.112 |
| 4 | 10,000 | 100 | 0.01 | 0 | −0.281 | −0.269 (−0.292, −0.248) | 0.461 | 0.448 (0.419, 0.477) | 0.090 |
Mutation rate Generation 1000 multiallelic loci. 95% confidence intervals from bootstrapping over loci. For Model 2, using Bayescan 2.1,
Model 2. Continent-island model:
In this scenario we have an infinite continent supplying a proportion of the alleles independently to populations 1 and 2, still with sizes There is no migration between the two populations, so Table 6 shows that the predicted values are contained in the confidence intervals of their estimated values. For this situation, the F-model is suitable, and at the bottom of Table 6 we report with their 95% credible intervals. slightly overestimates and underestimates
Model 3. Migrant-pool island model:
In this model, each population contributes to a migrant pool, from which migrant alleles are drawn. Among the migrant alleles in the case of two populations, half of the “migrant alleles” will in fact be resident alleles if the gametic pool is composed of the same proportion of alleles from each island, independent of its size. With otherwise the same parameter values, the predicted values, and our estimates after 400 generations, are shown in Table 6, and are in good agreement.
Model 4. Different population sizes, different migration rates:
We return to the two-populations model described by Equation 5, but now with and different migration rates and Predicted values after 400 generations are given in Table 6.
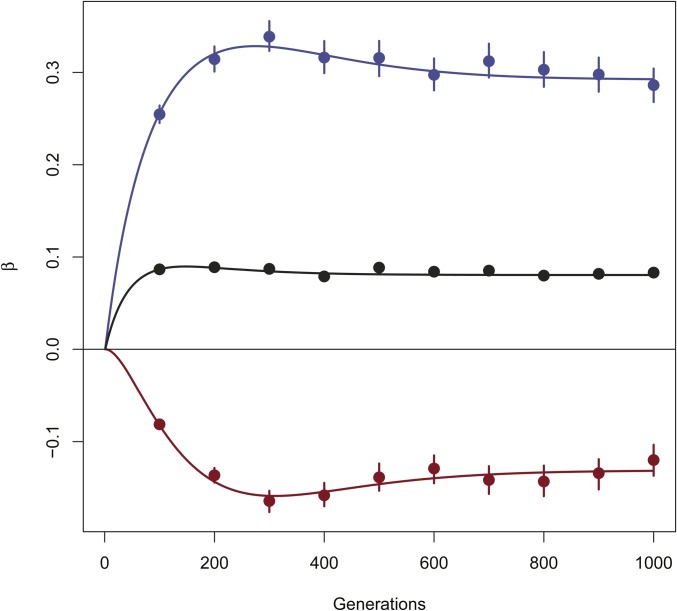
The results in Table 5 and Table 6 show good behavior of estimates with low bias. In Figure 3 we show the estimates for 10 different time points (independent replicates) for Model 4 with a mutation rate of in order to maintain sufficient levels of polymorphism. Again, expected values and estimates are in good agreement throughout the simulations.
Figure 3.
Estimated for independent simulations of the two-population model described by the system of Equation 5, at different times. Population sizes Migration rates Mutation rate in red, in blue, in black. Lines are expectations, points are estimates, and bars represent the 95% confidence intervals obtained by bootstrapping over loci.
Rare alleles:
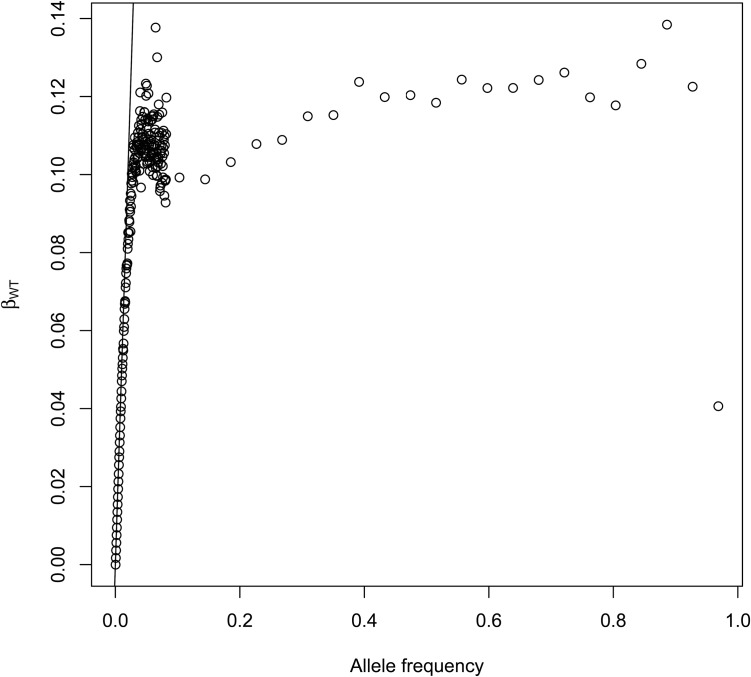
For r populations with total sample size and with copies of an allele private to population 1, the total count for this alleles is and so assuming similar sample sizes for each sample. In Figure 4 we display as a function of allele frequencies for SNPs located on chromosome 2 in the 1000 Genomes project. Individuals were grouped by regions (Africa, Europe, South Asia, East Asia and the Americas). The drawn line corresponds to The initial linear segment corresponds to alleles that are present in one continent only. starts departing from this line for allele counts >80, or equivalently, for worldwide sample frequencies > given the sampled chromosome number of
Figure 4.
as a function of allele frequencies () for SNPs located on chromosome 2. Data from the 1000 genomes project, individuals were grouped by regions (Africa, Europe, South Asia, East Asia, and Americas). The drawn line corresponds to The initial linear segment corresponds to alleles that are present in one continent only. starts departing from this line for allele counts >80, or equivalently, for worldwide frequencies > given the sampled chromosome number of 2426.
When a new allele appears, it will be present in one population only. We expect most, if not all, rare alleles to be private alleles, and thus the expected values for () for these rare alleles are their own-population frequencies. When starts departing from the allele frequency, it implies that some scattering has been happening. In species with a lot of migration, this will happen at low frequencies, whereas the species that are more sedentary should show a 1:1 relation between subpopulation allele frequencies and for a larger range of their site frequency spectrum.
Reference populations:
In Buckleton et al. (2016) we gave population-specific estimates for a set of 446 populations, using published data for 24 microsatellite loci collected for forensic purposes. We showed in that paper how the choice of a reference set of populations can affect results. Here, we illustrate this point with data from the 1000 genomes, using 1,097,199 SNPs on chromosome 22. For the samples originating from Africa, there is a larger with Africa as a reference set than there is, with the world as a reference set. African populations tend to be more different from each other on average than do any two populations in the world on average. The opposite was found for the collection of East Asian populations: there is a smaller with East Asia as a reference set than there is, with the world as a reference set. East Asian populations are more similar to each other than are any pair of populations in the world.
Inbreeding and coancestry
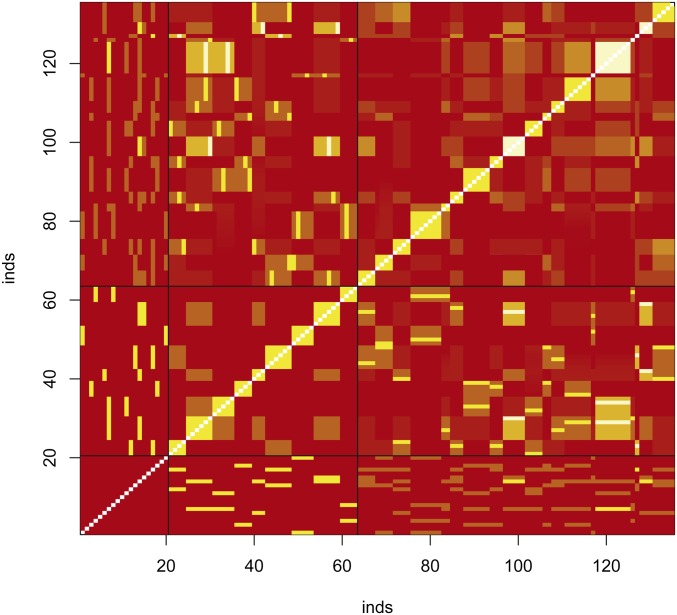
To check on the validity of our estimators of individual inbreeding and coancestry coefficients, we simulated data for a range of nine coancestries: Using the ms software (Hudson 2002), we generated data from an island model with two populations exchanging migrant per generation. We simulated 5000 independent loci, read either as haplotypes (5000) or as SNPs (∼80,000 polymorphic sites for the founders). We then chose 20 individuals from one of these populations and let them mate at random, without selfing. We did not assign or consider sex for these 20 founders. In order to generate a sufficient number of pairs of related individuals, we drew the number of offspring per mating from a Poisson distribution with a mean of five. These offspring were also allowed to mate at random, without selfing, and produced families with sizes drawn from a Poisson distribution with mean three. By keeping records of all matings we could generate the pedigree-based inbreeding and coancestry values for all 135 individuals: founders, their offspring, and their grand-offspring. The pedigree-based coancestries for all 9045 pairs of individuals are shown in Figure 5, although we note (Hill and Weir 2011) that the actual values have variation about expected or pedigree values.
Figure 5.
Pedigree-based coancestry coefficients for simulated data for 135 individuals with 20 founders. Red correspond to low values, yellow to high values of coancestry, white are missing data (unknown inbreeding coefficient of the founders). Black horizontal and vertical lines separate generations in the pedigree. The yellow blocks along the main diagonal correspond to sibships.
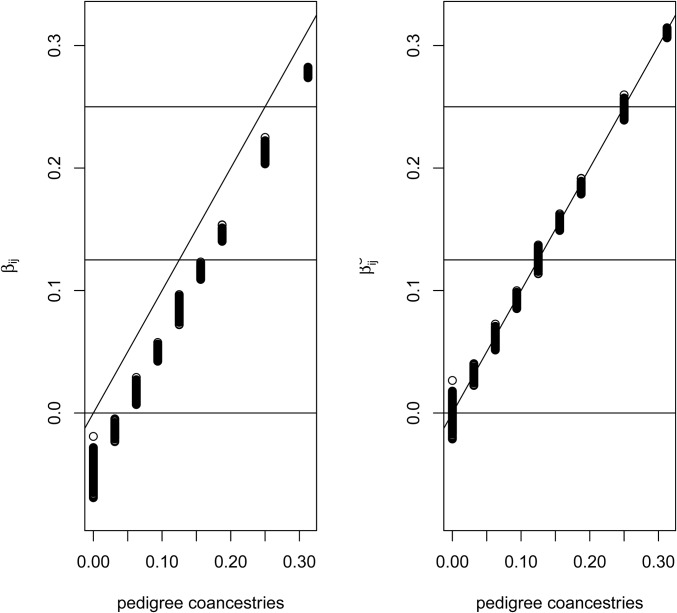
The left-hand plot of Figure 6 compares the coancestry estimates with the pedigree values for all pairs of individuals in the pedigree, and reflects the summing to zero by construction of the coancestries, whereas the pedigree coancestries are necessarily non-negative. The right-hand plot shows a “correction” of the estimates: we took the set of smallest values in the left-hand plot to represent the unrelated (relative to the assumed-unrelated) founders. If we write as the average value of the set of least-related pairs of individuals then our corrected values are
| (11) |
The corrected estimates are clearly close to the pedigree values. However, we are not sure if it is necessary, in general, to undertake this correction process. Whether or not it is applied, the values are still relative to those among all pairs of individuals in a study sample. In general, we will not have any individuals identified for which it is justified to assume zero relatedness or zero inbreeding, and we note the comment by Thompson (2013) “in most populations IBD within individuals is at least as great as IBD between.”
Figure 6.
Comparison of estimated and pedigree coancestries. Uncorrected estimates (Equation 6) on left, corrected estimates (Equation 11) on right.
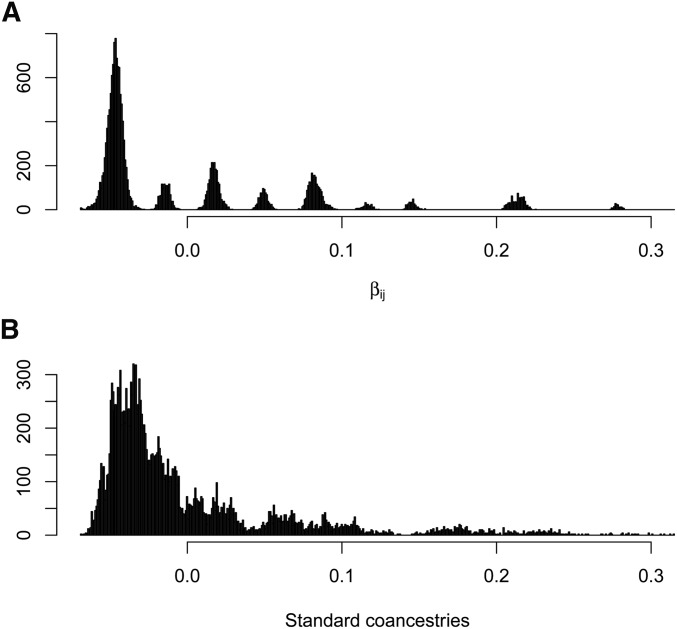
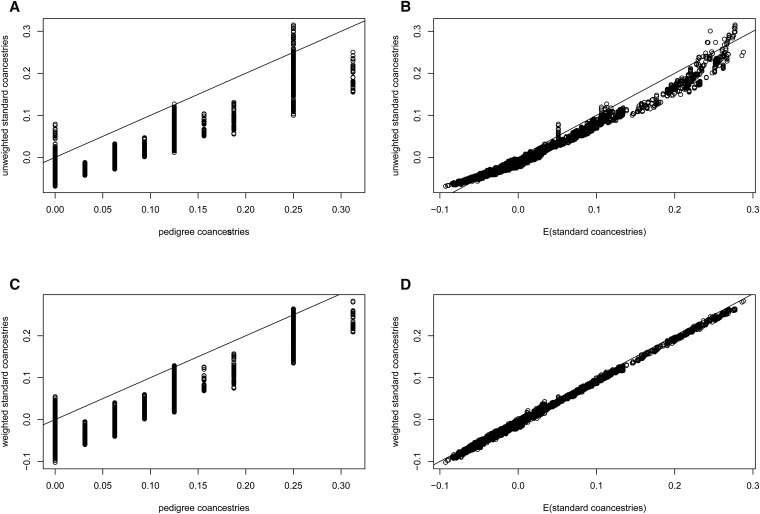
The distributions of estimates in Figure 7A are tightly clustered around nine values, corresponding to the nine distinct pedigree values A contrasting result is shown in Figure 7B, for the standard estimates (Equation 7), calculated as weighted averages over loci (i.e., taking the ratio of the sums over loci of the single-locus estimator numerators and denominators).
There is a current tendency in genome wide association studies (GWAS) to restrict the SNPs used in relatedness estimation to having a minor allele frequency (MAF) above some threshold. For example, the KING manual (http://people.virginia.edu/wc9c/KING/manual.html) lists a parameter minMAF to specify the minimum minor allele frequency to select SNPs for relationship inference in homogeneous populations. The thought is that lesser frequencies give rise to biased values, but that is not likely the case if “ratio of averages” estimates are used. To illustrate the effect of MAF filtering, we applied four different thresholds to our simulated data, and we show the means and SDs for estimates for each of nine pedigree values in Table 7. The estimates are the corrected values – i.e., relative to an assigned value of zero for the least-related class. There is clear evidence for the merits of retaining all SNPs, both in terms of bias and variance: all filtered estimates are downwardly biased, and the stronger the filter, the stronger the downward bias.
Table 7. Effects of filtering to L SNPs on coancestry estimate means (and SDs ).
| Pedigree value | ||||
|---|---|---|---|---|
| All SNPs | MAF | MAF | MAF | |
| 0 | 0.000 (0.50) | 0.000 (1.00) | 0.000 (1.99) | 0.000 (2.43) |
| 0.03125 | 0.031 (0.30) | 0.026 (0.30) | 0.010 (0.89) | 0.003 (1.45) |
| 0.06750 | 0.061 (0.34) | 0.056 (0.35) | 0.041 (1.13) | 0.036 (1.79) |
| 0.09375 | 0.092 (0.27) | 0.087 (0.27) | 0.069 (0.72) | 0.061 (1.13) |
| 0.12500 | 0.124 (0.41) | 0.120 (0.46) | 0.112 (1.90) | 0.109 (2.69) |
| 0.15625 | 0.156 (0.29) | 0.151 (0.29) | 0.133 (0.65) | 0.122 (1.15) |
| 0.18750 | 0.184 (0.26) | 0.179 (0.27) | 0.157 (1.07) | 0.144 (1.64) |
| 0.25000 | 0.249 (0.42) | 0.245 (0.45) | 0.241 (1.87) | 0.239 (2.62) |
| 0.31250 | 0.311 (0.20) | 0.307 (0.20) | 0.285 (0.77) | 0.271 (1.23) |
We continued a comparison of our proposed coancestry estimates by applying the estimates described by Wang (2014), listed in Table 8, and computed using the related R package (Pew et al. 2015). Additionally, related offers maximum likelihood estimators, derived by Milligan (2003) and Wang and Santure (2009). They are not computed here, because they require substantial computing time, which may rule them out for genomic data.
Table 8. Other estimates of relatedness.
| Method | Description |
|---|---|
| ped | The pedigree based relatedness |
| bij | developed here (Equation 4). These values are relative to the mean of the population and hence the mean of these relatedness must be 0 |
| stand.u | The standard estimator, Equation 7 average of ratios |
| Identical to the estimator derived by Ritland (1996) [Equation (4) in Wang (2014)] and also used in GCTA Yang et al. (2011) | |
| stand.w | Equation 7, ratio of averages |
| wang | The estimator developed by Wang (2002) |
| lynchli | The estimator derived by Lynch (1988) and improved by Li et al. (1993), Equation (7)in Wang (2014) |
| lynchrd | The estimator derived by Lynch and Ritland (1999) [Equations (5 and 6) in Wang (2014)] |
| quellergt | The estimator derived by Queller and Goodnight (1989) [Equations (2 and 3) in Wang (2014)] |
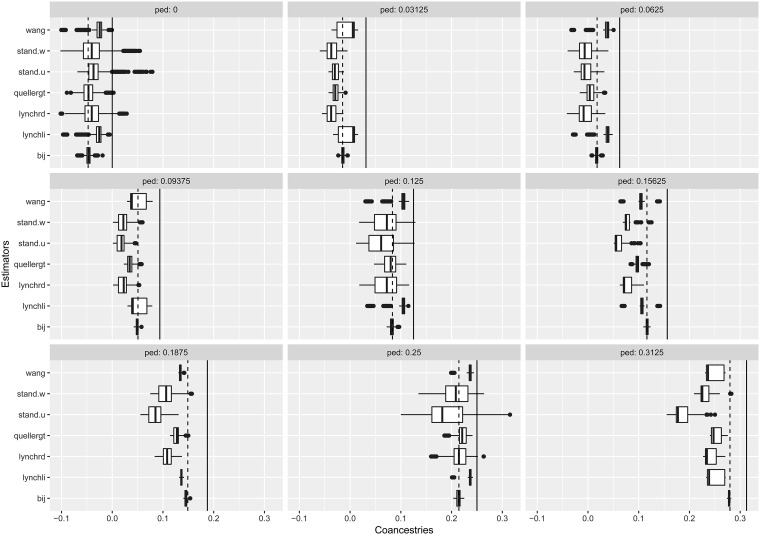
In Figure 8 we display box plots of coancestry estimates for seven alternative estimates, displayed according to nine pedigree values. The solid line for each panel corresponds to the pedigree value. The dashed line corresponds to an adjusted pedigree value, where the adjustment is obtained by subtracting the mean pedigree coancestry from the pedigree values, and dividing this by 1 – the mean pedigree value to insure that the range of possible values are covered. In Figure 6, we used estimates from the least related individuals to adjust the estimates, whereas here we adjusted the pedigree values to have an overall mean of zero.
Figure 8.
Boxplots of coancestry estimates for seven alternative estimates, displayed according to nine pedigree values. Vertical solid black line on each panel shows the pedigree coancestry, and vertical dashed line shows the mean-adjusted pedigree coancestry (see text). Estimators are defined in Table 6. shows very good statistical properties for all mean-adjusted pedigree coancestries.
All the estimates are negatively biased when compared to the pedigree values. When compared to the adjusted pedigree value, the β estimates show extremely good properties, with no bias, and very small variances. Other estimates, while also closer to these adjusted values, mostly underestimate, but sometimes overestimate (e.g., wang, lynchli) the adjusted pedigree values. The standard estimators (weighted or unweighted) consistently underestimate the adjusted pedigree values, except for the unrelated class.
Next, we illustrate how we can recover the average from the individual coancestries. For this, we use the pedigree described above, but take as founders 10 individuals from each of the two populations (mean between these two populations is ). Figure 9 illustrates the accuracy of our β estimates (Equation 6) compared with the standard estimates (Equation 7), for the coancestries of pairs of founders (but with the whole pedigree as the reference population). The values for pairs of founders from the same population (Boxplot A in Figure 9) are tightly distributed around 0.016, while ’s for pairs of individuals one from each population (boxplot B) are tightly distributed around The distribution for the same two categories for the standard estimator (boxplots C and D) is wider, in particular for pairs of individuals originating from the same population.
Figure 9.
Boxplots of coancestry estimates β (A and B) and the standard estimates (C and D) when the founders come from two populations. Coancestries were estimated for all the individuals in the pedigree shown in Figure 5, but only coancestries between founders are shown. (A and C): pairs of founders from the same population (B and D) pairs when the two members come from different populations.
The i.e., the average for the two populations from which the founders originated, is recovered from the individual coancestries as follows: each individual pair coancestry is calculated as (Table 3; the superscript p highlights that the estimates are taken over all pairs in the pedigree). We are seeking the overall among the founders only. The mean coancestry of founders from the same population in Figure 9 (boxplot A) corresponds to and the mean coancestry of founders, one from each population in the same figure (boxplot B) corresponds to Subtracting from and dividing by allows elimination of and recovery of the expression of For our situation, this gives as expected.
Discussion
A unified approach
Although there has been general recognition that family and evolutionary relatedness are just two ends of a continuum, we are not aware of previous moment estimates of population structure quantities such as or individual-pair coancestries that rest on this common framework. We have presented estimates that apply equally well to populations and individuals. While their statistical properties remain to be fully explored, it is reassuring to see how well they performed in the few simulations presented here.
Although individual-specific inbreeding coefficient, and individual-pair-specific coancestry coefficient moment estimates, are used routinely in association studies, we have not seen widespread adoption of population-specific moment estimates in evolutionary studies. We have shown here, theoretically and empirically, that these values can differ substantially among populations. This may simply reflect population size and migration rate differences, but different values for specific loci may also provide signatures of natural selection: see Balding and Nichols (1995), Beaumont and Balding (2004), Foll and Gaggiotti (2008) and Weir et al. (2005) for example. There is a growing literature for Bayesian analyses that address population-specific parameters (e.g., Karhunen and Ovaskainen 2012; Günther and Coop 2013), although these may not be amenable to analyses of genome-wide variant data.
There is also general understanding that identity by descent is a relative concept, rather than an absolute concept. This understanding has not led to an apparent recognition that the standard estimates of inbreeding and kinship are not unbiased for expected or pedigree values. Replacing population allele frequencies by sample values leads to bias in the usual estimates, regardless of sample size. Whenever sample allele frequencies from a study are used to estimate inbreeding or coancestry coefficients, the estimators are affected by the inbreeding and coancestry values for all study individuals. We will come back to this point in the section containing Equation 13
We also stress that all allelic variants, whatever their frequencies, need to be included in the estimation of population structure and inbreeding or relatedness. The estimates certainly depend on the allele frequencies, and restricting the range of frequencies used may reveal features of interest, but the underlying ibd parameters do not depend on the frequencies (see Equation 1 with the ibd interpretation). Exclusion of some alleles based on their frequencies will lead to biased estimates of the parameters as shown in Table 7.
Previous estimates
Weir and Cockerham estimates of :
The estimate of WC84 has been widely adopted, and it performs well for the model stated in that paper: data from a series of independent populations with equivalent histories and sizes. In the present notation, WC84 assumed for all populations i and all The estimate was designed to be unbiased for any number of sampled populations, any sample sizes and any number of alleles per locus. The analysis was a weighted one over populations: the average allele frequencies for a study had sample size weights, for alleles sampled from population i. Although our β estimates do not make explicit mention of allele frequencies, there is implicit use of sample frequencies that are unweighted averages over populations.
Weighting over populations has been discussed by Tukey (1957) and Robertson (1962). Those authors were concerned with bias and variance, and they used the language of variance components, within and between populations. For allele u, these components were given as and respectively, by WC84. Tukey said “In practice, we select two quadratic functions by some scheme involving intuition, find how their average values are expressed linearly in terms of the variance components, and then form two linear combinations of the original quadratics whose average values are the variance components. These linear combinations are then our estimates. Much flexibility is possible.” The estimates of WC84, Weir and Hill (2002) and Bhatia et al. (2013) all have this structure, although ratios of linear combinations are taken to remove the allele frequency parameters. Tukey went on to say that the weights (in the present notation) “gives the customary analyses, which treat observations as important and columns [i.e. populations] as unimportant.” Further, “the choice … treat the columns as important. This [unweighted] approach is appropriate when the column variance component is large compared with the within variance component.” Robertson (1962) also pointed to sample-size weights for small between-population variance components and equal weights for large values. Bhatia et al. (2013) were concerned with unequal values so their use of equal weights is consistent with Turkey’s statements. Their work provides simple averages of the different values as opposed to averages weighted by sample sizes. For unequal and unequal sample sizes, Weir and Hill (2002) said “the usual moment estimate [with sample-size weights] is of a complex function [of the ’s].” In our current model of unequal and nonzero we agree that unweighted analyses (population weights of 1) are appropriate, and that is what we have used in this paper. We note that Tukey’s “flexibility” in the choice of moment estimators, phrased in terms of weights, does not arise with maximum likelihood approaches. If sample allele frequencies are taken to be approximately normally distributed, then REML methods give appropriate and unique estimates.
What are the consequences of using the WC84 estimates when the current model of unequal and nonzero is more appropriate? We can show that the expected value of the Weir and Cockerham estimate is
This expression uses three functions of sample sizes: and The two weighted averages are and
The quantity Q is For equal sample sizes, or, for equal values of and Under these circumstances and we find the WC84 estimator performs well unless and/or values are quite different. We stress though that it is being estimated.
Nei estimates of :
Although we have phrased estimates in terms of matching proportions, we note that they are the complements of “heterozygosities” Our approach uses the average population-pair allele matching, whereas most previous treatments, from Nei (1973) onwards, use total heterozygosities where is the average sample allele frequency over populations: For large sample sizes, and Nei’s quantity and its expectation, in our notation, are
| (12) |
which reduce to and as r becomes large. Otherwise, the expectation of depends on the number r of populations. This expectation is bounded above by one, contrary to the claim of Bhatia et al. (2013). Nei and Chesser (1983) and Nei (1987) modified Nei’s earlier approach to remove the effects of the number of populations. Bounds on when that is defined as were given by Jakobsson et al. (2013).
Jost (2008) pointed out that does not provide a good measure of differentiation among populations, where differentiation reflects the collection of allele frequencies or their sample values We regard θ as an indicator of evolutionary history, rather than of allele frequencies, and we interpret it as probabilities of pairs of alleles being identical by descent. Jost introduced or as a measure of differentiation among populations. For the two-population drift scenario without mutation, D, unlike does not have a simple dependence on time, and so does not serve as a measure of evolutionary distance.
Standard coancestry estimates:
The expressions in Equation 7 provide unbiased estimates of and when the allele frequencies are known. When study sample allele frequencies are used, however, the expectations of these expressions, for one locus, are
| (13) |
where is the average of all within-individual coancestries is the average coancestry of individual j to all other individuals, and These expectations also hold for both the average over loci of the ratios for each locus, and for the ratio of averages when each locus has the same values of Note the difference with the expected values of shown in Table 2.
The differences diminish for studies with large numbers of individuals:
They diminish further for low average coancestries of the target individuals with other study individuals. An equivalent expression was given by Ochoa and Storey (2016b). The extent of bias of depends on the number of individuals in the sample, and how different the average coancestry of a target individual with all other study individuals is from the average coancestry of all pairs of study individuals. However, the standard estimates are not unbiased for This is illustrated in Figure 10, which displays, for the pedigree discussed previously (Figure 5) with founders originating from one population, the relation between the unweighted or weighted standard coancestry estimates (Equation 7) and pedigree coancestries in the left column, and the relation between the unweighted or weighted standard coancestries estimates and their expectation given by Equation 13 in the right column (B and D). The estimated standard coancestries do not match well the pedigree coancestries (Figure 10, A and C), contrary to the good match for (see Figure 6), which leads to the overdispersion of standard coancestry estimates seen in Figure 7B. But, standard coancestries match very well their expected values given by Equation 13, particularly so for the weighted standard coancestries (Figure 10D).
Figure 10.
Comparison of standard coancestry estimates (Equation 7) against pedigree coancestries (A and C) or against their expected values from Equation 13 (B and D), using the pedigree shown in Figure 5 and genotypes derived from founders originating from a single population. (A and B): unweighted standard coancestries; (C and D): weighted standard coancestries.
The standard estimates of Equation 7 appear as elements of the Genetic Relatedness Matrix (GRM) of Yang et al. (2011). We are grateful to P. Visscher (personal communication) for pointing out that the GRM was not designed for the purpose of kinship estimation, but was for estimating genetic variances in association mapping.
Population history
We commented earlier that can serve as a measure of genetic distance among populations in the sense that, for the genetic drift model, it depends on the time since the sampled populations diverged from an ancestral population. We see the need for further exploration of the role of population-specific estimates in evolutionary genetic studies, given the generally unrecognized prevalence of negative expected values for populations with correlated allele frequencies shown in Figure 1, and the relationship of estimates with the site-frequency spectrum suggested in Figure 4.
Conclusion
We have presented moment estimators for the probabilities that pairs of alleles, taken from individuals or from populations, are ibd relative to the ibd probabilities for alleles from all pairs of individuals or populations in a study. By identifying the reference set of alleles as those in the current study, we allow for negative values of measures of population structure or relatedness and their estimates. Alleles may have smaller ibd probabilities within some populations than between all pairs of populations in a study, for example. Some pairs of individuals in a study will be less related than the average for all pairs. Our estimates are phrased in terms of the proportions of pairs of alleles, within and between populations or individuals, that are of the same type (ibs).
For sets of populations, we advocate the use of population-specific values, as these more accurately reflect population history. For sets of individuals, our estimates seem to behave at least as well as those given previously. We note that our estimates have the same logical basis, and algebraic expressions, for populations and for individuals. The chief novelty of our method-of-moments approach is in allowing for allele frequencies to be correlated among populations when characterizing population structure, and correlated among all individuals when characterizing individual-pair relatedness.
Acknowledgments
We have benefited from discussions with Bill Hill, Peter Visscher, Loic Yengo Dimbou, and Oscar Gaggiotti. We also appreciate the helpful comments made by the reviewers and Associate Editor Graham Coop, and we are grateful for the encouragement of Senior Editor Lauren McIntyre. This work was supported in part by grants GM 075091, GM 099568, HL 120393, and contract HHSN268201300005C from the United States National Institutes of Health (NIH) and by grants 31003A_138180 and IZK0Z3_157867 from the Swiss National Science Foundation.
Footnotes
Communicating editor: G. Coop
Literature Cited
- 1000 Genomes Project Consortium, Abecasis, G. R., D. Altshuler, A. Auton, L. D. Brooks, R. M. Durbin et al., 2010 A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 (erratum: Nature 473: 544). [DOI] [PMC free article] [PubMed]
- Astle W., Balding D. J., 2009. Population structure and cryptic relatedness in genetic association studies. Stat. Sci. 24: 451–471. [Google Scholar]
- Balding D. J., 2003. Likelihood-based inference for genetic correlation coefficients. Theor. Popul. Biol. 63: 221–230. [DOI] [PubMed] [Google Scholar]
- Balding D. J., Nichols R. A., 1995. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica 96: 3–12. [DOI] [PubMed] [Google Scholar]
- Beaumont M. A., 2005. Adaptation and speciation: what can tell us? Trends Ecol. Evol. 20: 435–440. [DOI] [PubMed] [Google Scholar]
- Beaumont M. A., Balding D. J., 2004. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 13: 969–980. [DOI] [PubMed] [Google Scholar]
- Bhatia G., Patterson N., Sankararaman S., Price A. L., 2013. Estimating and interpreting the impact of rare variants. Genome Res. 23: 1514–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., Weir B. S., 2010. Population structure with localized haplotype clusters. Genetics 185: 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckleton J. S., Curran J. M., Goudet J., Taylor D., Thiery A., et al. , 2016. Population-specific values for forensic STR markers: a worldwide survey. Forensic Sci. Int. Genet. 23: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham C. C., 1969. Variance of gene frequencies. Evolution 23: 72–84. [DOI] [PubMed] [Google Scholar]
- Cockerham C. C., Weir B. S., 1983. Variance of actual inbreeding. Theor. Popul. Biol. 23: 85–109. [DOI] [PubMed] [Google Scholar]
- Epperson B. K., 1999. Gustave Malécot, 1911–1998: population genetics founding father. Genetics 152: 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M., Gaggiotti O., 2008. A genome scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180: 977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R., Gelfand A. E., Holsinger K. E., 2003. Exact moments calculations for genetic models with migration, mutation and drift. Theor. Popul. Biol. 63: 231–243. [DOI] [PubMed] [Google Scholar]
- Fu R., Dey D. K., Holsinger K. E., 2005. Bayesian models for the analysis of genetic structure when populations are correlated. Bioinf. 21: 1516–1529. [DOI] [PubMed] [Google Scholar]
- Gaggiotti O. E., Foll M., 2010. Quantifying population structure using the F-model. Mol. Ecol. Resour. 10: 821–830. [DOI] [PubMed] [Google Scholar]
- Goudet J., 2005. hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5: 184–186. [Google Scholar]
- Goudet J., Raymond M., De-Meeus T., Rousset F., 1996. Testing differentiation in diploid populations. Genetics 144: 1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther T., Coop G., 2013. Robust identification of local adaptation from allele frequencies. Genetics 195: 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D. L., Clark A. G., 1997. Principles of Population Genetics, Ed. 3 Sinauer Associates, Sunderland, MA. [Google Scholar]
- Hill W. G., Weir B. S., 2004. Moment estimation of population diversity and genetic distance from data on recessive markers. Mol. Ecol. 13: 895–908 (erratum: Mol. Ecol. 13: 3617). [DOI] [PubMed] [Google Scholar]
- Hill W. G., Weir B. S., 2011. Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genet. Res. 93: 47–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Weir B. S., 2012. Variation in actual relationship among descendants of inbred individuals. Genet. Res. 94: 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger K. E., Lewis P. O., Dey D. K., 2002. A Bayesian approach to inferring population structure from dominant markers. Mol. Ecol. 11: 1157–1164. [DOI] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Slatkin M., Maddison W. P., 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M., Edge M. D., Rosenberg N. A., 2013. The relationship between and the frequency of the most frequent allele. Genetics 193: 515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L., 2008. G(ST) and its relatives do not measure differentiation. Mol. Ecol. 17: 4015–4026. [DOI] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S. Y., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhunen M., Ovaskainen O., 2012. Estimating population-level coancestry coefficients by an admixture F model. Genetics 192: 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. C., Weeks D. E., Chakravarti A., 1993. Similarity of DNA fingerprints due to chance and relatedness. Hum. Hered. 43: 45–52. [DOI] [PubMed] [Google Scholar]
- Lynch M., 1988. Estimation of relatedness by DNA fingerprinting. Mol. Biol. Evol. 5: 584–599. [DOI] [PubMed] [Google Scholar]
- Lynch M., Ritland K., 1999. Estimation of pairwise relatedness with molecular markers. Genetics 152: 1753–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A., Mychaleckyj J. C., Rich S. S., Daly K., Sal M., et al. , 2010. Robust relationship inference in genome-wide association studies. Bioinformatics 26: 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., 1970. Effective number of alleles in a subdivided population. Theor. Popul. Biol. 1: 273–306. [DOI] [PubMed] [Google Scholar]
- Mathieson I., McVean G., 2012. Differential confounding of rare and common variants in spatially structured populations. Nat. Genet. 44: 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTavish E. J., Hillis D. M., 2015. How do SNP ascertainment schemes and population demographics affect inferences about population history? BMC Genomics 16: 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan B. G., 2003. Maximum-likelihood estimation of relatedness. Genetics 163: 1153–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., 1973. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York. [Google Scholar]
- Nei M., Chesser R. M., 1983. Estimation of fixation indices and gene diversities. Ann. Hum. Genet. 47: 253–259. [DOI] [PubMed] [Google Scholar]
- Nicholson G., Smith A. V., Johnson F., Gustafsson O., Stefansson K., et al. , 2002. Assessing population differentiation and isolation from single-nucleotide polymorphism data. J. Roy. Stat. Soc. B. Statistical Methodology 64: 695–715. [Google Scholar]
- Ochoa, A., and J. Storey, 2016a and kinship for arbitrary population structures I: generalized definitions. bioRxiv DOI: 10.1101/083915 [DOI] [PMC free article] [PubMed]
- Ochoa, A., and J. Storey, 2016b and kinship for arbitrary population structures II: method of moments estimators. bioRxiv DOI: 10.1101/083923 [DOI] [PMC free article] [PubMed]
- Peter B. M., 2016. Admixture, population structure, and F-statistics. Genetics 202: 1485–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pew J., Muir P. H., Wang J., Frasier T. R., 2015. Related: an R package for analysing pairwise relatedness from codominant molecular markers. Mol. Ecol. Resour. 15: 557–561. [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queller D. C., Goodnight K. F., 1989. Estimating relatedness using molecular markers. Evolution 43: 258–275. [DOI] [PubMed] [Google Scholar]
- Reich D., Thangaraj K., Patterson N., Price A. L., Singh L., 2009. Reconstructing Indian population history. Nature 461: 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J., Weir B. S., Cockerham C. C., 1983. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105: 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritland K., 1996. Estimators for pairwise relatedness and individual inbreeding coefficients. Genet. Res. 67: 175–185. [Google Scholar]
- Robertson A., 1962. Weighting in the estimation of variance components in the unbalanced single classification. Biometrics 18: 3–17. [Google Scholar]
- Rousset F., 1996. Equilibrium values of measures of population subdivision for stepwise mutation processes. Genetics 142: 1357–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver M. D., Kennedy G. C., Parra E. J., Lawson H. A., Sonpa V., et al. , 2004. The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum. Genomics 41: 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., 1985. Rare alleles as indicators or gene flow. Evolution 39: 53–65. [DOI] [PubMed] [Google Scholar]
- Song S., Dey D. K., Holsinger K. E., 2006. Differentiation among populations with migration, mutation and drift: implications for genetic inference. Evolution 60: 1–12. [PMC free article] [PubMed] [Google Scholar]
- Speed D., Balding D. J., 2015. Relatedness in the post-genomic era: is it still useful? Nat. Rev. Genet. 16: 33–44. [DOI] [PubMed] [Google Scholar]
- Thompson E. A., 1975. Estimation of pairwise relationships. Ann. Hum. Genet. 39: 173–188. [DOI] [PubMed] [Google Scholar]
- Thompson E. A., 2013. Identity by descent: variation in meiosis, across genomes, and in populations. Genetics 194: 301–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey J. W., 1957. Variances of variance components: II. The unbalanced single classification. Ann. Math. Stat. 28: 43–56. [Google Scholar]
- Wang B., Sverdlov S., Thompson E., 2017. Efficient estimation of realized kinship from SNP genotypes. Genetics 205: 1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., 2002. An estimator for pairwise relatedness using molecular markers. Genetics 160: 1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., 2014. Marker-based estimates of relatedness and inbreeding coefficients: an assessment of current methods. J. Evol. Biol. 27: 518–530. [DOI] [PubMed] [Google Scholar]
- Wang J., Santure A. W., 2009. Parentage and sibship inference from multilocus genotype data under polygamy. Genetics 181: 1579–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. S., 1996. Genetic Data Analysis II. Sinauer, Sunderland, MA. [Google Scholar]
- Weir B. S., Cockerham C. C., 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Weir B. S., Hill W. G., 2002. Estimating F-statistics. Annu. Rev. Genet. 36: 721–750. [DOI] [PubMed] [Google Scholar]
- Weir B. S., Cardon L. R., Anderson A. D., Nielsen D. M., Hill W. G., 2005. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 15: 1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1922. Coefficients of inbreeding and relationship. Am. Nat. 56: 330–338. [Google Scholar]
- Wright S., 1931. Evolution in Mendelian populations. Genetics 16: 97–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1943. Isolation by distance. Genetics 28: 114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1951. The genetical structure of populations. Ann. Eugen. 15: 323–354. [DOI] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., Visscher P. M., 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W. H., Bi I. V., Yamasaki M., et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.