ABSTRACT
Surfaces of food processing premises are exposed to regular cleaning and disinfection (C&D) regimes, using biocides that are highly effective against bacteria growing as planktonic cells. However, bacteria growing in surface-associated communities (biofilms) are typically more tolerant toward C&D than their individual free-cell counterparts, and survival of pathogens such as Listeria monocytogenes may be affected by interspecies interactions within biofilms. In this study, Pseudomonas and Acinetobacter were the most frequently isolated genera surviving on conveyor belts subjected to C&D in meat processing plants. In the laboratory, Pseudomonas, Acinetobacter, and L. monocytogenes dominated the community, both in suspensions and in biofilms formed on conveyor belts, when cultures were inoculated with eleven-genus cocktails of representative bacterial strains from the identified background flora. When biofilms were exposed to daily C&D cycles mimicking treatments used in food industry, the levels of Acinetobacter and Pseudomonas mandelii diminished, and biofilms were instead dominated by Pseudomonas putida (65 to 76%), Pseudomonas fluorescens (11 to 15%) and L. monocytogenes (3 to 11%). The dominance of certain species after daily C&D correlated with high planktonic growth rates at 12°C and tolerance to C&D. In single-species biofilms, L. monocytogenes developed higher tolerance to C&D over time, for both the peracetic acid and quaternary ammonium disinfectants, indicating that a broad-spectrum mechanism was involved. Survival after C&D appeared to be a common property of L. monocytogenes strains, as persistent and sporadic subtypes showed equal survival rates in complex biofilms. Biofilms established preferentially in surface irregularities of conveyor belts, potentially constituting harborage sites for persistent contamination.
IMPORTANCE In the food industry, efficient production hygiene is a key measure to avoid the accumulation of spoilage bacteria and eliminate pathogens. However, the persistence of bacteria is an enduring problem in food processing environments. This study demonstrated that environmental bacteria can survive foam cleaning and disinfection (C&D) at concentrations used in the industrial environment. The phenomenon was replicated in laboratory experiments. Important characteristics of persisting bacteria were a high growth rate at low temperature, a tolerance to the cleaning agent, and the ability to form biofilms. This study also supports other recent research suggesting that strain-to-strain variation cannot explain why certain subtypes of Listeria monocytogenes persist in food processing environments while others are found only sporadically. The present investigation highlights the failure of regular C&D and a need for research on improved agents that efficiently detach the biofilm matrix.
KEYWORDS: Listeria monocytogenes, conveyor belt, biofilm, cleaning, disinfection
INTRODUCTION
Food production premises are regularly subjected to cleaning and disinfection (C&D) regimes designed to reduce bacterial load and eliminate pathogens. Peracetic acid (PAA) and quaternary ammonium compounds (QAC) such as benzalkonium chloride are widely used as disinfectants in the food industry and in health care facilities. Disinfectants are agents that have multiple targets in the cell, and typically kill bacteria by disruption of the bacterial membrane (1). The use of chemical disinfectants in food processing environments is usually based on their efficacy in tests performed with planktonic bacteria (2). However, in natural and industrial environments, bacteria often grow as biofilms, which are complex and structured microbial communities encased in a self-produced protective extracellular matrix composed of polysaccharides, proteins, and/or extracellular DNA. The formation of biofilms is important for microbial survival in the food industry, and cells in biofilms typically exhibit increased tolerance toward antimicrobial agents compared with that of their planktonic counterparts (3, 4). Possible mechanisms contributing to the low efficacy of conventional biocides on biofilms include diffusion-reaction limitation associated with the biofilm matrix, slow growth, and development of persister cell subpopulations (4).
The microbiota found in food processing plant surfaces after C&D are commonly reported to be diverse and include foodborne pathogens and food spoilage bacteria. Predominant genera in meat processing plants after C&D include Pseudomonas, Acinetobacter, Staphylococcus, and Serratia (5–7). One of the pathogens regularly encountered in such environments is Listeria monocytogenes, which causes the life-threatening disease listeriosis. This bacterium poses a significant food safety challenge given its wide distribution in nature and its abilities to grow at refrigeration temperatures and to survive and persist on equipment in food processing environments. Contamination of food products with L. monocytogenes mainly occurs in the food production environment and is a concern especially with regard to ready-to-eat (RTE) products, such as cold meat cuts. The transfer of L. monocytogenes from food contact surfaces such as conveyor belts to processed food products has been documented and, in some cases, shown to result in outbreaks of listeriosis (8, 9).
Certain strains of L. monocytogenes can establish in the production environment and persist for months or even years, especially in humid areas and areas where C&D is difficult. Persistent strains of L. monocytogenes often belong to certain molecular subtypes, while other subtypes are found only sporadically (10–14). Several studies have investigated whether phenotypic traits such as the ability to form biofilms and survive biocide action may be responsible for the prolonged persistence of certain strains on food processing plant surfaces (15–18). Individual strains of L. monocytogenes have been shown to vary in their ability to form biofilms (19, 20) and differ in their tolerance toward disinfectants (21, 22). However, no single genetic determinant or individual trait responsible for L. monocytogenes persistence has been identified, and it is now generally thought that the perceived persistence of certain subtypes of L. monocytogenes is due to a complex combination of factors (13, 14).
The resident background microflora is recognized to play an important role with respect to protecting and sheltering pathogenic strains within food processing environments. Weak biofilm formers can, for instance, improve their survival by joining a multispecies biofilm (23–25). Additionally, it appears that biofilms composed of multiple genera are generally less susceptible to biocide action than their single-species counterparts (4, 23, 26, 27). For example, under most conditions, dual species biofilms of L. monocytogenes and Lactobacillus plantarum were more tolerant to benzalkonium chloride and PAA than were the corresponding single species biofilms (28). Nevertheless, specific bacterial interactions, which include competition, coaggregation, and metabolic cross-feeding, may have variable effects on the survival of individual biofilm community members (23). The growth of L. monocytogenes in dual-species biofilms with representative strains from food production environments has, for instance, resulted in both enhanced and reduced cell numbers of L. monocytogenes (29). However, it is not clear to what extent these effects vary between strains or subtypes of L. monocytogenes or how different L. monocytogenes strains survive in more complex multigenus biofilms subject to conditions similar to those found in the food industry.
The purpose of this study was to examine the biofilm formation and survival of strains belonging to bacterial genera commonly isolated from conveyor belts in meat processing environments under conditions simulating those encountered in these environments. This included an assessment of the efficacy of C&D under relevant conditions and an examination of how the background microbiota may affect the growth and survival of persistent and sporadic L. monocytogenes subtypes in biofilms exposed to C&D. Initially, the microbiota surviving C&D of conveyor belts in meat processing plants were identified. An experimental biofilm model system was then set up using conditions realistic for the food industry, including growth on coupons cut from conveyor belt material and exposure to daily cycles of C&D. Biofilms composed of L. monocytogenes strains were compared with complex multigenus biofilms inoculated with both L. monocytogenes and selected strains dominating the bacterial flora identified in meat processing environments. The development of the biofilm microbiota was investigated using viability counting, amplicon sequencing, and imaging techniques.
RESULTS
Identification of microbiota on conveyor belts in meat processing plants.
Sampling of nine conveyor belts after sanitation in two meat processing plants resulted in the identification of a total of 121 isolates from a total of 22 genera (Table 1). Eight genera were common for both plants, but overall, the microbiota after sanitation differed between plants and between single conveyor belts. For two of the six conveyor belts sampled in plant A, the bacterial numbers were very low and four or fewer isolates were collected (conveyors 4 and 5). For conveyors with higher bacterial numbers, Pseudomonas was most frequently isolated and dominated alone in one sample, together with Psychrobacter in another, and with Acinetobacter on a third conveyor belt. For one conveyor belt, which was associated with a permanent L. monocytogenes (MF5377) reoccurrence, a diverse microbiota was found in which Microbacterium dominated together with Epilithonimonas. In plant B, Sphingomonas dominated together with Rhodococcus on one conveyor and with Acinetobacter on another. Only five isolates were collected from the third conveyor belt. A total of 16 isolates were selected for the present study, representing the most dominant bacteria (Table 2).
TABLE 1.
Microbiota found on conveyor belts in meat processing plants
| Genus | No. of colonies isolated from indicated conveyor belt at: |
Total no. of colonies | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Plant A |
Plant B |
|||||||||
| 1 | 2 | 3 | 4 | 5 | 6a | 1 | 2 | 3 | ||
| Pseudomonas | 7b | 5 | 24b | 1b | 37 | |||||
| Acinetobacter | 1 | 6 | 6b | 13 | ||||||
| Microbacterium | 10b | 1 | 11 | |||||||
| Sphingomonas | 1 | 10b | 11 | |||||||
| Epilithonimonas | 8b | 8 | ||||||||
| Micrococcus | 4 | 1b | 1 | 6 | ||||||
| Psychrobacter | 4 | 2b | 6 | |||||||
| Rhodococcus | 1 | 4b | 5 | |||||||
| Corynebacterium | 2b | 2 | 4 | |||||||
| Brevundimonas | 1 | 2 | 1 | 4 | ||||||
| Vagococcus | 3 | 3 | ||||||||
| Erysipelothrix | 2 | 2 | ||||||||
| Kocuria | 1b | 1b | 2 | |||||||
| Chryseobacterium | 1 | 1 | ||||||||
| Exiguobacterium | 1 | 1 | ||||||||
| Leucobacter | 1 | 1 | ||||||||
| Lysinibacillus | 1 | 1 | ||||||||
| Moraxella | 1 | 1 | ||||||||
| Paenibacillus | 1 | 1 | ||||||||
| Rhothia | 1 | 1 | ||||||||
| Roseomonas | 1 | 1 | ||||||||
| Variovorax | 1 | 1 | ||||||||
| Total | 18 | 12 | 24 | 4 | 2 | 25 | 20 | 11 | 5 | 121 |
L. monocytogenes was isolated from this conveyor belt on several other occasions (MF5377).
Sample with isolates used in biofilm experiments in this study.
TABLE 2.
Bacterial strains used in biofilm experiments
| Strain | Planta | Bacterial species, MLST sequence type, or MLVA profileb |
|---|---|---|
| Background microbiota from conveyor belts | ||
| MF4640 | B | Acinetobacter johnsonii |
| MF4642 | B | Acinetobacter johnsonii |
| MF4643 | B | Corynebacterium sputi |
| MF4645 | B | Corynebacterium sp. |
| MF6392 | A | Epilithonimonas sp. |
| MF4644 | B | Kocuria sp. |
| MF6395 | A | Kocuria rhizophila |
| MF4634 | B | Microbacterium sp. |
| MF6393 | A | Micrococcus sp. |
| MF6396 | A | Pseudomonas sp. (P. putida group) |
| MF6394 | A | Pseudomonas sp. (P. fluorescens subgroup) |
| MF4836 | A | Pseudomonas sp. (P. mandelii subgroup) |
| MF4641 | B | Psychrobacter sp. |
| MF4633 | B | Rhodococcus erythropolis |
| MF4637 | B | Rhodococcus fascians |
| MF4632 | B | Sphingomonas sp. |
| L. monocytogenes strains from meat processing environmentsc | ||
| MF4536 | C | ST9, MLVA 6-11-15-18-6 (persistent, qacH positive) |
| MF5376 | A | ST7, MLVA 7-7-10-10-6 (persistent) |
| MF5634 | B | ST121, MLVA 6-7-14-10-6 (persistent, qacH positive) |
| MF5377 | A | ST8, MLVA 6-9-18-16-6 (persistent) |
| MF4565 | C | ST18, MLVA 8-8-17-21-6 (sporadic) |
| MF5630 | C | ST19, MLVA 6-9-18-10-6 (sporadic) |
| MF5378 | A | ST394, MLVA 6-9-19-10-6 (sporadic) |
Plants A, B, and C correspond to plants M2, M4, and M1, respectively, described by Møretrø et al. (22).
Listed MLVA profiles correspond to variable number tandem repeat (VNTR) loci LMV6-LMV1-LMV2-LMV7-LMV9 described by Lindstedt et al. (66). Persistent or sporadic MLVA profiles are determined using the criteria described in Materials and Methods.
L. monocytogenes strains were isolated by Møretrø et al. (22).
Three Pseudomonas sp. and two Acinetobacter sp. were subjected to whole-genome sequencing and phylogenetic analysis to further determine their taxonomic status. This analysis showed that strain MF6396 belonged to the Pseudomonas putida group and that strains MF6394 and MF4836 belonged to the Pseudomonas fluorescens and Pseudomonas mandelii subgroups, respectively, within the P. fluorescens complex (see Fig. S1 in the supplemental material). Thus, all three strains belong to the P. fluorescens lineage. For simplicity, these strains are referred to as P. putida MF6396, P. fluorescens MF6394, and P. mandelii MF4836 in the remainder of this text. Both Acinetobacter strains included in the experiments (MF4640 and MF4642) were determined to belong to the species A. johnsonii using in silico multilocus sequence typing (MLST) (see Fig. S2).
Pseudomonas and Acinetobacter dominated in laboratory multigenus biofilms.
A biofilm model system was set up to examine biofilm formation and survival under conditions simulating food production environments. Biofilms were grown on conveyor belt coupons placed vertically in 24-well plates with brain heart infusion (BHI) broth at 12°C, which is a temperature typically found in Norwegian meat processing facilities. In addition to the 16 strains from the background microbiota found on conveyors in meat processing plants (described above), seven L. monocytogenes strains belonging to different phylogenetic clusters were selected for inclusion in biofilm experiments (according to selection criteria in Materials and Methods). Four belonged to MLST sequence types (STs) responsible for persistent contaminations in Norwegian food processing plants, while three strains belonged to STs which were only sporadically encountered in the Norwegian food industry (Table 2) (22). Coupons were inoculated with a suspension of either the 16 background microbiota strains plus the seven L. monocytogenes strains (referred to as multigenus biofilms) or only the seven L. monocytogenes strains (L. monocytogenes biofilms). The biofilms were allowed to develop for 4 days and were subsequently subjected to C&D on days 4 to 7 using a chlorinated alkaline cleaning agent (Alkalifoam) and disinfection with either a QAC- or PAA-based disinfectant at concentrations recommended by the manufacturers. Wells containing multigenus biofilms usually contained a floating pellicle that was attached to the coupon at the air-liquid interface. Visible biofilm deposits were generally observed in this zone of the coupons after C&D.
The development of the microbiota in the multigenus biofilms was investigated using 16S rRNA amplicon sequencing. The results showed that after 4 days of biofilm growth, one of the A. johnsonii strains (MF4640) dominated the biofilm, while after 7 days of growth, the P. putida strain (MF6396) had taken over as the dominant strain. The proportion of L. monocytogenes in the multigenus biofilms was higher on day 7 than on day 4 (Fig. 1A). To investigate whether the shift in microbiota from day 4 to day 7 was due only to the establishment of a more mature biofilm or was also affected by the C&D cycles, new experiments were conducted in which only the three dominating Pseudomonas species strains and A. johnsonii strain MF4640 were included. Here, coupons that were rinsed daily with H2O were included in addition to coupons treated with C&D agents. The results presented in Fig. 1B show that the bacterial strain compositions identified on coupons subjected to C&D in these two additional experiments were similar to those obtained in the first three experiments in which biofilms were inoculated with all 16 background microbiota strains (Fig. 1A; see also Fig. S3). However, in the absence of C&D, no significant shift in the microbiota composition was observed from day 4 to day 7, and the A. johnsonii strain dominated, followed by P. putida. This indicated that daily exposure to C&D selected for P. putida, P. fluorescens, and L. monocytogenes and almost eliminated the P. mandelii and A. johnsonii strains.
FIG 1.
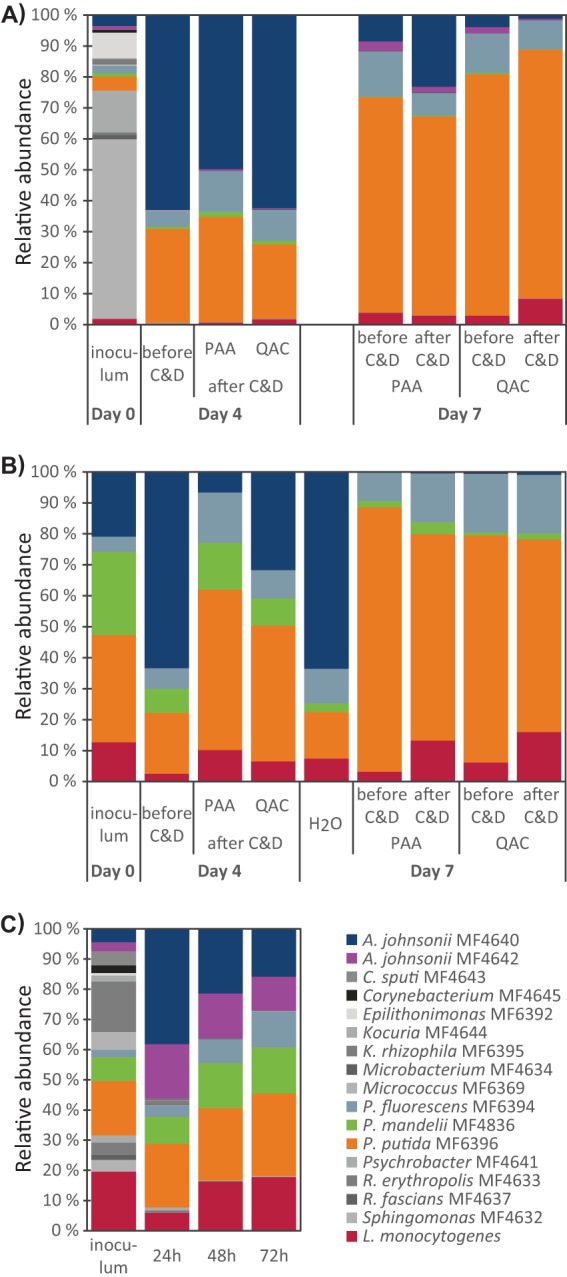
Development of the microbiota in multigenus biofilms on conveyor belt coupons (A, B) and planktonic cultures (C) at 12°C. Inocula were composed of L. monocytogenes plus either 16 (A, C) or four (B) background microbiota strains. The frequencies of different bacterial strains were determined by 16S rRNA amplicon sequencing. (A, B) Biofilms were allowed to develop for 4 days before being subjected to either daily cleaning with Alkalifoam and treatment with QAC or PAA disinfection agent or a daily rinse in H2O (B). Coupons were harvested either before or after C&D on the day of harvest, as indicated. (C) Development of microbiota in planktonic cultures grown with shaking in Erlenmeyer flasks for a total of 72 h. Presented results are averages from three (A, C) or two (B) independent experiments. Results for individual experiments are shown in Fig. S3 in the supplemental material.
No selection between different L. monocytogenes strains was observed in biofilms.
To determine whether the different L. monocytogenes strains had different fitness during growth in biofilms subjected to C&D, strain identification of single colonies collected after day 7 of biofilm growth was performed by sequencing of the dapE MLST allele (Fig. 2A; see also Fig. S4). The frequencies of each strain across all tested samples ranged from 5% for MF5378 to 28% for MF5360. The four strains belonging to persistent subtypes had an overall frequency of 51% across all samples, indicating that these strains did not have a greater ability to survive in biofilms exposed to C&D than strains belonging to sporadic subtypes. No evidence for selection between different L. monocytogenes strains was observed, either in the multigenus biofilms where L. monocytogenes was grown in the presence of 16 background flora strains or in biofilms containing L. monocytogenes only.
FIG 2.
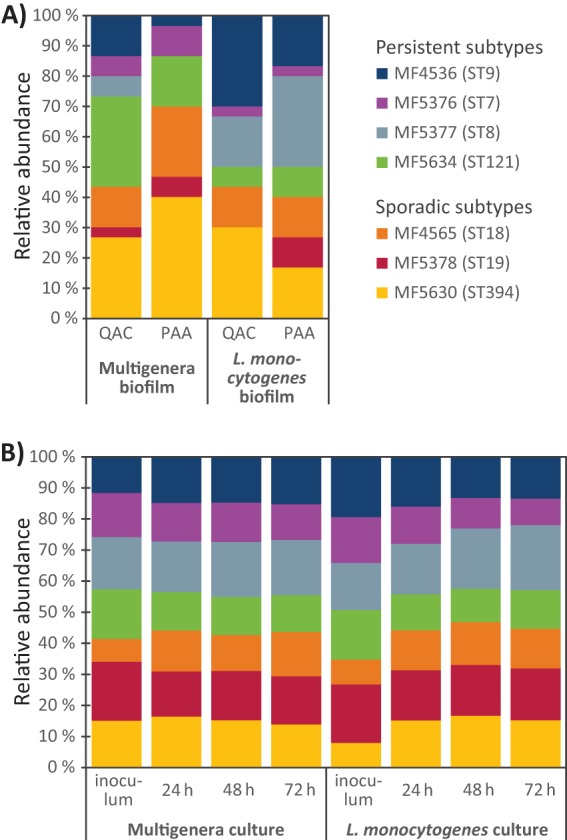
Competition between L. monocytogenes strains during biofilm (A) and planktonic (B) growth in BHI at 12°C. Inoculated suspensions were composed of equal amounts of seven different L. monocytogenes strains, grown either together with 16 background microbiota strains (multigenus biofilm/culture) or alone (L. monocytogenes biofilm/culture). (A) Frequencies of L. monocytogenes strains surviving in biofilms on conveyor belt coupons. Biofilms were allowed to develop for 4 days and then subjected to daily cleaning with Alkalifoam and disinfection with either a QAC- or PAA-based disinfectant on four consecutive days before harvest. The identities of single colonies of L. monocytogenes were determined by Sanger sequencing of the dapE allele. Results are pooled from three replicate experiments in which 10 single L. monocytogenes colonies were identified from each of the four conditions analyzed; 120 colonies in total. (B) Time course experiment showing the relative abundance of L. monocytogenes strains in planktonic cultures grown with shaking in Erlenmeyer flasks for a total of 72 h. The frequencies of different L. monocytogenes strains were determined by dapE amplicon sequencing. Presented results are averages from three independent experiments. Results for individual experiments are shown in Fig. S4.
Strains dominating in the multigenus biofilm showed high growth rates in planktonic culture.
The relative amounts of each bacterial strain in planktonic cultures inoculated with the same bacteria used in the multigenus biofilm experiments are shown in Fig. 1C. As in the biofilm experiments, the bacterial composition developed toward A. johnsonii, Pseudomonas spp., and L. monocytogenes. However, in contrast to the results seen during growth under biofilm conditions, the two A. johnsonii strains (MF4640 and MF4642) seemed to compete equally well under planktonic culture conditions. Similarly, the three Pseudomonas species strains were in approximately equal proportions in the planktonic cultures, while in the biofilms, there was significantly more P. putida MF6396 than P. fluorescens MF6394 and, especially, P. mandelii MF4836.
All seven L. monocytogenes strains were retained in approximately equal amounts when grown together in planktonic culture, both when they were grown alone and when they were grown with the 16 background microbiota strains (Fig. 2B; see also Fig. S4). The proportions of each strain present in the cultures containing only L. monocytogenes, determined using dapE amplicon sequencing, ranged from an average of 9% (MF5376/ST7) to 21% (MF5377/ST8) after 72 h of growth. When the seven L. monocytogenes strains were grown with the 16 background microbiota strains, the proportions of each L. monocytogenes strain after 72 h ranged from 12% (MF5376/ST7 and MF5634/ST121) to 18% (MF5377/ST8). These results indicated that during planktonic growth at 12°C, none of the seven L. monocytogenes strains appeared to have a growth advantage allowing them to outcompete any of the other strains.
When the individual strains were grown in separate wells in a Bioscreen C instrument (Fig. 3; see also Table S1), the largest maximal growth rate during the exponential phase of growth was attained by P. fluorescens MF6394, followed by P. mandelii MF4836, the two A. johnsonii strains, and then P. putida MF6396. The cultures containing Pseudomonas strains ultimately reached higher values of optical density at 600 nm (OD600) than the Acinetobacter cultures. Other strains with high growth rates were the seven L. monocytogenes strains (which all had similar growth curves) and Epilithonimonas strain MF6392, followed by the Psychrobacter and Microbacterium strains (MF4641 and MF4634). It thus seems that the strains showing rapid planktonic growth at 12°C in BHI culture medium are highly competitive in the biofilms grown on conveyor belt coupons.
FIG 3.
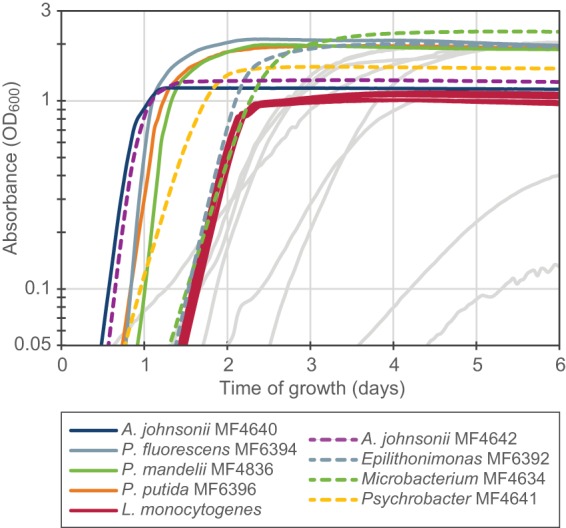
Growth of the 16 background microbiota strains and the seven L. monocytogenes strains in BHI medium at 12°C. The experiment was performed in a Bioscreen C instrument, with measurement of the absorbance at 600 nm once every hour. Results shown are averages from triplicate wells in one representative experiment out of three experiments performed. Strains included in the biofilm experiments with four background microbiota strains (Fig. 1B) are represented by solid colored lines. Dashed lines represent selected strains that also showed good growth properties. Gray lines represent the remaining eight strains.
The Pseudomonas strains contained different sets of known biofilm-associated genes.
The difference in competitiveness between the three Pseudomonas strains and between the two A. johnsonii strains in planktonic culture compared with that during growth in the conveyor belt biofilm model (Fig. 1) could possibly be due to differences in the ability to form biofilms. Therefore, the genomes of the Pseudomonas spp. and A. johnsonii strains were screened for known biofilm-associated genes using a BLAST analysis. All three Pseudomonas strains contained the alg operon required for alginate synthesis and homologs to the lapABCD and lapG genes required for the expression of the large surface protein LapA on the cell surface. Each of the three strains contained only one of the genes responsible for Pel, Psl, and cellulose synthesis: P. fluorescens MF6394 contained a psl operon, P. mandelii MF4836 contained a pel operon, while a homolog to the wss operon required for cellulose synthesis was present in P. putida MF6396. Thus, it appears that all three strains harbor genetic factors enabling biofilm formation (see Table S2). With respect to the two A. johnsonii strains, not much is known about biofilm formation in non-baumannii Acinetobacter strains, and no homologs to genes shown to be involved in biofilm formation in Acinetobacter baumannii were identified in the genomes of the two A. johnsonii strains employed in the current study. The two strains did however have different genome sizes, as the genome of A. johnsonii MF4640 was 13% larger than the 3.36-Mbp large genome of strain MF4642. A large portion of the additional genetic material in MF4640 appears to constitute plasmids and other mobile genetic elements.
The sanitation regime was inefficient at killing bacteria in conveyor belt biofilms.
To assess sanitation efficacy in the biofilm model system, the total numbers of CFU in biofilms growing on conveyor belt coupons were determined both before and after coupons were subjected to C&D.
After the initial 4 days of biofilm development, the cell densities in multigenus biofilms reached approximately 1 × 108 CFU per coupon (3 cm2 surface area). Coupons were then subjected to daily cycles of C&D for 3 days and were sampled again on day 7 after allowing 24 h of regrowth after the last C&D cycle. Control coupons were rinsed with sterile deionized water every day. There was no significant difference in cell densities on coupons with multigenus biofilms sampled on days 4 and 7 prior to C&D, regardless of whether coupons had been treated with QAC, PAA, or rinsed in H2O (P > 0.05) (Fig. 4, gray bars). Thus, neither the QAC- nor PAA-based C&D regimes altered the total amounts of biofilm on conveyor belt materials present 24 h after C&D treatment.
FIG 4.
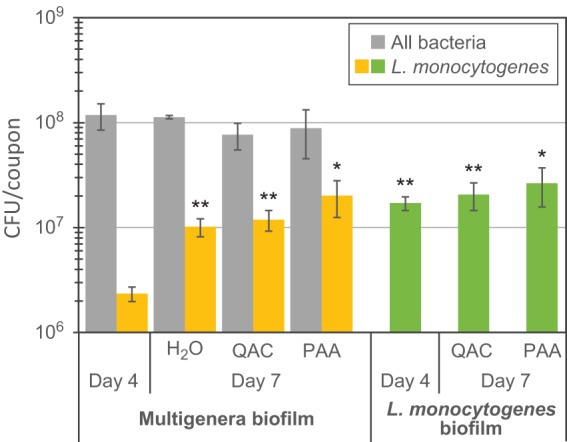
Total bacterial numbers in biofilms on conveyor belt coupons prior to C&D. Biofilms were allowed to develop undisturbed until day 4 before being rinsed in H2O or cleaned with Alkalifoam and disinfected with either a QAC- or a PAA-based disinfection agent on three consecutive days. The microbiota on coupons harvested on day 7 had been allowed to regrow for 24 h after the last disinfection step. The L. monocytogenes counts in multigenus biofilms were determined by plating on selective agar. Mean values from five replicates are shown, except for the sample labeled H2O, where the mean from two replicates is shown. Error bars show standard errors of the means. Asterisks represent differences in L. monocytogenes CFU per coupon relative to that in multigenus biofilms harvested on day 4 (two-tailed paired Student t tests). *, P = 0.08; **, P < 0.05.
However, while the total numbers of CFU on each coupon in the multigenus biofilms were similar in all tested samples, the amounts of L. monocytogenes in the biofilms increased approximately 10-fold from day 4 to day 7 (Fig. 4, yellow bars). The fractions (with standard errors) of L. monocytogenes in the multigenus biofilms increased from 2.3% (± 1.1%) on day 4 to 9% (± 2%), 18% (± 4%), and 32% (± 7%) on day 7 in the H2O-rinsed, QAC-treated, and PAA-treated biofilms, respectively. However, in the biofilms where L. monocytogenes bacteria were grown alone, there was no statistically significant difference in L. monocytogenes counts per coupon between days 4 and 7 (P > 0.05) (Fig. 4, green bars), with approximately 2 × 107 CFU per coupon on both days and across the different treatments.
When the total numbers of CFU per coupon before and after C&D were compared, between 0.6- and 0.9-log10 reductions in total CFU were observed on day 4 and day 7, respectively, for coupons harboring multigenus biofilms (Fig. 5, gray bars). The difference in log10 reductions between treatments or day of sampling was not statistically significant (P > 0.05).
FIG 5.
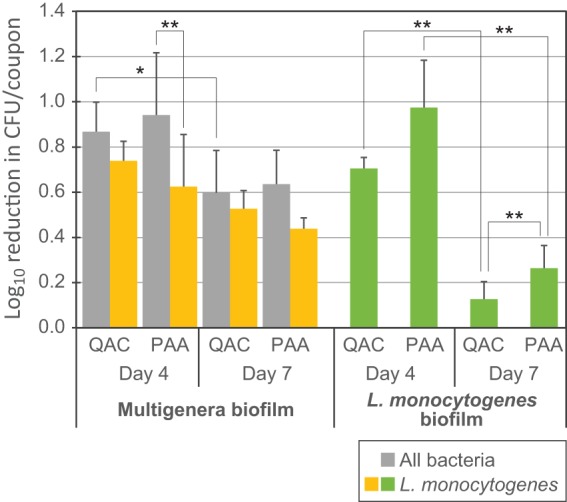
Tolerance of biofilms to C&D regimes relevant in the food industry. The log10 reduction upon C&D for multigenus biofilms and biofilms inoculated with seven L. monocytogenes strains grown on conveyor belt coupons is shown. Biofilms were allowed to develop undisturbed until day 4 before being cleaned with Alkalifoam and disinfected with either a QAC- or a PAA-based disinfection agent on four consecutive days. Calculated reductions are relative to control coupons rinsed in H2O only. The L. monocytogenes counts in multigenus biofilms were determined by plating on selective agar. Mean values from five experiments and standard errors of the means are shown. Asterisks represent comparisons of samples using the two-tailed paired Student t tests. *, P = 0.06; **, P < 0.05.
The log10 reduction for the L. monocytogenes component of the multigenus biofilm was significantly lower than the log10 reduction in total CFU per coupon when the day 4 coupons were treated with PAA disinfection (0.6- versus 0.9-log10 reduction; P = 0.04). For the other treatments (QAC treatment on days 4 and 7 and PAA treatment on day 7), there was no difference in survival of the flora strains and the L. monocytogenes strains in the multigenus biofilm upon C&D (P > 0.05) (Fig. 5, compare gray and yellow bars). This indicates that the proportion of L. monocytogenes cells in the biofilm was relatively stable during a cycle of C&D.
For the L. monocytogenes biofilms, on day 4, the reduction in CFU per coupon upon sanitation treatment was approximately the same as for the multispecies biofilms. On day 7, however, there was almost no reduction in bacterial numbers upon C&D, with average reductions in cell numbers of only 0.13 log10 and 0.26 log10 CFU per coupon upon QAC and PAA treatment of the biofilms, respectively (Fig. 5, green bars).
Overall, these experiments indicated that biofilms on conveyor belt materials were not eliminated when exposed to a C&D regime relevant for the food industry. Little or no development of tolerance to C&D agents was observed for the multigenus biofilms during the course of the experiments. However the L. monocytogenes biofilms did develop increased tolerance over time, as no significant reductions in CFU were observed during the C&D process after the coupons had been exposed to three daily cycles of cleaning followed by disinfection with either PAA or QAC.
All strains were susceptible to the sanitation agents in suspension tests.
To examine whether any of the strains included in the multigenus biofilms had a specific tolerance toward the employed C&D agents that could explain survival, bactericidal suspension tests were performed on each strain, using both QAC and PAA disinfection agents as well as the Alkalifoam cleaning agent. For all strains, the bacterial reductions were >4 log10 units after exposure to the recommended user concentrations of the QAC and PAA disinfectants for 5 min at 12°C (see Table S3). Most strains also showed the same levels of tolerance to the cleaning agent alone. The exceptions were the two Corynebacterium species strains and the Micrococcus species strain, which showed only between 10 and 100-fold reductions in CFU upon treatment with the cleaning agent, and the two Kocuria species strains, P. putida strain MF6396, and the Psychrobacter species strain, which showed 3-log10 to 4-log10 reductions in CFU per ml upon treatment with the cleaning agent. These results indicate that all strains were susceptible to the C&D treatment when grown in suspension.
CLSM analysis showed that biofilms predominantly settle on the underside of the conveyor belt material.
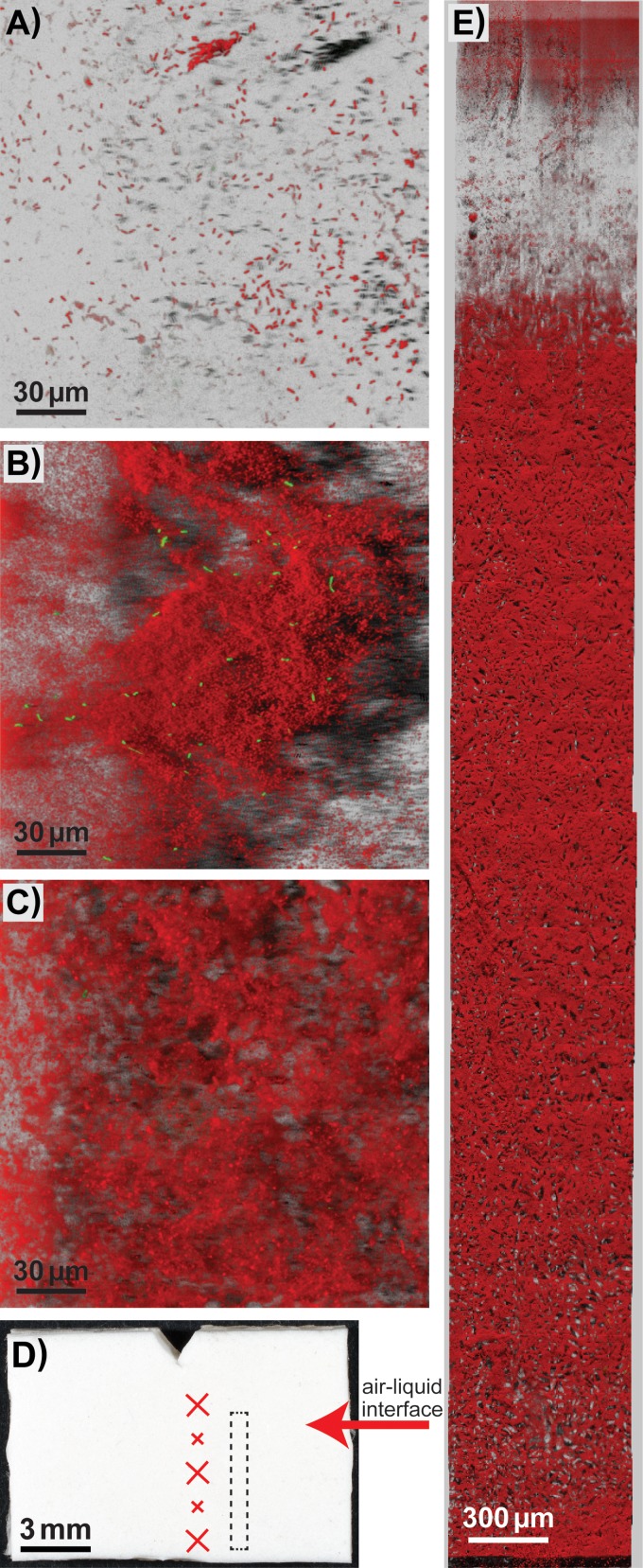
Confocal laser scanning microscopy (CLSM) was employed to examine the spatial organization of biofilms formed on the conveyor belt coupons. The three-dimensional image reconstructions shown in Fig. 6 and Fig. 7 were obtained by scans of several predefined location patterns on each coupon (see Fig. 6D), and were selected from 174 acquired confocal Z-stack scans (see Table S4). In the majority of captured scans, relatively few sparse cells—attached singly or as small clusters—were observed on the coupon surface. However, a significant number of images showed the presence of large heterogeneous three-dimensional biofilms. These were also observed on some of the coupons examined immediately after cleaning and disinfection with QAC or PAA. In the multigenus biofilms, green fluorescent protein (GFP)-expressing L. monocytogenes cells were often absent despite observations of significant numbers of background flora cells. When present, L. monocytogenes was spatially organized as single cells mixed in between the cells of the background flora strains. In some of the images, the biofilm also appeared to have a slightly layered structure, with L. monocytogenes cells found closer to the bottom layer of the biofilms (Fig. 7K). No separate L. monocytogenes monospecies microcolonies were observed on the coupons in which multigenus biofilms were grown.
FIG 6.
Biofilms on the top coating of conveyor belt material examined using CLSM. Green represents GFP-expressing L. monocytogenes cells, while Acinetobacter and Pseudomonas background flora strains are shown in red. (A to C and E) CLSM images shown are Easy3D shadow projection reconstructions obtained from the confocal Z-stack series using the IMARIS software. Coupons with biofilms were imaged either on day 4 (B) or on day 7 (A, C, E) after initiation of biofilm growth and harvested after rinsing in H2O (C, E) or after C&D with QAC (A, B). (D) Photograph of the front side of a conveyor belt coupon. The arrow indicates the approximate location of the air-liquid interface during biofilm development, and crosses show the approximate locations on each coupon where the CLSM images were acquired (when only three images were acquired, the locations indicated by the large crosses were used). (E) Mosaic 3D meta-image obtained using a motorized stage that automatically moves the sample between scans and tiles the adjacent fields. The box drawn with dashed lines in panel D approximately corresponds to the area of the coupon covered by the CLSM image in panel E.
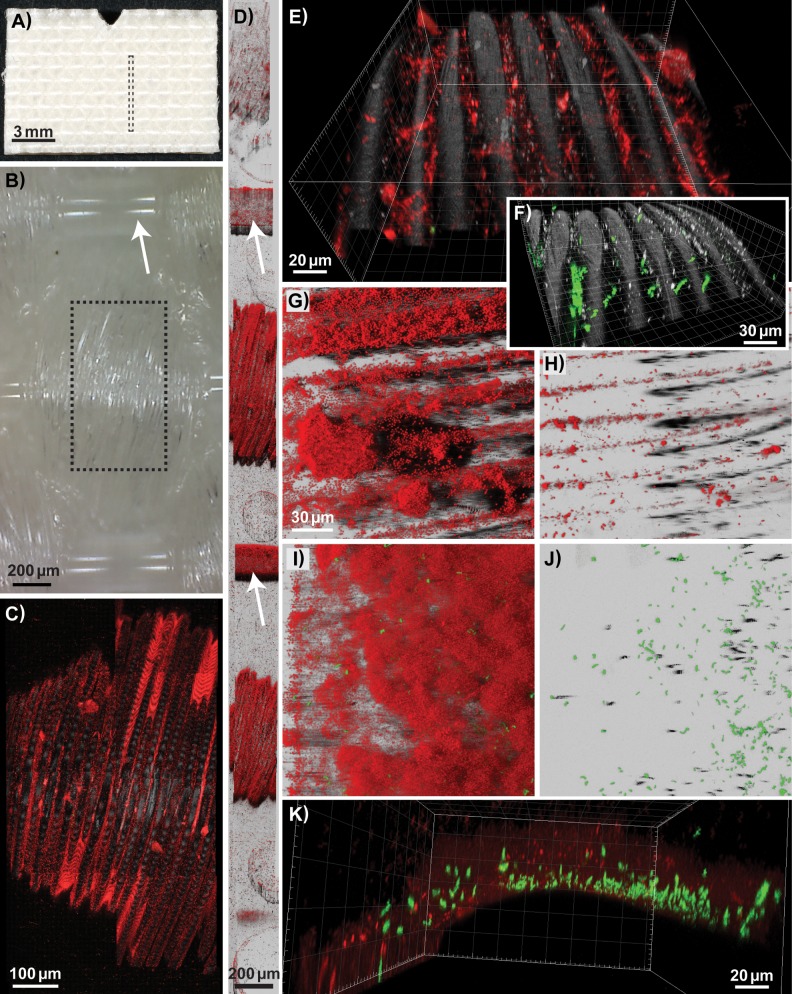
FIG 7.
Biofilms formed on the underside of the conveyor belt examined using CLSM. Pseudomonas and Acinetobacter background flora strains (shown in red) and GFP-expressing L. monocytogenes strains (in green) were used as inoculum, except in (F), where biofilms were inoculated with L. monocytogenes strains only. (A, B) Photographs showing the underside of a clean conveyor belt coupon. (B) Photograph taken with a USB microscope lens showing the weave pattern of the fabric, with the smooth warp thread indicated by an arrow. Biofilms formed on the weft thread of the woven fabric, on coupons rinsed in H2O on days 4 to 7 (C, D, and F), on a coupon rinsed in H2O on day 7 after treatment with QAC on days 4 to 6 (E), and on coupons harvested on day 4 for after treatment with PAA (G) or QAC (H). (C, E, and F) The smaller fibers constituting the weft thread are represented by the reflection signal (in gray). Mosaic 3D meta-images are shown in section view mode (C) and Easy3D blend representation (D), with warp treads indicated by arrows. The boxes drawn with dashed lines in panels A and B show the size and location of the area of a coupon depicted by the CLSM images shown in panels D and C, respectively. (I to K) Different representations of the same scan, showing a biofilm harvested on day 4 after rinsing in H2O, formed on the warp thread of the woven fabric. Images are shown as three-dimensional (E, F, and K) and Easy 3D (G, H, I, and J) IMARIS representations. The scales are the same in the Easy3D blend images in panels G to J. The red channel is not shown in panels F and J.
The top face of the conveyor belt is coated with polyvinyl chloride (PVC) and is a matt antistatic surface (Fig. 6D). The underside of the conveyor belt is a urethane-impregnated woven polyester fabric. The photomicrograph in Fig. 7B, taken of the underside of a coupon, shows the linen weave pattern with single smooth warp threads and weft threads composed of bundles of smaller fibers. The difference between the flat top face and the heterogeneous topography of the rear face of the conveyor belt coupons can be seen in the overview images obtained by stitching together multiple CLSM scans—acquired across the length of the coupon from top to bottom—shown in Fig. 6E and 7D. Notably, for the rear side of the coupons, scans were only obtained for the most elevated parts of the fabric, since the microscope was not able to focus in the areas constituting the “valleys” in the fabric surface. Most striking were images acquired for biofilms formed on weft threads composed of bundles of smaller fibers on the rear side of the conveyor belt coupons, as shown in Fig. 7E and F. Both the background flora and L. monocytogenes cells are predominantly found in the gap between these fibers. In the multigenus biofilms, mushroom-shaped biofilm structures could be observed to protrude upwards from the cleft harboring bacterial cells (Fig. 7G).
Quantitative analysis of the biovolume of GFP-expressing L. monocytogenes cells in the biofilms was performed by an analysis of the green channel of the acquired CLSM image stacks (Table 3; see also Fig. S5). The calculated biovolume of L. monocytogenes cells was higher prior to C&D than after treatment with QAC or PAA. Also, the results suggest that the total L. monocytogenes biovolumes were higher in biofilms harvested on day 7 than in biofilms harvested on day 4, both in multigenus biofilms and in L. monocytogenes single-species biofilms. Finally, the analysis strongly indicates that significantly more L. monocytogenes cells were attached to the woven-structured underside of the conveyor belt than on the PVC-coated top surface. The strongest effect was seen for L. monocytogenes biofilms rinsed in H2O daily from days 4 to 7 and harvested on day 7, in which areas (standard errors) of L. monocytogenes cells of 14 (± 11) μm3 and 2,841 (± 1,439) μm3 were found on the top and bottom faces of the conveyor belt coupons, respectively.
TABLE 3.
Biovolumes of L. monocytogenes on conveyor belt couponsa
| Biofilm type | Day | Vol (μm3 [± SE]) |
|||
|---|---|---|---|---|---|
| Before C&D |
After C&D |
||||
| H2O | QAC | QAC | PAA | ||
| Front of coupon | |||||
| Multigenus | 4 | 149 (32) | NAb | 39 (23) | 0.06 (0.03) |
| 7 | 35 (22) | 1.3 (1.2) | 0.67 (0.61) | NTc | |
| L. monocytogenes | 4 | 3.4 (1.1) | NA | 0.18 (0.11) | 0.1 (0.08) |
| 7 | 14 (11) | NT | NT | NT | |
| Back of coupon | |||||
| Multigenus | 4 | 331 (141) | NA | 11 (5) | 0 (0) |
| 7 | 62 (14) | 68 (66) | 0.1 (0.06) | NT | |
| L. monocytogenes | 4 | 172 (73) | NA | 0.61 (0.61) | NT |
| 7 | 2,841 (1,439) | 11 (3.6) | 6.3 (3.9) | NT | |
Values were obtained from CLSM Z-scans using the ICY image analysis software (64). The numbers of coupons tested for each condition are given in Table S4 in the supplemental material.
NA, not applicable.
NT, not tested.
In summary, the microscopy showed that L. monocytogenes cells were spatially intermixed with background flora species in the multigenus biofilms. Furthermore, bacteria appeared to be predominantly situated in the gaps between filament fibers on the undersides of the conveyor belts.
DISCUSSION
This study aimed to decipher the growth and survival of L. monocytogenes on conveyor belts in the food industry using conditions relatively realistically representative of those found in meat production environments. This included growing strains of L. monocytogenes in multigenus biofilms with strains from the background microbiota isolated in these environments. An initial investigation of the microbiota on conveyor belts after C&D in two RTE-meat processing plants resulted in the isolation of a relatively small number of bacteria, but nevertheless, a high diversity was found between as well as within samples (Table 1). A relatively diverse microbiota was therefore used in the initial biofilm experiments (Table 2). Similar to what has been found in other studies, Pseudomonas was relatively common after C&D (5–7, 30–32). Enterobacteriaceae have also been reported to be common in meat processing environments (5–7, 30, 31, 33, 34) but were absent in our study. Instead, microbiota of conveyor belts were dominated by bacteria less frequently reported in previous studies, such as Acinetobacter, Microbacterium, Sphingomonas, and Epilithonimonas (Table 1). The composition of the microbiota is dependent on a number of factors, such as the sanitation regime, the temperature, and the humidity. Biofilm formation reflecting all these various conditions would not be possible in in vitro laboratory studies. In this study, we chose to simulate conditions with high humidity and nutrient content at a temperature relevant for meat processing environments (12°C) and apply C&D cycles similar to those found in the food industry.
The compositions of the biofilms formed on conveyor belt coupons under these conditions were largely stable—with a dominance of Pseudomonas and Acinetobacter strains—regardless of whether four or 16 background strains were used as inocula and regardless of whether coupons had been treated with QAC, PAA, or rinsed in H2O (Fig. 1A and B). Stable coexistence of Acinetobacter and Pseudomonas strains in biofilms has been reported previously (35, 36). In the present study, the compositions of the biofilms shifted from Acinetobacter-dominated biofilms in the day 4 samples to P. putida-dominated biofilms in the day 7 samples subjected to daily C&D (Fig. 1A and B). This transition was not seen in biofilms subjected instead to daily rinses in H2O on days 4 to 7 (Fig. 1B; column labeled H2O). Furthermore, in suspension, P. putida MF6396 had a higher tolerance toward the lethal effect of the chloralkali cleaning agent than A. johnsonii and the other included Pseudomonas strains. This suggests that the dominance of the P. putida strain in biofilms subjected to daily C&D could be a consequence of the C&D treatments and tolerance of the P. putida strain toward the cleaning agent. However, the relative levels of Acinetobacter also decreased over time in the planktonic competition experiments performed in this study (Fig. 1C) and in multigenus biofilm experiments performed in a previous study, in which the effect of C&D was not assessed (37). Potentially, interspecies interactions, such as competition for limiting nutrient sources, may also have contributed to the observed transition in microbial composition between the day 4 and day 7 biofilms.
Specific bacteria may show enhanced survival in biofilms challenged by biocides by means of interspecies interactions, such as coaggregation and metabolic cross-feeding (27). Interactions with other bacteria in biofilms may potentially explain the persistence of pathogens such as L. monocytogenes in food production environments. In this study, the proportion of L. monocytogenes in the multigenus biofilms increased during the course of the experiment, concomitant with the shift toward a P. putida-dominated biofilm. This is consistent with L. monocytogenes specifically interacting with the P. putida strain. Interestingly, this specific strain (MF6396) was isolated from a conveyor belt which was persistently contaminated with L. monocytogenes and from which the persistent ST8 strain L. monocytogenes MF5377 was isolated (see Table 1) (38). It is therefore likely that MF6396 and MF5377 may have originated from the same microhabitat in the meat production plant. An examination of biofilms using CLSM in this study showed that cells of L. monocytogenes were found intermixed with background flora cells, with no spatially segregated L. monocytogenes microcolonies observed within the multigenus biofilms (Fig. 6 and 7). Such spatial distribution patterns in multispecies biofilms are indicative of interspecies coaggregation and cooperation (39, 40), further suggesting that L. monocytogenes cells may directly interact with one or more of the other species found in the biofilm. Previous studies have shown that coculture of L. monocytogenes and resident apathogenic bacteria from food production environments resulted in both positive and negative effects on the biomass of L. monocytogenes (29, 41). Potential specific interactions between the individual strains examined in this study are subject to further examination in our laboratory.
The observation that certain subtypes of L. monocytogenes are more likely than others to persist in food processing environments has prompted several investigators to examine whether genetic determinants or various phenotypic traits could be associated with this ability. One of the aims of this study was to examine whether this perceived persistence may be linked to strain-specific differences in the ability of L. monocytogenes to interact with the resident microflora in biofilms. Few studies have addressed this point specifically, although in a recent study, Overney et al. (42) found that the survival rates of two reference strains of L. monocytogenes (EGD-e and LO28) did not differ when they were grown in dual culture biofilms with a P. fluorescens strain and when biofilms were subject to daily cycles of C&D and desiccation. A similar result was obtained in this study, where seven L. monocytogenes strains—four of which belonged to subtypes linked to persistent contaminations in food production facilities—were shown to be equally capable of growth and survival in biofilms exposed to C&D (Fig. 2). This result was obtained with both monospecies and multigenus biofilms and is consistent with the growing consensus that individual genetic traits linked to specific subtypes do not account for the existence of persistent subtypes of L. monocytogenes (13, 14).
It is widely acknowledged that the efficacy of C&D agents is lower for biofilms than for bacteria growing in planktonic culture (3, 4, 27). A high level of tolerance to C&D was also observed for biofilms in this study, with less than 1-log10 reductions in total CFU per coupon obtained across treatments when the C&D agents were applied at the concentrations recommended by the manufacturers (Fig. 5). A similar level of efficacy of C&D agents applied at recommended user concentrations was seen in a study by Pan et al. (43), where L. monocytogenes biofilms grown on stainless steel or Teflon coupons and subjected to daily cycles of sanitation followed by starvation and incubation in dilute culture medium were followed over a period of 3 weeks. In their study, treatments of biofilms with the minimum recommended user concentrations of peroxide, QAC, or chloride disinfectants resulted in less than 0.3 log10 CFU · cm−2 after the first week of their simulated food processing regimen. However, not all studies find the efficacy of C&D agents against biofilms to be this low. In some studies, the disinfection agents have to be diluted below recommended user concentrations to maintain sufficient numbers of cells above the detection threshold after the disinfection of biofilm coupons (42). Also, the results obtained in this study do not support the previous observation of PAA being more effective against L. monocytogenes biofilms than QAC (44). In any case, the low efficacy of C&D seen in the present study cannot be attributed to the commonly observed greater tolerance of multispecies biofilms than of their single-species counterparts toward biocides (4, 23, 26), because the opposite was actually observed; L. monocytogenes biofilms were shown to become more tolerant to daily C&D than the multigenus biofilms (Fig. 5). Since no significant difference in survival of bacteria was observed between treatments with QAC or PAA disinfectants, the low efficacy of C&D was not likely to be a result of specific resistance mechanisms, such as the presence of efflux pumps conferring resistance toward chemical agents. This is supported by the observation that no selection between different L. monocytogenes strains was seen despite two of the strains possessing the qacH gene encoding an efflux pump conferring increased tolerance to low concentrations of QAC compounds (Table 2) (45).
The explanation for the low efficacy of the C&D treatment could instead, at least partly, be ascribed to features of the coupon material on which the biofilms were grown. Within food processing plants, conveyor belts have been shown to be favorable for contamination by L. monocytogenes that is difficult to remove (9, 46). Furthermore, cracks or scratches in the surfaces of materials used in the food industry have been shown to support the development of L. monocytogenes biofilms deeply rooted in microscopic sutures and ridges (47). The underside of the conveyor belt used as the surface for biofilm growth in this study had a woven surface with filament fiber threads. When coupons were viewed using CLSM, bacteria could be seen to shelter in the clefts between these fibers (Fig. 7), and a quantitative biovolume analysis further suggested that significantly more L. monocytogenes cells were attached to the underside of the conveyor belt than were on the smooth top coating (Table 3). Bacteria could also be expected to find harborage sites on the cut edges of the conveyor belt coupons, which, though likely to be sealed when initially installed in food production plants to prevent the penetration by soil and bacteria, could model situations where conveyers in a production facility become worn or frayed.
The observed increases in tolerance to C&D by L. monocytogenes biofilms over time for both the QAC and PAA disinfectants (Fig. 5) concur with results obtained in the study by Pan et al. (43), in which L. monocytogenes appeared to develop similar levels of biofilm-specific resistance to disinfection with peroxide, QAC, and chloride during the course of the experiment. This indicates that a broad-spectrum mechanism, probably related to the biofilm mode of growth, was responsible for the increased tolerance seen in both studies. This increase may also potentially be linked to attributes of the coupon surface on which the biofilms were grown. When biofilms were examined using CLSM in this study, larger L. monocytogenes biofilm aggregates were always seen confined to the clefts and surface structures on the underside of the conveyor belt material, while the multigenus biofilms were regularly observed to protrude outwards from the crevices in which they were rooted. Conceivably, spatial growth patterns and/or a relatively modest growth rate could account for L. monocytogenes biofilms not extending beyond the shelter of the crevices during the 24 h separating the two cycles of C&D, thereby resulting in the observed lower reduction in L. monocytogenes numbers upon C&D on day 7 than on day 4 (Fig. 5).
In summary, the results from this study showed that L. monocytogenes can grow and survive in multigenus biofilms formed from bacteria belonging to the background microbiota isolated in meat industry environments, even after several rounds of C&D. Furthermore, the results suggest that regular C&D agents used in the food industry fail to remove biofilms from heterogeneous surfaces harboring cracks or crevices. Although the underside of a conveyor belt is not intended to be in direct contact with food, it may confer harborage sites from which bacteria can shelter and cross-contaminate food-contact surfaces during processing. Further research into more efficient methods for the removal of biofilms and a greater focus on hygienic design of food processing equipment are warranted.
MATERIALS AND METHODS
Isolation of bacteria from conveyor belts in meat processing plants.
Two plants processing RTE meats were visited. Samples from a total of nine conveyors, including six from plant A and three from plant B, were taken after C&D, before the start of production. The daily sanitation included a chloralkali agent for cleaning followed by disinfections using QAC in plant A and PAA in plant B. In addition, the conveyors in plant B were disinfected with 70% ethanol several times during the production day, between the processing of different products and before breaks. An area of approximately 900 cm2 was sampled with neutralizing sampling cloths (Sodibox, Nevez, France). The cloths were stored at 4°C and analyzed within 36 h. Ten milliliters of peptone water (1 g · l−1 peptone [Oxoid], 0.85% NaCl, pH 7.2) was added to the plastic bag containing the cloth, and after a 30-s treatment in a Stomacher, 1-ml samples were plated to blood agar directly and after dilution to obtain single well-separated colonies for identification. The agar plates were incubated at 20°C for 5 days. Up to 20 colonies were picked at random, restreaked for purification, and subjected to 16S rRNA sequencing (V3 to V4 region) for identification using the universal 16S rRNA primers TCCTACGGGAGGCAGCAGT and GGACTACCAGGGTATCTAATCCTGTT (48), as previously described (37). The taxonomy of each strain was assigned using the SeqMatch tool of the Ribosomal Database Project (RDP), with database v.11.5 (https://rdp.cme.msu.edu).
Selection criteria for background microbiota strains included in biofilm experiments.
A total of 16 strains isolated from conveyor belts in meat processing plants A and B were selected for inclusion in multigenus biofilm experiments (Table 2). Of these, 14 isolates represented the nine most frequently found genera after C&D. All these were among the dominating genera (>20% of the colonies) in at least one sample. More than one isolate was chosen from genera with diversity in the 16S rRNA amplicon sequences. In addition, Kocuria isolates were included, because Kocuria spp. were reported to promote biofilm formation of L. monocytogenes in an earlier study (29). All strains included in the experiments had unique 16S rRNA amplicon sequences, enabling their differentiation by 16S rRNA sequencing.
Selection criteria for L. monocytogenes strains.
Seven L. monocytogenes strains from three different meat processing facilities (plants A, B, and C, corresponding to plants M2, M4, and M1, respectively, from Møretrø et al. [22]) were selected for inclusion in experiments in this study (Table 2). These strains had been collected as part of two research projects where nine Norwegian food processing plants were sampled, resulting in the isolation of a total of 680 L. monocytogenes strains subsequently typed using multiple locus variable-number tandem-repeat analysis (MLVA) (22). Of the seven strains used in this study, four were from MLVA profiles that were identified as persistent in the said projects using the statistical approach described by Malley et al. (49) (results to be published separately), and which were detected after C&D in more than one of the nine sampled Norwegian facilities. The four included persistent strains were further selected from individual strains isolated after C&D at sampling points in which the same MLVA genotype had been found on several occasions. Three L. monocytogenes strains were selected from so-called sporadic MLVA profiles. These strains were selected based on the criteria that they should be isolated during production and that their MLVA profiles were not commonly found after C&D. All seven selected strains had different dapE alleles, enabling their differentiation by MLST sequencing (50). Alleles and sequence types for MLST were compared with those available in the Institute Pasteur's L. monocytogenes MLST database (http://bigsdb.web.pasteur.fr/listeria/listeria.html).
Whole-genome sequencing.
DNA isolation, whole-genome sequencing and de novo genome assembly were performed essentially as previously described (38), with 300-bp paired-end sequencing on a MiSeq instrument (Illumina), except that genome assembly was performed with v3.10.0 of SPAdes (51) and six k-mer sizes were included (21, 33, 55, 77, 99, and 127). Contigs with sizes of <500 bp and with coverage of <35 were removed from the assemblies. The sequences were annotated using the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) server (http://www.ncbi.nlm.nih.gov/genome/annotation_prok/).
Sequence search for known biofilm genes.
The Pseudomonas and Acinetobacter genome assemblies from this study were analyzed for the presence of genes known to be involved in biofilm formation in these genera (52–54) using BLAST+ v2.2.30 (55). The following genes were used as queries in the analysis: pslA to pslR (PA2231 to PA2246), pelA to pelG (PA3064 to PA3058), the alg operon (PA3540 to PA3551), and cdrA (PA4625) from P. aeruginosa PAO1 (accession number AE004091); wssA to wssJ from P. fluorescens SBW25 (accession number AY074776); lapA to lapG (PP0168 to PP0164), lapF (PP0806), the bcs operon (PP2629 to PP2638), peaA to peaI (PP3133 to PP3141), and the peb locus (PP1795 to PP1788) from P. putida KT4220 (accession number AE015451); csuA to csuE, encoding the pilus usher-chaperone assembly system from A. baumannii 19606 (accession number AY241696); pgaA to pgaD (A1S_2160 to A1S_2162 and A1S_3792) from A. baumannii ATCC 17978 (accession number CP000521); and the gene encoding Bap from A. baumannii 307-0294 (accession number EU117203). Genome comparisons were performed using Mauve (56).
Phylogenetic analysis.
The sequences of single genes or whole genomes from reference strains used in the phylogenetic analyses were downloaded from GenBank, and their accession numbers are listed in Table S5 in the supplemental material. Acinetobacter strains were typed in silico using the MLST scheme described by Diancourt et al. (57), while Pseudomonas strains were analyzed using the multilocus sequence analysis (MLSA) scheme described by Mulet et al. (58). The concatenated sequences of the seven MLST alleles (for Acinetobacter) or the four MLSA alleles (for Pseudomonas) were aligned using CLC Main Workbench 7 (CLC bio). Phylogenetic trees were then inferred from the alignments in MEGA7 (59) using the neighbor-joining method. The evolutionary distances were computed using the Jukes-Cantor method, and bootstrap confidence values were generated using 1,000 replicates.
C&D agents.
The C&D agents used in this study were selected to represent products with concentrations of active ingredients typical of industrial formulations. The industrial chlorinated alkaline cleaning agent ISS Alkalifoam 27 (Ecolab, Norway), referred to as “Alkalifoam” throughout the text, was used at a 1% concentration, which is the minimum recommended user concentration indicated by the manufacturer. At this concentration, the solution contains a minimum of 0.02% NaOH and 0.03% sodium hypochlorite. Two industrial disinfection agents were used. One was Aco Hygiene Des QA (Aco Kjemi, Norway), which is a formulation based on quaternary ammonium compounds, referred to as “QAC” throughout the text. The second was Diverfoam active (Lilleborg, Norway), which is based on peracetic acid and is referred to as “PAA” throughout the text. Both were used at the indicated minimum user concentrations, which are 1% for QAC and 1.5% for PAA. At these concentrations, the QAC solution contains a minimum of 0.05% benzalkonium chloride, while the PAA solution contains a minimum of 0.02% peracetic acid, 0.05% acetic acid, and 0.15% hydrogen peroxide.
Growth conditions in planktonic culture.
Bacteria were grown in BHI broth (Oxoid) throughout all experiments. Overnight cultures and precultures were grown in 5-ml volumes in culture tubes and 50-ml Nunc tubes, respectively, with shaking at 30°C, except for Sphingomonas sp. MF4632, which was grown at 20°C. All biofilm and growth experiments were carried out at 12°C. For plating, RAPID'L.mono (RLM) agar (Bio-Rad) and BHI agar (Oxoid) plates were used.
For the generation of growth curves for single strains, overnight cultures were diluted to approximately 105 CFU · ml−1 and inoculated in volumes of 250 μl in 100-well polystyrene microwell plates (Oy Growth Curves Ab, Ltd.). The plates were incubated for 7 days at 12°C in a Bioscreen C instrument (MTX Lab Systems, Inc.), with continuous shaking and recording of OD600 every hour. Blank wells contained BHI broth only, and values for blanks were subtracted from sample values to obtain actual absorbance measurements. Triplicate wells were used for each sample, and each strain was tested three or four times.
For the planktonic competition experiment, overnight cultures were mixed in roughly equal CFU numbers in an inoculum diluted to a final total concentration of 105 CFU · ml−1. Fifty-milliliter culture volumes were incubated in 500-ml baffled Erlenmeyer bottles at 12°C with shaking at 200 rpm. Every 24 h, samples were withdrawn and plated to determine the CFU count, and cells were pelleted by centrifugation and stored at −20°C for use in amplicon sequencing analysis (see below).
Construction of GFP-labeled L. monocytogenes.
L. monocytogenes strains were transformed with plasmid pNF8, from which the GFP is constitutively expressed (60). The pNF8 plasmid was a kind gift from Hanne Ingmer at the University of Copenhagen. Transformation was performed using the procedure described by Monk et al. (61). Erythromycin at a concentration of 10 μg · ml−1 was used for the selection of cells harboring pNF8. The identity of all strains after transformation was confirmed by PCR amplification and sequencing of the dapE MLST allele (50) using primers GTTTTCCCAGTCACGACGTTGTACGACTAATGGGCATGAAGAACAAG and TTGTGAGCGGATAACAATTTCATCGAACTATGGGCATTTTTACC for PCR (overhangs underlined) and primers GTTTTCCCAGTCACGACGTTGTA and TTGTGAGCGGATAACAATTTC for sequencing.
Biofilm experiments with C&D.
Precultures of each strain were inoculated from glycerol stocks prepared from exponential-phase cultures and were maintained at −80°C, grown separately to logarithmic phase, and mixed in roughly equal CFU numbers in an inoculum diluted to a final total concentration of ∼106 CFU · ml−1. The bacterial suspensions were inoculated in 24-well plates containing coupons of food-grade PVC conveyor belt material (E 8/2 U0/V5 MT white FDA; Forbo-Siegling Transilon) cut to 1.0 cm by 1.5 cm, autoclaved, and placed vertically in each well. One-milliliter of inoculum was added to each well so that wells were half-filled with culture broth, resulting in the air/liquid interface crossing the length of the coupon (see Fig. 6D). The plates were incubated at 12°C with gentle orbital shaking, and the culture medium was refreshed on day 3.
Control coupons not subjected to C&D were harvested after 4 days of biofilm development. Sets of coupons subjected to C&D (see below) on day 4 were either harvested after treatment or, for coupons to be harvested on day 7, placed in a new 24-well tray containing 1 ml BHI in each well and were incubated as before for 24 h. The cycles of C&D followed by incubation in BHI were repeated on days 5 and 6. On day 7, sets of coupons treated with either QAC or PAA on days 4 to 6 were harvested prior to and after C&D treatment. Coupons sampled prior to C&D (both on days 4 and 7) were rinsed three times in ∼10 ml H2O (in 15-ml Falcon tubes) to remove nonadherent bacteria before harvest. Control coupons subjected to rinsing in H2O instead of treatment with C&D agents on days 4 to 7 were included in selected experiments.
Treatment with C&D agents was performed as follows. C&D agents were applied as foam (as intended by the manufacturers) produced in foam pump bottles (Sunvita, Norway). Each coupon was rinsed three times in ∼10 ml H2O (in 15-ml Falcon tubes) and placed vertically in wells of a clean 24-well tray. The wells were filled with 1% Alkalifoam and coupons were incubated 5 min, rinsed as before in H2O, and placed in a second clean 24-well plate. The wells were then filled with 1% QAC or 1.5% PAA, and the coupons were incubated 5 min and finally rinsed as before in H2O. The average weight of foam applied to each well was ∼350 mg.
Cells attached to coupons were harvested as follows. Each coupon was transferred to a glass tube containing 4.5 ml peptone water and 2 g glass beads of ∼2-mm diameter (no. 1401/2; Assistent). Tubes were then vortexed for 30 s and sonicated for 10 min (Branson 3510 ultrasonic cleaner) to dislodge attached cells and disperse cell aggregates. After withdrawing 45.5 μl or 500 μl for plating dilutions on agar plates (to determine total and L. monocytogenes CFU count per coupon), the remaining cells were pelleted by centrifugation (16,000 × g for 5 min) and stored at −20°C. The identity of single L. monocytogenes colonies from dilutions plated after harvesting coupons subjected to sanitation on day 7 was determined by PCR amplification and sequencing of the dapE MLST allele (50) as described above.
Biofilms analyzed using CLSM were grown and subjected to rinsing or C&D as described above, with the following exceptions. For L. monocytogenes, the strains labeled with GFP were used, and overnight cultures for these were grown in the presence of 10 μg · ml−1 erythromycin. The biofilm inoculum was prepared from overnight cultures diluted to an OD600 of 0.01. These were mixed so that the inoculum contained 12.5% (vol/vol) of each of the four background microbiota strains, Acinetobacter MF4640 and Pseudomonas strains MF4836, MF6394, and MF6396, and 50% of a mixture of equal amounts of the seven L. monocytogenes strains (Table 2). Biofilms were grown under static conditions. The rinsing of coupons in H2O before and after treatment with C&D agents was performed three times in 2.5-ml volumes of H2O in 24-well plates. After coupons were either subjected to C&D or rinsed in H2O (to remove nonadherent bacteria from control coupons), coupons were left in BHI until imaging the same day.
DNA isolation and amplicon sequencing.
For the purification of genomic DNA for amplicon sequencing analyses, cells were lysed using Lysing Matrix B and a FastPrep-24 instrument (both MP Biomedicals), and DNA was isolated using either the PowerSoil-htp 96-well soil DNA isolation kit (Mo Bio) (biofilm experiment no. 1 to 3) or the DNeasy blood and tissue kit (Qiagen) (biofilm experiment no. 4 and 5 and planktonic competition experiments). Libraries for amplicon sequencing to analyze microbial compositions were prepared following the 16S Metagenomic sequencing library preparation protocol from Illumina (62). Briefly, amplicon PCR was performed with primers targeting either the V3 to V4 region of the 16S rRNA gene or the dapE gene of L. monocytogenes, followed by an index PCR performed using the Nextera XT index kit (Illumina). The primers used to amplify the 16S rRNA gene were TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG and GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC, and those used to amplify dapE were TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCGACTAATGGGCATGAAGAACAAG and GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCATCGAACTATGGGCATTTTTACC (overhangs underlined; N = A, C, G, or T; W = A or T; H = A, C, or T; V = A, C, or G). PCR products were purified using the AMPure XP system (Agencourt) after each PCR and after pooling. Purified indexed PCR products and the pooled samples were quantified using the Quant-iT PicoGreen double-stranded DNA (dsDNA) kit (Invitrogen). The library was spiked with 10% PhiX control and sequenced using MiSeq v3 reagents using paired 300-bp reads on a MiSeq instrument (Illumina).
Metagenomic analysis using QIIME software.
Demultiplexed raw reads from the MiSeq run were processed with the QIIME (Quantitative Insights Into Microbial Ecology) software package v1.9.1 (63). After paired-end reads were joined, they were quality filtered on q20. Then, samples amplified with dapE primers were assigned to their respective dapE allele using a closed reference operational taxonomic unit (OTU) picking protocol against a custom reference file containing the dapE allele sequences of the seven L. monocytogenes strains (dapE allele numbers 4, 6, 7, 8, 9, 18, and 21 as listed at the Institute Pasteur's L. monocytogenes MLST database). The OTU picking script was run with default parameters except that the sequence similarity threshold was set to 1. For samples amplified with 16S rRNA primers, samples were analyzed using an open reference OTU picking protocol, in which reads were first matched against a custom reference file containing the 16S rRNA allele sequences of L. monocytogenes plus those of the 16 background flora strains included in the experiments. The 16S rRNA reference file is included as Table S6 in the supplemental material.
Bactericidal suspension test.
Overnight cultures were diluted to approximately 108 CFU · ml−1 in peptone water, and 1 ml of the diluted culture was added directly to 9 ml of H2O (control) or user concentrations of Alkalifoam (1%), QAC (1%), or PAA (1.5%), resulting in a final cell concentration of approximately 107 CFU · ml−1. After 5 min, 0.5 ml of the solution was transferred to Dey-Engley (D/E) neutralizing broth (Difco) and dilutions were plated on BHI agar plates. The tests were performed with all solutions at 12°C. The experiment was performed three to four times for each strain.
Confocal laser scanning microscopy.
Surface-associated bacteria on conveyor belt coupons were stained with the cell-permeant Syto 61 red fluorescent nucleic acid strain (Life Technologies), diluted to 5 mM in dimethyl sulfoxide (DMSO) and used at a 1:2,000 dilution. L. monocytogenes was pinpointed in the complex biofilm through specific emission of GFP expression. Images were acquired using a Leica SP8 confocal laser scanning microscope (Leica Microsystems) and the MIMA2 microscopy platform. Images were obtained using an HC PL APO 63× long-distance water objective with a numerical aperture of 1.2. The emitted GFP fluorescence signal from L. monocytogenes cells was collected on a hybrid detector in the range 500 to 550 nm after excitation at 488 nm with an Argon laser set at 20% of its maximal intensity. The red fluorescence emitted by the bacteria labeled with Syto 61 was collected on a photomultiplier in the range 645 to 675 nm after excitation with a 633-nm-wavelength HeNe laser. To contrast the surface topography, the reflected signal from the 633-nm-wavelength HeNe laser was collected. Samples were scanned at 600 Hz every micron with 246 μm by 246 μm images to acquire multicolor three-dimensional (3D) stacks.
Representative CLSM images from each coupon were acquired by scanning Z-stacks at different locations of the sample with a fixed pattern on the coupon surface. In most cases, three or five stacks were taken from both the top face and underside of the conveyor belt coupon at even intervals following a line from the bottom to the top of the coupon, as marked with crosses in the photograph of the coupon in Fig. 6D. The numbers of obtained Z-stack CLSM scans for the different samples and treatments are summarized in Table S4. On a few coupons, a larger view of the coupon was obtained thanks to a mosaic 3D meta-image, obtained using a motorized stage that automatically moves the sample between scans and tiles the adjacent fields. All CLSM stacks were processed using IMARIS (Bitplane) to produce projection images of the biofilms. Quantitative analysis of each Z-stack to quantify the green biovolumes corresponding to the L. monocytogenes subpopulations of the biofilms was performed using the ICY image analysis software (64) using a homemade script previously described, with some adaptations (65). The biomass software algorithm did not reliably score low cell numbers correctly; therefore, the biovolume of scans where ≤5 cells were observed in the green channel of the image projection was set to 0, when the scored biovolume was above a threshold of 800 μm3. The biovolume of the red channel was not extracted, since red fluorescent background interfered with the Syto 61-specific signal.
Accession number(s).
Data from this whole-genome shotgun project have been deposited at DDBJ/ENA/GenBank under the accession numbers MVOJ00000000, MVOK00000000, MVOL00000000, MVOM00000000, and MVON00000000. The versions described in this paper are versions XXXX01000000. The raw reads are available from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession numbers SRR5273317, SRR5273318, SRR5273319, SRR5273320, and SRR5273321.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bjørn C. T. Schirmer for isolation of microbiota strains from meat processing plants and Merete Rusås Jensen, Anette Wold Åsli, Tove Maugesten, Signe M. Drømtorp, and Julien Deschamps for excellent technical assistance.
This work was supported by Norwegian Research Funding for Agriculture and Food Industry (grant no. 234355). The collection of strains used in this work was funded by Norwegian Research Funding for Agriculture and Food Industry (grant no. 207765) and The Norwegian Seafood Research Fund (grant no. FHF-900521).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01046-17.
REFERENCES
- 1.Maillard JY. 2002. Bacterial target sites for biocide action. J Appl Microbiol 92(Suppl):1–21. [PubMed] [Google Scholar]
- 2.Coughlan LM, Cotter PD, Hill C, Alvarez-Ordóñez A. 2016. New weapons to fight old enemies: novel strategies for the (bio)control of bacterial biofilms in the food industry. Front Microbiol 7:1641. doi: 10.3389/fmicb.2016.01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Azizi M, Farag N, Khardori N. 2016. Efficacy of selected biocides in the decontamination of common nosocomial bacterial pathogens in biofilm and planktonic forms. Comp Immunol Microbiol Infect Dis 47:60–71. doi: 10.1016/j.cimid.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Bridier A, Briandet R, Thomas V, Dubois-Brissonnet F. 2011. Resistance of bacterial biofilms to disinfectants: a review. Biofouling 27:1017–1032. doi: 10.1080/08927014.2011.626899. [DOI] [PubMed] [Google Scholar]
- 5.Brightwell G, Boerema J, Mills J, Mowat E, Pulford D. 2006. Identifying the bacterial community on the surface of Intralox belting in a meat boning room by culture-dependent and culture-independent 16S rDNA sequence analysis. Int J Food Microbiol 109:47–53. doi: 10.1016/j.ijfoodmicro.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Møretrø T, Langsrud S, Heir E. 2013. Bacteria on meat abattoir meat production process surfaces after sanitation: characterisation of survival properties of Listeria monocytogenes and the commensal bacterial flora. Adv Microbiol 3:255–264. doi: 10.4236/aim.2013.33037. [DOI] [Google Scholar]
- 7.Mettler E, Carpentier B. 1998. Variations over time of microbial load and physicochemical properties of floor materials after cleaning in food industry premises. J Food Prot 61:57–65. doi: 10.4315/0362-028X-61.1.57. [DOI] [PubMed] [Google Scholar]
- 8.Stephan R, Althaus D, Kiefer S, Lehner A, Hatz C, Schmutz C, Jost M, Gerber N, Baumgartner A, Hächler H, Mäusezahl-Feuz M. 2015. Foodborne transmission of Listeria monocytogenes via ready-to-eat salad: a nationwide outbreak in Switzerland, 2013-2014. Food Control 57:14–17. doi: 10.1016/j.foodcont.2015.03.034. [DOI] [Google Scholar]
- 9.Miettinen MK, Björkroth KJ, Korkeala HJ. 1999. Characterization of Listeria monocytogenes from an ice cream plant by serotyping and pulsed-field gel electrophoresis. Int J Food Microbiol 46:187–192. doi: 10.1016/S0168-1605(98)00185-8. [DOI] [PubMed] [Google Scholar]
- 10.Ferreira V, Wiedmann M, Teixeira P, Stasiewicz MJ. 2014. Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J Food Prot 77:150–170. doi: 10.4315/0362-028X.JFP-13-150. [DOI] [PubMed] [Google Scholar]
- 11.Larsen MH, Dalmasso M, Ingmer H, Langsrud S, Malakauskas M, Mader A, Møretrø T, Mozina SS, Rychli K, Wagner M, Wallace RJ, Zentek J, Jordan K. 2014. Persistence of foodborne pathogens and their control in primary and secondary food production chains. Food Control 44:92–109. doi: 10.1016/j.foodcont.2014.03.039. [DOI] [Google Scholar]
- 12.Morganti M, Scaltriti E, Cozzolino P, Bolzoni L, Casadei G, Pierantoni M, Foni E, Pongolini S. 2015. Processing-dependent and clonal contamination patterns of Listeria monocytogenes in the cured ham food chain revealed by genetic analysis. Appl Environ Microbiol 82:822–831. doi: 10.1128/AEM.03103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stasiewicz MJ, Oliver HF, Wiedmann M, den Bakker HC. 2015. Whole-genome sequencing allows for improved identification of persistent Listeria monocytogenes in food-associated environments. Appl Environ Microbiol 81:6024–6037. doi: 10.1128/AEM.01049-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carpentier B, Cerf O. 2011. Review–persistence of Listeria monocytogenes in food industry equipment and premises. Int J Food Microbiol 145:1–8. doi: 10.1016/j.ijfoodmicro.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Lundén JM, Miettinen MK, Autio TJ, Korkeala HJ. 2000. Persistent Listeria monocytogenes strains show enhanced adherence to food contact surface after short contact times. J Food Prot 63:1204–1207. doi: 10.4315/0362-028X-63.9.1204. [DOI] [PubMed] [Google Scholar]
- 16.Martínez-Suárez JV, Ortiz S, López-Alonso V. 2016. Potential impact of the resistance to quaternary ammonium disinfectants on the persistence of Listeria monocytogenes in food processing environments. Front Microbiol 7:638. doi: 10.3389/fmicb.2016.00638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bridier A, Sanchez-Vizuete P, Guilbaud M, Piard JC, Naitali M, Briandet R. 2015. Biofilm-associated persistence of food-borne pathogens. Food Microbiol 45:167–178. doi: 10.1016/j.fm.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Ortiz S, López-Alonso V, Rodríguez P, Martínez-Suárez JV. 2015. The connection between persistent, disinfectant-resistant Listeria monocytogenes strains from two geographically separate Iberian pork processing plants: evidence from comparative genome analysis. Appl Environ Microbiol 82:308–317. doi: 10.1128/AEM.02824-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borucki MK, Peppin JD, White D, Loge F, Call DR. 2003. Variation in biofilm formation among strains of Listeria monocytogenes. Appl Environ Microbiol 69:7336–7342. doi: 10.1128/AEM.69.12.7336-7342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadam SR, den Besten HM, van der Veen S, Zwietering MH, Moezelaar R, Abee T. 2013. Diversity assessment of Listeria monocytogenes biofilm formation: impact of growth condition, serotype and strain origin. Int J Food Microbiol 165:259–264. doi: 10.1016/j.ijfoodmicro.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 21.Poimenidou SV, Chrysadakou M, Tzakoniati A, Bikouli VC, Nychas GJ, Skandamis PN. 2016. Variability of Listeria monocytogenes strains in biofilm formation on stainless steel and polystyrene materials and resistance to peracetic acid and quaternary ammonium compounds. Int J Food Microbiol 237:164–171. doi: 10.1016/j.ijfoodmicro.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 22.Møretrø T, Schirmer BCT, Heir E, Fagerlund A, Hjemli P, Langsrud S. 2017. Tolerance to quaternary ammonium compound disinfectants may enhance growth of Listeria monocytogenes in the food industry. Int J Food Microbiol 241:215–224. doi: 10.1016/j.ijfoodmicro.2016.10.025. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez-Vizuete P, Orgaz B, Aymerich S, Le Coq D, Briandet R. 2015. Pathogens protection against the action of disinfectants in multispecies biofilms. Front Microbiol 6:705. doi: 10.3389/fmicb.2015.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giaouris E, Heir E, Hébraud M, Chorianopoulos N, Langsrud S, Møretrø T, Habimana O, Desvaux M, Renier S, Nychas GJ. 2014. Attachment and biofilm formation by foodborne bacteria in meat processing environments: causes, implications, role of bacterial interactions and control by alternative novel methods. Meat Sci 97:298–309. doi: 10.1016/j.meatsci.2013.05.023. [DOI] [PubMed] [Google Scholar]
- 25.Røder HL, Raghupathi PK, Herschend J, Brejnrod A, Knøchel S, Sørensen SJ, Burmølle M. 2015. Interspecies interactions result in enhanced biofilm formation by co-cultures of bacteria isolated from a food processing environment. Food Microbiol 51:18–24. doi: 10.1016/j.fm.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Burmølle M, Webb JS, Rao D, Hansen LH, Sørensen SJ, Kjelleberg S. 2006. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol 72:3916–3923. doi: 10.1128/AEM.03022-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert P, Maira-Litran T, McBain AJ, Rickard AH, Whyte FW. 2002. The physiology and collective recalcitrance of microbial biofilm communities. Adv Microb Physiol 46:202–256. [PubMed] [Google Scholar]
- 28.van der Veen S, Abee T. 2011. Mixed species biofilms of Listeria monocytogenes and Lactobacillus plantarum show enhanced resistance to benzalkonium chloride and peracetic acid. Int J Food Microbiol 144:421–431. doi: 10.1016/j.ijfoodmicro.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 29.Carpentier B, Chassaing D. 2004. Interactions in biofilms between Listeria monocytogenes and resident microorganisms from food industry premises. Int J Food Microbiol 97:111–122. doi: 10.1016/j.ijfoodmicro.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 30.Møretrø T, Sonerud T, Mangelrød E, Langsrud S. 2006. Evaluation of the antimicrobial effect of a triclosan-containing industrial floor used in the food industry. J Food Prot 69:627–633. doi: 10.4315/0362-028X-69.3.627. [DOI] [PubMed] [Google Scholar]
- 31.Hood S, Zottola E. 1997. Isolation and identification of adherent gram-negative microorganisms from four meat-processing facilities. J Food Prot 60:1135–1138. doi: 10.4315/0362-028X-60.9.1135. [DOI] [PubMed] [Google Scholar]
- 32.Stellato G, La Storia A, De Filippis F, Borriello G, Villani F, Ercolini D. 2016. Overlap of spoilage-associated microbiota between meat and the meat processing environment in small-scale and large-scale retail distributions. Appl Environ Microbiol 82:4045–4054. doi: 10.1128/AEM.00793-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marouani-Gadri N, Augier G, Carpentier B. 2009. Characterization of bacterial strains isolated from a beef-processing plant following cleaning and disinfection—influence of isolated strains on biofilm formation by Sakai and EDL 933 E. coli O157:H7. Int J Food Microbiol 133:62–67. doi: 10.1016/j.ijfoodmicro.2009.04.028. [DOI] [PubMed] [Google Scholar]
- 34.Hultman J, Rahkila R, Ali J, Rousu J, Björkroth KJ. 2015. Meat processing plant microbiome and contamination patterns of cold-tolerant bacteria causing food safety and spoilage risks in the manufacture of vacuum-packaged cooked sausages. Appl Environ Microbiol 81:7088–7097. doi: 10.1128/AEM.02228-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhargava N, Sharma P, Capalash N. 2014. Pyocyanin stimulates quorum sensing-mediated tolerance to oxidative stress and increases persister cell populations in Acinetobacter baumannii. Infect Immun 82:3417–3425. doi: 10.1128/IAI.01600-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen SK, Haagensen JA, Gjermansen M, Jørgensen TM, Tolker-Nielsen T, Molin S. 2007. Characterization of a Pseudomonas putida rough variant evolved in a mixed-species biofilm with Acinetobacter sp. strain C6. J Bacteriol 189:4932–4943. doi: 10.1128/JB.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langsrud S, Moen B, Møretrø T, Løype M, Heir E. 2016. Microbial dynamics in mixed culture biofilms of bacteria surviving sanitation of conveyor belts in salmon-processing plants. J Appl Microbiol 120:366–378. doi: 10.1111/jam.13013. [DOI] [PubMed] [Google Scholar]
- 38.Fagerlund A, Langsrud S, Schirmer BC, Møretrø T, Heir E. 2016. Genome analysis of Listeria monocytogenes sequence type 8 strains persisting in salmon and poultry processing environments and comparison with related strains. PLoS One 11:e0151117. doi: 10.1371/journal.pone.0151117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elias S, Banin E. 2012. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev 36:990–1004. doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- 40.Liu W, Røder HL, Madsen JS, Bjarnsholt T, Sørensen SJ, Burmølle M. 2016. Interspecific bacterial interactions are reflected in multispecies biofilm spatial organization. Front Microbiol 7:1366. doi: 10.3389/fmicb.2016.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodríguez-López P, Saá-Ibusquiza P, Mosquera-Fernández M, López-Cabo M. 2015. Listeria monocytogenes-carrying consortia in food industry. Composition, subtyping and numerical characterisation of mono-species biofilm dynamics on stainless steel. Int J Food Microbiol 206:84–95. doi: 10.1016/j.ijfoodmicro.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Overney A, Jacques-André-Coquin J, Ng P, Carpentier B, Guillier L, Firmesse O. 2017. Impact of environmental factors on the culturability and viability of Listeria monocytogenes under conditions encountered in food processing plants. Int J Food Microbiol 244:74–81. doi: 10.1016/j.ijfoodmicro.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Pan Y, Breidt F Jr, Kathariou S. 2006. Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Appl Environ Microbiol 72:7711–7717. doi: 10.1128/AEM.01065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saá Ibusquiza P, Herrera JJ, Cabo ML. 2011. Resistance to benzalkonium chloride, peracetic acid and nisin during formation of mature biofilms by Listeria monocytogenes. Food Microbiol 28:418–425. doi: 10.1016/j.fm.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 45.Müller A, Rychli K, Zaiser A, Wieser C, Wagner M, Schmitz-Esser S. 2014. The Listeria monocytogenes transposon Tn6188 provides increased tolerance to various quaternary ammonium compounds and ethidium bromide. FEMS Microbiol Lett 361:166–173. doi: 10.1111/1574-6968.12626. [DOI] [PubMed] [Google Scholar]
- 46.Tolvanén R, Lunden J, Korkeala H, Wirtanen G. 2007. Ultrasonic cleaning of conveyor belt materials using Listeria monocytogenes as a model organism. J Food Prot 70:758–761. doi: 10.4315/0362-028X-70.3.758. [DOI] [PubMed] [Google Scholar]
- 47.Doijad SP, Barbuddhe SB, Garg S, Poharkar KV, Kalorey DR, Kurkure NV, Rawool DB, Chakraborty T. 2015. Biofilm-forming abilities of Listeria monocytogenes serotypes isolated from different sources. PLoS One 10:e0137046. doi: 10.1371/journal.pone.0137046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nadkarni MA, Martin FE, Jacques NA, Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 49.Malley TJ, Stasiewicz MJ, Gröhn YT, Roof S, Warchocki S, Nightingale K, Wiedmann M. 2013. Implementation of statistical tools to support identification and management of persistent Listeria monocytogenes contamination in smoked fish processing plants. J Food Prot 76:796–811. doi: 10.4315/0362-028X.JFP-12-236. [DOI] [PubMed] [Google Scholar]
- 50.Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog 4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mann EE, Wozniak DJ. 2012. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev 36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nilsson M, Chiang WC, Fazli M, Gjermansen M, Givskov M, Tolker-Nielsen T. 2011. Influence of putative exopolysaccharide genes on Pseudomonas putida KT2440 biofilm stability. Environ Microbiol 13:1357–1369. doi: 10.1111/j.1462-2920.2011.02447.x. [DOI] [PubMed] [Google Scholar]
- 54.Longo F, Vuotto C, Donelli G. 2014. Biofilm formation in Acinetobacter baumannii. New Microbiol 37:119–127. [PubMed] [Google Scholar]
- 55.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mulet M, Lalucat J, García-Valdés E. 2010. DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol 12:1513–1530. doi: 10.1111/j.1462-2920.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- 59.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fortineau N, Trieu-Cuot P, Gaillot O, Pellegrini E, Berche P, Gaillard JL. 2000. Optimization of green fluorescent protein expression vectors for in vitro and in vivo detection of Listeria monocytogenes. Res Microbiol 151:353–360. doi: 10.1016/S0923-2508(00)00158-3. [DOI] [PubMed] [Google Scholar]
- 61.Monk IR, Gahan CG, Hill C. 2008. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl Environ Microbiol 74:3921–3934. doi: 10.1128/AEM.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Illumina. 2013. 16S metagenomic sequencing library preparation guide. http://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf.
- 63.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Chaumont F, Dallongeville S, Chenouard N, Hervé N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Le Montagner Y, Lagache T, Dufour A, Olivo-Marin JC. 2012. Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods 9:690–696. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- 65.Sanchez-Vizuete P, Le Coq D, Bridier A, Herry JM, Aymerich S, Briandet R. 2015. Identification of ypqP as a new Bacillus subtilis biofilm determinant that mediates the protection of Staphylococcus aureus against antimicrobial agents in mixed-species communities. Appl Environ Microbiol 81:109–118. doi: 10.1128/AEM.02473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lindstedt BA, Tham W, Danielsson-Tham ML, Vardund T, Helmersson S, Kapperud G. 2008. Multiple-locus variable-number tandem-repeats analysis of Listeria monocytogenes using multicolour capillary electrophoresis and comparison with pulsed-field gel electrophoresis typing. J Microbiol Methods 72:141–148. doi: 10.1016/j.mimet.2007.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.