Abstract
How essential, regulatory genes originate and evolve is intriguing because mutations of these genes not only lead to lethality in organisms, but also have pleiotropic effects since they control the expression of multiple downstream genes. Therefore, the evolution of essential, regulatory genes is not only determined by genetic variations of their own sequences, but also by the biological function of downstream genes and molecular mechanisms of regulation. To understand the origin of essential, regulatory genes, experimental dissection of the complete regulatory cascade is needed. Here, we provide genetic evidences to reveal that PhoP-PhoQ is an essential two-component signal transduction system in the gram-negative bacterium Xanthomonas campestris, but that its orthologs in other bacteria belonging to Proteobacteria are nonessential. Mutational, biochemical, and chromatin immunoprecipitation together with high-throughput sequencing analyses revealed that phoP and phoQ of X. campestris and its close relative Pseudomonas aeruginosa are replaceable, and that the consensus binding motifs of the transcription factor PhoP are also highly conserved. PhoPXcc in X. campestris regulates the transcription of a number of essential, structural genes by directly binding to cis-regulatory elements (CREs); however, these CREs are lacking in the orthologous essential, structural genes in P. aeruginosa, and thus the regulatory relationships between PhoPPae and these downstream essential genes are disassociated. Our findings suggested that the recruitment of regulatory proteins by critical structural genes via transcription factor-CRE rewiring is a driving force in the origin and functional divergence of essential, regulatory genes.
Keywords: essential gene, transcription-cis regulatory element rewiring, two-component signal transduction system, PhoP-PhoQ, Xanthomonas campestris pv. campestris
ESSENTIAL genes play critical roles in the vitality of an organism, such as in cell division, macromolecule synthesis, ribosome assembly, and embryo development. Genetic inactivation of these genes leads to lethality or infertility (Wilson et al. 1977). Consequently, null mutations in essential genes would be promptly erased by the overwhelming force of purifying selection (Jordan et al. 2002; Bergmiller et al. 2012). Based on this, it was predicted that essential genes would evolve much slower than nonessential genes. However, recent theoretical and experimental studies have revealed that essential genes can also originate from novel genes during speciation, implying that the molecular mechanisms determining the origins of essential genes are diverse (Hurst and Smith 1999; Chen et al. 2010). Among them, the evolution of essential, regulatory genes is more intriguing, since they modulate the expression of various downstream genes, which complicates the evolutionary impact of mutations in these regulatory genes. However, to date, how essential, regulatory genes originate and evolve remains largely unclear (Peter and Davidson 2011; Harms and Thornton 2013).
One of the difficulties in studying the evolution of essential regulatory genes is that the biological functions of these genes depend substantially on the genetic context during regulation and growth conditions of the organism (DʼElia et al. 2009). In fact, the theoretical prediction of which regulatory gene is essential per se is a challenge. For example, a number of genes that are essential in one organism have been experimentally confirmed to be nonessential in closely related organisms (Dussurget et al. 1996; Rodriguez et al. 2002; Ng et al. 2004; Bisicchia et al. 2007). Bioinformatic studies that predicted essential regulatory genes merely through homology searching inevitably introduced bias by including nonessential genes or ignoring the genetic context of the regulatory genes in the analyses. Even in genome-wide mutational studies, the failure to obtain a viable gene mutant is also weak evidence to support the essentiality of the gene. For instance, in bacteria, the polar effect of an insertional mutation within an operon and the growth conditions complicate the subsequent genetic analysis (Glass et al. 2006; Xu et al. 2011). Furthermore, the evolution of the biological function of regulatory genes may not be caused by genetic polymorphisms in themselves but instead may be determined by their downstream genes or even their mode of molecular regulation. Therefore, to solve these problems, a “bottom-up” strategy, which dissects the evolutionary changes in the complete regulatory cascade based on genetic and biochemical analyses, would not only avoid the problematic prediction of essential regulatory genes but could also reconstruct the possible molecular steps of genetic variations during their evolution (Perez and Groisman 2009a; Harms and Thornton 2013). Together with theoretical studies, a more complete view of the origin of essential regulatory genes could be achieved by multidisciplinary investigations.
In prokaryotes, PhoP-PhoQ represents a typical two-component signal transduction system (TCS), which is the predominant sense-and-response mechanism that is usually compared with the “nervous system” of eukaryotic cells (Goulian 2010). PhoP-PhoQ is also one of the most extensively studied signaling systems in all organisms (Groisman 2001; Ohl and Miller 2001). PhoQ is an inner membrane-bound receptor histidine kinase (HK). After detecting environmental signals of Mg2+, Ca2+, low pH, or cationic antimicrobial peptides, PhoQ activates its autokinase activity by phosphorylating a conserved histidine residue (His) and then transfers the phosphoryl group to PhoP, a cytosolic response regulator (RR) with transcription factor activity. Eventually, the activated PhoP modulates the transcription of downstream genes by binding directly to their cis-regulatory elements (CREs) (Kato and Groisman 2008; Prost and Miller 2008). PhoP-PhoQ is a pleiotropic TCS, regulating the virulence of a number of pathogenic bacteria, especially those belonging to Proteobacteria (Oyston et al. 2000; Minagawa et al. 2003; Groisman et al. 2013). The inactivation of this system generally leads to a remarkable attenuation of bacterial virulence. The evolution of the PhoP-PhoQ regulon has been studied in bacteria belonging to Enterobacteriaceae, which revealed extensive genetic polymorphisms and species-specific regulation in different bacterial species, suggesting that PhoP-PhoQ is an evolutionarily active system in bacterial adaptation to various ecological niches (Zwir et al. 2005; Perez and Groisman 2009b; Perez et al. 2009; Ram and Goulian 2013).
Interestingly, during our previous study to systematically knockout all genes encoding RRs in the gram-negative bacterium Xanthomonas campestris pathovar (pv). campestris, a species belonging to the γ-Proteobacteria class and the causative agent of black rot disease of crucifers, repeated efforts to inactivate the orthologs of phoP and phoQ failed, suggesting that phoPXcc and phoQXcc are essential (Qian et al. 2008a,b). However, as mentioned above, previously reported phoP and phoQ orthologs in other bacteria, such as Escherichia coli, Salmonella enterica, and Yersinia pestis, are undoubtedly nonessential (Kato and Groisman 2008). Even in closely related species that belong to the Pseudomonadaceae family, such as Pseudomonas aeruginosa and P. fluorescens (Macfarlane et al. 2000; Yan et al. 2009), phoP and phoQ are also mutable. These results prompted a hypothesis that the functional divergence of phoP-phoQ during bacterial evolution resulted in the essentiality of their orthologs in X. campestris, which provided a research case to study the molecular mechanism responsible for the origin of essential, regulatory genes.
In the present study, we demonstrate that phoP and phoQ of X. campestris pv. campestris are bona fide essential genes. The conditional mutants of phoP and phoQ exhibited abnormalities in daughter cell separation during cytogenesis when the transcription of the two genes was repressed. Based on the identified PhoPXcc-binding consensus sequence, evolutionary and molecular analyses revealed that PhoPXcc controls the transcription of a number of essential, structural genes by directly binding to their CREs. However, in the closely related bacterium P. aeruginosa, PhoPPae did not regulate the transcription of the orthologs of the aforementioned essential, structural genes, due to the lack of the PhoPPae-binding sites in these downstream genes because of transcription factor (TF)-CRE rewiring. Our study suggests that TF-CRE rewiring is the genetic force that drove the functional divergence of phoP-phoQ essentiality in X. campestris.
Materials and Methods
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Supplemental Material, Table S1 in File S1. All X. campestris pv. campestris mutants were derived from the wild-type (WT) strain X. campestris pv. campestris 8004. These strains were grown at 28° in nutrient broth, yeast extract and glycerol (NYG)-rich medium (peptone, 5.0 g liter–1; yeast extract, 3.0 g liter–1; and glycerol, 20.0 g liter–1; pH 7.0) or minimal medium for Xanthomonas campestris (MMX) minimal medium (glucose, 2.0 g liter–1; (NH4)2SO4, 2.0 g liter–1; sodium citrate, 1.0 g liter–1; K2HPO4, 4.0 g liter–1; KH2PO4, 6.0 g liter–1; and MgSO4·7H2O, 0.2 g liter–1, pH 7.0). All P. aeruginosa mutants were derived from the strain PAO1. The P. aeruginosa strains were grown in rich nutrient yeast broth (NYB) (25 g liter–1 Oxoid nutrient broth no. 2 and 5 g liter–1 yeast extract, pH 7.0) or minimal M9 medium (glucose 10 mM, Na2HPO4 48 mM, KH2PO4 22 mM, NaCl 9 mM, NH4Cl 19 mM, MgSO4 2 mM, and CaCl2 100 µM, pH 7.2). The E. coli DH5α strain was used as the host for the preparation of all recombinant vectors. Antibiotics were added at the following concentrations for bacterial selection: kanamycin (50 μg ml−1), gentamycin (100 μg ml−1), spectinomycin (100 μg ml−1), ampicillin (100 μg ml−1), and rifamycin (25 μg ml−1). Where stated, L-arabinose and IPTG were added to the media at a concentration of 0.05% and 2 mM, respectively. X. campestris pv. campestris and P. aeruginosa competent cells were prepared according to previous reports (Qian et al. 2008b; Wang et al. 2015). The transformation of X. campestris pv. campestris competent cells was conducted on a Bio-Rad Gene Pulser XCell (Bio-Rad, Hercules, CA), set at 18 kV cm−1 and a pulse time of 0.35–0.55 msec. The transformation of P. aeruginosa competent cells was performed according to a previous report (Wang et al. 2015).
Construction of in-frame deletion mutants of the chromosomal phoP and phoQ genes
General molecular biology methods, such as PCR, Southern blotting, DNA ligation, restriction enzyme treatment, transformation, and plasmid isolation, were carried out according to the methods described by Sambrook et al. (1989). To mutate an essential gene in the bacterial chromosome of X. campestris pv. campestris, a conditional regulatable recombinant strain (WT-pphoQ or WT-pphoP), which contained an additional gene copy (phoQXcc or phoPXcc) provided in trans in a pHM2 vector, was constructed before inactivation (Table S1 in File S1). This plasmid-borne gene copy was under the control of a tightly regulated arabinose promoter (pBAD-araC). The full-length phoQ or phoP genes with engineered SalI and KpnI restriction sites, and the arabinose promoter (pBAD-araC) fragment cloned from E. coli S17-1 with EcoRI and SalI restriction sites, were amplified, sequenced, and ligated into the promoterless pHM2 broad-host-range vector. These recombinant pHM2 vectors were extracted from E. coli and electroporated into X. campestris pv. campestris 8004 competent cells to generate the strains WT-pphoQ and WT-pphoP. The primers used to amplify the above full-length genes and CREs are listed in Table S2 in File S1.
The initiation of homologous double-crossover recombination events was used to mutate the chromosomal copies of phoQXcc and phoPXcc (Schafer et al. 1994). The primer sequences used to amplify the phoP and phoQ gene fragments from X. campestris pv. campestris 8004 are listed in Table S2 in File S1. In-frame gene deletion mutants (including ΔphoP-pphoP and ΔphoQ-pphoQ) were constructed by homologous double-crossover recombination using the selection/counterselection suicide vector pk18mobsacB (Schafer et al. 1994). In brief, 5′ and 3′ DNA fragments of the target gene were amplified, sequenced, and ligated using introduced restriction sites to generate an insert with a reading frame deletion. The insert was then ligated into the pK18mobsacB suicide vectors. These recombinant vectors were electroporated into the previously constructed recombinant strains WT-pphoP and WT-pphoQ and screened on NYG plates containing kanamycin (50 μg ml−1) to obtain a single-crossover mutant. MMX agar plates supplemented with 10% sucrose and 0.05% L-arabinose were then used to select the second double-crossover recombinants, using a previously described method (Schafer et al. 1994). The resulting constructs (ΔphoP-pphoP and ΔphoQ-pphoQ) were verified by PCR, sequencing, and Southern blotting. To inactivate the chromosomal copy of phoPXoo and phoQXoo of X. oryzae pv. oryzae PXO99A, a similar strategy as above was conducted. To in-frame delete the chromosomal copy of phoPPae and phoQPae of P. aeruginosa PAO1, a homologous, double-crossover method was used as reported (Wang et al. 2015). The primers with appropriate restriction sites, used to construct the above mutants, are listed in Table S2 in File S1.
To construct the recombinant strain used in chromatin immunoprecipitation sequencing (ChIP-seq), the His6 epitope coding sequence was fused at the 3′-end of phoPXcc by PCR. This DNA insert was used to substitute the chromosomal phoPXcc copy of the ΔphoQ-pphoQ strain by double crossover using the suicide vector pK18mobsacB. The primers used to construct this strain are listed in Table S2 in File S1.
RT-PCR and quantitative real-time RT-PCR (qRT-PCR)
Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA), following the manufacturer’s instructions. The concentration of total RNA was determined using a NanoDrop 1000 Spectrophotometer (Thermo Fisher). One microgram of RNA from each sample was treated with Turbo DNA-free DNase (Ambion) before RT-PCR, to exclude DNA contamination. First strand cDNA was synthesized using SuperScriptIII reverse transcriptase (Invitrogen) and random hexamer primers. To verify the phoP-phoQ operon structure, seven pairs of primers (P1–P10) were designed to amplify the potential intergenic transcripts by RT-PCR. The PCR reactions were performed in a MyCycler thermocycler (Bio-Rad). The primers used in the RT-PCR assays are listed in Table S2 in File S1.
To quantify mRNA expression levels in various bacterial cells, qRT-PCR was performed in a CFX96 Real-Time System (Bio-Rad), using Maxima SYBR Green/Fluorescein dye (Fermentas) and the primers listed in Table S2 in File S1. The results were analyzed in Excel (Microsoft), and the expression levels were normalized to tmRNA levels as internal controls.
Protein expression, purification, and western blotting
PhoPXcc-His6, PhoQXcc-His6, PhoPXoo-His6, PhoQXoo-His6, PhoPPae-His6, and PhoPPae-His6 recombinant proteins were expressed by constructing the corresponding recombinant pET30a (Novagen) expression vectors and transforming them into E. coli BL21(DE3) cells. The primers used to produce these constructs are listed in Table S2 in File S1. For protein expression, overnight cultures of each strain were reinoculated into 1 liter of Luria-Bertani medium and grown until they reached OD600 = 0.6, when protein expression was induced by adding 1 mM IPTG for 6 hr at 28° or 18 hr at 16°.
His6-tagged proteins were purified using affinity chromatography with nickel- nitrilotriacetic acid (Ni-NTA) agarose beads (Millipore, Bedford, MA), according to the manufacturer’s instructions. The following buffers were used to purify the recombinant proteins: lysis buffer [50 mM Tris-HCl, pH 7.5; 0.5 M NaCl; 10% glycerol; 0.1% Triton X-100; 1 mM PMSF; and 1 × protease inhibitor cocktail (GE Healthcare, UK)]; binding buffer (300 mM NaCl; 50 mM sodium phosphate buffer, pH 7.0; and 10 mM imidazole, pH 8.0); Ni-NTA wash buffer (300 mM NaCl; 50 mM sodium phosphate buffer, pH 7.0; and 30 mM imidazole, pH 8.0); and Ni-NTA elution buffer (300 mM NaCl, 250 mM imidazole, and 50 mM sodium phosphate buffer, pH 7.0). Approximately 3 g of cells was harvested and resuspended in 10 ml of lysis buffer, sonicated, and centrifuged at 15,000 × g to clear the supernatant. His6-tagged proteins were bound to 0.5–1.0 ml of a Ni-NTA slurry for 4 hr, washed six times with 10 ml of wash buffer, and then eluted with 2.0 ml of elution buffer. The purified proteins were then concentrated with Centricon YM-10 or YM-30 columns (Millipore) and maintained in storage buffer (50 mM Tris-HCl, pH 8.0; 0.5 mM EDTA; 50 mM NaCl; and 5% glycerol) at −80° until use. The protein purity was determined by 12% SDS-PAGE followed by Coomassie Brilliant Blue G-250 staining.
Inverted membrane vesicles (IMVs) containing full-length PhoQ proteins were prepared according to a previous study (Rosen and McClees 1974). In brief, 0.5 liter of IPTG-induced E. coli BL21(DE3) cells was collected and lysed by sonication. The membrane fraction, containing the PhoQ of various bacteria, was collected by ultracentrifugation at 80,000 × g for 60 min at 4° and washed twice in high-salt buffer (20 mM sodium phosphate, pH 7.0; 2 M KCl; 10% glycerol; 5 mM EDTA; 5 mM DTT; 1 mM PMSF; and protease inhibitor cocktail) and twice in high-salt buffer without DTT and protease inhibitor. The purified membrane fraction was pelleted by ultracentrifugation (120,000 × g for 10 min at 4°) and resuspended in 0.5 ml of final storage buffer (20 mM Tris-HCl, pH 7.5, 10% glycerol). The total protein was quantified using a Bio-Rad Protein Assay kit after the lysis of the membrane fraction with 2 × SDS-PAGE loading buffer without dye. The membranes were used immediately in autokinase, phosphotransferase, and phosphatase assays.
In vitro protein phosphorylation assays
PhoQ kinase, phosphotransferase, and phosphatase activity assays were performed as previously described (Jung et al. 1997). To detect the autokinase activity of PhoQ in vitro, 2–10 μM (final concentrations) of purified PhoQ-His6 protein or 20 μg of IMVs was phosphorylated with 5 μCi γ-[32P]-ATP in reaction buffer (50 mM Tris-HCl, pH 7.8; 2 mM DTT; 25 mM NaCl; 25 mM KCl; and 5 mM MgCl2) at room temperature. Aliquots of the reaction mixture were removed at different times, and the reaction was stopped by adding 5 μl of 5 × SDS-PAGE loading buffer (250 mM Tris-HCl, pH 6.8; 10% SDS; 50% glycerol; 0.5% bromophenol blue; and 5% β-mercaptoethanol). The samples were loaded onto 12% SDS-PAGE gels without preheating and electrophoresed at room temperature for 60 min at 120 V. Bovine serum albumin was substituted for the HK PhoQ protein in negative control reactions. Following electrophoresis, the gel was separated and sealed in a plastic bag. Autoradiographic signals were detected with a Phosphor Imager and scanned by the Typhoon 9400 system (Amersham, Piscataway, NJ).
To detect the phosphotransferase activity from PhoQ to PhoP in vitro, 20 μg of IMVs was auto-phosphorylated as described above. Then, 10 μM purified PhoPXcc-His6 protein was added to initiate the phosphotransfer reaction. Twenty-microliter aliquots were removed at required time points and mixed with 5 × SDS-PAGE loading buffer to stop the reaction. The samples were kept on ice until SDS-PAGE analysis. Following electrophoresis, signals were detected by autoradiography, as described above.
For phosphatase assays, 20 μg of IMVs containing PhoQ was incubated with 30 μCi γ-[32P]-ATP in 300 μl of reaction buffer for 20 min at room temperature. Then, 10 μM purified PhoP-His protein was added, and the reaction mixture was incubated for 10 min to saturation. A 20-μl aliquot was removed and mixed with 5 × SDS-PAGE loading buffer and kept as a reference sample for the phosphor-PhoQ and phosphor-PhoP proteins after the phosphotransfer reaction. The IMVs were then removed by centrifugation (16,000 × g for 3 min at 4°) and washed. The supernatant containing the PhoPXcc-P protein was transferred to a Centricon YM-10 column (Millipore) for centrifugation (12,000 × g, 2 min at 4°). The column was washed with reaction buffer three times to remove free γ-[32P]-ATP. A 280-μl volume of reaction buffer containing 1 mM ADP was added to the column to dissolve and recover the phosphor-PhoP protein fraction. The mixture was then divided equally into two parts. One part was incubated with 15 μg of IMVs containing unphosphorylated PhoQ protein, and the other was incubated with an equal volume of final storage buffer (20 mM Tris-HCl, pH 7.5 and 10% glycerol). Aliquots were removed from the respective mixtures at different times and mixed with 5 × SDS-PAGE loading buffer to stop the reaction. The samples were kept on ice until SDS-PAGE. Following electrophoresis, the signals were detected by autoradiography, as mentioned above.
Electrophoretic mobility shift assay (EMSA)
DNA duplex fragments corresponding to the sequences upstream of targeted genes were PCR-amplified and purified. The PCR products were labeled with α-[32P]-dCTP by DNA polymerase Klenow fragment (Promega, Madison, WI). The primers used to amplify this product are listed in Table S2 in File S1. For the EMSA, ∼2 fmol of 32P-labeled DNA in a 20-μl volume was incubated at room temperature for 30 min with purified PhoP-His6 protein. The binding buffer used for protein–DNA incubations contained 10 mM Tris-HCl, pH 7.5; 50 mM KCl; 1 mM DTT; 5 mM MgCl2; 0.05% NP-40; 50 ng μl−1 poly(dI·dC); and 2.5% glycerol. For competition, an unlabeled DNA probe was co-incubated with the labeled probe and the purified PhoP-His6 protein. The reactions were stopped by adding 10 × DNA loading buffer, and the samples were run on a 5% nondenaturing PAGE gel in 0.5 × TBE buffer at room temperature. The gel was dried, and the shifted bands were detected by autoradiography. If short DNA probes (< 30 nt) were used, the DNA sequences were chemically synthesized, end-labeled by T4 Polynucleotide Kinase with [γ-32P]ATP, and used in EMSA.
Two-dimensional electrophoresis (2-DE) of proteins
Bacterial strains were grown in NYG medium overnight to OD600 = 0.4; the cells were then washed and resuspended with the same volume of MMX medium three times. The cells were harvested and treated with MMX medium with 2 mM IPTG or 0.05% L-arabinose for 1 hr. The bacterial cells were harvested by centrifugation at 10,000 × g for 5 min at 4°. Total proteins were extracted and dissolved in isoelectric focusing (IEF) buffer [7 M urea, 2 M thiourea, 4% CHAPS, 40 mM DTT, and 2% immobilized pH gradient (IPG) buffer, pH 4–7]. The protein concentration was determined using a 2-DE Quant kit (GE Healthcare), followed by protein purification by a 2-DE clean-up kit (GE Healthcare). The procedures of the first-dimension IEF and 2-DE were performed. To achieve the best resolution, 500 μg of total protein was loaded onto 24 cm of Immobiline DryStrips (pH 4–7; GE Healthcare) with rehydration solution (7 M urea, 2 M thiourea, 2% CHAPS, 15 mM DTT, and 1% IPG buffer, pH 4–7) for gel rehydration. The IEF was performed using an Ettan IPGphor III system (GE Healthcare), according to the manual supplied. After equilibration, the protein samples were separated on 12.5% SDS-PAGE gels for 2-DE, which was performed using an Ettan DALT six system (GE Healthcare). The gels were run at a constant current in two steps at 15°: 1 W/gel for 1 hr and then at 8 W/gel for 5 hr. Finally, the 2-DE gels were stained with colloidal Coomassie staining. Each sample pair was analyzed three times. The gels were scanned at a 300 dots per inch resolution using an ImageScanner III scanner, and the data were analyzed using ImageMaster 2D Platinum 7.0 software (GE Healthcare).
Protein spots were detected in automatic analysis mode, and those showing significant differences (P value < 0.05) were selected and excised manually from gels. The excised portions of the gel were subjected to in-gel digestion, using trypsin for subsequent protein identification with the Autoflex II matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) system. The details of the procedures for in-gel digestion and MS analysis are given in our previous work (Deng et al. 2014).
ChIP-seq analysis
The protocol used for ChIP followed that of a previous study (Wang et al. 2014). In brief, bacterial strains were grown in MMX medium plus IPTG or L-arabinose until the OD600 nm was 0.4. The cells were cross-linked with 1% formaldehyde and subsequently quenched by 0.5 M glycine for 5 min. Bacterial cultures (4 ml), harvested by centrifugation, were washed twice in 10 ml of cold TBS buffer (20 mM Tris-HCl, pH 7.5 and 150 mM NaCl) and then resuspended in 1 ml of lysis buffer (10 mM Tris, pH 8.0; 20% sucrose; 50 mM NaCl; 10 mM EDTA, and 10 mg/ml lysozyme). Immunoprecipitation (IP) buffer (50 mM HEPES-KOH pH 7.5; 150 mM NaCl; 1 mM EDTA; 1% Triton X-100; 0.1% sodium deoxycholate; 0.1% SDS; and 1 mM phenylmethylsulfonyl fluoride) was added to the bacterial cell suspension, and the cells were sonicated with a Diagenode Bioruptor UCD-300 sonicator (Diagenode, Denville, NJ) to generate DNA fragments of ∼200 bp (on average). After centrifugation, the solution was precleared with 20 μl of protein A sepharose at 4° for 10 min on a slow rotator, and a 100-µl aliquot was kept as the loading control DNA (input sample). For the ChIP assays, 50 μl of protein A sepharose (50% slurry) and 2 µl of an anti-His6 antibody were added to an 800-μl aliquot, and the mixture was incubated at 4° overnight with slow rotation. The next day, the beads were collected by centrifugation and washed with IP buffer and wash buffer [10 mM Tris-HCl, pH 8.0; 250 mM LiCl; 1 mM EDTA; 0.5% Nonidet-P40 (equivalent to Triton X-100); and 0.5% sodium deoxycholate]. The immunoprecipitated chromatin was removed from the beads by adding 100 µl of elution buffer (50 mM Tris, pH 7.5; 10 mM EDTA; and 1% SDS), and the solution was incubated for 10 min at 65°. RNase A and proteinase K were used to remove RNA and protein, respectively. The DNA was purified using a PCR purification kit (QIAGEN, Valencia, CA) after an overnight incubation at 65° to reverse the protein–DNA cross-links. High-throughput sequencing was performed using the Illumina Highseq-2000 system by the Beijing Institute of Genomics genomic service.
The reads of high-throughput sequencing were treated using the Burrows-Wheeler Alignment tool (BWA) method (Li and Durbin 2009). The clean reads were aligned against the genomic sequence of X. campestris pv. campestris 8004. Peak calling was conducted by MACS2 (Zhang et al. 2008) and Perl scripts. The PhoP-binding motif analysis was completed using the MEME and FIMO tools from the MEME software suite (Bailey et al. 2015). We first determined the binding motif and then scanned the full genome for its presence. The motif identification was done using the MEME program on the set of sequences defined by the PhoP-binding regions. Using default settings, the previously determined PhoP motif was recovered and then tailored to the correct size by setting the width parameter to 16 bp. We then used these motifs and the position-specific probability matrix generated by MEME to rescan the entire genome using the FIMO program.
DNase I footprinting assay
To characterize the PhoP-binding sites on PftsA, PphoP, and POar, DNase I footprinting experiments were performed. DNA probes for PftsA, PphoP, and POar were generated using a pair of 5′-end-labeled primers for the gene of interest (forward or reverse) and an unlabeled primer (reverse or forward). Binding reaction mixtures were prepared in binding buffer [10 mM Tris-HCl, pH 7.5; 50 mM KCl; 1 mM DTT; 5 mM MgCl2; 0.05% NP-40; 50 ng μl−1 poly(dI·dC); and 2.5% glycerol]. The 32P-end-labeled DNA probes were mixed with purified PhoP-His6 protein at concentrations ranging from 20 to 200 μM in a 50-μl total reaction volume. The binding reactions were carried out at 25° for 1 hr and subsequently subjected to treatment with 0.375 U DNase I (Promega) at 25° for 3 min. The digestion reactions were stopped by the addition of 50 μl of DNase I stop solution (20 mM EGTA, pH 8.0). The digested DNA samples were isolated by phenol–chloroform extraction and ethanol precipitation. Pellets containing DNA were air-dried and then resuspended in formamide containing loading dye (10 mM Tris-HCl, pH 8.0; 20 mM EDTA, pH 8.0; 0.05% bromophenol blue; and 0.05% xylene cyanol). The samples were heated at 95° for 3 min and loaded into an 8% polyacrylamide gel containing 7 M urea. The dried gels were exposed to a phosphor screen and scanned using a Typhoon Trio system (GE Healthcare). The sequencing ladder was generated using an Accupower DNA sequencing kit (Bioneer, Korea) with the same 32P-end-labeled oligonucleotide primer (Table S2 in File S1).
Microscale thermophoresis (MST) assays
The MST method was used to assay the binding interactions between proteins and DNA, and between proteins and metal ions. For protein–DNA interaction, 5′-FAM (fluorescein)-labeled oligonucleotide primers were synthesized by the Genscript Technology Company (China) and were annealed with an unlabeled complementary primer to form double-stranded (ds) DNA. A series of 16 PhoP-His6 protein solutions with different concentrations were prepared by consecutive twofold dilutions from the highest concentration. Protein at different concentrations and double-stranded DNA (dsDNA) at 20 nM were mixed at a volume ratio of 1:1. The samples were loaded into silica capillaries after incubating at room temperature for 20 min. Measurements were performed at 25° in MST-optimized buffer [50 mM Tris-HCl buffer (pH 7.4) with 150 mM NaCl, 10 mM MgCl2, and 0.05% (V/V) Tween-20] using 40–90% light emitting diode (LED) power and 80% infrared (IR)-laser power for comparison. For protein–metal interaction, PhoP was labeled by Protein Labeling Kit RED-NHS (NanoTemper Technologies, München, Germany) and dissolved in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 10 mM MgCl2, and 0.05% (v/v) Tween-20. Both protein–DNA and protein–metal binding affinity were measured by a Monolith NT.115T (NanoTemper Technologies). The data analyses were performed using Origin v.8.5 analysis software.
Data availability
All data are included in the figures and tables. Bacterial strains are available upon request.
Results
TCS system XC4031–XC4030 of X. campestris is the ortholog of PhoP-PhoQ
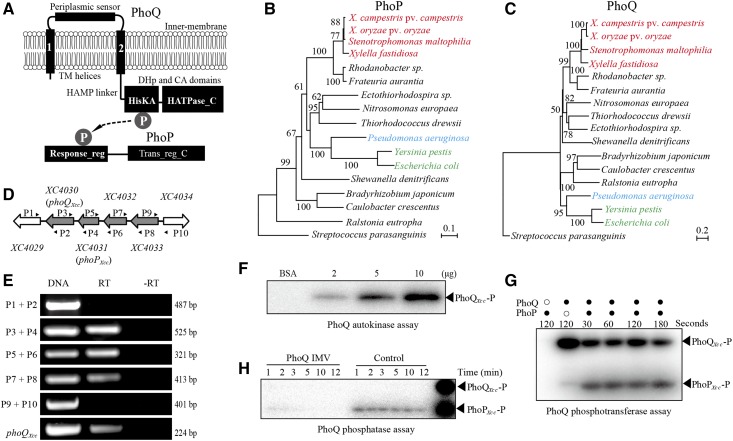
Phylogenetic analysis and similarity search revealed that XC4031–XC4030 of X. campestris pv. campestris strain 8004 are the PhoP-PhoQ orthologs of Enterobacteriaceae (Figure 1A, B, and C). Among the proteins of X. campestris pv. campestris, XC4031–XC4030 share the highest similarity with the PhoP-PhoQ sequences of E. coli, S. enterica, or P. aeruginosa. These PhoQ-PhoP orthologs have similar structures. PhoQ usually contains an N-terminal periplasmic sensor region surrounded by two transmembrane helices. The C-terminal dimerization and histidine phosphotransfer and catalytic ATP-binding domains are linked to the N-terminal input region by an ∼140-aa HAMP (present in HKs, adenylate cyclases, methyl-accepting proteins, and phosphatases) domain (Figure 1A). PhoP is an OmpR-family TF with an N-terminal receiver domain as the phosphoryl acceptor and a C-terminal helix-turn-helix region (Trans_reg_C) that binds dsDNA (Figure 1A). The identities of PhoP and PhoQ between X. campestris pv. campestris (belongs to Xanthomonadaceae) and its close relative P. aeruginosa PAO1 (belongs to Pseudomonadaceae) are 56 and 35% (e-values are 2 × 10−89 and 4 × 10−51), respectively (Figure S1 in File S1). A RT-PCR assay revealed that in X. campestris pv. campestris, phoQXcc-phoPXcc-XC4032-XC4033 constitute an operon because they were cotranscribed as a polycistronic mRNA (Figure 1, D and E). Among them, XC4032 encodes a conserved hypothetical protein, while XC4033 encodes a putative dihydrouridine synthase that is involved in tRNA synthesis. This tetra-cistronic operon structure differs from the bicistronic phoP-phoQ operon generally encoded by enterobacterial species (Kasahara 1992).
Figure 1.
Phylogeny and signal transduction of the two-component signaling system PhoP-PhoQ. (A) Schematic view of the PhoP and PhoQ secondary structures and cellular locations. TM: transmembrane; protein domain names are indicated according to the pfam database. HisKA (PF00512); HAMP (PF00672); HPt (PF01627); HATPase_c (PF02518); Response_Reg (PF00072); and Trans_reg_C (PF00486). “P” in gray circles represents the phosphoryl group that is transferred from PhoQ to PhoP. (B and C) Phylogenetic relationships of PhoP and PhoQ, respectively. Orthologous protein sequences from representative species of α-, β- and γ-Proteobacteria were used to construct the trees by the Neighbor-Joining method. The scale bar indicates the nucleotide substitutions per site. Orthologous PhoP and PhoQ from the gram-positive bacterium Streptococcus parasanguinis were used as outgroups. Phylogenetic trees were evaluated by bootstrapping (500 duplicates). Bacteria belonging to Xanthomonadaceae, Pseudomonadaceae, and Enterobacteriaceae are listed in red, blue, and green, respectively. (D) Schematic view of the phoPXcc-phoQXcc operon in X. campestris pv. campestris strain 8004. The large arrows represent the location and transcriptional direction of genes; the small black arrows (P1–P10) indicate the primers used in the RT-PCR reactions. (E) Verification of the structure of the phoPXcc-phoQXcc operon by RT-PCR. DNA: positive control using X. campestris pv. campestris 8004 DNA as the PCR template. −RT: no reverse transcriptase negative control. RT: reverse transcriptase added during cDNA synthesis. Bacterial total RNA was used as the template for cDNA synthesis. The experiments were repeated three independent times and a representative is shown. (F) Full-length PhoQXcc has autokinase activity. PhoQXcc-P represents phosphorylated PhoQXcc. Inverted membrane vesicles (IMVs) of PhoQXcc (2–10 µg) were co-incubated with [γ-32P]ATP for 20 min. (G) Full-length PhoQXcc phosphorylates PhoPXcc. PhoQXcc IMVs were autophosphorylated as in (D), and 10 μM of PhoPXcc was then added to the reaction mixtures. Aliquots were taken at the indicated times. Each sample contained 10 μg of PhoQXcc IMVs. (H) PhoQXcc has phosphatase activity toward PhoPXcc. PhoQXcc IMVs were autophosphorylated, and the phosphoryl groups were transferred to PhoPXcc. The IMVs were removed by centrifugation, and the free [γ-32P]ATP was removed by ultrafiltration. The reaction mixture was equally separated into two parts. Unphosphorylated PhoQXcc IMVs and ADP (1 mM) were added to one of the mixtures. Phosphorylated PhoPXcc and PhoQXcc were used as the indicators (the last lane). In (F–H), the reactions were stopped by adding 5 μl of 5 × SDS-PAGE loading buffer. The experiments were repeated three times, and a representative is shown.
To biochemically verify that PhoPXcc-PhoQXcc is a TCS, the full-length PhoP-PhoQ sequences of X. campestris pv. campestris, X. oryzae pv. oryzae, and P. aeruginosa PAO1 were expressed with C-terminal His6 epitope tags and purified. In vitro phosphorylation assays showed that the IMVs of PhoQXcc could be autophosphorylated and that the phosphoryl groups are transferred onto PhoPXcc within a short period of time (30 sec, Figure 1, F and G). PhoQXcc IMVs also have phosphatase activity to completely dephosphorylate PhoPXcc-P within 5 min (Figure 1H). In addition, both PhoQXoo and PhoQPae could reciprocally phosphorylate PhoPXcc with similar kinetics (Figure S2 in File S1). These results demonstrated that PhoPXcc-PhoQXcc is a bona fide TCS, and that the enzymatic activity of PhoQ seems to be conserved in the tested bacteria.
phoP-phoQ of X. campestris pv. campestris are essential genes and can be genetically complemented by their orthologs from P. aeruginosa
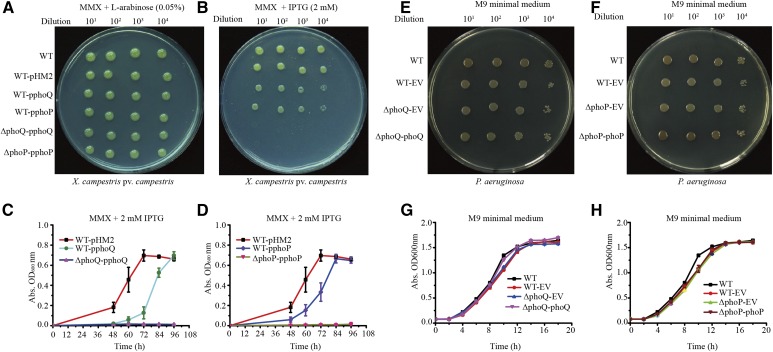
To define stringently whether phoPXcc and phoQXcc are essential genes, we constructed two merodiploid strains (WT-pphoP and WT-pphoQ) in which an additional copy of phoPXcc or phoQXcc, respectively, was supplied in trans by the broad-host-range vector pHM2 (Table S1 in File S1). The plasmid-borne copies were controlled by a tightly regulated arabinose promoter (pBAD-araC) from E. coli. Starting from the two recombinant strains, the chromosomal phoPXcc and phoQXcc (ΔphoP-pphoP and ΔphoQ-pphoQ) genes were successfully deleted in-frame, and Southern blotting and PCR amplification were used to detect whether the targeted DNA sequences were correctly mutated (Figure S3 in File S1). The recombinant rate in the construction of the phoPXccmutant was quite low, since only one correct clone was obtained from > 6000 candidate colonies in selecting the second round of homologous crossover. Note that if we started from the WT strain without a plasmid-borne copy of phoPXcc or phoQXcc, or when L-arabinose was absent during the double-crossover selection, no correct mutant could be obtained. In addition, when the bacterial strains were grown on minimal MMX medium containing 0.05% L-arabinose to activate the transcription of the vector-borne phoPXcc or phoQXcc genes, the growth rates of the mutants ΔphoP-pphoP and ΔphoQ-pphoQ were similar to that of the WT strain (Figure 2A and Figure S4, A and B in File S1). However, when 2 mM IPTG was added to repress the pBAD-araC promoter activity that controls the transcription of plasmid-borne phoPXcc or phoQXcc, the growth of the phoP and phoQ mutants was completely arrested (Figure 2, B–D). Taken together, the above results genetically demonstrated that phoPXcc and phoQXcc are bona fide essential genes. The inactivation or transcriptional repression of them led to the nonviability of bacterial cells.
Figure 2.
Growth of bacterial strains in minimal media. (A) Bacterial growth of X. campestris pv. campestris on an MMX plate containing L-arabinose (0.05%). (B) Bacterial growth of X. campestris pv. campestris on an MMX plate containing IPTG (2 mM). In both (A and B), 2 μl of a serially diluted bacterial culture was inoculated on the plates and cultured at 28° for 96 hr. (C) Growth curve of the phoQXcc mutant of X. campestris pv. campestris in MMX with 2 mM IPTG. (D) Growth curve of the phoPXcc mutant of X. campestris pv. campestris in MMX with 2 mM IPTG. (E) Bacterial growth of P. aeruginosa on an M9 plate. (F) Bacterial growth of P. aeruginosa on an M9 plate. In both (E and F), 2 μl of a serially diluted bacterial culture was inoculated on the plates and cultured at 28° for 96 hr. (G) Growth curve of the phoQPae mutant of P. aeruginosa in M9 medium. (H) Growth curve of the phoPPae mutant of P. aeruginosa in M9 medium. Vertical bars represent SD (n = 4). EV, empty vector; MMX, minimal medium for Xanthomonas campestris.
We also successfully constructed phoPPae and phoQPae mutants of P. aeruginosa PAO1 without needing to provide additional copies (Figure 2, E and F). The growths of phoPPae and phoQPae mutants were similar to the WT strain (Figure 2, G and H). Although there is a study in which phoPXoo and phoQXoo genes of the rice bacterial blight disease pathogen X. oryzae pv. oryzae were mutated (Lee et al. 2008), our repeated efforts to construct phoPXoo and phoQXoo mutants of X. oryzae pv. oryzae PXO99A failed. This result corresponds to a recent report indicating that phoPXoo and phoQXoo could not be inactivated in the same strain (Zheng et al. 2016). Thereafter, by providing additional copies through recombinant vectors, phoPXoo and phoQXoo mutants were successfully constructed using a strategy similar to that used to inactivate phoPXcc and phoQXcc as described above (Figure S5, C and D in File S1).
To investigate the functional replaceability of the phoP-phoQ genes in various bacteria, the phoP and phoQ genes from X. oryzae pv. oryzae PXO99A and P. aeruginosa PAO1 were provided in trans to X. campestris pv. campestris to construct heterogeneous merodiploid strains. The subsequent inactivation of the chromosomal copies of phoPXcc or phoQXcc revealed that phoPPae, phoQPae, and phoQXoo complemented their essential functions, since correct phoPXcc or phoQXcc mutants were obtained (Figure S5, E–G in File S1). However, in the presence of plasmid-borne phoPXoo, a correct phoPXcc mutant could not be obtained after various efforts. Since the protein sequences of PhoPXcc and PhoPXoo are identical, the most likely reason is that the recombination rate during the selection of the second round of phoP homologous cross-over is extremely low. Collectively, these results suggest that, although X. campestris pv. campestris and P. aeruginosa belong to Xanthomonadaceae and Pseudomonadaceae, respectively, the genetic functions of their phoP and phoQ genes are highly conserved. Therefore, the functional divergence between the phoP-phoQ genes in the two bacteria is unlikely to be caused by the regulatory properties of their protein products.
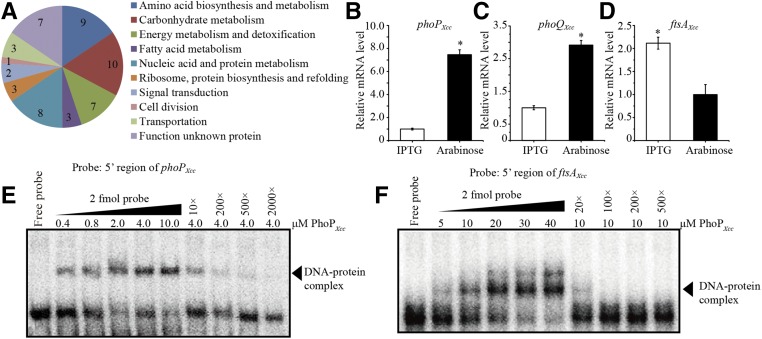
Comparative proteomic analysis reveals that PhoPXcc regulates cell division
Based on the above results, we reasoned that the PhoP regulons in X. campestris pv. campestris and P. aeruginosa are remarkably different. However, the PhoP regulon and its consensus binding motif has never been studied in Xanthomonas spp. before; therefore, a comparative proteomic analysis was used to screen the differentially expressed proteins that are directly or indirectly controlled by PhoPXcc. Bacterial cells of the ΔphoP-pphoP mutant grown in MMX medium containing 0.05% L-arabinose were collected, washed, and separated into two portions, which were allowed to grow for 60 min in fresh MMX medium containing either 2 mM IPTG or 0.05% L-arabinose. The total proteins from the two samples were analyzed by 2-DE together with MALDI-TOF-MS/MS. As shown in Figure S6 in File S1, 53 differentially expressed proteins were identified. These proteins were classified into 10 functional categories (Figure 3A and Table S3 in File S1), representing a set of genes directly or indirectly regulated by PhoPXcc. Among these proteins, PhoPXcc itself was identified because its expression level was downregulated by 2.19 times in the ΔphoP-pphoP mutant (Table S3 in File S1). This was verified by qRT-PCR analysis, which revealed that in the ΔphoP-pphoP mutant, the transcriptional level of the plasmid-borne phoPXcc gene was substantially repressed by the IPTG stimulation (to 13.4% of the level observed in the sample induced by L-arabinose, Figure 3B). Meanwhile, the amount of mRNA transcribed from the chromosomal phoQXcc gene decreased to 34.5% of that observed in the sample induced by L-arabinose (Figure 3C). An EMSA confirmed that PhoPXcc specifically binds to the 5′ sequence upstream of phoPXcc (Figure 3E). These results demonstrate that PhoPXcc directly and positively controls its own transcription via an autoregulatory loop, as reported in other bacteria (Soncini et al. 1995; Mitrophanov et al. 2010).
Figure 3.
Comparative proteomic and molecular analyses reveal that PhoP is subject to autoregulation and controls ftsA transcription. (A) Functional classification of differentially expressed proteins in the phoPXcc mutant. The ΔphoP-pphoP mutant was grown in MMX medium with 0.05% L-arabinose or 2 mM IPTG. A comparative proteomic analysis was carried out using 2-DE with MALDI-TOF-MS/MS. The 2-DE analysis was repeated three independent times, each with two technical replicates. (B–D) phoPXcc regulates the transcription of its own operon and ftsAXcc. qRT-PCR was used to measure the mRNA levels of phoPXcc (plasmid-borne copy), phoQXcc (chromosomal copy) and ftsAXcc. Total RNA was extracted from the phoP mutant (ΔphoP-pphoP) that was grown in MMX medium containing IPTG (2 mM, for repression of the phoPXcc expression) or L-arabinose (0.05%, for induction of the phoPXcc expression). tmRNA transcripts were used as an internal control. The experiments were replicated three independent times, each with three technical repeats. A representative experiment is shown. (E and F) PhoPXcc binds to the 5′ promoter regions upstream of phoPXcc and ftsAXcc. Increasing amounts of the PhoPXcc protein were tested in an EMSA. Increasing amounts of unlabeled probes were used as competitors to bind to PhoPXcc protein extracts. The experiments were repeated three times, and a representative is shown. 2-DE, two-dimensional electrophoresis; MALDI-TOF-MS, matrix-assisted laser desorption/ionization time of flight mass spectrometry; MMX, minimal medium for Xanthomonas campestris; qRT-PCR, quantitative real-time RT-PCR.
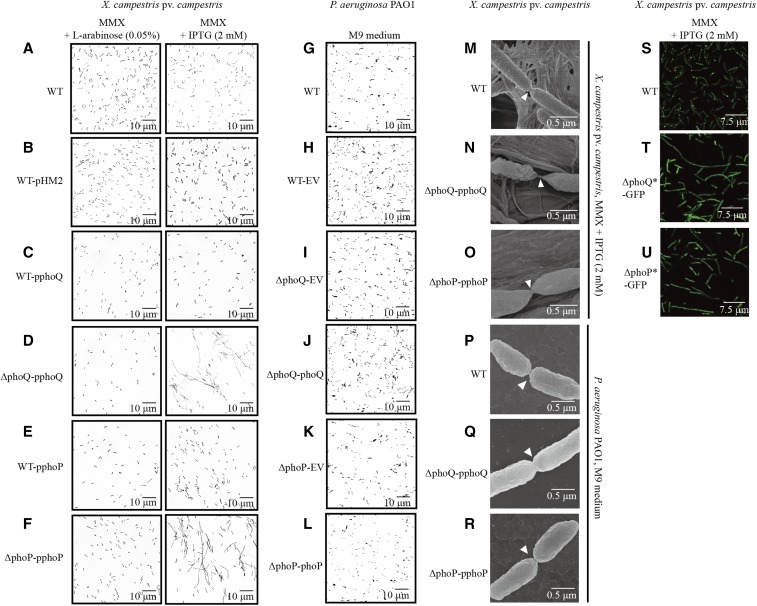
We noticed that the IPTG-mediated repression of phoPXcc expression caused an upregulation in the expression of the FtsAXcc protein (XC3506), which belongs to the actin/HSP 70 protein family and is involved in cell division. qRT-PCR quantification verified that, in the ΔphoP-pphoP mutant, the addition of 2 mM IPTG resulted in a 2.1 times increase in the mRNA level compared with that of the ΔphoP-pphoP cells treated with 0.05% L-arabinose (Figure 3D). In addition, an EMSA also confirmed that PhoPXcc directly and specifically binds to the 5′ promoter region of ftsAXcc (Figure 3F). Since FtsA controls the Z-ring formation during bacterial cytogenesis by binding to the membrane and recruiting a tubulin superfamily GTPase, FtsZ (Adams and Errington 2009), the fact that PhoPXcc negatively regulates ftsAXcc transcription implied that the cell division of X. campestris pv. campestris is subject to control by PhoPXcc. Subsequently, microscopic observation revealed that under L-arabinose (0.05%) induction, the cellular morphologies of all examined bacterial strains were similarly rod-shaped (Figure 4, A–F, left panels). However, under IPTG repression, bacterial cells of the mutants ΔphoP-pphoP and ΔphoQ-pphoQ changed into long, aseptate cell filaments, indicating that the separation of daughter cells is abnormal in these strains (Figure 4, D and F, right panels). In addition, high-resolution field emission scanning electron microscopy showed that in the phoQXcc-repressed state, while the WT strain displayed a smooth, circular arc septum between daughter cells (Figure 4M), the conditional mutants ΔphoP-pphoP and ΔphoQ-pphoQ exhibited a sharp connection between cells (Figure 4, N and O). Moreover, the cells of the two mutants, especially ΔphoP-pphoP, had different surface morphology compared to that of the WT strain (Figure 4, N and O). In contrast, in P. aeruginosa, phoPPae and phoQPae mutations did not impact the morphology of bacterial cells during cytogenesis (Figure 4, G–L and P–R). Therefore, the PhoP-PhoQ system plays a remarkably different role in the regulation of cell division in X. campestris pv. campestris and P. aeruginosa.
Figure 4.
Inactivation of phoPXcc and phoQXcc causes abnormalities in bacterial cell division. (A–F) Morphology of X. campestris pv. campestris cells observed under light microscopy. The bacterial strains were grown in MMX plus L-arabinose (0.05%) or MMX plus IPTG (2 mM). (G–I) Morphology of P. aeruginosa cells observed under light microscopy. The bacterial strains were grown in M9 medium. (M–O) Morphology of X. campestris pv. campestris cells observed under field emission scanning electron microscopy. The bacterial strains were grown in MMX plus IPTG (2 mM). (P–R) Morphology of P. aeruginosa cells observed under field emission scanning electron microscopy. The bacterial strains were grown in M9 medium. (S–U) Localization of FtsZ-eGFP in X. campestris pv. campestris cells. The bacterial strains were grown in MMX plus IPTG (2 mM). Scale bars are provided in the photographs. The genotypes of the bacterial strains are listed in Table S1 in File S1. eGFP, enhanced GFP; EV, empty vector; MMX, minimal medium for Xanthomonas campestris; WT, wild-type.
To observe the Z-ring formation in X. campestris pv. campestris, we produced an FtsZ-eGFP (enhanced GFP) fusion construct and integrated the DNA sequence into an unrelated locus (between XC1972 and XC1973) in the bacterial chromosome of different strains. These recombinants were not disrupted at the native ftsZXcc gene and did not exhibit morphological differences compared with the donor strains. As Figure 4S shows, in the WT background, the FtsZ-eGFP signals of 95% of the bacterial cells (n = 200) were localized to the growth poles of cells or in the septation sites between daughter cells, which was similar to previous reports in other γ-Proteobacteria. However, in the ΔphoP-pphoP and ΔphoQ-pphoQ mutants, after IPTG treatment, the FtsZ-eGFP signal of the majority of bacterial cells was dispersed throughout the whole cell (only 2.0 and 1.5% of the cells, respectively, exhibited a polar GFP signal, n = 200, Figure 4, T and U). These results supported the view that the inactivation of phoPXcc-phoQXcc results in abnormal daughter cell separation, which is possibly caused by disrupted ftsAXcc transcription and the subsequent mislocalization or degradation of FtsZXccduring Z-ring formation.
The consensus DNA-binding motifs of PhoP are highly conserved between X. campestris and P. aeruginosa
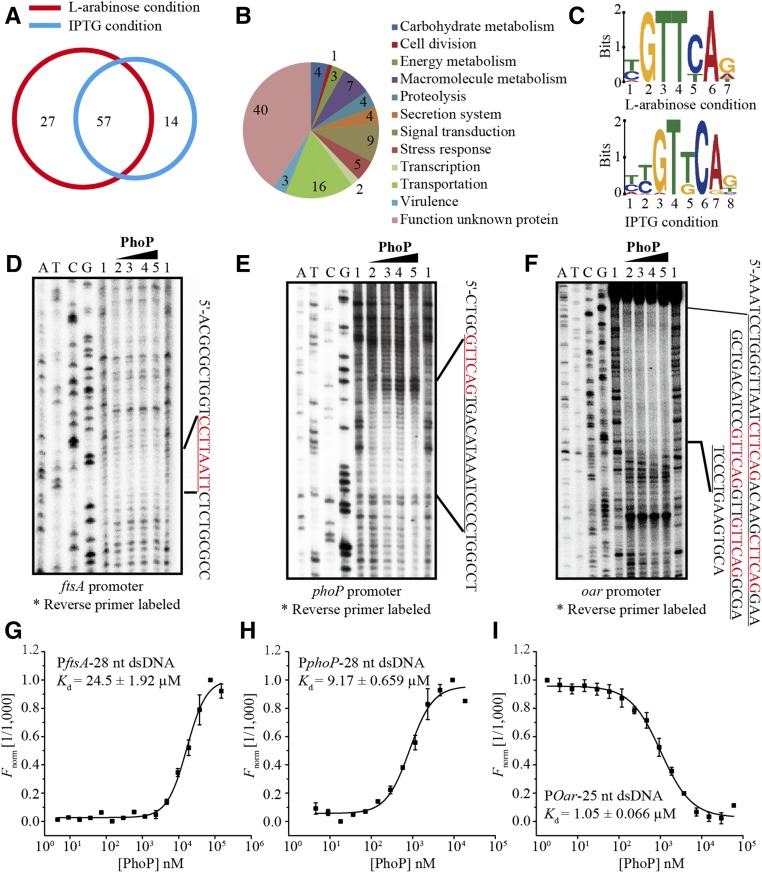
To dissect the consensus binding motif of PhoPXcc and compare it to the other PhoP-boxes, ChIP-seq was employed to screen for PhoPXcc-binding sequences (CREs) throughout the genome. We constructed a recombinant strain based on the HK gene phoQXcc mutant (ΔphoQ-pphoQ). In this strain (ΔphoQ-phoP-his6), a sequence encoding a His6 epitope tag was fused at the 3′-end of the coding sequence of phoPXcc in the bacterial chromosome. To control the transcription of the HK phoQXcc, this strain was grown overnight, washed, and then subcultured in MMX medium containing either 2 mM IPTG or 0.05% L-arabinose for 60 min. After using a monoclonal anti-His6 antibody to retrieve the PhoPXcc protein together with its bound DNA, high-throughput sequencing was conducted. Peak calling of the ChIP-seq data revealed that, under the states of phoQXcc induction (by L-arabinose) or repression (by IPTG), PhoPXcc binds to the upstream regions of 84 and 71 genes, respectively (Table S4 in File S1). There were 27 and 14 genes, respectively, that were specifically identified in the phoQXcc activation or repression conditions (Figure 5A). These unique genes represent different binding sites that associate with PhoPXcc at various phosphorylation levels that are regulated by the HK PhoQXcc. The ChIP-seq analysis identified that PhoPXcc modulates 98 coding genes whose products belong to various functional categories (Figure 5B and Table S4 in File S1).
Figure 5.
Dissection of the consensus binding motif of PhoPXcc. (A) Venn diagram showing the number of PhoPXcc-binding promoter regions identified by the ChIP-seq analysis. (B) Functional categories of all genes with promoters bound by PhoPXcc under PhoQXcc induction and repression conditions. (C) Consensus PhoPXcc-binding DNA motifs predicted by ChIP-seq and MEME. The height of each nucleotide is proportional to the level of conservation at that site. Upper and lower panels: motifs predicted under PhoQXcc induction and repression conditions, respectively. (D–F) Binding sites of PhoPXcc on the promoter regions of (D) oar, (E), phoP, and (F) ftsA. DNase I footprinting assays were conducted. The amounts of PhoPXcc protein used in the reactions were 1: zero; 2: 40 μM, 3: 80 μM, 4: 120 μM, and 5: 160 μM. The DNA regions protected by PhoPXcc are shown on the right of the footprinting results, with the PhoPXcc-binding sites shown in red. (G–I) PhoPXcc–DNA-binding affinities were quantified by MST. (G) PhoPXcc–PftsAXcc. (H) PhoPXcc–PphoPXcc. (I) PhoPXcc–PoarXcc. DNA probes were labeled with FAM and used for MST. The FAM-labeled DNA (20 nM) was incubated with the PhoPXcc protein in an NT standard capillary in the MST assay. Titrations of the PhoPXcc protein ranged from 1.5 to 50 μM. The solid curve is the fit of the data points to the standard KD-Fit function. The black bars represent SD (n = 3). Kd = dissociation constant. ChIP-seq, chromatin immunoprecipitation sequencing; FAM, fluorescein; MST, microscale thermophoresis; NT, NanoTemper.
Based on the ChIP-seq results, we searched for common recognition elements that physically contact PhoPXcc, which led to the identification of two highly similar binding motifs under the phoQXcc-induced and phoQXcc-repressed conditions (Figure 5C). The consensus PhoPXcc-binding motif, (T/C)(T/C)GT(T/G)CA(G/A), is highly similar to the known PhoPPae-box of P. aeruginosa (CGGTCAG) (McPhee et al. 2006), but quite different from the PhoPSal-box [(G/T)GTTTA(A/T)] found in enterobacteria (Zwir et al. 2005). To experimentally verify the predicted PhoPXcc-binding motif, we selected the promoter regions of three representative genes based on proteomics and ChIP-seq information, oar, phoP, and ftsA, and performed DNase I footprinting to map the dsDNA sequences protected by PhoPXcc. As anticipated, PhoPXcc protected the 5′ sequences upstream of the oar, phoP, and ftsA coding regions (Figure 5, D–F). These PhoPXcc protected sequences (in red in Figure 5, D–F) are in accordance with the DNA motif revealed by ChIP-seq. A close examination revealed that, within the protected regions, the promoters of ftsA and phoP contain only one PhoP-binding motif each (Figure 5, D and E). However, the promoter of oar has four motif sequences (Figure 5F). In addition to this, MST detected that PhoPXcc binds to 25–28-bp-long, FAM-labeled oligonucleotides synthesized according to the PhoPXcc-binding motifs from Poar (disassociation constant Kd = 1.05 ± 0.066 μM), PphoP (9.17 ± 0.659 μM), and PftsA (24.5 ± 1.920 μM) (Figure 5, G–I). These results verified the DNA motif bound by PhoPXcc, which is highly conserved between X. campestris pv. campestris and P. aeruginosa.
High level of genetic polymorphisms in PhoP-binding sites identified between bacterial genomes
In all organisms, transcription regulation consists of three major units: TFs, CREs, and downstream genes (Wilson and Odom 2009; Li and Johnson 2010). Since PhoPPae of P. aeruginosa can genetically substitute for the regulatory roles of PhoPXcc of X. campestris pv. campestris (Figure S5, E and F in File S1), and the sequences of their CREs are highly conserved (Figure 5C), the essentiality of PhoPXcc in X. campestris pv. campestris may be due to the properties of downstream regulated genes. That is, PhoPXcc may control the expression of essential, structural genes, while PhoPPae does not control these genes. Therefore, we hypothesized that the difference in the essentiality between PhoPXcc and PhoPPae in their regulatory roles may be caused by rewiring between the TFs and CREs of downstream genes.
Since the PhoPXcc-binding motif was identified, we used this consensus sequence to search the genome of X. campestris pv. campestris 8004 and identified putative binding sites in the promoter regions of 198 genes (Table S5 in File S1). This provides an opportunity to compare and evaluate the results from comparative proteomic and ChIP-seq analyses. Considering the operon organization of the genes in the genome of X. campestris pv. campestris, 22 genes (39.3%) whose protein products were identified by 2-DE were also obtained by PhoP-binding motif search (including those in the same operons, Figure S7A in File S1). Meanwhile, 59 genes (60.2%) identified by the ChIP-seq analysis contain a PhoP-binding motif in their promoter region (including those in the same operon, Figure S7B in File S1). This comparison revealed that in dissecting the regulon of PhoPXcc, ChIP-seq is more efficient than the comparative proteomic analysis in identifying genes directly regulated by PhoPXcc. This result is anticipated because PhoPXcc-binding DNA sequences were enriched during IP before sequencing, while comparative proteomic analysis identified proteins directly or indirectly regulated by the transcription factor. Especially, IPTG stimulation is toxic to bacterial cells, and some proteins identified by proteomic analysis, such as GroEL, XC0333, and XC2653, may be involved in the IPTG stress response (Raivio 2005; Maleki et al. 2016). Their coding sequences lack PhoP-binding sites in the promoter regions. In addition, it is noticeable that though ftsA was identified as a differently expressed protein by 2-DE (Table S3 in File S1), the gene was absent in the results of the ChIP-seq analysis (Table S4 in File S1). However, a binding-motif search revealed that ftsA is a PhoPXcc-regulated gene because it contains a binding motif (Table S5 in File S1). Therefore, the fact that ftsA is missed in ChIP-seq analysis is possibly caused by low PhoP-PftsA binding affinity, which is in accordance with the result of the MST assay (Figure 5G).
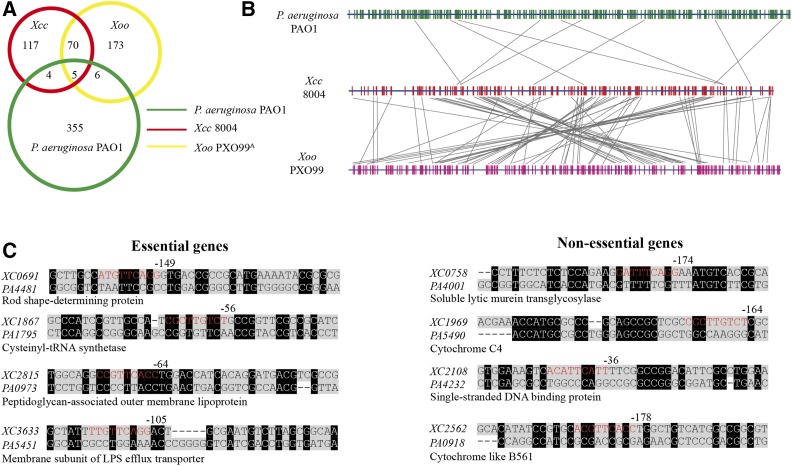
To determine the genome-wide variation caused by PhoP-CRE rewiring, the conserved PhoP consensus binding motif was also used to search the complete genomic sequences of X. oryzae pv. oryzae PXO99A and P. aeruginosa PAO1. As shown in Figure 6A, there are 255 and 370 genes of the two genomes, respectively, that are putatively regulated by PhoP, as they contain conserved PhoP-binding motifs in their promoter regions. A remarkable number of genetic polymorphisms were identified in the genes regulated by PhoP, which was caused by PhoP-CRE rewiring. Although X. campestris pv. campestris and X. oryzae pv. oryzae belong to the same genus, and their PhoP protein sequences are identical, only 75 orthologous, PhoP-regulated genes were shared between them (Figure 6, A and B). For P. aeruginosa, there are 9 or 11 PhoP-regulated genes shared by it and X. campestris pv. campestris or X. oryzae pv. oryzae, respectively (Figure 6, A and B). These three genomes share only five genes whose expressions are putatively regulated by PhoP, including orthologs of XC1128 (encoding aspartyl-tRNA synthetase), XC1739 (protein-export membrane protein SecF), XC2267 (flagellar motor switch protein FliM), XC3652 (β-ketoacyl-synthase I), and XC3986 (protease Do). As Figure 6, A and B shows, a large number of species-specific, PhoP-regulated genes were identified in the above three genomes. Collectively, these results revealed a very high level of genetic polymorphisms of TF-CRE relationships among the three bacterial species, indicating that the PhoQ-PhoP regulon has extensively differentiated during bacterial speciation to adapt to various ecological niches.
Figure 6.
Genome-wide polymorphisms of cis-regulatory elements of PhoP between X. campestris pv. campestris, X. oryzae pv. oryzae, and P. aeruginosa. (A) Venn diagram exhibiting the number of cis-regulatory elements regulated by PhoP in the three bacterial species. Xcc: X. campestris pv. campestris 8004; Xoo: X. oryzae pv. oryzae PXO99A. (B) Comparison of PhoP-binding motifs between bacterial species. The vertical red lines and green lines represent putative PhoP-binding motifs distributed in the genomes of X. campestris pv. campestris 8004, X. oryzae pv. oryzae PXO99A, and P. aeruginosa PAO1. The gray lines link the orthologous genes with these PhoP-binding motifs in the three bacterial genomes. (C) Alignment of the promoter sequences of candidate genes of X. campestris pv. campestris and P. aeruginosa. Eight pairs of orthologous genes from the two bacteria were selected for further genetic and molecular investigations. The PhoP-binding motifs are in red. Locations of the nucleotides from the translational initiation sites are labeled above the panels. LPS, lipopolysaccharide.
Functional divergence of PhoPXcc and PhoPPae in regulating the expression of essential, structural genes and sensing stimuli
The essentiality of genes is a complex biological trait that is determined by various factors. Previous studies have revealed that the essentiality of regulatory genes is usually polygenic, that is, they control the expression of multiple genes critical in viability (Dubrac et al. 2008). Based on this notion and the high level of genetic polymorphism detected in PhoP-CRE relationships (Figure 6), we reason that PhoPXcc regulates multiple cellular processes that are essential to bacterial survival, while PhoQPae does not modulate these processes.
To challenge this hypothesis, eight PhoPXcc-regulated, candidate genes of X. campestris pv. campestris were selected for further investigation. These genes contain PhoP-binding motifs in their promoter regions and are involved in fundamental physiological pathways that are associated with essentiality, including cell wall construction (XC0758 and XC2815), lipopolysaccharide transportation (XC3633), cytogenesis (XC0691), tRNA synthetase (XC1867), cytochrome (XC1969 and XC2562), and DNA replication (XC2108, Figure 6C). First, insertion inactivation by homologous, single crossover was used to rapidly inactivate these genes. It showed that XC0758, XC1969, XC2108, and XC2562 can be disrupted, indicating that they are nonessential genes (Figure S4, C–F in File S1). The inactivation of XC0758 caused the growth deficiency of bacteria grown in the MMX medium (Figure S4F in File S1). However, the other four genes, which encode a rod shape-determining protein (XC0691), a cysteinyl-tRNA synthetase (XC1867), a peptidoglycan-associated outer membrane lipoprotein (XC2815), and a membrane subunit of lipopolysaccharide efflux transporter (XC3633), failed to be inactivated after repeated efforts, indicating that they are essential genes. Thereafter, plasmid-borne copies of these genes were transformed in trans into the WT strain by recombinant pHM1 vectors. Under these genetic backgrounds, correct mutants of the four genes were obtained in the presence of the additional, plasmid-borne copy, supporting the view that they are essential, structural genes in X. campestris pv. campestris. Recently, Turner et al. (2015) reported the identification of essential genes in P. aeruginosa by a Monte Carlo simulation-based, high-throughput transposon sequencing method, revealing that the orthologs of these four genes, PA4481, PA1795, PA0973, and PA5451 (Figure 6C), respectively, are also bona fide essential genes in P. aeruginosa (Turner et al. 2015).
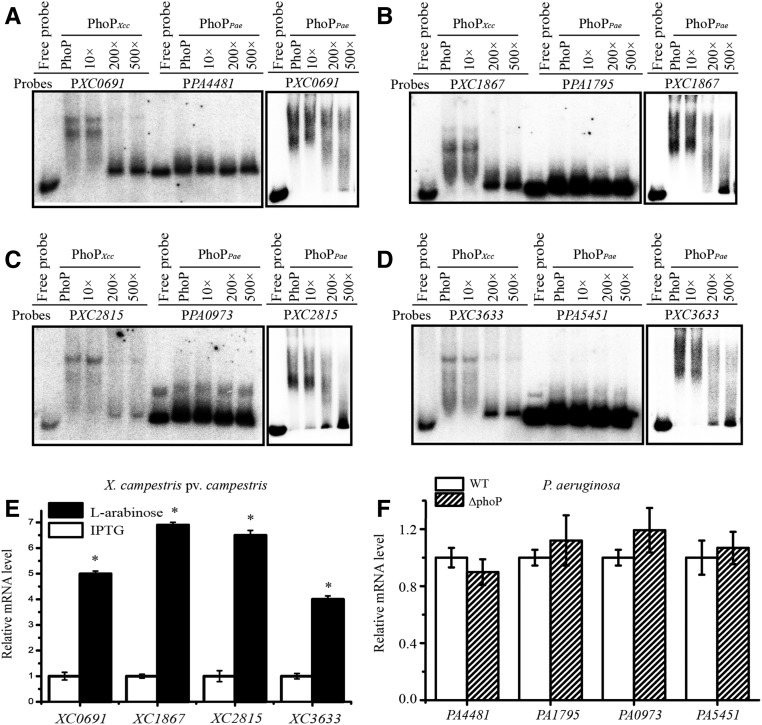
Next, EMSA and qRT-PCR analyses were employed to investigate the regulatory roles of PhoP on the four essential genes in X. campestris pv. campestris and P. aeruginosa. As Figure 7, A–D show, both PhoPXcc and PhoPPae specifically bind to the promoter sequences of all four essential genes of X. campestris pv. campestris in vitro, but PhoPPae did not bind to any of the promoter regions of the orthologous genes from P. aeruginosa. In addition, qRT-PCR analysis revealed that in the phoPXcc mutant of X. campestris pv. campestris, the repression of phoP transcription by IPTG stimulation resulted in a significant downregulation of XC0691, XC1867, XC2815, and XC3633 (Figure 7E). Compared to this, in the phoPPae mutant of P. aeruginosa, the mRNA levels of the four essential genes were similar to those of the WT strain (Figure 7F). Consequently, these results revealed that PhoPXcc of X. campestris pv. campestris is a positive regulator of these four essential genes, whereas PhoPPae of P. aeruginosa does not regulate the expression of their orthologs. In addition, the regulatory role of PhoP in binding to the four nonessential genes of X. campestris pv. campestris and P. aeruginosa was also analyzed by EMSA, and the results are similar to those of the essential genes described above (Figure S8 in File S1). As the sequence alignments show (Figure 6C), the promoter regions of the four genes in P. aeruginosa generally lack the conserved CRE of PhoP, which might be caused by point mutations. It also revealed that even in the genus of Xanthomonas and close-relative species, substantial genetic polymorphism was detected in the promoter regions of the afore-mentioned four essential genes. For example, X. campestris pv. vesicatoria, X. axonopodis pv. citri, and X. oryzae pv. oryzae usually have PhoP-binding motifs in the upstream sequences of these genes (Figure S9 in File S1), but X. perforans and X. gardneri exhibited remarkable genetic variation in the promoter regions and lack PhoP-binding sites (Figure S9 in File S1). This high level of genetic polymorphism indicates that the essentiality of PhoP-PhoQ system is phylogenetic clade-dependent. Before mutational evidence is available, caution must be taken to avoid predicting that phoP-phoQ are essential genes in Xanthomonas spp. or their close relatives.
Figure 7.
PhoP of X. campestris pv. campestris and P. aeruginosa controls the transcription of downstream essential genes differently. (A–D) PhoP-promoter binding assay. EMSA revealed that both PhoPXcc and PhoPPae bind directly to the promoter regions of four essential genes (XC0691, XC1867, XC2815, and XC3633) of X. campestris pv. campestris, but PhoPPae did not bind the promoter regions of their orthologs in P. aeruginosa (PA4481, PA1795, PA0973, and PA5451). The DNA probes were labeled with [γ-32P]ATP. Increasing amounts of unlabeled probes were used as competitors. The experiments were repeated three times, and a representative is shown. (E) mRNA levels of the essential genes in the phoPXcc mutant of X. campestris pv. campestris. (F) mRNA levels of the essential genes in the phoPPae mutant of P. aeruginosa. qRT-PCR was used to estimate the transcription levels of the four aforementioned essential genes. In both (E and F), tmRNA transcripts were used as an internal control. The experiments were replicated three independent times, each with three technical repeats. A representative experiment is shown. The vertical bars represent SD. * indicates significant differences (Student’s t-test, P < 0.05). EMSA, electrophoretic mobility shift assay; qRT-PCR, quantitative real-time RT-PCR; WT, wild-type.
PhoQ of S. enterica and P. aeruginosa bind and detect Ca2+ and Mg2+ (Kato and Groisman 2008). To investigate whether PhoQXcc also senses metal ions, MST analysis was used to determine the possible binding events between PhoQXcc and Ca2+, Mg2+, and Mn2+. As Figure S10 in File S1 shows, neither the soluble, truncated PhoQXcc (containing the cytoplasmic region) nor the full-length PhoQXcc embedded in the IMVs bind to these metal ions. As a positive control, the known iron sensor of X. campestris pv. campestris, VgrS, directly binds Fe3+ with a Kd value of 23.21 ± 2.05 μM (Figure S10G in File S1). These results demonstrated that unlike PhoQ of S. enterica or P. aeruginosa, PhoQXcc does not have the capability to sense these metals, which is in parallel to the fact that the sensor regions of these PhoQ contain substantial genetic polymorphisms (Figure S1 in File S1). Furthermore, qRT-PCR analyses revealed that when the phoPXcc expression was induced by L-arabinose, stimulation by various concentrations of Mg2+, Mn2+, and Ca2+ resulted in decreased mRNA levels of XC0691, a PhoPXcc regulated gene (Figure S11, A–C in File S1). The expression of XC0691 remained stable when phoPXcc expression was repressed by IPTG stimulation (Figure S11, A–C in File S1). As for XC1867, another PhoPXcc-regulated gene, stimulation by Mg2+ and Ca2+ did not affect its transcription levels (Figure S11, D and E in File S1). However, stimulation by 10 mM Mn2+ resulted in a significant increase of XC1867 mRNA levels in the background of phoP activation, while repression of phoPXcc expression generally caused a decrease of the XC1867 mRNA amounts under the metal treatment (Figure S11, D and E in File S1). Collectively, these results suggest that PhoPXcc regulates the expression of XC0691 and XC1867 under the treatment of some conditions of metal stresses. However, the regulation is likely indirect since PhoQXcc of X. campestris pv. campestris cannot bind these metals (Figure S10 in File S1).
Taken together, our study strongly supports the view that in X. campestris pv. campestris, the essentiality of PhoPXcc can be attributed to the fact that the TF controls the transcription of a number of essential, structural genes. Because of TF-CRE rewiring during evolution, these regulatory relationships between PhoPPae and the downstream essential genes were disassociated in P. aeruginosa. In addition, PhoQXcc has functional divergence from its orthologs in sensing metal ions.
Discussion
In prokaryotes, the majority of TCSs are nonessential and promote the capability of cells to adapt to various ecological niches (Hoch 2000; Salazar and Laub 2015). Only a few bacterial TCSs are essential, such as DivK, CckA-CtrA, MtrA-MtrB, and YycG-YycF (Zahrt and Deretic 2000; Laub et al. 2007; Dubrac et al. 2008; Winkler and Hoch 2008). Most of these essential TCS genes take part in the regulation of the cell cycle or in cell wall development. In the present study, we demonstrated that phoPXcc-phoQXcc, which encode a canonical, nonessential TCS that modulates the expression of virulence factors of many other pathogenic bacteria (Prost and Miller 2008), are bona fide essential regulatory genes in the gram-negative bacterium X. campestris pv. campestris (Figure 2 and Figure S3 in File S1). The regulon and consensus DNA-binding motif of PhoPXcc was dissected by comparative proteomics, ChIP-seq, and molecular analyses (Figure 3 and Figure 5). These analyses revealed that PhoPXcc modulates the transcription of at least four essential, structural genes by directly binding to their promoter regions, indicating that the essentiality of this TF can be attributed to its polygenic manner of regulation, through which it controls the expression of multiple genes that are involved in fundamental pathways of bacterial cells (Figure 7). Although PhoPPae-PhoQPae of P. aeruginosa are genetically equivalent to their orthologs in X. campestris pv. campestris and their PhoP-binding motifs are highly conserved, due to the TF-CRE rewiring, PhoPPae does not control the aforementioned essential genes in P. aeruginosa, since there are no PhoP-binding sites in the promoter regions of these genes (Figure 7). We believe that the PhoP-CRE rewiring event that occurred during bacterial speciation resulted in the essentiality of PhoP-PhoQ in X. campestris pv. campestris.
In the past decade, along with the rapid development of high-throughput sequencing, genomic approaches to the study of TF-CRE binding landscapes have allowed genome-wide binding variations to be investigated between orthologous TFs from different prokaryotes and eukaryotes (Zwir et al. 2005; Hershberg and Margalit 2006; Perez and Groisman 2009a; Siepel and Arbiza 2014). In yeast and fruit flies, the genetic variation of TF-CRE rewiring has been estimated to be as high as > 50% (Wittkopp and Kalay 2012). Our comparative analyses revealed that, even for the closely related X. campestris pv. campestris and X. oryzae pv. oryzae, ∼70% of the PhoP-CRE relationships were different between them. Furthermore, only five downstream genes (< 2%) were putatively shared by the PhoP regulon of the three bacterial species tested in this study (Figure 6). Together with previous studies in Enterobacteriaceae (Zwir et al. 2005; Perez et al. 2009; Perez and Groisman 2009b), these studies suggest that extensive PhoP-CRE rewiring events have occurred during bacterial evolution. Previous studies in different organisms have also proposed that rewiring between TFs and CREs plays a fundamental role during the evolution of morphological traits and the origin of species, such as the organ development of fruit flies (Koshikawa et al. 2015), the mating system of yeasts (Sorrells et al. 2015), animal transcription networks (Schmidt et al. 2010; Stefflova et al. 2013; Vierstra et al. 2014), and the virulence regulation of bacterial pathogens (Perez and Groisman 2009b). In the present study, because the phoP-phoQ orthologs proved to be mutable in other bacterial species, especially those from close relatives of Proteobacteria, the essentiality of the phoP-phoQ genes in X. campestris pv. campestris is likely a phylogenetic clade-specific apomorphy that accompanied bacterial speciation. We hypothesized that the existing essential, structural genes recruited the regulatory TF PhoP by genetic recombination and mutational events that occurred in their promoter regions. Therefore, the result of our study not only provides an additional case but also defines a limit of the driving force of TF-CRE rewiring during evolution; if essential regulatory genes whose inactivation will lead to nonviability can originate from TF-CRE rewiring, then in all probability this kind of genetic variation could drive the evolution of regulatory relationships between TFs and all kinds of downstream genes.
Although the molecular mechanisms responsible for evolving novel functions (neofunctionalization) of structural genes have been investigated extensively (Long et al. 2003), the molecular steps by which a novel essential gene could originate from ancient or newly arisen genes are less studied. Ross et al. (2013) reported that an essential gene of Drosophila melanogaster, Umbrea, acquired its essential function in a stepwise manner during speciation, which was driven by the deletion of ancestral sequences encoding a heterochromatin localizing domain (Ross et al. 2013). As the sequence alignment shows (Figure 6C), TF-CRE rewiring caused by the accumulation of point mutations or indels (insertion/deletions) is also a molecular mechanism that drives the evolution of essentiality. In addition, our results support the established notion that mutations within CRE sequences, rather than those in the coding sequence of trans-regulatory TFs, accelerate the evolutionary innovation of regulatory gene function (Zwir et al. 2005; Perez et al. 2009; Wittkopp and Kalay 2012; Visweswariah and Busby 2015). In the study of PhoP-PhoQ evolution in 10 bacterial species belong to Enterobacteriaceae, a substantial number of genetic polymorphisms were also detected in the PhoP regulon, which suggest that TF-CRE rewiring plays an important role in bacterial speciation (Perez et al. 2009).
Although a transcriptional rewiring event was established between PhoP and its CRE in essential, structural genes, a noticeable result of the present study is that the cognate HK PhoQ was also proven to be essential (Figure 2 and Figure S3 in File S1), suggesting that a strong coevolutionary tendency exists between the TCS proteins. In the prototypical TCS, the sensor HKs encode various N-terminal domains to detect specific environmental stimuli (Hoch 2000). The coevolution of PhoP-PhoQ implies that the environmental cues sensed by PhoQ might be the selective forces that drove the evolution of the PhoP-CRE interaction in X. campestris pv. campestris. We propose that PhoP-PhoQ-dependent regulation in reaction to these environmental stimuli must confer adaptive advantages to this bacterium. In enterobacteria, PhoQ senses the concentrations of Mg2+, Ca2+, and antimicrobial peptides, as well as low pH levels (Groisman et al. 2013). However, MST and qRT-PCR analyses showed that PhoQXcc did not bind to Mg2+ and Ca2 and that the change of gene expression levels under the metal treatments were indirectly regulated by PhoQ-PhoP (Figure S10 and Figure S11 in File S1). Therefore, the environmental stimuli sensed by PhoQXcc remain unclear. A comparison of the signal input region of PhoQ revealed a substantial number of sequence polymorphisms. For example, the residues constituting the acidic region, which is important for interacting with the bacterial inner membrane through metal bridges during the detection of Mg2+ or Ca2+ (Molnar et al. 2014; Matamouros et al. 2015), are changed in the PhoQ of X. campestris pv. campestris (Figure S1 in File S1). This information suggested that the PhoQ of X. campestris pv. campestris detects different environmental stimuli compared with those of S. enterica or P. aeruginosa. In addition, these results suggest that the loss of the metal-sensing function of the PhoQXcc did not involve in the evolution of its essentiality, especially considering that phoQPae can substitute for phoQXcc during mutagenesis. Based on these analyses, further investigation is necessary to identify these stimuli and compare the signaling processes between PhoP-PhoQ TCSs in different bacteria. This will increase our understanding of the impact of selective forces on the development of regulatory networks that are controlled by TF-CRE rewiring.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.200204/-/DC1.
Acknowledgments
We thank Lei Wang from the Department of Biology, Beijing City University, for help with constructing the partial recombinant vectors. Luyan Z. Ma of our institute kindly provided the P. aeruginosa PAO1 strains and pEXGm vector. This work was financially supported by the National Science Foundation of China (grants 31370127, 31400071, and 31671989), the Ministry of Science and Technology of China (grant 2016YFD0100602), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB11040700), and the State Key Laboratory of Plant Genomics.
Footnotes
Communicating editor: J. F. Miller
Literature Cited
- Adams D. W., Errington J., 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7: 642–653. [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Johnson J., Grant C. E., Noble W. S., 2015. The MEME suite. Nucleic Acids Res. 43: W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmiller T., Ackermann M., Silander O. K., 2012. Patterns of evolutionary conservation of essential genes correlate with their compensability. PLoS Genet. 8: e1002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisicchia P., Noone D., Lioliou E., Howell A., Quigley S., et al. , 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65: 180–200. [DOI] [PubMed] [Google Scholar]
- Chen S., Zhang Y. E., Long M., 2010. New genes in Drosophila quickly become essential. Science 330: 1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Elia M. A., Pereira M. P., Brown E. D., 2009. Are essential genes really essential? Trends Microbiol. 17: 433–438. [DOI] [PubMed] [Google Scholar]
- Deng C. Y., Deng A. H., Sun S. T., Wang L., Wu J., et al. , 2014. The periplasmic PDZ domain-containing protein Prc modulates full virulence, envelops stress responses, and directly interacts with dipeptidyl peptidase of Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 27: 101–112. [DOI] [PubMed] [Google Scholar]
- Dubrac S., Bisicchia P., Devine K. M., Msadek T., 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70: 1307–1322. [DOI] [PubMed] [Google Scholar]
- Dussurget O., Rodriguez M., Smith I., 1996. An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative-stress response. Mol. Microbiol. 22: 535–544. [DOI] [PubMed] [Google Scholar]
- Glass J. I., Assad-Garcia N., Alperovich N., Yooseph S., Lewis M. R., et al. , 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 103: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulian M., 2010. Two-component signaling circuit structure and properties. Curr. Opin. Microbiol. 13: 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A., 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183: 1835–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A., Hollands K., Kriner M. A., Lee E. J., Park S. Y., et al. , 2013. Bacterial Mg2+ homeostasis, transport, and virulence. Annu. Rev. Genet. 47: 625–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M. J., Thornton J. W., 2013. Evolutionary biochemistry: revealing the historical and physical causes of protein properties. Nat. Rev. Genet. 14: 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R., Margalit H., 2006. Co-evolution of transcription factors and their targets depends on mode of regulation. Genome Biol. 7: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A., 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3: 165–170. [DOI] [PubMed] [Google Scholar]
- Hurst L. D., Smith N. G., 1999. Do essential genes evolve slowly? Curr. Biol. 9: 747–750. [DOI] [PubMed] [Google Scholar]
- Jordan I. K., Rogozin I. B., Wolf Y. I., Koonin E. V., 2002. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 12: 962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Tjaden B., Altendorf K., 1997. Purification, reconstitution, and characterization of KdpD, the turgor sensor of Escherichia coli. J. Biol. Chem. 272: 10847–10852. [DOI] [PubMed] [Google Scholar]
- Kasahara M., 1992. Molecular analysis of the Escherichia coli phoP-phoQ operon. J. Bacteriol. 174: 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Groisman E. A., 2008. The PhoQ/PhoP regulatory network of Salmonella enterica. Adv. Exp. Med. Biol. 631: 7–21. [DOI] [PubMed] [Google Scholar]
- Koshikawa S., Giorgianni M. W., Vaccaro K., Kassner V. A., Yoder J. H., et al. , 2015. Gain of cis-regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 112: 7524–7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub M. T., Shapiro L., McAdams H. H., 2007. Systems biology of Caulobacter. Annu. Rev. Genet. 41: 429–441. [DOI] [PubMed] [Google Scholar]
- Lee S. W., Jeong K. S., Han S. W., Lee S. E., Phee B. K., et al. , 2008. The Xanthomonas oryzae pv. oryzae PhoPQ two-component system is required for AvrXA21 activity, hrpG expression, and virulence. J. Bacteriol. 190: 2183–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Johnson A. D., 2010. Evolution of transcription networks–lessons from yeasts. Curr. Biol. 20: R746–R753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M., Betran E., Thornton K., Wang W., 2003. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 4: 865–875. [DOI] [PubMed] [Google Scholar]
- Macfarlane E. L., Kwasnicka A., Hancock R. E., 2000. Role of Pseudomonas aeruginosa PhoP-phoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146(pt. 10): 2543–2554. [DOI] [PubMed] [Google Scholar]
- Maleki F., Khosravi A., Nasser A., Taghinejad H., Azizian M., 2016. Bacterial heat shock protein activity. J. Clin. Diagn. Res. 10: BE01–BE03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamouros S., Hager K. R., Miller S. I., 2015. HAMP domain rotation and tilting movements associated with signal transduction in the PhoQ sensor kinase. MBio 6: e00616-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee J. B., Bains M., Winsor G., Lewenza S., Kwasnicka A., et al. , 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 188: 3995–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa S., Ogasawara H., Kato A., Yamamoto K., Eguchi Y., et al. , 2003. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J. Bacteriol. 185: 3696–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov A. Y., Hadley T. J., Groisman E. A., 2010. Positive autoregulation shapes response timing and intensity in two-component signal transduction systems. J. Mol. Biol. 401: 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar K. S., Bonomi M., Pellarin R., Clinthorne G. D., Gonzalez G., et al. , 2014. Cys-scanning disulfide crosslinking and bayesian modeling probe the transmembrane signaling mechanism of the histidine kinase, PhoQ. Structure 22: 1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W. L., Kazmierczak K. M., Winkler M. E., 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 53: 1161–1175. [DOI] [PubMed] [Google Scholar]
- Ohl M. E., Miller S. I., 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52: 259–274. [DOI] [PubMed] [Google Scholar]
- Oyston P. C., Dorrell N., Williams K., Li S. R., Green M., et al. , 2000. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68: 3419–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez J. C., Groisman E. A., 2009a Evolution of transcriptional regulatory circuits in bacteria. Cell 138: 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez J. C., Groisman E. A., 2009b Transcription factor function and promoter architecture govern the evolution of bacterial regulons. Proc. Natl. Acad. Sci. USA 106: 4319–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez J. C., Shin D., Zwir I., Latifi T., Hadley T. J., et al. , 2009. Evolution of a bacterial regulon controlling virulence and Mg(2+) homeostasis. PLoS Genet. 5: e1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter I. S., Davidson E. H., 2011. Evolution of gene regulatory networks controlling body plan development. Cell 144: 970–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost L. R., Miller S. I., 2008. The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell. Microbiol. 10: 576–582. [DOI] [PubMed] [Google Scholar]
- Qian W., Han Z. J., He C., 2008a Two-component signal transduction systems of Xanthomonas spp.: a lesson from genomics. Mol. Plant Microbe Interact. 21: 151–161. [DOI] [PubMed] [Google Scholar]
- Qian W., Han Z. J., Tao J., He C., 2008b Genome-scale mutagenesis and phenotypic characterization of two-component signal transduction systems in Xanthomonas campestris pv. campestris ATCC 33913. Mol. Plant Microbe Interact. 21: 1128–1138. [DOI] [PubMed] [Google Scholar]
- Raivio T. L., 2005. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 56: 1119–1128. [DOI] [PubMed] [Google Scholar]
- Ram S., Goulian M., 2013. The architecture of a prototypical bacterial signaling circuit enables a single point mutation to confer novel network properties. PLoS Genet. 9: e1003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez G. M., Voskuil M. I., Gold B., Schoolnik G. K., Smith I., 2002. ideR, An essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70: 3371–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen B. P., McClees J. S., 1974. Active transport of calcium in inverted membrane vesicles of Escherichia coli. Proc. Natl. Acad. Sci. USA 71: 5042–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross B. D., Rosin L., Thomae A. W., Hiatt M. A., Vermaak D., et al. , 2013. Stepwise evolution of essential centromere function in a Drosophila neogene. Science 340: 1211–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar M. E., Laub M. T., 2015. Temporal and evolutionary dynamics of two-component signaling pathways. Curr. Opin. Microbiol. 24: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual, Ed. 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schafer A., Tauch A., Jager W., Kalinowski J., Thierbach G., et al. , 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145: 69–73. [DOI] [PubMed] [Google Scholar]
- Schmidt D., Wilson M. D., Ballester B., Schwalie P. C., Brown G. D., et al. , 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Arbiza L., 2014. Cis-regulatory elements and human evolution. Curr. Opin. Genet. Dev. 29: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncini F. C., Vescovi E. G., Groisman E. A., 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177: 4364–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells T. R., Booth L. N., Tuch B. B., Johnson A. D., 2015. Intersecting transcription networks constrain gene regulatory evolution. Nature 523: 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefflova K., Thybert D., Wilson M. D., Streeter I., Aleksic J., et al. , 2013. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell 154: 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner K. H., Wessel A. K., Palmer G. C., Murray J. L., Whiteley M., 2015. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. USA 112: 4110–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J., Rynes E., Sandstrom R., Zhang M., Canfield T., et al. , 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346: 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswariah S. S., Busby S. J., 2015. Evolution of bacterial transcription factors: how proteins take on new tasks, but do not always stop doing the old ones. Trends Microbiol. 23: 463–467. [DOI] [PubMed] [Google Scholar]
- Wang F. F., Deng C. Y., Cai Z., Wang T., Wang L., et al. , 2014. A three-component signalling system fine-tunes expression kinetics of HPPK responsible for folate synthesis by positive feedback loop during stress response of Xanthomonas campestris. Environ. Microbiol. 16: 2126–2144. [DOI] [PubMed] [Google Scholar]
- Wang S., Liu X., Liu H., Zhang L., Guo Y., et al. , 2015. The exopolysaccharide Psl-eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environ. Microbiol. Rep. 7: 330–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. C., Carlson S. S., White T. J., 1977. Biochemical evolution. Annu. Rev. Biochem. 46: 573–639. [DOI] [PubMed] [Google Scholar]
- Wilson M. D., Odom D. T., 2009. Evolution of transcriptional control in mammals. Curr. Opin. Genet. Dev. 19: 579–585. [DOI] [PubMed] [Google Scholar]
- Winkler M. E., Hoch J. A., 2008. Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J. Bacteriol. 190: 2645–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P. J., Kalay G., 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13: 59–69. [DOI] [PubMed] [Google Scholar]
- Xu P., Ge X., Chen L., Wang X., Dou Y., et al. , 2011. Genome-wide essential gene identification in Streptococcus sanguinis. Sci. Rep. 1: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Wu X. G., Wei H. L., Wang H. M., Zhang L. Q., 2009. Differential control of the PcoI/PcoR quorum-sensing system in Pseudomonas fluorescens 2P24 by sigma factor RpoS and the GacS/GacA two-component regulatory system. Microbiol. Res. 164: 18–26. [DOI] [PubMed] [Google Scholar]
- Zahrt T. C., Deretic V., 2000. An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182: 3832–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Yao X., Duan M., Luo Y., Liu B., et al. , 2016. Two overlapping two-component systems in Xanthomonas oryzae pv. oryzae contribute to full fitness in rice by regulating virulence factors expression. Sci. Rep. 6: 22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwir I., Shin D., Kato A., Nishino K., Latifi T., et al. , 2005. Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc. Natl. Acad. Sci. USA 102: 2862–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the figures and tables. Bacterial strains are available upon request.