Males and females exhibit marked differences in phenotypes and gene expression, particularly in the gonads. Genes with male- or testisbiased expression..
Keywords: ovary-biased genes, gonads, Aedes aegypti, molecular evolution, protein sequence divergence, Genetics of Sex
Abstract
Males and females exhibit highly dimorphic phenotypes, particularly in their gonads, which is believed to be driven largely by differential gene expression. Typically, the protein sequences of genes upregulated in males, or male-biased genes, evolve rapidly as compared to female-biased and unbiased genes. To date, the specific study of gonad-biased genes remains uncommon in metazoans. Here, we identified and studied a total of 2927, 2013, and 4449 coding sequences (CDS) with ovary-biased, testis-biased, and unbiased expression, respectively, in the yellow fever mosquito Aedes aegypti. The results showed that ovary-biased and unbiased CDS had higher nonsynonymous to synonymous substitution rates (dN/dS) and lower optimal codon usage (those codons that promote efficient translation) than testis-biased genes. Further, we observed higher dN/dS in ovary-biased genes than in testis-biased genes, even for genes coexpressed in nonsexual (embryo) tissues. Ovary-specific genes evolved exceptionally fast, as compared to testis- or embryo-specific genes, and exhibited higher frequency of positive selection. Genes with ovary expression were preferentially involved in olfactory binding and reception. We hypothesize that at least two potential mechanisms could explain rapid evolution of ovary-biased genes in this mosquito: (1) the evolutionary rate of ovary-biased genes may be accelerated by sexual selection (including female–female competition or male–mate choice) affecting olfactory genes during female swarming by males, and/or by adaptive evolution of olfactory signaling within the female reproductive system (e.g., sperm-ovary signaling); and/or (2) testis-biased genes may exhibit decelerated evolutionary rates due to the formation of mating plugs in the female after copulation, which limits male–male sperm competition.
SEX-BIASED genes, that is, genes whose expression is upregulated in one sex relative to the other, are believed to underlie sexual dimorphism and have been extensively reported in anisogamous eukaryotes including mammals, birds, fish, insects, worms, fungi, higher plants, and algae (Meiklejohn et al. 2003; Ranz et al. 2003; Zhang et al. 2004; Cutter and Ward 2005; Yang et al. 2006; Ellegren and Parsch 2007; Haerty et al. 2007; Whittle et al. 2007; Small et al. 2009; Assis et al. 2012; Parsch and Ellegren 2013; Whittle and Johannesson 2013; Lipinska et al. 2015; Wang et al. 2015). The number of genes exhibiting sex bias can represent a majority of the genome, with estimates from 50 to >75% of the genome being sex-biased in insect genera such as Drosophila and Nasonia (Ranz et al. 2003; Ellegren and Parsch 2007; Assis et al. 2012; Wang et al. 2015). The sex biases in expression appear to have molecular evolutionary consequences. For example, in animals studied to date, male-biased genes typically evolve more rapidly at the protein sequence level, exhibiting elevated ratios of nonsynonymous to synonymous substitutions (dN/dS), than female-biased or unbiased genes [reviewed by Ellegren and Parsch (2007); see also Mank et al. (2007) and Parsch and Ellegren (2013)]. Evidence suggests that sex biases in expression between whole individuals might largely result from the sex-limited organs, namely gonads and their contained sex cells (Arbeitman et al. 2002, 2004; Parisi et al. 2004; Ellegren and Parsch 2007; Small et al. 2009; VanKuren and Vibranovski 2014). This is consistent with the fact that the gonads comprise highly differentiated organs, with extremely divergent transcriptomes, between males and females (Parisi et al. 2004; Harrison et al. 2015). In this regard, gonadal gene expression may largely shape the evolution of many protein-coding genes.
Genes expressed in the gonads are believed to play a central role in evolution, contributing toward reproductive success and fitness, reproductive isolation, and speciation. These genes are also most apt to be influenced by major evolutionary processes such as sexual conflict, mate-choice, and intrasexual competition (Civetta and Singh 1998; Swanson and Vacquier 2002; Jagadeeshan and Singh 2005; Ellegren and Parsch 2007; Turner and Hoekstra 2008). With respect to molecular evolution of reproductive and gonad genes, studies in animals including mammals and insects have shown that certain seminal protein genes (Swanson et al. 2001), testis (Good and Nachman 2005; Khaitovich et al. 2005; Ellegren and Parsch 2007; Haerty et al. 2007), and sperm genes (Torgerson et al. 2002; Good and Nachman 2005; Ellegren and Parsch 2007; Haerty et al. 2007) typically evolve rapidly as compared to the rest of the genome, and thus agree with the global pattern of rapid sequence evolution of male-biased genes. Specific testis and ovary genes directly involved in fertilization have also been demonstrated to evolve rapidly in various animal systems (Civetta and Singh 1998; Swanson and Vacquier 2002). Nonetheless, observed effects of expression within sex and reproductive genes varies among studies and with the type of tissue examined. For example, seminal fluid protein sequences evolve fast in numerous models including Drosophila species (Haerty et al. 2007), but not in others such as A. albopictus (Boes et al. 2014), and while spermatogenesis genes typically evolve rapidly (including in Drosophila) (Ellegren and Parsch 2007; Haerty et al. 2007), some fly research suggests that sperm genes may be more conservative in their rates of evolution (Dorus et al. 2006). In this regard, while male reproductive genes typically evolve rapidly, this may not be universal to all male sexual tissues, and the signal varies to some extent among organisms and studies.
At present, studies in animals that specifically compare whole testis- vs. ovary-biased expression and its effect on the evolution of protein-coding DNA remain few, with the exception of some examples from organisms such as guppies (Poecilia: Sharma et al. 2014), various birds (Harrison et al. 2015), sea urchins (Allocentrotus and Strongylocentrotus: Oliver et al. 2010), and the greatest focus on the insect genus Drosophila (Jagadeeshan and Singh 2005; Ellegren and Parsch 2007; Haerty et al. 2007; Meisel 2011; Assis et al. 2012). In each of these example taxa, higher dN/dS was reported for the testis-linked genes than for the ovary-linked genes [with the exception of guppies, where testis- and ovary-biased genes both evolved rapidly compared to unbiased genes (Sharma et al. 2014)]. Such research is an important step in deciphering the role of gonads in evolution (Jagadeeshan and Singh 2005), and particularly in DNA sequence evolution.
In addition to protein sequence divergence (Ellegren and Parsch 2007; Parsch and Ellegren 2013), another facet of protein-coding DNA that may be related to sex-biased expression is optimal codon usage (Hambuch and Parsch 2005; Ellegren and Parsch 2007; Whittle et al. 2007), that is, those codons that promote efficient and/or accurate translation (Sharp et al. 1986; Duret and Mouchiroud 1999; Akashi 2001). Synonymous codon usage appears to often be nonarbitrary, and is believed to arise either from natural selection promoting biochemically efficient and accurate protein synthesis (Duret and Mouchiroud 1999; Duret 2000; Stoletzki and Eyre-Walker 2007) or from mutational bias (Osawa et al. 1988; Sueoka 1988; Sharp et al. 1995). A role of natural selection concurs with data showing that tRNA abundance and tRNA gene copy number match the most common codons in the genome (Ikemura 1981, 1985; Duret 2000), and that a subset of codons are preferentially used in highly transcribed genes (e.g., Sharp et al. 1986; Stenico et al. 1994; Duret and Mouchiroud 1999; Cutter et al. 2006; Ingvarsson 2008; Whittle et al. 2011; Whittle and Extavour 2016a). Optimization of codon usage has been shown in diverse eukaryotes including fungi, plants, worms, and insects such as Tribolium and Drosophila [Duret and Mouchiroud 1999; reviewed by Hershberg and Petrov (2008); Behura and Severson (2011, 2013); and Williford and Demuth (2012)]. However, signals have been weak or absent in other taxa, including the insect Bombyx (Jia et al. 2015; Whittle and Extavour 2016b). As optimal codons typically have been linked to selective pressures, the level of optimal codon usage may be influenced by sex-biased expression (Hambuch and Parsch 2005; Whittle et al. 2007), including selective pressures arising from the testes and ovaries.
Available data to date suggests that protein-coding sequence (CDS) and optimal codon usage might evolve in concert in some eukaryotes. For instance, in Drosophila, wherein male- (or testis-) biased genes evolve rapidly (Zhang et al. 2004; Ellegren and Parsch 2007; Haerty et al. 2007), it has been reported that male-biased genes exhibit lower frequency of optimal codons as compared to female-biased and unbiased genes (Hambuch and Parsch 2005). In addition, fast evolving sex-biased genes have been suggested to have reduced codon usage bias in other groups outside the metazoans. For example, in the hermaphrodite fungus Neurospora, a rare case of a fungal species wherein protein-coding genes with biased expression in female-limited organs (protoperithecia) evolve rapidly, the female genes also exhibit reduced optimal codon usage as compared to male organ- (conidia-) biased and unbiased genes (Whittle and Johannesson 2013). Similarly, rapidly evolving sex-biased genes show less optimal codon use in brown algae (Ectocarpus: Lipinska et al. 2015), and male-biased genes from the sex organs (anthers) in certain plants exhibit lower optimal codon usage than ovary-biased genes (Whittle et al. 2007). This frequently reported inverse correlation between rates of protein sequence evolution and optimal codon usage in sex-biased genes might result from adaptive evolution in proteins driving fixation of linked nonoptimal codons (selective sweeps) and/or from relaxed selection (Betancourt and Presgraves 2002; Kim 2004; Hambuch and Parsch 2005; Whittle and Johannesson 2013). In this regard, an understanding of the dynamics shaping the evolution of DNA coding for sex-biased gonad genes should include its relationship to both protein divergence and optimal codon usage.
To gain further insight into the evolution of sex-biased gonadal genes, research needs to be expanded to nontraditional models, including within insects, which comprise the largest and most diverse class of animals on earth, representing up to 90% of all known animal species (Erwin 1982). The yellow fever mosquito Aedes aegypti has gained momentum as a model in biology, and extensive large-scale genomic and transcriptome resources have recently been made available (Nene et al. 2007; Akbari et al. 2013; Chen et al. 2015), making it suitable for molecular evolutionary research. A. aegypti is a holometabolous insect that serves as a vector for human pathogens including yellow fever (Jentes et al. 2011), Dengue (Simmons et al. 2012), and chikungunya (Leparc-Goffart et al. 2014), which are transmitted by a female bite (Hall et al. 2015). Originating from Africa, over the last 40 years this insect has spread across all continents excluding Antarctica (Kraemer et al. 2015). It is a short-lived organism with a typical life cycle of 15–30 days (Christophers 1960). The reproductive biology of A. aegypti has been well-established. Mating competency typically begins in males that are at least 24-hr old, and mating usually results during male swarming, but also by single-pair mating (Oliva et al. 2014). While males are polygynous, they limit fertilization by other males both by transferring sufficient sperm to fertilize all eggs a female will lay in her lifetime, and by transmitting accessory gland secretions that act as physical and chemical barriers to insemination by other males [reviewed by Oliva et al. (2014)]. After mating, egg maturation generally occurs after blood feeding, but can occur in its absence in a small portion (3–4%) of females (Ariani et al. 2015). A. aegypti does not have heteromorphic sex chromosomes among its three pairs of chromosomes (Juneja et al. 2014). Instead, sex determination is governed by a dominant male-determining factor (M-factor) located on a homomorphic (sex-determining) chromosome (with a genotype of Mm in males and mm in females). In fact, sex may largely depend on a single gene in this region named Nix, which is found only in males and initiates male development, and its absence causes feminization (Hall et al. 2015). With respect to the genome, the complete sequence for A. aegypti is ∼1.38 GB, a relatively large size for an insect, and fivefold larger than the model mosquito Anopheles gambiae (Nene et al. 2007). The large genome size of A. aegypti is thought to result from an abundance of repetitive sequences (Nene et al. 2007; Juneja et al. 2014). Together, given its genomic and trancriptomic resources and its well-understood sexual biology, A. aegypti comprises a suitable system for the study of the molecular evolution of sex-biased gonad genes.
In the present study, we investigate the connection between sex-biased gonad expression and protein-CDS evolution in A. aegypti. First, using large-scale embryo transcriptomes, we identify a list of optimal codons in this mosquito, and observe clear signals that natural selection favors codons ending in G or C (GC3). Second, using testis and ovary transcriptomes, we identify CDS with sex-biased gonad expression. The data show that, as compared to testis-biased CDS, ovary-biased and unbiased CDS exhibit higher dN/dS and lower frequency of optimal codons (Fop; Wright 1990) in this insect. Surprisingly, these trends are opposite to the faster evolution of male-biased genes (and low Fop) reported in most organisms studied to date (Ellegren and Parsch 2007; Parsch and Ellegren 2013). Evaluation of ovary- and testis-specific gene expression as compared to embryos reveals that female gonad expression is consistently linked to higher dN/dS, even when these genes are additionally coexpressed in other tissues, and those patterns are connected to positive selection. Further, ovary genes were preferentially involved in olfactory functions. We compare sex-biased expression in nonreproductive male and female carcasses (defined as whole bodies without the gonads) and find that somatic female-biased genes evolve faster than somatic male-biased genes. It is shown that this effect largely results from coexpression of ovary-biased genes in the female soma. Together, we propose that the fast evolution of ovary-biased genes may result from sexual selection acting to accelerate evolution of female gonadal genes, including those involved in olfactory functions, and/or by reduced male–male sperm competition due to the formation of mating barriers in the female reproductive tract by male seminal fluids in A. aegypti.
Results and Discussion
Of the 15,796 protein-coding genes described for A. aegypti (Table S1 in File S1, assembly v3.29), we assessed expression in those CDS that started with an ATG start codon, had no unknown or ambiguous nucleotides, and comprised the longest CDS per gene when multiple isoforms were described. These filters yielded a total of 14,678 CDS for analysis. In addition to dN/dS, we wished to study if sex-biased gonadal expression shaped codon usage. For this, we first had to determine whether A. aegypti was a species where selection shaped its codon usage, which we assessed using comparative analysis of the large-scale transcriptome data set from embryos (see File S1 and specifically Table S1 contained within it).
Identification of optimal codons in A. aegypti
We report that A. aegypti exhibits strong signals showing that natural selection shapes optimal codon usage in this taxon (Table 1). As described in detail in Supplemental Note 1 in File S1, for the identification of optimal codons in this mosquito, we conducted contrasts of codon usage in highly (highest 5%) vs. lowly expressed (lowest 5%) genes, which included assessment of changes in ∆RSCU (Sharp et al. 1986) and correspondence analysis. Both of these approaches have precedent for the identification of codons preferentially used under high expression (optimal codons) (Duret and Mouchiroud 1999; Cutter et al. 2006; Ingvarsson 2008; Wang et al. 2011; Whittle et al. 2011; Whittle and Extavour 2016a). For details, see Supplemental Note 1 in File S1. The codon with the largest statistically significant and positive ∆RSCU per amino acid was taken as the primary optimal codon per amino acid. As shown in Table 1, we identified 18 primary optimal codons in A. aegypti, which ended in C or G.
Table 1. The ∆RSCU between highly (high) and lowly (low) expressed genes in A. aegypti.
| Amino acid | Codon | RSCU (High) | RSCU (Low) | ∆RSCU | P value | Correspondence analysis |
|---|---|---|---|---|---|---|
| Ala | GCT | 0.9704 | 0.9909 | −0.020 | ||
| Ala | GCC | 1.8235 | 1.2438 | +0.579 | ** | GCC |
| Ala | GCA | 0.6085 | 0.9658 | −0.357 | ** | |
| Ala | GCG | 0.5976 | 0.7994 | −0.201 | ** | |
| Arg | CGT | 1.4934 | 1.0031 | +0.490 | ** | |
| Arg | CGC | 1.6334 | 0.9633 | +0.670 | ** | CGC |
| Arg | CGA | 0.9562 | 1.3965 | −0.440 | ** | |
| Arg | CGG | 0.9879 | 1.1382 | −0.150 | * | |
| Arg | AGA | 0.5099 | 0.8639 | −0.354 | ** | |
| Arg | AGG | 0.3541 | 0.6256 | −0.271 | ** | |
| Asn | AAT | 0.6486 | 0.9013 | −0.252 | ** | |
| Asn | AAC | 1.3266 | 1.0987 | +0.227 | ** | AAC |
| Asp | GAT | 1.0194 | 1.1458 | −0.126 | ** | |
| Asp | GAC | 0.9433 | 0.8542 | +0.089 | ** | GAC |
| Cys | TGT | 0.5477 | 0.9489 | −0.401 | ** | |
| Cys | TGC | 1.1112 | 1.0227 | +0.088 | * | TGC |
| Gln | CAA | 0.6219 | 0.9189 | −0.297 | ** | |
| Gln | CAG | 1.3192 | 1.0811 | +0.238 | ** | CAG |
| Glu | GAA | 1.0727 | 1.1491 | −0.076 | ** | |
| Glu | GAG | 0.9118 | 0.8509 | +0.060 | ** | GAG |
| Gly | GGT | 1.095 | 1.0235 | +0.071 | * | |
| Gly | GGC | 1.1659 | 0.8714 | +0.294 | ** | GGC |
| Gly | GGA | 1.4366 | 1.5836 | −0.147 | ** | |
| Gly | GGG | 0.2838 | 0.5214 | −0.237 | ** | |
| His | CAT | 0.7198 | 0.9272 | −0.207 | ** | |
| His | CAC | 1.1469 | 1.0602 | +0.086 | * | CAC |
| Ile | ATT | 0.9168 | 1.076 | −0.159 | ** | |
| Ile | ATC | 1.8039 | 1.3602 | +0.443 | ** | ATC |
| Ile | ATA | 0.2421 | 0.5638 | −0.321 | ** | |
| Leu | TTA | 0.2848 | 0.4383 | −0.153 | ** | |
| Leu | TTG | 1.1941 | 1.4717 | −0.277 | ** | |
| Leu | CTT | 0.5594 | 0.7806 | −0.221 | ** | |
| Leu | CTC | 0.8132 | 0.7709 | +0.042 | ||
| Leu | CTA | 0.4033 | 0.6167 | −0.213 | ** | |
| Leu | CTG | 2.708 | 1.922 | +0.786 | ** | CTG |
| Lys | AAA | 0.6329 | 0.9424 | −0.309 | ** | |
| Lys | AAG | 1.3515 | 1.0419 | +0.309 | ** | AAG |
| Phe | TTT | 0.4731 | 0.7328 | −0.259 | ** | |
| Phe | TTC | 1.499 | 1.2673 | +0.231 | ** | TTC |
| Pro | CCT | 0.461 | 0.7075 | −0.246 | ** | |
| Pro | CCC | 0.9911 | 0.6824 | +0.308 | ** | CCC or CCG |
| Pro | CCA | 1.0912 | 1.1852 | −0.094 | * | |
| Pro | CCG | 1.4071 | 1.4248 | −0.017 | ||
| Ser | TCT | 0.5852 | 0.6641 | −0.078 | * | |
| Ser | TCC | 1.6615 | 1.0864 | +0.575 | ** | TCC or TCG |
| Ser | TCA | 0.4453 | 0.7651 | −0.319 | ** | |
| Ser | TCG | 1.5813 | 1.3423 | +0.238 | ** | |
| Ser | AGT | 0.6007 | 1.0136 | −0.412 | ** | |
| Ser | AGC | 1.098 | 1.1286 | −0.030 | ||
| Thr | ACT | 0.6453 | 0.8135 | −0.168 | ** | |
| Thr | ACC | 1.8154 | 1.2705 | +0.544 | ** | ACC |
| Thr | ACA | 0.5641 | 0.7742 | −0.210 | ** | |
| Thr | ACG | 0.9379 | 1.1417 | −0.203 | ** | |
| Tyr | TAT | 0.5447 | 0.7627 | −0.217 | ** | |
| Tyr | TAC | 1.3839 | 1.2342 | +0.149 | ** | TAC |
| Val | GTT | 0.995 | 1.08 | −0.085 | * | |
| Val | GTC | 1.2726 | 0.8775 | +0.395 | ** | GTC |
| Val | GTA | 0.4135 | 0.634 | −0.220 | ** | |
| Val | GTG | 1.2939 | 1.4084 | −0.114 | * |
Gene expression was measured using embryos. Putative optimal codons are in bold and underlined. The primary optimal codon per amino acid predicted by correspondence analysis is also shown. A total of 18 primary optimal codons were identified. * P < 0.05, ** P < 0.001 using t-tests. ∆RSCU, change in relative synonymous codon usage (rounded down to the nearest 1/1000); RSCU, relative synonymous codon usage (mean values); note that standard errors ranged between 0.010 and 0.050 and are not shown for presentation purposes.
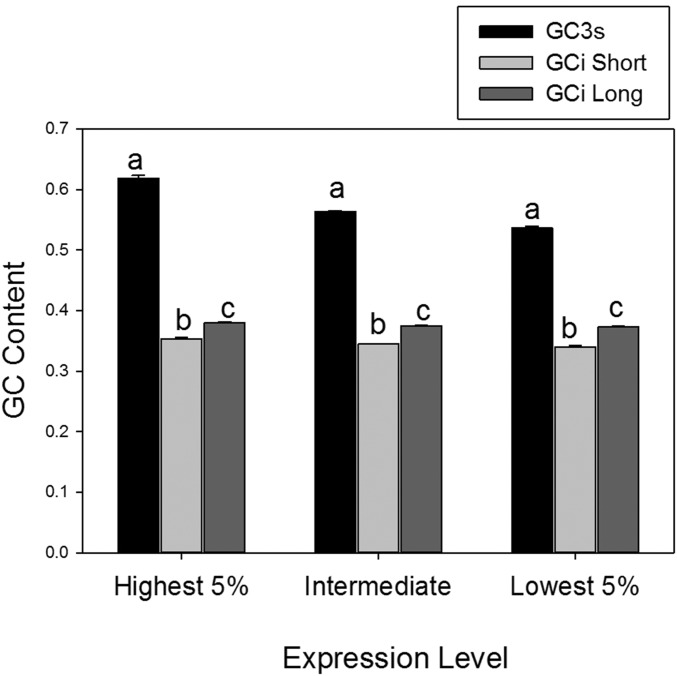
The strong link observed between expression level and C- and G-ending codons, as found here, is a good indicator that selection drives the use of those codons (Sharp et al. 1986; Stenico et al. 1994; Duret and Mouchiroud 1999; Cutter et al. 2006; Ingvarsson 2008; Whittle et al. 2011). Nonetheless, for an additional layer of stringency, we wanted to fully exclude that mutational bias may have caused differences between expression classes (Beletskii and Bhagwat 1996; Comeron 2004). For instance, it is possible that highly transcribed genes could have a mutational bias (such as reported for C to T mutations) or a transcription-coupled mutational asymmetry (Beletskii and Bhagwat 1996; Green et al. 2003) causing disproportionately greater usage of C3 or G3 codons in A. aegypti. To further confirm optimal codon usage was due to natural selection and not mutational bias, one can compare the GC content of synonymous GC3s to selectively neutral regions such as introns (GCi), where the latter should reflect mutational pressures [note that GC3s is an effective proxy of optimal codon usage, as the frequency of optimal codons (Fop, Ikemura 1981) correlates strongly to GC3s: R = 0.93 and P < 2.0 × 10−7; Figure S1 in File S1]. For this, as described in detail in File S1, we examined 36,272 introns and GC3 content in the genes under study. The introns were divided by length [short (≤130 bp) and long introns (>130 bp) (Farlow et al. 2012)], and for the long introns, 50 bp were removed from the ends, which may contain regulatory regions (Williford and Demuth 2012). The results showed that the GC3s for the CDS with the highest 5% expression in embryos (average GC3s = 0.618 ± 0.005), intermediate 5th to 95th expression level (0.564 ± 0.001), and the CDS with lowest 5% expression (GC3s = 0.536 ± 0.003), were each markedly and statistically significantly higher than the GCi content of their associated introns [for both short and long introns], which averaged between 0.340 and 0.380 (Figure 1; Mann-Whitney U (MWU)-test P < 0.001 for all contrasts). Further, unlike GC3s, which increased with expression, GCi was not elevated with respect to transcription level (and slightly decreased for short introns, Figure 1), also showing that mutational biases could not explain optimal codon usage in this taxon.
Figure 1.
The average GC content at synonymous third codon positions (GC3s) and the GC content of introns (GCi). Results are shown for the set of coding sequences (CDS) with high (highest 5%), intermediate (between the 5th and 95th percentile), or low (lowest 5%) expression in A. aegypti embryos. GCi for short (≤130 bp) and long (>130 bp) introns (after removal of 50 bp at each end) per expression class are shown. Different letters (a, b, or c) above the bars showing mean values of GC3s, GCi (short) and GCi (long) within each expression class indicate a statistically significant difference (P < 0.05) using paired Mann-Whitney U (MWU) tests (the P-value was <0.001 in each contrast). Note that the GC3s values were also statistically significant different between the three expression classes using paired contrasts (MWU-test P < 0.001). Error bars are standard errors.
Together, we conclude that the preferential use of 18 optimal codons under high expression in A. aegypti (Table 1) is caused by natural selection. Thus, in addition to dN/dS, we were able to study how sex-biased gonadal expression affects selection on optimal codon usage (see below).
Expression profiles of testis- and ovary-biased genes in A. aegypti
We identified CDS with sex-biased expression in testes (plus accessory glands; henceforward denoted as testes) and ovaries in A. aegypti using large-scale transcriptome data sets (Akbari et al. 2013) and studied their molecular evolution, including protein sequence divergence and optimal codon usage. To measure interspecies divergence (Yang 2007), we used a species from the same genus, A. albopictus (cf. Zhang et al. 2004; Proschel et al. 2006; Assis et al. 2012), whose genome sequence has recently become available (Chen et al. 2015). A total of 9389 putative orthologs were identified between A. aegypti and A. albopictus (see Materials and Methods). For consistency across all results, this CDS list was used for all of our subsequent sex-biased expression and molecular evolutionary analysis. Sex-biased genes were defined as those with FPKM > 1 in at least one tissue, at least a twofold difference between testis and ovary expression, and P < 0.05 using differential expression analysis in A. aegypti (see Materials and Methods). All CDS not matching these criteria were defined as unbiased. A total of 2927, 2013, and 4449 CDS were identified for study as testis-biased, ovary-biased, and unbiased, respectively, suggesting a greater propensity for CDS to be involved in testis than ovary function.
Fold sex bias in the gonads is linked to expression level
We assessed the relationship between the degree of sex-biased gonadal expression (fold bias) and expression level in A. aegypti. The results showed that greater ovary and testis bias were each linked to elevated expression levels in A. aegypti. For this analysis, the testis- and ovary-biased CDS were each classified into three nonoverlapping fold bias classes: (1) ≥2-fold bias and <5-fold bias; (2) ≥5-fold bias and <10-fold bias; and (3) ≥10-fold bias. This classification approach is comparable to previous male/female sex bias studies, and allows observations of clear demarcations with respect to bias level (Lipinska et al. 2015; Perry et al. 2015). In addition, for rigor, given that prior data in eukaryotes indicate that expression level can be inversely related to CDS length, and particularly elevated for the shortest CDS in eukaryotic genomes (Akashi 2001, 2003; Urrutia and Hurst 2003; Comeron 2004; Lemos et al. 2005; Williford and Demuth 2012; Whittle and Extavour 2016a), the testis- and ovary-biased genes were divided into short (≤250 aa) and long (>250 aa) length classes. The CDS classes were separated using the 25th percentile approximated across all 9389 CDS under study (cf. Bloom et al. 2006).
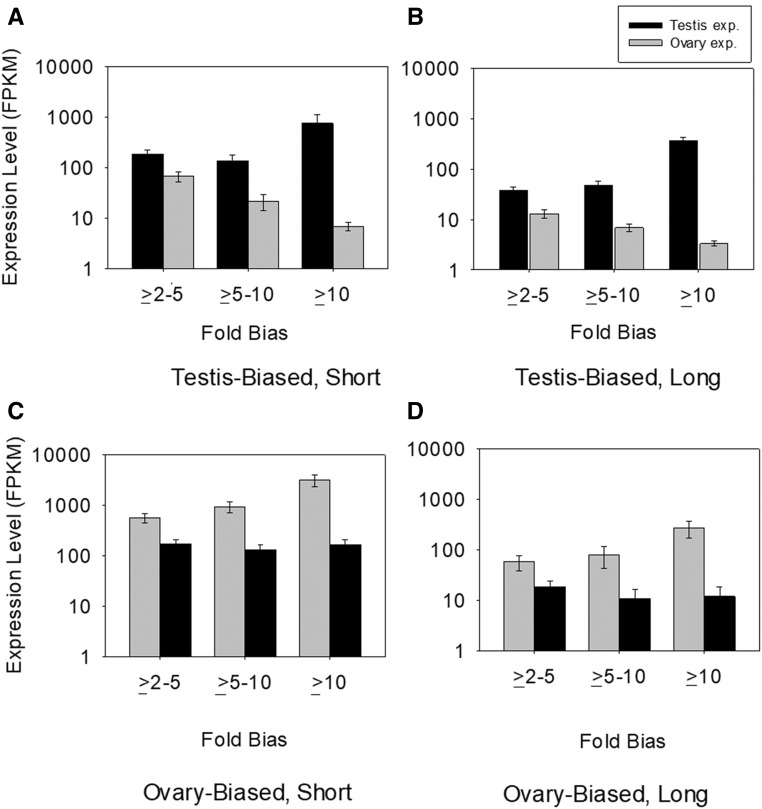
As shown in Figure 2, an increase in testis-fold bias resulted from both elevated testis expression and reduced ovary expression. Specifically, as testis-fold bias progressively increased from ≥2-5 fold bias, to ≥5-10 fold bias, and to the ≥10-fold bias classes, there was elevated average (and median) expression level in testes, and reduced expression in ovaries, for both short and long CDS (MWU-tests P < 0.001 for all contrasts between fold bias classes). In fact, CDS with ≥10-fold bias had the highest expression in testis (average = 771.6 ± 347.1 FPKM for short CDS and 368.2 ± 57.7 FPKM for long CDS), and the lowest expression in ovaries (6.9 ± 1.2 FPKM for short CDS and 3.3 ± 0.4 for long CDS) among all of the 3-fold bias categories (MWU-tests P < 0.001 for all contrasts, Figure 2, A and B). This trend concurs with findings in whole (early-stage) male/females in Drosophila and from sexual structures (gametophytes) in brown algae (Ectocarpus), where greater male bias resulted from both elevated expression in male tissues and decreased expression in female tissues (Lipinska et al. 2015; Perry et al. 2015).
Figure 2.
The average expression level in testes and ovaries of CDS with sex-biased expression in A. aegypti. The testis-biased and ovary-biased CDS were each subdivided with respect to fold bias, and the expression level in each tissue type for the biased genes is shown. (A and B) testis-biased short and long CDS. (C and D) ovary-biased short and long CDS. Within (A and B), all paired contrasts between fold class categories for testis-expressed and for ovary-expressed CDS were statistically significant (P < 0.05) with Mann-Whitney U tests P < 0.001 in each contrast. For (C), all paired contrasts between fold bias class were statistically significant for ovary expression (P < 0.001 for each contrast), but not for testis expression (all P > 0.86). For (D), all paired contrasts between fold classes were statistically significant for ovary expression, and between the ≥ 2 to 5 vs. the ≥ 5 to 10 and ≥ 10 classes for testis expression (P was < 0.001). Note the y-axes are in log scale. CDS, coding sequences; FPKM, frequency per kilobase million.
For the ovary-biased CDS, a different trend was found. For short ovary-expressed genes, higher sex bias was linked to elevated expression in ovaries and no notable change in testis expression (Figure 2C). However, for long CDS, greater bias in ovary expression was linked to both a marked increase in ovary expression and a relatively mild reduction in testis expression in the ≥5- to 10-fold bias and ≥10-fold bias classes as compared to the ≥2- to 5-fold bias class (Figure 2D) (MWU-test P < 0.001 for each paired contrast). This finding differs from Drosophila and algae, wherein high bias in female tissues did not result from upregulation in females (which were largely unchanged across fold bias classes) but rather arose from downregulation in males (Lipinska et al. 2015; Perry et al. 2015). Thus, unlike those systems, in A. aegypti, ovary bias arises specifically from upregulation in the female gonad and is not determined by male expression levels.
It is notable that the expression of sex-biased gonad genes was higher for short than for long CDS for all expression classes (MWU-test P < 0.001 for all paired contrasts of each tissue type per fold biased category). This trend is consistent with observations of shorter CDS in highly expressed genes reported in various eukaryotic systems, which might involve selection to minimize synthesis costs of abundant proteins (Akashi 2001, 2003; Urrutia and Hurst 2003; Comeron 2004; Lemos et al. 2005; Williford and Demuth 2012; Whittle and Extavour 2016a). Our results extend this phenomenon to include genes with sex-biased gonad expression in A. aegypti.
High dN/dS and low optimal codon usage in ovary-biased genes
To study protein sequence divergence, we assessed dN/dS (Yang 2007) of A. aegypti and its sister species A. albopictus, an approach consistent with paired species sequence contrasts previously employed to reveal evolutionary patterns of sex-biased genes (Zhang et al. 2004; Proschel et al. 2006; see Ellegren and Parsch 2007 Assis et al. 2012). Values were determined using the Nei–Gojobori method (Nei and Gojobori 1986) in MEGA (Kumar et al. 2012) for all sex-biased and unbiased CDS [PAML was also utilized to estimate these values (Yang 2007); see below in this section, and Supplemental Note 2 in File S1]. These Aedes taxa have adequately diverged to accumulate effects on dN (average = 0.075 ± 0.001). All CDS had dN < 1 and nearly all CDS (97.0%) had dS < 1.5 (73.9% were <1.0), which comprises an effective range to study dN, dS, and dN/dS (cf. Castillo-Davis et al. 2004).
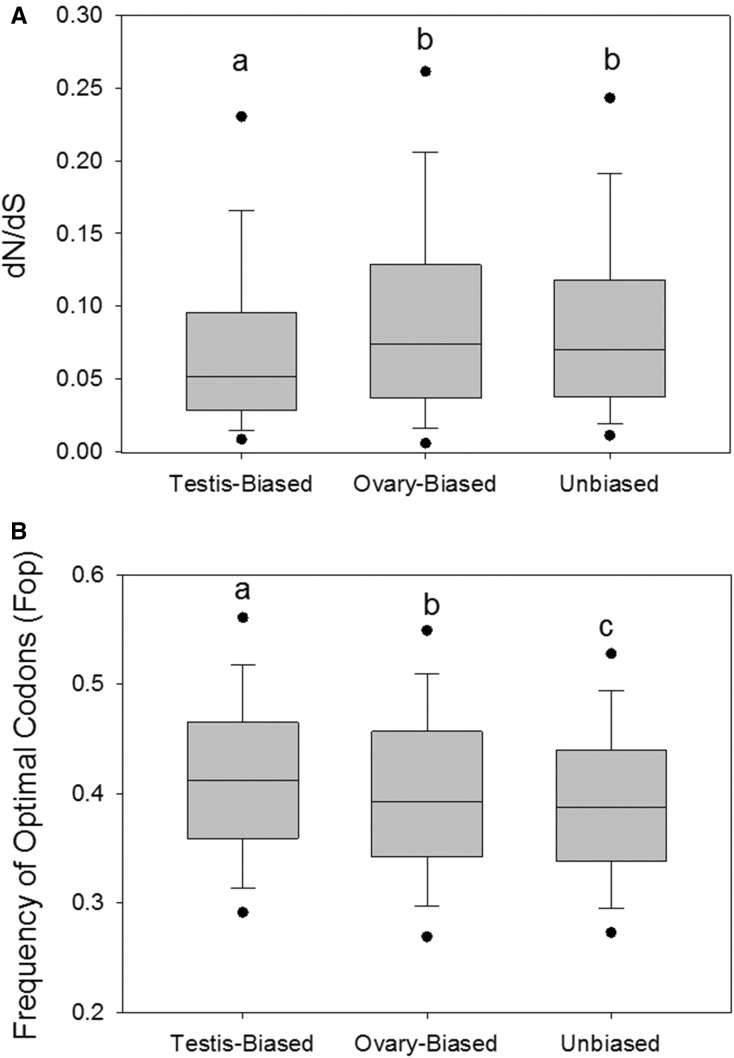
As shown in Figure 3, comparison of all testis-biased, ovary-biased, and unbiased CDS defined in A. aegypti revealed that dN/dS was statistically significantly higher for ovary-biased CDS than testis-biased CDS in this mosquito (MWU-tests P < 0.001). However, no differences were observed between ovary-biased and unbiased CDS (MWU-test P > 0.05). This represents an unusual pattern for metazoans, including other insects such as Drosophila, wherein male- (and/or testis-) biased genes typically evolve rapidly (Zhang et al. 2004; Ellegren and Parsch 2007; Haerty et al. 2007; Assis et al. 2012; Ellegren 2013; Harrison et al. 2015). Faster evolution in the female gonad genes might be related to the fact that ovary-biased expression results from active upregulation in the ovary in this taxon (Figure 2), rather than decreased testis expression (Lipinska et al. 2015; Perry et al. 2015), suggesting a distinct mechanism of sex-biased expression. Further, the higher dN/dS in unbiased genes, which were in a similar range as ovary-biased genes, might indicate that ovary expression accelerates the evolution of the unbiased gene set (as compared to testis-biased), even when the genes are coexpressed in testis (see below section: Ovary and testis expression relative to embryos and dN/dS).
Figure 3.
Box and whisker plots of dN/dS (A) and Fop (B) showing the distribution of all testis-biased, ovary-biased, and unbiased CDS in Aedes. Different letters above any two bars for the testis-biased, ovary-biased, and unbiased genes within each figure indicate a statistically significant (P < 0.05) difference in values using Mann-Whitney U tests (P was < 0.001 in all contrasts). NTestis-Biased = 2927; NOvary-Biased = 2013, and NUnbiased = 4449. Fop was measured using A. aegypti, and dN/dS measured using A. albopictus as a reference. CDS, coding sequences; dN/dS, nonsynonymous to synonymous substitution rates; Fop, frequency of optimal codons.
In addition to the Nei–Gojobori method (Nei and Gojobori 1986) in MEGA (Kumar et al. 2012), we also used PAML to measure dN, dS, and dN/dS, based on a substitution model that specifically includes codon usage bias (Yang 2007). Using this method, we found nearly identical results for all figures and tables (see Supplemental Note 2 in File S1). Further, values of dN/dS across all 9389 CDS under study were highly correlated between the two estimation methods (Figure S2 in File S1), with Spearman’s R = 0.95, P < 2.0 × 10−6, as were dN (R = 0.84, P < 2.0 × 10−7) and dS (R = 1.0, P < 2.0 × 10−7) values.
With respect to codon usage, Fop was statistically significantly higher for testis- than ovary-biased CDS and unbiased CDS (Figure 3B), trends that also contrast with prior findings in other insects, where male-biased genes typically exhibited lower optimal codon usage (Zhang et al. 2004; Hambuch and Parsch 2005). While Fop has been proposed to be lower in longer CDS due to reduced selective constraint or genetic interference (Duret and Mouchiroud 1999; Hambuch and Parsch 2005), a gene length effect per se cannot explain the female effect herein because, as a group, ovary-biased CDS had shorter average lengths than testis-biased CDS (average = 416.6 ± 8.5 and 627.7 ± 6.7 codons, respectively; MWU-test P < 0.001). These trends in dN/dS and Fop in this mosquito resemble findings from a nonmetazoan, the fungus Neurospora, where genes with biased expression in female sex organs had higher dN/dS combined with lower Fop as compared to their male counterparts (Whittle and Johannesson 2013).
Fold bias, CDS length, and expression level and dN/dS
To evaluate in finer detail the relationship between sex-biased gonadal expression on molecular evolution in A. aegypti, we examined dN, dS, dN/dS, and Fop for each sex combination of the three classes of fold bias and the two classes of CDS lengths defined in our above section, Fold sex bias in the gonads is linked to expression level. The assessment is presented in detail in Figure S3, Supplemental Note 2, and Table S2 in File S1. In brief, the results showed that ovary-biased CDS exhibited higher dN in five of six categories of fold bias and CDS lengths, and higher dN/dS in four of six classes. Fop was lower for ovary-biased CDS than testis-biased genes for all six combinations of fold bias and CDS length (see also Figure S3 and Table S3 in File S1), indicating a persistently reduced Fop in the ovary-biased genes (compared to testis-biased genes) irrespective of the level of sex bias and CDS length in A. aegypti. Nevertheless, we noted that dN/dS was higher for testis-biased CDS in short CDS with 5-fold or higher bias. Thus, while the primary signal is of high dN/dS in A. aegypti, a subset of short testis-biased genes appear to have evolved rapidly.
As a further analysis, we examined dN/dS and Fop in the most highly expressed testis-biased, ovary-biased, and unbiased genes (≥1000 FPKM). An extended description of the findings is provided in Supplemental Note 3 in File S1. In summary, we found that within the relatively small subset of sex-biased CDS that were extremely highly transcribed in this taxon (N = 220, or 2.34% of 9389 genes, Table 2), testis-biased and ovary-biased genes exhibited higher dN/dS and lower Fop as compared to unbiased genes (Table 2; note that dS varied among these gene sets and is discussed in Supplemental Note 4 in File S1). We conducted functional annotation targeted to the extremely highly expressed CDS using the gene ontology (GO) tool DAVID (database for annotation, visualization, and integrated discovery) (Huang da et al. 2009), which revealed that these testis- and ovary-biased CDS were highly represented by ribosomal protein genes (Table S4 in File S1). Accordingly, we propose that ribosomal protein genes, in addition to their core role as an essential component of the ribosome needed for translation, may also play a role in sexual differentiation in the sexual organs in A. aegypti. This notion is consistent with findings that ribosomal protein gene copies exhibit differential expression across tissues in plants, yeast, and certain animals, and suggestions that they are involved in tissue-specific gene regulation (Uechi et al. 2002; Komili et al. 2007; Whittle and Krochko 2009).
Table 2. Mean dN/dS, dN, dS, and Fop for the small subset of the most extremely high expressed (≥ 1000 FPKM) testis-biased and ovary-biased CDS in Aedes.
| Parameter | Testis-biased short | Ovary-biased short | Testis-biased long | Ovary-biased long | Unbiased short | Unbiased long |
|---|---|---|---|---|---|---|
| dN/dS | 0.098 (0.023)a | 0.061 (0.007)b | 0.118 (0.014)a | 0.083 (0.016)b | 0.034 (0.016)a | 0.083 (0.030) |
| dN | 0.065 (0.017)a | 0.040 (0.008)b | 0.084 (0.013)a | 0.053 (0.017)b | 0.016 (0.009)a | 0.021 (0.006) |
| dS | 0.577 (0.116)a | 0.566 (0.036)a | 0.631 (0.049)a | 0.503 (0.078)b | 0.285 (0.034)a | 0.279 (0.040) |
| Fop | 0.499 (0.027)a | 0.507 (0.011)a | 0.448 (0.0169)a | 0.507 (0.020)b | 0.617 (0.014)a | 0.621 (0.011) |
| N | 21 | 107 | 49 | 17 | 23 | 3 |
Standard errors are in parenthesis. Different letters for each parameter within each length class (a versus b) indicate a statistically significant difference using randomization tests (P < 0.05). As unbiased long CDS had N = 3, the parameter values were included for completeness, but were not used in randomization tests. dN, nonsynonymous substitution rate; dS, synonymous substitution rate; Fop, frequency of optimal codons.
The unbiased short CDS exhibit a statistically significant difference from testis- and ovary-biased CDS (short and long), P < 0.05.
Taken together, the data show that fold bias, CDS length, and expression level are each relevant factors in the evolution of ovary-biased and testis-biased genes in A. aegypti, and that multiple approaches and analyses show a consistent and predominant effect of high dN/dS in ovary-biased genes.
Ovary and testis expression relative to embryos and dN/dS
To better understand the effects of sex-biased expression on molecular evolution, we sought to assess whether effects of testis or ovary expression on dN/dS and Fop may be influenced by coexpression (or lack thereof) in a nonsexual somatic structure, namely the embryos [noting that precursors to the germ cells are a minor part (<2%) of the population of cells therein] or by having expression specifically restricted to the ovaries or testes. Embryos exhibit a highly diverse transcriptome, expressing a majority of the genes in the genome across all embryonic stages, and are comprised of an array of cell types and tissues (e.g., Diez-Roux et al. 2011; Mittmann and Wolff 2012; Combs and Eisen 2013; Schwager et al. 2015; Donoughe and Extavour 2016). Therefore, we considered that mixed-stage embryos (8–12 hr) would provide a suitable reference as a somatic nonsexual tissue for assessing genes with gonad-specific expression and gonad-somatic coexpression. We note that expression breadth is believed to influence molecular evolution, where genes expressed more broadly across tissues/structures are thought to be more pleiotrophic and constrained, and thus to evolve more slowly, than those that are tissue-specific (Mank and Ellegren 2009; Assis et al. 2012; Lipinska et al. 2015).
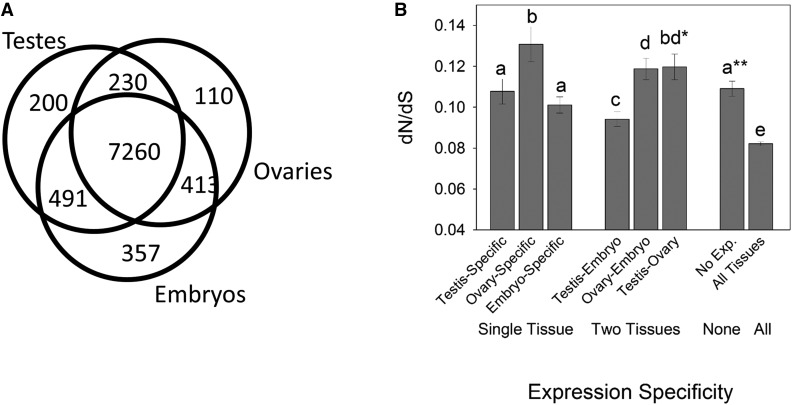
The Venn diagram in Figure 4A shows the number of the 9389 genes under study with specific or shared gene expression in the testis, ovaries, and embryos. We examined each of the seven nonoverlapping gene sets, including tissue-specific (testis-specific, ovary-specific, and embryo-specific), those expressed in two tissues (testis-ovaries, testis-embryos, and ovaries-embryos), and those expressed in all three tissues or none of the three tissues. For this, expression was defined as FPKM > 0 and nonexpression as FPKM = 0, or presence/absence of expression per tissue type. The results showed that, among all gene sets, dN/dS was highest for ovary-specific genes (Figure 4B). The fact that ovary-specific CDS had higher dN/dS than both testis-specific and embryo-specific CDS is consistent with accelerated evolution of genes from the female gonad (MWU-tests P < 0.001; Figure 4B), supporting our conclusions that ovary expression is typically connected to accelerated evolution in Aedes. The CDS lengths were shortest for ovary-specific CDS among all three categories (ovary-, testis-, and embryo-specific CDS lengths were 335.3 ± 32.3, 419.8 ± 32.4, and 449.9 ± 18.6 codons, respectively). As shorter CDS may be expected to be more conserved at the molecular evolutionary level (Akashi 2003; Lemos et al. 2005), CDS length cannot explain the faster evolution of these ovary-biased genes in Aedes. Thus, specific expression in ovaries per se appears to enhance the rate of protein evolution.
Figure 4.
(A) The number of genes expressed in testes, ovaries, and embryos in A. aegypti among the 9389 genes under study (328 CDS had no expression). (B). The mean dN/dS of genes for tissue-specific (single tissue) and genes coexpressed (two tissue types or all three), and those not expressed in any tissues (none). Standard errors are shown in error bars. Different letters above any two bars indicate a statistically significant difference (P < 0.05) between those dN/dS values, using a paired Mann-Whitney U (MWU)-test (note that P was < 0.01 for all paired contrasts). *The ovary-specific and testis-ovary dN/dS did not differ across all genes per group (MWU-test P > 0.05), but the former had higher values using contrasts of values above the median (P < 0.001). No difference was observed between CDS coexpressed in ovary–embryo vs. testis–ovary as denoted by the same letter (d) in both categories. **No expression (No exp.) exhibited no difference in dN/dS as compared to testis-specific CDS, but was different from embryo-specific CDS. CDS, coding sequences; dN/dS, nonsynonymous to synonymous substitution rates.
The results in Figure 4B suggest that the reason unbiased genes evolve faster than testis-biased genes in Aedes (Figure 3) is due to ovarian expression in the unbiased gene set. For instance, among genes expressed in two of the three tissue types, dN/dS was higher for testis–ovary and ovary–embryo genes than for testis–embryo genes. From this, we infer that coexpression in ovaries accelerates evolution, regardless of the other tissue in which the gene is expressed (i.e., testis or embryos, Figure 4B). Moreover, testis–ovary coexpressed genes exhibited higher dN/dS than either testis-specific genes or testis–embryo coexpressed genes. These findings suggest that testis expression does not accelerate divergence, but rather that the high dN/dS values in the ovary–testis coexpressed gene set result from ovary expression, not testis expression. Together, it may be inferred that the elevated dN/dS observed for unbiased genes herein (Figure 3) likely results from gene expression in the ovaries. In other words, the reason why unbiased genes have more elevated dN/dS values than testis-biased genes (Figure 3) is likely due to ovary expression per se. Notably, this conclusion concurs with the data shown in Figure 2, A and B, which show that greater testis bias is linked to lower ovary expression, while unbiased genes by definition show no difference in expression with respect to testis. Therefore, ovary expression, assuming a link to higher dN/dS (Figure 4B), would be more apt to accelerate divergence of unbiased genes than testis expression. It can be concluded that unbiased expression between the ovaries and testes does not imply that the genes are not affected by sexual expression, but rather that their evolution may in fact be accelerated from ovarian transcription in Aedes.
Ovary-specific CDS also evolved faster than those CDS not expressed in any of the three tissue types (Figure 4B). We would anticipate that genes not expressed in any tissues under study might be subject to reduced purifying selective forces (or relaxed selection) as compared to all of the other gene expression categories we consider here (Pal et al. 2001; Subramanian and Kumar 2004), particularly since embryos express genes from a large portion of the genome. Since ovary-specific genes in this mosquito evolve relatively rapidly, this finding suggests that the main mechanism of faster evolution of ovary-specific genes at least partly involves forces other than relaxed purifying selection (see below section: Positive selection is more common in ovary- than testis- and embryo-specific genes). Among all gene sets in Figure 4B, the slowest rates of evolution, or lowest dN/dS, were for those genes expressed in all three tissues (Figure 4B). Although the study of even more tissues is beyond the scope of our objectives here, and would be needed to fully conclude such a pleiotropic effect, we suggest that our results of embryonic gene expression analysis are consistent with enhanced pleiotropy and purifying selection in more broadly expressed genes (Mank and Ellegren 2009). For the findings on Fop for gene sets described in Figure 4, see Supplemental Note 5 in File S1.
Positive selection is more common in ovary- than testis- and embryo-specific genes
Given that positive selection is unlikely to be detected using whole CDS (dN/dS > 1) unless most sites in a CDS exhibit positive selection, we assessed whether sex-related genes exhibited signals of episodic positive selection at specific sites in A. aegypti. We assessed those genes with testis-, ovary-, and embryo-specific expression in Figure 4A, which are those genes most apt to indicate the effect of tissue type expression on events of positive selection. For this analysis, we studied trees of three for A. aegypti, A. albopictus, and their outgroup relative from the same family C. quinquefasciatus (Culicidae). This taxon was chosen due to availability of CDS data and its phylogenetic proximity to Aedes (Arensburger et al. 2010). We found that 63.6, 76.5, and 80.1% of the Aedes CDS list for ovary-specific, testis-specific, and embryo-specific, respectively, had putative orthologs in C. quinquefasciatus. The fact that we found fewer orthologs of genes belonging to the ovary-specific data set is consistent with the faster rates of evolution observed for these genes (Ellegren and Parsch 2007). Further, as Culex diverged from Aedes ∼50 MYA (Arensburger et al. 2010), in those CDS with orthologs we observed substantial saturation for whole-branch dS (usually in the Culex branch) in some genes (e.g., dS > 3, between 28.2 and 35.3% of each data set under study using a CODEML branch-model) (Yang 2007). Nevertheless, a majority of genes were not highly saturated for dS using an outgroup, and thus were suitable for analysis [note that, as described above, for the paired species within the Aedes genus, nearly all CDS (97%) had dS < 1.5 in our main analysis, but we permitted greater flexibility of dS < 3 when using the outgroup Culex in this present assessment].
For our assessment, we conducted branch site analysis using A. aegypti, our main target species under study that was used in all sex-related expression analyses (Figure 4A), as the foreground branch. The results showed that the highest proportion of genes with statistically significant signals of positive selection [with 2 × ∆ lnLikelihood χ2 P < 0.05 (Yang 2007)] (see Materials and Methods) in the A. aegypti branch was for ovary-specific genes (23.7%), nearly double that of testis-specific (12.2%) and embryo-specific genes (11.9%). Nonetheless, we observed a nonnegligible level of positive selection in the latter two tissue types. A follow-up assessment using A. albopictus as the foreground branch also revealed greater incidences of positive selection in the ovary-specific gene set (28.9% of genes) than the testis- (19.1%) and embryo-specific (19.5%) gene sets (note that these gene sets showing signs of positive selection were largely nonoverlapping, and <6% per gene set had signs of positive selection in both taxa). While A. albopictus shows a greater tendency than A. aegypti for positive selection in all three tissue-specific data sets, as in A. aegypti, A. albopictus ovary-specific genes still exhibited more frequent instances of adaptive evolution than testis- or embryo-specific genes. Together, these results indicate a putative role of positive selection in the faster evolution of ovary-related genes in Aedes (Figure 3). Further, the results suggest that selective sweeps might contribute at least partly toward the inverse link between dN/dS and Fop (Figure 3).
Causes of fast evolution of ovary genes and GO
We used DAVID (Huang da et al. 2009) to conduct a targeted GO assessment of the ovary-specific and ovary–embryo coexpressed genes, as well as the testis-specific and testis–embryo coexpressed genes identified in Figure 4. These gene sets were chosen because we thought it was highly likely that they would be involved in gonad-specific functions, and would reflect the effect of gonadal expression on molecular evolution, such as that observed in Figure 4B. The results are provided in Table 3 and have been clustered by functionality, with the top three clusters shown per gene set. We found that the ovary-specific genes were strongly preferentially involved in olfactory functions, including odorant binding and odorant receptor activity (cluster 1 for that gene set, Table 3). Similarly, olfactory signaling genes were prevalent in the ovary–embryo coexpressed gene set (see cluster number 2 for that gene set), but were not a major functional cluster found for the testis-specific and testis–embryo coexpressed genes (Table 3). In A. aegypti, olfactory signals have been shown to be involved in male swarming and attraction of females, and thus are directly linked to mate recognition (Fawaz et al. 2014). Additionally, olfactory molecules have also been suggested to facilitate chemotaxis between sperm and oocytes within female reproductive structures to ensure fertilization in various organisms (Parmentier et al. 1992; Goto et al. 2001), including vector mosquitoes such as A. aegypti (Pitts et al. 2014). Further, after insemination, chemotaxis between sperm and female structures may direct the sperm into the spermathecae for their storage until fertilization (Degner and Harrington 2016). In fact, some authors have hypothesized that olfactory signaling may have evolutionarily originated from the reproductive system (Pitts et al. 2014) and later been co-opted for use in the antennae and other sensing systems. Our data suggest that such olfactory molecules are prevalent and rapidly evolving in ovary genes, raising the possibility that their rapid rates of evolution may be related to mating and fertilization processes. We speculate that such olfactory genes are absent as a top functional class in the testis-specific or testis–embryo coexpressed set because olfactory signals in female ovaries are continuously emitted, perhaps to maximize mating or fertilization opportunities, but may be restricted in male testis or sperm to brief periods directly linked to mating, and thus not observed for male gonads herein.
Table 3. Functional annotation of ovary-specific, testis-specific, ovary–embryo coexpressed, and testis–embryo coexpressed genes for A. aegypti.
| Gene type | Role | P value |
|---|---|---|
| Ovary-specific | ||
| Cluster 1: enrichment score: 4.41 | Odorant binding | 6.50E−10 |
| Olfactory receptor activity | 4.10E−05 | |
| Plasma membrane | 5.10E−05 | |
| Olfaction | 5.90E−05 | |
| Olfactory receptor | 6.20E−05 | |
| Sensory transduction | 1.00E−04 | |
| Transducer | 1.40E−03 | |
| Receptor | 7.60E−03 | |
| Cluster 2: enrichment score: 2.08 | Integral component of membrane | 5.00E−04 |
| Transmembrane helix | 2.00E−02 | |
| Transmembrane | 2.00E−02 | |
| Membrane | 2.50E−02 | |
| Cluster 3: enrichment score: 1.24 | Sensory perception of taste | 1.90E−02 |
| 7TM chemoreceptor | 6.30E−02 | |
| Cell membrane | 1.60E−01 | |
| Testis-specific | ||
| Cluster 1: enrichment score: 2.36 | Leucine-rich repeat | 3.60E−04 |
| LRR_TYP (Leucine-rich repeats, typical) | 9.50E−03 | |
| Leucine-rich repeat, typical subtype | 2.40E−02 | |
| Cluster 2: enrichment score: 1.49 | Ves allergen | 1.60E−02 |
| SCP (SCP domain proteins) | 3.10E−02 | |
| Allergen V5/Tpx-1-related | 4.60E−02 | |
| CAP domain | 4.60E−02 | |
| Cluster 3: enrichment score: 1.24 | FBG (Fibrinogen-related domains) | 4.40E−02 |
| Fibrinogen, α/β/γ chain, C-terminal globular domain | 6.60E−02 | |
| Fibrinogen, α/β/γ chain, C-terminal globular, subdomain 1 | 6.60E−02 | |
| Ovary–embryo coexpressed (absent testis) | ||
| Cluster 1: enrichment score: 3.96 | Integral component of membrane | 2.90E−05 |
| Membrane | 1.60E−04 | |
| Transmembrane helix | 1.70E−04 | |
| Transmembrane | 1.80E−04 | |
| Cluster 2: enrichment score: 2.37 | Plasma membrane | 5.10E−05 |
| Odorant binding | 7.10E−05 | |
| Olfactory receptor activity | 1.10E−02 | |
| Olfactory receptor | 1.10E−02 | |
| Olfaction | 1.30E−02 | |
| Transducer | 1.60E−02 | |
| Sensory transduction | 2.10E−02 | |
| Receptor | 5.80E−02 | |
| Cluster 3: enrichment score: 1.96 | Gustatory receptor protein | 3.90E−03 |
| Gustatory receptor | 1.80E−02 | |
| Taste receptor activity | 1.80E−02 | |
| Testis–embryo coexpressed (absent ovary) | ||
| Cluster 1: enrichment score: 3.45 | Secreted | 7.90E−07 |
| Lipid metabolic process | 1.30E−05 | |
| Dol/Ves 1 allergen | 5.50E−04 | |
| Phosphatidylcholine 1-acylhydrolase activity | 6.30E−04 | |
| Lipase, N-terminal | 7.30E−04 | |
| Lipase | 7.30E−04 | |
| Carboxylic ester hydrolase activity | 4.00E−01 | |
| Cluster 2: enrichment score: 2.69 | ||
| Homeobox | 9.00E−05 | |
| Sequence-specific DNA binding | 9.40E−05 | |
| Homeodomain | 1.70E−04 | |
| HOX (Homeodomain and Homeodomain-like) | 2.60E−04 | |
| Homeobox, conserved site | 3.30E−04 | |
| Homeodomain, metazoa | 1.20E−03 | |
| DNA-binding | 1.80E−03 | |
| Homeodomain-like | 3.60E−03 | |
| Helix-turn-helix motif | 5.30E−03 | |
| Regulation of transcription, DNA-templated | 4.10E−02 | |
| Nucleus | 4.60E−02 | |
| Cluster 3: enrichment score: 1.92 | CAP domain | 1.00E−02 |
| Allergen V5/Tpx-1-related | 1.00E−02 | |
| SCP (SCP domain proteins) | 1.60E−02 |
The gene ontology was determined using the DAVID (database for annotation, visualization, and integrated discovery) system (Huang da et al. 2009), and the three clusters with the greatest enrichment score are shown per category. P-values are from a modified Fisher’s test, wherein lower values indicate greater enrichment. The number of genes per category is provided in Figure 4A.
We propose that a hypothesis potentially explaining our findings is that sexual selection drives the rapid evolution of ovary-biased and ovary-specific genes. For instance, positive selection mediated by female–female competition acting to attract swarming males, or resulting from male mate choice among females (Oliva et al. 2014) [assuming some coexpression between sensilli and ovaries, as suggested in Foret and Maleszka (2006) and Matthews et al. (2016)], could cause fast evolution of ovary genes. Alternatively, it is possible that chemotaxis between ovaries and sperm is involved in attracting sperm movement into the reproductive tract after mating, which may promote the observed rapid evolution. For example, while the chemical activation underlying sperm movement into the spermathecae in mosquitoes (and other insects) is largely unknown, it has been suggested that secretions from one reproductive tissue might regulate sperm motility in another, such as the spermathecae (Degner and Harrington 2016). Thus, sperm movement could be driven, at least in part, by ovarian signals, and thus individuals containing ovaries with stronger olfactory reception or signaling may be more successful in compelling sperm to move into the spermathecae for storage (after mating), or triggering a high number of sperm to be released from the spermathecae for fertilization [a phenomenon that would be adaptive when sperm are plentiful (Degner and Harrington 2016)], thereby exerting an adaptive selective pressure on ovary-biased genes. In fact, evidence suggests that up to one-third of available sperm do not reach the spermatheceae after mating in A. aegypti [reviewed by Oliva et al. (2014)]. Putative signaling from ovary to sperm may be crucial for ensuring sperm storage, and thus supply, over a female’s life span and reproductive success. Other speculative, yet conceivable, mechanisms include competition among eggs to attract incoming sperm, or male (sperm) choice among eggs [as numerous sperm may be released from the spermathecae for fertilization at a time; see Degner and Harrington 2016)], based on the strength or favorability of olfactory signaling. To determine the feasibility of these latter possibilities, it will be important for further studies to assess whether olfactory binders and receptors are observed not only in ovaries containing previtellogenic eggs (Raikhel and Dhadialla 1992), but also in the reproductive tract of mated females where fertilization ultimately occurs, including the mature eggs and transferred sperm (Degner and Harrington 2016). Together, to distinguish between these possible scenarios, extensive further study will be needed to assess the causes of rapid evolution of ovary-expressed genes, including olfactory proteins and receptors, in A. aegypti.
Our finding that A. aegypti exhibits fast evolution of ovary-biased genes differs from most other models studied to date, including its Dipteran insect relative Drosophila melanogaster, which has been commonly shown to exhibit faster evolution of whole male-biased than female-biased genes, and testis-biased than ovary-biased genes (e.g., Zhang et al. 2004, 2007; Proschel et al. 2006; Ellegren and Parsch 2007; Haerty et al. 2007; Meisel 2011; Assis et al. 2012; Parsch and Ellegren 2013; Perry et al. 2015). There are several factors that could underlie the observed differences. We note that, unlike Drosophila, ovary-biased expression in A. aegypti was caused by upregulation of genes in ovaries (Figure 2), whereas in D. melanogaster, greater ovary-biased expression (or preovary and pretestis depending on developmental stage) has been reported to result from downregulation in testes (Perry et al. 2015), suggesting that different mechanisms of gene regulation account for ovary-biased expression in the two insects. Further, the ovary gene sets in A. aegypti (Figure 4) include a high representation of olfactory signaling genes (Table 3), a finding not, to our knowledge, reported to be prevalent in D. melanogaster, and which might reflect a key difference in the biology of sperm attraction and storage between these two insect systems. We also note that unusually long sperm lengths and formation of sperm bundles in the female tract (which are thought to improve velocity) have been reported in D. melanogaster, features not believed to be inherent to A. aegypti [reviewed in Degner and Harrington (2016)]. Such sperm features might potentially reduce the need or effectiveness of ovarian or egg-based signaling in D. melanogaster to attract sperm to the spermatheceae, or the release of sperm from the spermatheceae for fertilization, and perhaps make Drosophila ovary-biased genes less likely to be subjected to adaptive evolutionary pressure than in A. aegypti. Based on these observations from our own work and that of others, we speculate that ovary-biased genes in A. aegypti, partly including olfactory signaling genes, might play an essential role in sperm attraction, storage, and fertilization, thus potentially exerting adaptive evolutionary pressures on mutations on these genes in the mosquito.
We propose that a secondary mechanism that may contribute toward the faster evolution of ovary-biased genes than testis-biased genes in A. aegypti is putatively reduced male–male sperm competition. In this mosquito, males ensure monogamy of females by transferring sufficient sperm to fertilize all eggs, and the accessory glands produce physical (and chemical) barriers in females that are normally formed and functional within 40 min after copulation (Oliva et al. 2014). This rapid postcopulation barrier to further mating could greatly limit sexual selection via male–male sperm competition (albeit not perfectly, given barriers are not established immediately Oliva et al. 2014), a feature differing from insects such as Drosophila, which exhibit strong intermale sperm competition after mating (Ellegren and Parsch 2007). In D. melanogaster, females mate with multiple males, and sperm from several males may become mixed in the female storage organs, resulting in strong sperm competition (Price et al. 1999). While various types of mating plugs form in D. melanogaster, they primarily ensure sperm are stored (and not lost from the female) in the storage organs, but do not prevent mating and sperm transfer from competing males (Avila et al. 2015). This differs from postmating barriers (initially physical barriers, followed by chemical barriers) that form after a first mating in Aedes females, which greatly limit or impede subsequent reinsemination by other males, and act to ensure female monogamy [reviewed by Oliva et al. (2014)]. As sperm competition between males is believed to contribute toward adaptive evolution of genes (Ellegren and Parsch 2007), this form of sexual selection might be highly limited or absent in A. aegypti, possibly decelerating the rate of evolution of testis-biased genes as compared to the ovaries in this insect.
Importantly, a prior report has shown that seminal fluid proteins with orthologs between A. aegypti and A. albopictus did not exhibit elevated dN/dS as compared to control housekeeping genes (Boes et al. 2014). In addition, no evidence of enhanced positive selection was observed across whole CDS or at specific codon sites in these Aedes species (Boes et al. 2014), findings in striking contrast to the typically fast and adaptive evolution of such genes observed in other organisms studied to date (Swanson and Vacquier 2002; Clark et al. 2006; Haerty et al. 2007). Together with our data (wherein testes samples included accessory glands; Akbari et al. 2013), this suggests that at least two male reproductive gene sets, namely testes and seminal fluid proteins, which typically exhibit fast sequence evolution in studied metazoan systems, evolve slowly in species of Aedes.
We speculate that an additional factor that might contribute to fast evolution of ovary genes is infection. For instance, a bacterial infection of the germ lines (by Wolbachia), which has been shown to enhance female germ line fertility, has been linked to accelerated gene divergence in D. melanogaster (Flores et al. 2015). Thus, similar types of infections specifically affecting the ovaries, and fertility, is another potential avenue contributing to fast evolution of ovary-biased genes in A. aegypti. We note that the specific infection of Wolbachia might not naturally occur in or have this effect in A. aegypti (Segoli et al. 2014), but a similar symbiotic effect is plausible from other bacterial endosymbionts. Finally, we cannot exclude the possibility that ovary-biased CDS might be dispensable for reproductive success or fitness (Mank and Ellegren 2009), and thus some genes may exhibit relaxed selection, accelerating protein sequence evolution and reducing Fop. However, our finding of positive selection in ovary-biased genes suggests that this possibility cannot explain the findings. Therefore, we suggest that our findings are better explained by sexual selection, as described above. Further studies aimed toward addressing these possible causes may provide insights into why ovary-biased genes evolve fast in this mosquito.
Sex biases in whole males vs. females
As a final part of our assessment, we were interested in the connection between the gonads and somatic male and female sex biases in expression (Ellegren and Parsch 2007), and thus compared these two scales of sex-biased expression in A. aegypti. We assessed genes with ≥2-fold sex-biased expression from adult male and female carcasses, which excluded the sex organs (denoted hereafter as somatic-, or SOM-male and SOM-female, respectively) (Akbari et al. 2013), in A. aegypti (Table S1 in File S1). We found that SOM-male-biased genes (N = 3743) were more common than SOM-female-biased genes (N = 774), consistent with prior reports of sex-biased expression in the somatic tissues of this taxon (Akbari et al. 2013). Further, SOM-male-biased CDS had lower dN/dS than SOM-female-biased CDS and unbiased CDS (N = 4872; MWU-test P < 0.001, Figure S4A in File S1), and Fop was higher for the SOM-male- (than female-) biased genes (MWU-test P < 0.001; Figure S4B in File S1). Thus, these trends suggest that somatic female-biased genes exhibit faster evolution than somatic male-biased genes.
A total of 55.4% of the testis-biased gene set defined for our main analysis (Figure 3) were also SOM-male-biased (N = 1621 of 2927), while only 13.1% of ovary-biased genes were SOM-female-biased (N = 264 of 2013), indicating disparity in the gene sets exhibiting sex biases in the gonads as compared to the somatic tissues. Those genes that were both SOM-female- and ovary-biased had markedly higher median dN/dS as compared to those that were both SOM-male- and testis-biased (medians of 0.088 and 0.050, respectively, MWU-test P < 0.007, Figure S4C in File S1). However, importantly, after excluding genes with gonad-biased expression (including those only sex-biased in the soma), no difference in dN/dS profiles was observed between the SOM-female- (N = 510) and SOM-male-biased genes (N = 2122) (median = 0.069 and 0.068, respectively, MWU-test P = 0.400, Figure S4D in File S1). Collectively, the results suggest that the faster evolution of SOM-female- compared to SOM-male-biased genes (Figure S4, A and C in File S1) likely results from gene coexpression in the gonads (and gonad-biased expression, Figure S4D in File S1), rather than from sex-biased expression in the soma. Data from specific nonsexual organs and tissues in species of Aedes will help further ascertain whether or how the individual somatic structures contribute to whole-sex differences in expression and sequence divergence in Aedes (Matthews et al. 2016)
Together, these findings demonstrate that: (1) as ovary-biased genes were not necessarily SOM-female-biased (N = 2013 and 775, respectively), the sex-limited organs alone are linked to expression and sequence divergence in Aedes, in a manner that cannot be explained by biases in whole males vs. females; and (2) faster evolution of somatic female-biased genes than somatic male-biased genes (Figure S4A in File S1) may be explained by the presence of gonad-biased genes in each data set (Figure S4, C and D in File S1).
It is worth noting that a recent study in preprint for four species of Anopheles, a related mosquito from the same family as Aedes (Culicidae), has indicated that female-biased genes identified after blood feeding evolve faster than male-biased genes in that genus (Papa et al. 2016) [see also a recent study by of testis vs. male carcass genes (Cassone et al. 2017)]. Both female carcass- (excluding the gonads) and ovary-biased genes showed elevated dN/dS as compared to their male counterparts (Papa et al. 2016). Papa et al. (2016) hypothesized that blood feeding contributes to rapid evolution of female (carcass)-biased genes, as high dN/dS genes of genes upregulated in somatic carcasses of blood-fed females were preferentially involved in functions of salivary glands, digestion, and immunity. We do not consider blood feeding to be a primary candidate to explain our findings on expression (Figure 2), dN/dS, (Figure 3 and Figure 4), or GO functions (Table 3) in ovary genes in A. aegypti, as first, the females and thus ovaries were not blood fed herein, and second, ovaries are not directly involved in the somatic physical mechanisms of blood feeding. Nonetheless, as blood feeding is perhaps the most obvious biological novelty of vectors such as mosquitoes, an indirect role is possible, for instance, via coexpression of some genes in non-blood-fed and blood-fed ovaries. As described above, our findings point toward a primary role of selection during mating and fertilization, including a possible role of olfactory genes, in the fast evolution of ovary genes in A. aegypti (Figure 4 and Table 3), and are not consistent with a major role of blood feeding.
The findings from our present study, and those of Papa et al. (2016), suggest that rapid evolution of ovary-biased genes as compared to testis-biased genes might be a common feature of the mosquitoes, spanning both blood-fed and nonblood-fed ovaries. Mating plugs have historically been thought to limit sperm competition in many metazoans, particularly mammals and insects (Parker 1970). As described above (see section: Causes of fast evolution of ovary genes and GO), the employment of mating plugs in A. aegypti promotes female monogamy and may greatly limit sperm competition in this mosquito (Oliva et al. 2014), a possibility that has also been noted for at least some species of Anopheles (Rogers et al. 2009; Oliva et al. 2014; Papa et al. 2016), and might contribute to slowed evolution of testis-biased genes in both genera. Additional research across more genera of mosquitoes will be essential to assess whether fast evolution of ovary-biased genes is a ubiquitous feature of this taxonomic group.
Conclusions
Here, we have provided an uncommon example in metazoans showing that ovary-biased genes evolve rapidly as compared to testis-biased genes in A. aegypti. We propose that the fast evolution of ovary-biased and ovary-specific genes, including those involved in olfactory functions, may partly result from sexual selection (positive selection) during courtship/mating and/or fertilization. In addition, we propose that higher dN/dS in ovary- than testis-biased genes may partly arise from the formation of post-mating barriers in the female reproductive tract, resulting in low male-male sperm competition. Further behavioral research in mating in this taxon, including male and female choice, and gene expression analysis in post-mating sperm and eggs in the female reproductive tract, including olfactory genes, will help further discern the factors contributing to the evolution of sex-biased gonad genes in this mosquito.
Materials and Methods
Expression in A. aegypti testes, ovaries, embryos (8–12 hr), and whole male and female carcasses was determined by mapping all reads to the list of 14,678 CDS under study from A. aegypti using Geneious 9 (www.Geneious.com: Kearse et al. 2012). Expression was determined as frequency per kilobase million (FPKM, or RPKM since all were single reads; Supplemental Material, Table S1 in File S1). These expression values were utilized for subsequent expression analyses.
Optimal codon usage
To determine optimal codon usage, we used embryo tissues and relative synonymous codon usage (∆RSCU) values between the 5% least and most transcribed in the CDS list for A. aegypti across the full CDS list described in the Results and Discussion. For the correspondence analysis of codon usage, we used CodonW (Peden 1999) and the complete CDS list for A. aegypti. The correspondence analysis separates the codons into discrete classes on the principle axis based on the effective number of codons (ENC, Wright 1990) and predicts the list of putative optimal codons for a taxon (those linked to high expression), which should be followed-up with expression data analysis for confirmation, as conducted herein using ∆RSCU (Table 1) (Peden 1999). We note that for A. aegypti, a partial list of seven optimal codons had been suggested previously, where the list included codons ending in G, C, A, or T (Behura and Severson 2011). However, this comprises an atypical and small optimal codon list (Sharp et al. 1986; Stenico et al. 1994; Duret and Mouchiroud 1999; Cutter et al. 2006; Ingvarsson 2008; Whittle et al. 2011), and was obtained for different goals than herein. Thus, we conducted an assessment to identify optimal codons herein using gene expression data and correspondence analysis, yielding the list of 18 optimal codons in Table 1. Further details are provided Supplemental Note 1 in File S1.
Orthology and expression analysis
In our analysis of sex-biased genes, A. aegypti and A. albopictus orthologs were determined using reciprocal BLASTX (https://blast.ncbi.nlm.nih.gov). Only those CDS having a top match (with e < 10−6) in both reciprocal searches between the two species were retained for further sex bias analysis for a total of 9489 complete CDS. Sex-biased CDS were identified as those with at least twofold difference in expression, with P < 0.05, and FPKM ≥ 1 in at least one of the compared tissues. P-values were determined using expression values normalized to the median in Geneious 9, which is based on the same method used in DeSeq (Anders and Huber 2010), and its two-sample expression contrast method employing the Binomial distribution (http://www.geneious.com/ Kearse et al. 2012). We presented results for all genes with P < 0.05 rather than corrected values as we had a large cutoff for definition as sex-biased [≥2-fold and P < 0.05; rather than 1.25- or 1.5-fold with 10% FDR used in other studies (Mank et al. 2007)], and we aimed to study all available genes exhibiting signals of sex-biased expression. Nonetheless, we repeated all our molecular evolutionary analysis of testis- and ovary-biased genes using only the gene sets identified after Bonferroni correction, which classified an additional 11.0% of the 9389 genes as unbiased (N = 5484 vs. N = 4449), resulting mostly from the weakest (<10 FPKM) expressed genes. This approach yielded nearly identical results for all tables and figures (data not shown). In the calculation of expression level of unbiased CDS (Table 2), we averaged the expression in testis and ovaries.
Alignments and dN/dS
To measure dN, dS, and dN/dS, orthologs for A. aegypti and A. albopictus were aligned by codons using MUSCLE (Edgar 2004) as implemented in MEGA (Kumar et al. 2012) and gaps removed. The dN and dS (and dN/dS) values were calculated using the Nei–Gojobori method (Jukes–Cantor) in MEGA (Kumar et al. 2012) with γ variation among sites. Values were also calculated using maximum likelihood and yn00 in PAML (Yang 2007), which includes codon bias as a parameter. Results in all tables and figures are for Nei–Gojobori (both methods yielded highly similar results), and PAML findings are cited in the Results and Discussion and shown in Figure S2 in File S1.
Pairwise contrasts of dN, dS, dN/dS, or Fop using randomization tests (Ludbrook and Dudley 1998) were conducted as follows: for each comparison, genes from the two compared categories were pooled. The pooled data set was resampled with replacement 1000 times. Each sample was randomly divided into two bins of size N1 and N2, corresponding to the gene sample sizes of category one and two, respectively. For each of the 1000 samples, we determined ∆X = mean XCategory 1- mean XCategory2, where X = dN/dS, dN, dS, or Fop depending on the parameter under study. This gives the sampling distribution of differences in X under the null hypothesis. P-values were calculated as the fraction of samples in the distribution wherein ∆X exceeded the observed ∆X.
Intron analysis
Introns were extracted using the annotation files in Table S1 in File S1. Evidence suggests that short introns are nearly entirely selectively neutral while longer introns may be subjected to some selection, particularly in regulatory flanking regions (Farlow et al. 2012). Thus, we divided introns into short and long classes approximately divided by the median (130 bp), and removed the 50-bp ends from the long introns prior to analysis of GC content (Williford and Demuth 2012). All introns <20 bp were excluded from analysis.
Positive selection
For assessing episodic positive selection in A. aegypti for the tissue-specific genes in Figure 4, we used trees of three closely related species, A. aegypti, A. albopictus, and the outgroup species Culex quinquefasciatus, a species that represents a reasonably close mosquito relative (from the same subfamily as Aedes) with whole genomic sequence available for study (available at http://metazoa.ensembl.org/). Orthologous CDS in all three species were identified using reciprocal BLASTX (https://blast.ncbi.nlm.nih.gov) with CDS sequences of our main study organism A. aegypti as the query and A. albopictus (as described above) or C. quinquefasciatus CDS as the target CDS with a cutoff e-value of e < 10−6. Alignments were conducted for CDS of the three species in MEGA as described above (in Materials and Methods) and gaps removed (aligned by codons). Prior to branch-site analysis, branch analysis was conducted using CODEML in PAML (Yang 2007) for the three taxa using the free-ratios model, and poor or very saturated alignments (due to distance of Culex to Aedes) measured by dS > 3 (cf. Lloyd et al. 2015) in the (typically longest) Culex branch were discarded. The proportion of CDS without orthologs or with high saturation is cited in the Results and Discussion. Subsequently, branch-site analysis was conducted using CODEML (Yang and Swanson 2002; Yang 2007), which allows dN/dS to vary among branches and sites. A. aegypti or A. albopictus was used as the foreground branch in an unrooted tree of the three species. Positive selection was inferred for all CDS with a statistically significant value for 2 × ∆ln Likelihood using a χ2-test (P < 0.05) (Yang and Swanson 2002; Yang 2007). The number of CDS under study for ovary-specific, testis-specific, and embryo-specific data sets is provided in Figure 4A.
Data availability
Sources for all data used for analysis are provided in Table S1 in File S1 and were downloaded between August and November 2015 (A. aegypti) and in March 2016 (A. albopictus). No custom scripts or tools were created to perform the present analysis.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.201343/-/DC1.
Acknowledgments
We are grateful to the anonymous reviewers for valuable comments on the manuscript. This work was supported by funds from Harvard University and National Institutes of Health grant 5R01H D0-73499 to C.G.E.
Footnotes
Communicating editor: B. P. Lazzaro
Literature Cited
- Akashi H., 2001. Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 11: 660–666. [DOI] [PubMed] [Google Scholar]
- Akashi H., 2003. Translational selection and yeast proteome evolution. Genetics 164: 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O. S., Antoshechkin I., Amrhein H., Williams B., Diloreto R., et al. , 2013. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 (Bethesda) 3: 1493–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman M. N., Furlong E. E., Imam F., Johnson E., Null B. H., et al. , 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270–2275. [DOI] [PubMed] [Google Scholar]
- Arbeitman M. N., Fleming A. A., Siegal M. L., Null B. H., Baker B. S., 2004. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development 131: 2007–2021. [DOI] [PubMed] [Google Scholar]
- Arensburger P., Megy K., Waterhouse R. M., Abrudan J., Amedeo P., et al. , 2010. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science 330: 86–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariani C. V., Smith S. C., Osei-Poku J., Short K., Juneja P., et al. , 2015. Environmental and genetic factors determine whether the mosquito Aedes aegypti lays eggs without a blood meal. Am. J. Trop. Med. Hyg. 92: 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R., Zhou Q., Bachtrog D., 2012. Sex-biased transcriptome evolution in Drosophila. Genome Biol. Evol. 4: 1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila F. W., Wong A., Sitnik J. L., Wolfner M. F., 2015. Don’t pull the plug! the Drosophila mating plug preserves fertility. Fly (Austin) 9: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behura S. K., Severson D. W., 2011. Coadaptation of isoacceptor tRNA genes and codon usage bias for translation efficiency in Aedes aegypti and Anopheles gambiae. Insect Mol. Biol. 20: 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behura S. K., Severson D. W., 2013. Codon usage bias: causative factors, quantification methods and genome-wide patterns: with emphasis on insect genomes. Biol. Rev. Camb. Philos. Soc. 88: 49–61. [DOI] [PubMed] [Google Scholar]
- Beletskii A., Bhagwat A. S., 1996. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc. Natl. Acad. Sci. USA 93: 13919–13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A. J., Presgraves D. C., 2002. Linkage limits the power of natural selection in Drosophila. Proc. Natl. Acad. Sci. USA 99: 13616–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. D., Drummond D. A., Arnold F. H., Wilke C. O., 2006. Structural determinants of the rate of protein evolution in yeast. Mol. Biol. Evol. 23: 1751–1761. [DOI] [PubMed] [Google Scholar]
- Boes K. E., Ribeiro J. M., Wong A., Harrington L. C., Wolfner M. F., et al. , 2014. Identification and characterization of seminal fluid proteins in the Asian tiger mosquito, Aedes albopictus. PLoS Negl. Trop. Dis. 8: e2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone B. J., Kay R. G., Daugherty M. P., White B. J., 2017. Comparative transcriptomics of malaria mosquito testes: function, evolution, and linkage. G3 (Bethesda) 7: 1127–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis C. I., Bedford T. B., Hartl D. L., 2004. Accelerated rates of intron gain/loss and protein evolution in duplicate genes in human and mouse malaria parasites. Mol. Biol. Evol. 21: 1422–1427. [DOI] [PubMed] [Google Scholar]
- Chen X. G., Jiang X., Gu J., Xu M., Wu Y., et al. , 2015. Genome sequence of the Asian tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc. Natl. Acad. Sci. USA 112: E5907–E5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophers S. R., 1960. Aedes aegypti L. The Yellow Fever Mosquito, Its Life History, Bionomics and Structure. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Civetta A., Singh R. S., 1998. Sex-related genes, directional sexual selection, and speciation. Mol. Biol. Evol. 15: 901–909. [DOI] [PubMed] [Google Scholar]
- Clark N. L., Aagaard J. E., Swanson W. J., 2006. Evolution of reproductive proteins from animals and plants. Reproduction 131: 11–22. [DOI] [PubMed] [Google Scholar]
- Combs P. A., Eisen M. B., 2013. Sequencing mRNA from cryo-sliced Drosophila embryos to determine genome-wide spatial patterns of gene expression. PLoS One 8: e71820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., 2004. Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics 167: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., Ward S., 2005. Sexual and temporal dynamics of molecular evolution in C. elegans development. Mol. Biol. Evol. 22: 178–188. [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Wasmuth J. D., Blaxter M. L., 2006. The evolution of biased codon and amino acid usage in nematode genomes. Mol. Biol. Evol. 23: 2303–2315. [DOI] [PubMed] [Google Scholar]
- Degner E. C., Harrington L. C., 2016. A mosquito sperm’s journey from male ejaculate to egg: mechanisms, molecules, and methods for exploration. Mol. Reprod. Dev. 83: 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G., Banfi S., Sultan M., Geffers L., Anand S., et al. , 2011. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 9: e1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoughe S., Extavour C. G., 2016. Embryonic development of the cricket Gryllus bimaculatus. Dev. Biol. 411: 140–156. [DOI] [PubMed] [Google Scholar]
- Dorus S., Busby S. A., Gerike U., Shabanowitz J., Hunt D. F., et al. , 2006. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat. Genet. 38: 1440–1445. [DOI] [PubMed] [Google Scholar]
- Duret L., 2000. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 16: 287–289. [DOI] [PubMed] [Google Scholar]
- Duret L., Mouchiroud D., 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H., 2013. The evolutionary genomics of birds. Annu. Rev. Ecol. Evol. Syst. 44: 239–259. [Google Scholar]
- Ellegren H., Parsch J., 2007. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 8: 689–698. [DOI] [PubMed] [Google Scholar]
- Erwin T. L., 1982. Tropical forests: their richness in Coleoptera and other arthropod species. Coleopt. Bull. 26: 74–75. [Google Scholar]
- Farlow A., Dolezal M., Hua L., Schlotterer C., 2012. The genomic signature of splicing-coupled selection differs between long and short introns. Mol. Biol. Evol. 29: 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawaz E. Y., Allan S. A., Bernier U. R., Obenauer P. J., Diclaro J. W., II, 2014. Swarming mechanisms in the yellow fever mosquito: aggregation pheromones are involved in the mating behavior of Aedes aegypti. J. Vector Ecol. 39: 347–354. [DOI] [PubMed] [Google Scholar]
- Flores H. A., Bubnell J. E., Aquadro C. F., Barbash D. A., 2015. The Drosophila bag of marbles gene interacts genetically with Wolbachia and shows female-specific effects of divergence. PLoS Genet. 11: e1005453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foret S., Maleszka R., 2006. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 16: 1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Nachman M. W., 2005. Rates of protein evolution are positively correlated with developmental timing of expression during mouse spermatogenesis. Mol. Biol. Evol. 22: 1044–1052. [DOI] [PubMed] [Google Scholar]
- Goto T., Salpekar A., Monk M., 2001. Expression of a testis-specific member of the olfactory receptor gene family in human primordial germ cells. Mol. Hum. Reprod. 7: 553–558. [DOI] [PubMed] [Google Scholar]
- Green P., Ewing B., Miller W., Thomas P. J., Green E. D., 2003. Transcription-associated mutational asymmetry in mammalian evolution. Nat. Genet. 33: 514–517. [DOI] [PubMed] [Google Scholar]
- Haerty W., Jagadeeshan S., Kulathinal R. J., Wong A., Ram K. R., et al. , 2007. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics 177: 1321–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. B., Basu S., Jiang X., Qi Y., Timoshevskiy V. A., et al. , 2015. SEX DETERMINATION. A male-determining factor in the mosquito Aedes aegypti. Science 348: 1268–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambuch T. M., Parsch J., 2005. Patterns of synonymous codon usage in Drosophila melanogaster genes with sex-biased expression. Genetics 170: 1691–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P. W., Wright A. E., Zimmer F., Dean R., Montgomery S. H., et al. , 2015. Sexual selection drives evolution and rapid turnover of male gene expression. Proc. Natl. Acad. Sci. USA 112: 4393–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R., Petrov D. A., 2008. Selection on codon bias. Annu. Rev. Genet. 42: 287–299. [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Ikemura T., 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151: 389–409. [DOI] [PubMed] [Google Scholar]
- Ikemura T., 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2: 13–34. [DOI] [PubMed] [Google Scholar]
- Ingvarsson P. K., 2008. Molecular evolution of synonymous codon usage in Populus. BMC Evol. Biol. 8: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeshan S., Singh R. S., 2005. Rapidly evolving genes of Drosophila: differing levels of selective pressure in testis, ovary, and head tissues between sibling species. Mol. Biol. Evol. 22: 1793–1801. [DOI] [PubMed] [Google Scholar]
- Jentes E. S., Poumerol G., Gershman M. D., Hill D. R., Lemarchand J., et al. , 2011. The revised global yellow fever risk map and recommendations for vaccination, 2010: consensus of the informal WHO working group on geographic risk for yellow fever. Lancet Infect. Dis. 11: 622–632. [DOI] [PubMed] [Google Scholar]
- Jia X., Liu S., Zheng H., Li B., Qi Q., et al. , 2015. Non-uniqueness of factors constraint on the codon usage in Bombyx mori. BMC Genomics 16: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja P., Osei-Poku J., Ho Y. S., Ariani C. V., Palmer W. J., et al. , 2014. Assembly of the genome of the disease vector Aedes aegypti onto a genetic linkage map allows mapping of genes affecting disease transmission. PLoS Negl. Trop. Dis. 8: e2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., et al. , 2012. Genious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P., Hellmann I., Enard W., Nowick K., Leinweber M., et al. , 2005. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309: 1850–1854. [DOI] [PubMed] [Google Scholar]
- Kim Y., 2004. Effect of strong directional selection on weakly selected mutations at linked sites: implication for synonymous codon usage. Mol. Biol. Evol. 21: 286–294. [DOI] [PubMed] [Google Scholar]
- Komili S., Farny N. G., Roth F. P., Silver P. A., 2007. Functional specificity among ribosomal proteins regulates gene expression. Cell 131: 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer M. U., Sinka M. E., Duda K. A., Mylne A. Q., Shearer F. M., et al. , 2015. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife 4: e08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Peterson D., Tamura K., 2012. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 28: 2685–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B., Bettencourt B. R., Meiklejohn C. D., Hartl D. L., 2005. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol. Biol. Evol. 22: 1345–1354. [DOI] [PubMed] [Google Scholar]
- Leparc-Goffart I., Nougairede A., Cassadou S., Prat C., de Lamballerie X., 2014. Chikungunya in the Americas. Lancet 383: 514. [DOI] [PubMed] [Google Scholar]
- Lipinska A., Cormier A., Luthringer R., Peters A. F., Corre E., et al. , 2015. Sexual dimorphism and the evolution of sex-biased gene expression in the brown alga ectocarpus. Mol. Biol. Evol. 32: 1581–1597. [DOI] [PubMed] [Google Scholar]
- Lloyd J. P., Seddon A. E., Moghe G. D., Simenc M. C., Shiu S. H., 2015. Characteristics of plant essential genes allow for within- and between-species prediction of lethal mutant phenotypes. Plant Cell 27: 2133–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludbrook J., Dudley H., 1998. Why permutation tests are superior to t and F tests in biomedical research. Am. Stat. 52: 127–132. [Google Scholar]
- Mank J. E., Ellegren H., 2009. Are sex-biased genes more dispensable? Biol. Lett. 5: 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank J. E., Hultin-Rosenberg L., Axelsson E., Ellegren H., 2007. Rapid evolution of female-biased, but not male-biased, genes expressed in the avian brain. Mol. Biol. Evol. 24: 2698–2706. [DOI] [PubMed] [Google Scholar]
- Matthews B. J., McBride C. S., DeGennaro M., Despo O., Vosshall L. B., 2016. The neurotranscriptome of the Aedes aegypti mosquito. BMC Genomics 17: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Parsch J., Ranz J. M., Hartl D. L., 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 100: 9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel R. P., 2011. Towards a more nuanced understanding of the relationship between sex-biased gene expression and rates of protein-coding sequence evolution. Mol. Biol. Evol. 28: 1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittmann B., Wolff C., 2012. Embryonic development and staging of the cobweb spider Parasteatoda tepidariorum C. L. Koch, 1841 (syn.: Achaearanea tepidariorum; Araneomorphae; Theridiidae). Dev. Genes Evol. 222: 189–216. [DOI] [PubMed] [Google Scholar]
- Nei M., Gojobori T., 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Nene V., Wortman J. R., Lawson D., Haas B., Kodira C., et al. , 2007. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 316: 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva C. F., Damiens D., Benedict M. Q., 2014. Male reproductive biology of Aedes mosquitoes. Acta Trop. 132: S12–S19. [DOI] [PubMed] [Google Scholar]
- Oliver T. A., Garfield D. A., Manier M. K., Haygood R., Wray G. A., et al. , 2010. Whole-genome positive selection and habitat-driven evolution in a shallow and a deep-sea urchin. Genome Biol. Evol. 2: 800–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S., Ohama T., Yamao F., Muto A., Jukes T. H., et al. , 1988. Directional mutation pressure and transfer RNA in choice of the third nucleotide of synonymous two-codon sets. Proc. Natl. Acad. Sci. USA 85: 1124–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C., Papp B., Hurst L. D., 2001. Highly expressed genes in yeast evolve slowly. Genetics 158: 927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa F., Windbichler N., Waterhouse R. M., Cagnetti A., D’Amato R., et al. , 2016. Rapid evolution of female-biased genes among four species of Anopheles malaria mosquitoes. bioRxiv DOI: https://10.1101/081620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M., Nuttall R., Edwards P., Minor J., Naiman D., et al. , 2004. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker G. A., 1970. Sperm competition and its evolutionary consequences in the insects. Biol. Rev. Camb. Philos. Soc. 45: 525–567. [Google Scholar]
- Parmentier M., Libert F., Schurmans S., Schiffmann S., Lefort A., et al. , 1992. Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature 355: 453–455. [DOI] [PubMed] [Google Scholar]
- Parsch J., Ellegren H., 2013. The evolutionary causes and consequences of sex-biased gene expression. Nat. Rev. Genet. 14: 83–87. [DOI] [PubMed] [Google Scholar]
- Peden J. F., 2000. Analysis of Codon Usage. University of Nottingham, UK. [Google Scholar]
- Perry J. C., Harrison P. W., Mank J. E., 2015. The ontogeny and evolution of sex-biased gene expression in Drosophila melanogaster. Mol. Biol. Evol. 31: 1206–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts R. J., Liu C., Zhou X., Malpartida J. C., Zwiebel L. J., 2014. Odorant receptor-mediated sperm activation in disease vector mosquitoes. Proc. Natl. Acad. Sci. USA 111: 2566–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. S. C., Dyer K. A., Coyne J. A., 1999. Sperm competition between Drosophila males involves both displacement and incapacitation. Nature 400: 449–452. [DOI] [PubMed] [Google Scholar]
- Proschel M., Zhang Z., Parsch J., 2006. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics 174: 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikhel A. S., Dhadialla T. S., 1992. Accumulation of yolk proteins in insect oocytes. Annu. Rev. Entomol. 37: 217–251. [DOI] [PubMed] [Google Scholar]
- Ranz J. M., Castillo-Davis C. I., Meiklejohn C. D., Hartl D. L., 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300: 1742–1745. [DOI] [PubMed] [Google Scholar]
- Rogers D. W., Baldini F., Battaglia F., Panico M., Dell A., et al. , 2009. Transglutaminase-mediated semen coagulation controls sperm storage in the malaria mosquito. PLoS Biol. 7: e1000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwager E. E., Meng Y., Extavour C. G., 2015. vasa and piwi are required for mitotic integrity in early embryogenesis in the spider Parasteatoda tepidariorum. Dev. Biol. 402: 276–290. [DOI] [PubMed] [Google Scholar]
- Segoli M., Hoffmann A. A., Lloyd J., Omodei G. J., Ritchie S. A., 2014. The effect of virus-blocking Wolbachia on male competitiveness of the dengue vector mosquito, Aedes aegypti. PLoS Negl. Trop. Dis. 8: e3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma E., Kunstner A., Fraser B. A., Zipprich G., Kottler V. A., et al. , 2014. Transcriptome assemblies for studying sex-biased gene expression in the guppy, Poecilia reticulata. BMC Genomics 15: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R., 1986. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 14: 5125–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Averof M., Lloyd A. T., Matassi G., Peden J. F., 1995. DNA sequence evolution: the sounds of silence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 349: 241–247. [DOI] [PubMed] [Google Scholar]
- Simmons C. P., Farrar J. J., Nguyen v. V., Wills B., 2012. Dengue. N. Engl. J. Med. 366: 1423–1432. [DOI] [PubMed] [Google Scholar]
- Small C. M., Carney G. E., Mo Q., Vannucci M., Jones A. G., 2009. A microarray analysis of sex- and gonad-biased gene expression in the zebrafish: evidence for masculinization of the transcriptome. BMC Genomics 10: 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenico M., Lloyd A. T., Sharp P. M., 1994. Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res. 22: 2437–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N., Eyre-Walker A., 2007. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol. Biol. Evol. 24: 374–381. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Kumar S., 2004. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics 168: 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N., 1988. Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci. USA 85: 2653–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W. J., Vacquier V. D., 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3: 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson W. J., Clark A. G., Waldrip-Dail H. M., Wolfner M. F., Aquadro C. F., 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98: 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson D. G., Kulathinal R. J., Singh R. S., 2002. Mammalian sperm proteins are rapidly evolving: evidence of positive selection in functionally diverse genes. Mol. Biol. Evol. 19: 1973–1980. [DOI] [PubMed] [Google Scholar]
- Turner L. M., Hoekstra H. E., 2008. Causes and consequences of the evolution of reproductive proteins. Int. J. Dev. Biol. 52: 769–780. [DOI] [PubMed] [Google Scholar]
- Uechi T., Maeda N., Tanaka T., Kenmochi N., 2002. Functional second genes generated by retrotransposition of the X-linked ribosomal protein genes. Nucleic Acids Res. 30: 5369–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia A. O., Hurst L. D., 2003. The signature of selection mediated by expression on human genes. Genome Res. 13: 2260–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanKuren N. W., Vibranovski M. D., 2014. A novel dataset for identifying sex-biased genes in Drosophila. J. Genomics 2: 64–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Shao Z. W., Xu Y., Liu J., Liu Y., et al. , 2011. Optimal codon identities in bacteria: implications from the conflicting results of two different methods. PLoS One 6: e22714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Werren J. H., Clark A. G., 2015. Genetic and epigenetic architecture of sex-biased expression in the jewel wasps Nasonia vitripennis and giraulti. Proc. Natl. Acad. Sci. USA 112: E3545–E3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C. A., Extavour C. G., 2016a Expression-linked patterns of codon usage, amino acid frequency and protein length in the basally branching arthropod Parasteatoda tepidariorum. Genome Biol. Evol. 8: 2722–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C. A., Extavour C. G., 2016b Refuting the hypothesis that the acquisition of germ plasm accelerates animal evolution. Nat. Commun. 7: 12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C. A., Johannesson H., 2013. Evolutionary dynamics of sex-biased genes in a hermaphrodite fungus. Mol. Biol. Evol. 30: 2435–2446. [DOI] [PubMed] [Google Scholar]
- Whittle C. A., Krochko J. E., 2009. Transcript profiling provides evidence of functional divergence and expression networks among ribosomal-protein gene paralogs in Brassica napus. Plant Cell 21: 2203–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C. A., Malik M. R., Krochko J. E., 2007. Gender-specific selection on codon usage in plant genomes. BMC Genomics 8: 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C. A., Sun Y., Johannesson H., 2011. Evolution of synonymous codon usage in Neurospora tetrasperma and Neurospora discreta. Genome Biol. Evol. 3: 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williford A., Demuth J. P., 2012. Gene expression levels are correlated with synonymous codon usage, amino acid composition, and gene architecture in the red flour beetle, Tribolium castaneum. Mol. Biol. Evol. 29: 3755–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F., 1990. The “effective number of codons” used in a gene. Gene 87: 23–29. [DOI] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z., Swanson W. J., 2002. Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol. Biol. Evol. 19: 49–57. [DOI] [PubMed] [Google Scholar]
- Yang X., Schadt E. E., Wang S., Wang H., Arnold A. P., et al. , 2006. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 16: 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sturgill D., Parisi M., Kumar S., Oliver B., 2007. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature 450: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Hambuch T. M., Parsch J., 2004. Molecular evolution of sex-biased genes in Drosophila. Mol. Biol. Evol. 21: 2130–2139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sources for all data used for analysis are provided in Table S1 in File S1 and were downloaded between August and November 2015 (A. aegypti) and in March 2016 (A. albopictus). No custom scripts or tools were created to perform the present analysis.