Abstract
Systems biology and systems medicine have played an important role in the last two decades in shaping our understanding of biological processes. While systems biology is synonymous with network maps and ‘-omics’ approaches, it is not often associated with mechanical processes. Here, we make the case for considering the mechanical and geometrical aspects of biological membranes as a key step in pushing the frontiers of systems biology of cellular membranes forward. We begin by introducing the basic components of cellular membranes, and highlight their dynamical aspects. We then survey the functions of the plasma membrane and the endomembrane system in signaling, and discuss the role and origin of membrane curvature in these diverse cellular processes. We further give an overview of the experimental and modeling approaches to study membrane phenomena. We close with a perspective on the converging futures of systems biology and membrane biophysics, invoking the need to include physical variables such as location and geometry in the study of cellular membranes.
1 Introduction
Systems biology, an emerging interdisciplinary field aimed at understanding biological systems holistically, is often associated with network maps of protein-protein interactions, proteomic, genomic, and bioinformatics approaches [1, 2, 3]. A quick Google search on systems biology leads us to the NIH definition, which includes contributions in computational modeling, proteomics, bioinformatics, genomics, and even immunology. These efforts have been used successfully to identify potential drug targets and genes related to various diseases [4, 5, 6].
One common feature of the network and gene maps and their interactions is that they occur in compartmentalized regions within the cell that are separated by a lipid bilayer. Systems biology approaches have made significant contributions to the different aspects of cellular membrane function. Lipidomics, the mapping of cellular lipids, heralded the era of ‘-omics’ into membrane biology [7, 8, 9, 10, 11]. Using methods such as liquid chromatography [12, 13, 14] and mass spectrometry [15, 16, 17], researchers have been able to extract lipids from distinct cellular compartments, and characterize and classify them into their specific molecular species. While each extraction and characterization methods has different strengths and weaknesses [18], these approaches announced the era of quantitative knowledge of lipids, lipid metabolites, and their role in the metabolome. Coupled with studies that highlighted the role of lipid signaling in health and disease [19] and signaling processes that regulate cellular membranes [20, 21, 22], it has become increasingly evident that systems biology of cellular membranes has come of age. This progression has also led to increasingly complex dynamical models that include compartments to represent the plasma membrane [23, 24, 25, 26, 27], calcium fluxes through the ER membrane [28, 29], and signaling at the Golgi membrane as well [30, 31, 32].
What then is the future of systems approaches for the study of cellular membranes? We propose that the next era of systems approaches in cellular membranes stems from combining the existing knowledge base of lipid diversity and function with spatial and mechanical aspects of cellular membranes, which impact diverse membrane processes from the assembly of trafficking vesicles to the function of organelles and cell invasion by pathogens. In this review, we make the case for an important role played by lipid bilayer mechanics in systems biology. Given the rather broad nature of this undertaking, we present a few important examples of membrane regulation of cellular processes and discuss experimental and modeling methods to study these processes. We propose that membrane curvature as a governing principle of biological function and suggest that the mechanics of lipid membranes cannot be neglected. We close with a perspective on the converging futures of systems biology and membrane biophysics, invoking the need to include physical variables such as location and geometry in the study of cellular membranes.
2 Lipid bilayers and cellular function
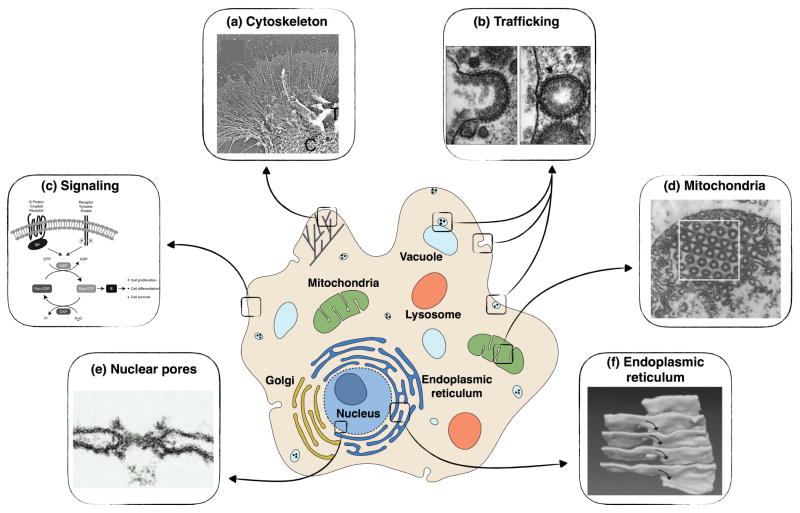
The function of cellular membranes is tied closely to their geometry. Membranes are important not only for protecting the cytosolic components but also for organelle function and cytoskeletal remodeling. (a) SEM image of lamellipodia and filopodia in Aplysia growth cone (adapted from [33]); (b) Micrographs of clathrin-mediated endocytosis at different stages of budding (adapted from [34]); (c) Membrane-bound receptors, G-protein coupled receptors, and receptor tyrosine kinases cause Ras GTPase activation at the plasma membrane and endomembranes [35]; (d) TEM images of mitochondrial membrane in amoebae Chaos carolinensis show triply periodic minimal structures (adapted from [36]); (e) TEM image of a nuclear membrane shaped as a catenoid by the nuclear pore complex (adapted from [37]); (f) 3D reconstruction from SEM endoplasmic reticulim in an acinar cells of mouse salivary gland, showing stacks of parallel membrane sheets connected by helicoidal ramps (adapted from [38]).
Lipid membranes are ubiquitous in the cellular environment, and not only mark cellular boundaries, but also separate the organelles from the cellular interior and from one another. Although compartmentalization of cellular features is one of the most recognized roles of the cellular membranes, membranes also perform essential biological functions through signaling, interaction with the cytoskeleton, and close regulation of organelle activity by modulating their shapes (Figure 1). Cell membranes also provide substrates for surface reactions in signaling, an important feature that uses the local surface-to-volume ratio to tune the signal strength [39, 40, 41]. Cellular membranes achieve this wide range of function through a heterogeneous lipid and protein composition and their range of functionality is further enhanced by dynamic rearrangements of lipids through trafficking and lipid synthesis [42].
Figure 1.
2.1 Dynamic membrane composition
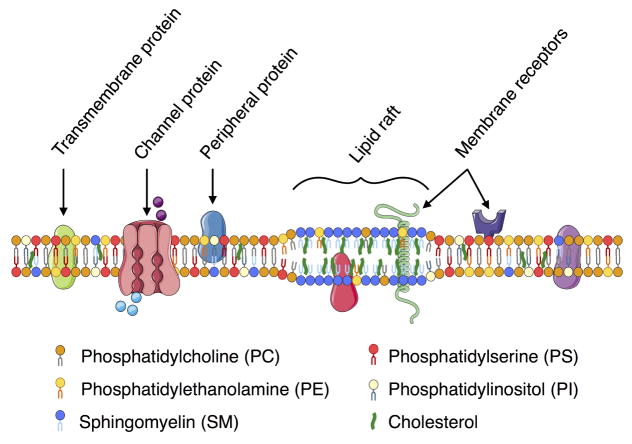
Biological membranes are complex in both lipid composition and protein inclusion, as illustrated by this schematic of a mammalian plasma membrane. Varying membrane composition can lead to a change in membrane thickness and other mechanical properties, which in turn affect their function.
Although about six nanometers thick, cellular membranes are fascinatingly complex in their heterogeneity (Figure 2). Phospholipids are the predominant type of lipid found in cellular membranes. In mammalian cells, lipid membranes are also enriched locally in cholesterol and sphingolipids. Phosphatidylcholine (PC) is one of the key components of cellular membranes, often occupying half of the entire phospholipid content. However, lipid distribution varies across the different organelle membranes, with different endo membranes containing different ratios of phospholipid to cholesterol and small amounts of signaling lipids. Membrane composition not only varies across organelles but also varies across organisms, cell types, differentiation and development stages, and in disease states [43, 44, 45, 46]. A comprehensive review of different membrane lipids, their role, and composition is given in [42], while a constantly updated database on lipids, lipidomics, and the role of lipids in different diseases can be found in the LIPID MAPS project (www.lipidmaps.org). These resources serve to highlight the complexity of cellular membranes, the role of lipids in regulating cellular function in health and disease, and the contributions of systems biology approaches to membrane biology.
Figure 2.
The rich heterogeneous composition of cellular membranes is constantly regulated in a dynamic manner. This dynamic in-plane architecture of the plasma membrane was first discussed in the ‘fluid-mosaic’ model [47], where the proteins were thought to randomly diffuse in a sea of lipids. Subsequently, the ‘lipid-raft’ hypothesis was proposed by Simons et al. [48]. This hypothesis suggested that the lipids organize laterally in the plane of the plasma membrane, and this organization can result in functional microdomains that are enriched in cholesterol and sphingolipids (Figure 2). While this hypothesis is still the subject of considerable debate nearly 20 years since it was first proposed, it has played an important role in our understanding of the dynamics of lipid membranes. We now acknowledge that lipids undergo dynamic rearrangement in both model and cellular membranes [49, 50, 51, 52, 53, 54, 55, 56, 57] and are not passive background components but active participants that promote the activity and functionality of membrane proteins.
The lateral organization of lipids in cellular membranes can be understood in terms of phase separation in synthetic vesicles. Recent analyses have shown that a synthetic vesicle made of multi-component membranes can phase separate into liquid-ordered and liquid-disordered domains in the presence of cholesterol [56]. The dynamics of phase separation depend on temperature and membrane stress [55, 56, 57, 54, 58, 59], and provide a biophysical and thermodynamic interpretation of the raft hypothesis. These studies show that microdomain formation and dissolution is an inherent part of membrane dynamics. Within cells, however, the dynamics of formation and dissipation of heterogeneities in the plasma membrane are driven by several complex inter-related processes such as the compositional variation of the lipids and proteins present in the membrane, curvature stresses that can be induced by membrane heterogeneity, and the out-of-equilibrium nature of the extracellular environment. Whether these heterogeneities form signaling corrals or pinning structures to the cytoskeleton, and what, if any, is their function in cells, is still a matter of debate and investigation.
2.2 Signaling at cellular membranes, geometry, and mechanics
Cellular membranes play a critical role in information transfer through signal transduction, which serves as a cornerstone of biological function [37]. Some examples of signal transduction at the plasma membrane include ligand-receptor binding and the binding of scaffolding molecules and coat proteins to the plasma membrane from the cytoplasm [22, 21, 60, 61, 62], and regulation of ion channels and pumps, many of which are known to be mechanosensitive [63, 64, 65, 66, 67, 68].
Canonical signaling through receptor-ligand pathways has been well studied in vitro and in vivo in many model systems [69, 70, 71, 72, 73]. Recent studies have shown that signal strength and lifetime at the plasma membrane are regulated by receptor levels, availability of receptors, and ligand accessibility [74, 75]. Live cell imaging and data analysis using simultaneous detection of the endocytic machinery and cargo have shown that the dynamics of the clathrin-coated pits depend on the cargo, which in turn regulate the formation and maturation of the endocytic pits [76]. These studies indicate that the time available for receptor clustering at the plasma membrane and for the initiation of productive signaling depends on the interplay between receptor-ligand interaction and the endocytic pathway [76]. For example, in EGFR signaling, clathrin-mediated endocytosis is associated predominantly with receptor recycling and sustained signaling whereas clathrin-independent endocytosis controls receptor degradation and signal extinction [77]. Endocytosis also controls the graded response of signaling intensity to EGF concentration through compartmental regulation [77]. Thus, the dynamic response of a signaling pathway at the plasma membrane is intricately tied to the geometry and mechanics of the cellular membranes.
Network analyses, mathematical modeling, and other systems approaches to signaling have complemented experimental investigations of signal transduction in cells. There have been many modeling efforts focused on developing large interaction networks of proteins for a given process [6, 78]; these approaches have helped identify the key players and their relation with one another in the regulation of cellular phenomena. Recent advances in spatio-temporal dynamics of signaling activity emphasized the crucial role of cell shape [79, 39, 41], showing in particular that local membrane curvature can govern the kinetics of biochemical reactions [80, 39, 81, 41]. These studies highlight for instance the effect of cell geometry on the signaling activity of second messengers like cAMP [40] and of kinases such as MAP kinase [39].
Signaling is not solely limited to the plasma membrane; indeed a large part of cell signaling occurs within the cell at the endomembranes. Using siRNA screens, [82] identified the kinases and phosphatases involved in trafficking [82, 83]. In particular, the Raf-MEK pathway was shown to regulate Sec16, which is an upstream regulator of COPII vesicle biogenesis. Growth factor signaling through endosomes is well known for the EGFR family of receptors [69, 70, 71] and dysregulation of endosomal signaling is implicated in several diseases [84, 85, 86]. siRNA screening and bioinformatics approaches were also used to identify the signaling networks that regulate the Golgi apparatus and revealed that a large proportion of signaling genes, in fact, regulated Golgi architecture [87, 88]. Another siRNA screen showed a previously unknown link between secretory pathways, actin cytoskeleton reorganization, growth-factor signaling, and small G-protein regulation [89]. Intracellular signaling also occurs between organelles; KDEL receptors on the Golgi membrane are activated by traffic chaperones released from the ER. The binding of chaperone molecules to the KDEL receptors on the Golgi membranes leads to the activation of the Src family of kinases [90, 91, 92]. Subsequently, the same group of researchers showed that the activation of Src kinase was through dynamin 2 [93], a protein that is implicated in vesiculation at the Golgi [92]. The dynamin family of proteins plays an important role in membrane fission in mammalian endocytosis [94, 60]. Thus, the spatial orchestration of Src activation through multiple intracellular compartments and its relation to dynamin suggests that signaling at intracellular membranes is closely tied to geometry and mechanical maintenance of the organelle equilibrium.
Interestingly, over the variety of approaches used to identify the signaling networks and the proteins involved in endomembrane signaling, a common thread exists – that is, the signaling proteins seem to closely regulate the secretory pathways and vesicle generation capability, indicating that coupling signaling with geometry and membrane mechanics is the next step toward putting these network maps in their proper biophysical context.
2.3 Membrane curvature generation
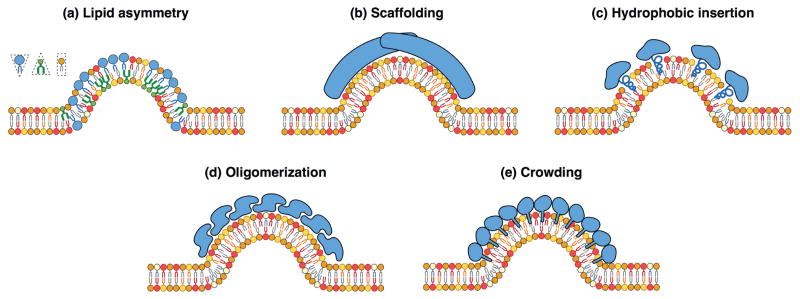
Membrane curvature can be induced by different mechanisms. (a) Due to the tail chemistry, lipids can have cylindrical, conic, or reversed conic shapes, therefore inducing curvature when the lipid composition is different between the two lipid layers. (b) Large intrinsically curved proteins, such as BAR domain family of proteins, can scaffold and bend the membrane. (c) Insertion of amphipathic α -helices in one leaflet induces membrane curvature. (d) Oligomerization of several monomers can scaffold and curve the membrane. (e) High surface concentration of membrane binding proteins produces a steric pressure that can bend the lipid bilayer.
Not only do membranes serve as platforms for cell signaling, but they also are capable of supporting curvature stresses due to their low bending resistance to external load. On the other hand, cellular membranes have high stretching modulus, and membrane area expansion is energetically expensive [95]. Cellular membranes use the curvature generating feature in many different biological functions such as trafficking, fission, fusion, and three-dimensional organization (e.g. caveolae) [96]. The mechanical aspect of membrane organization and conformation has motivated the development of many biophysical models over the years, experimental as well as theoretical, ranging from lipid to continuum scales. Theoretical contributions to the field are reviewed below in Section 4.2.
Many biological processes are regulated by membrane-protein interactions. An example that highlights the importance of protein-induced membrane bending is clathrin-mediated endocytosis, where a multicomponent protein coat forms to cluster cargo and bends the membrane into a budded morphology (see Figure 1b). Clathrin assembles into a lattice-like cage on the plasma membrane with the assistance of adaptor proteins that directly bind lipids, cargo proteins, other adaptors and clathrin itself [97, 98]. This assembly is generally thought to act as a scaffold that imposes its spontaneous curvature on the underlying membrane [99]. Recent work suggests that other components of the coat can also contribute to membrane bending via scaffolding by F-BAR domains, amphipathic helix insertion into the bilayer, and adaptor protein crowding [100, 97, 101, 102, 103]. The contributions from each of these membrane bending mechanisms, summarized in Figure 3, can be combined into a single measure of the curvature generating capability of the coat, or spontaneous curvature, with an effective strength that depends on its composition, density and area coverage [104, 105].
Figure 3.
In addition to bending in response to curvature-inducing proteins, curved membranes can also serve as interfaces that facilitate the binding of curvature-sensing proteins. This relation between protein-curvature generation and curvature-sensing, can be illustrated by the mechanism of binding of antimicrobial peptides (AMPs) to lipid membranes. AMPs are multi-functional molecules involved in the host defense system, which have been shown to generate negative Gaussian curvature in model bacterial membranes [106, 107]. On the other hand, certain AMPs such as TRP3, duramycin, and cinnamycin, bond preferentially to highly curved membranes [108, 109]. These curvature-mediated binding events can be utilized in therapeutic applications, for instance using nanoparticles coated with native RBC membranes that sequester and neutralize protein toxins in the blood stream as an effective drug delivery mechanism [110].
2.4 Cytoskeleton membrane interactions
Cell shape is controlled by the physical properties of the plasma membrane, the biochemical reactions involving membrane components, and the underlying cytoskeleton [111, 112, 113]. In particular, actin has long been known to be intimately associated with membranes. Upon activation by extracellular ligand binding to receptors in the plasma membrane, multiple signaling pathways containing small GTPases regulate the actin network dynamics and thus cell shape [27]. Actin has long been known to be intimately associated with membranes, and two major forms of actin regulation have been linked to the plasma membrane: (1) modulation of the actin monomer pool by phosphoinositides; and (2) modulation of actin assembly factors by membrane-associated small GTPases, membrane-associated proteins, and direct binding of assembly factors to the plasma membrane.
Phosphoinositide (PI) lipids associate with diverse types of actin-binding proteins, and either inhibit or stimulate their activity [114]. The actin nucleation promotion factors WAVE and WASP facilitate actin polymerization via the Arp2/3 complex upon binding PI(4,5)P2 and play important roles in endocytosis [115]. In contrast, actin-capping protein, the F-actin severing protein ADF/Cofilin, and the G-actin binding protein profilin, are all inhibited by binding to PI(4,5)P2 [114] and upstream signaling networks [27]. Membrane-cytoskeletal interactions also regulate critical biological processes such as clathrin mediated endocytosis, where F-BAR proteins recruit and modulate the activity of nucleation-promoting factors for the Arp2/3 complex[116, 117, 118, 119, 120, 121, 122]. These associations regulate WASP activity and myosin contractile activity in yeast and cytoskeleton assembly in endocytic patches [123, 124, 115]. Thus, the role of the cytoskeleton cannot be ignored while studying membrane processes and vice versa.
3 Experimental systems for the study of lipid bilayers
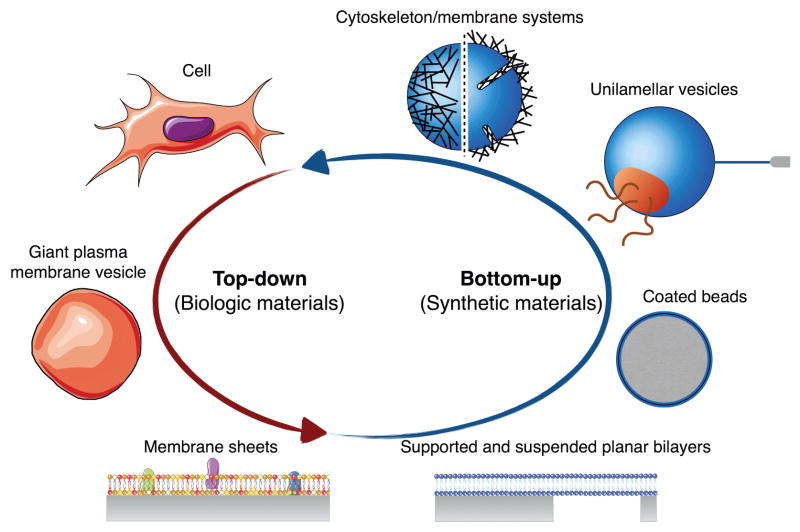
The overwhelming diversity, complexity, and variability of lipid membranes in mammalian cells make the study of their biophysical and biochemical properties particularly challenging. In order to study membranes in relatively simplified conditions, one can either follow a top-down approach, deconstructing the cellular membrane complexity into simpler systems, or adopt a bottom-up approach, building up increasingly complex membrane models (Figure 4). While we are quite far from building artificial cells, the past few decades have been rich in successes in constructing biomimetic model membranes which enable the study of structure, composition, dynamics, and interactions of lipid membranes. Here we review the main membrane systems from a biophysical perspective; reviews more focused on preparation protocols can be found, for instance, in [125], while reviews on lipid monolayer systems are available in [126, 127].
Figure 4.
Model systems for the study of cellular membranes. In the top-down approach, the cellular structural complexity is simplified to some or one of its part by extracting membrane components from the cell. In the bottom-up approach on the other hand, increasingly complex membrane systems are built-up from synthetic materials.
3.1 Top-down approach
Although some membrane chemical and physical properties can be directly studied in living cells [128, 129, 62], an approach that is especially popular in the study of endocytosis from the plasma membrane [130, 131], this approach presents several difficulties. First, there is an inherent lack of control of most biological processes that can influence membrane behavior. Second, acting on the plasma membrane, its inner leaflet, or endomembranes, is not feasible without altering several other cell functions, and therefore introducing additional doubt about the origin of the measured phenomena. Finally, most cellular membranes are highly curved [132], limiting the use of diffusion measurement such as single-particle tracking, which, with the exception of 3D particle tracking microscopes [133] cannot resolve the plasma membrane folds and is based on the assumption of a flat cell surface [134].
These difficulties can be partially overcome by isolating cellular material to facilitate detailed study. Giant plasma membrane vesicles (GPMVs) can be extracted from living cells by inducing chemical blebbing [135, 49]. This system enables the study of the plasma membrane in isolation from the cytoskeleton and organelles. GPMVs have been primary employed to investigate the formation of membranes heterogeneities such as lipid rafts in membranes of cell-like compositional complexity [49, 136, 137]. Interestingly, partitioning between two fluid membrane phases in cell-derived GPMVs has been observed at relatively low temperatures between 15 and 25°C [49, 50], suggesting that at physiological conditions, cell plasma membranes are in the vicinity of a critical point, allowing compositional fluctuations to maintain heterogeneities of approximately 20 nm in size [50, 138].
Alternatively, stable membrane patches originating from cells adhering to a support coated with adhesive proteins can be isolated by detaching the cell from the patch, either by sonication [139] or mechanically disrupting the plasma membrane [140, 141]. This technique provides direct access to the inner leaflet and its associated proteins. Moreover, it gives control on the geometry of membrane sheet, allowing the use of surface sensitive techniques to measure diffusion properties or lipid-protein interactions [142, 143].
3.2 Bottom-up approach to lipid bilayer study
An alternative approach to cell-based investigation is the use of reconstituted membrane systems, where many relevant parameters such as concentration, composition, mechanical, or geometrical properties can be controlled with precision. Such reconstituted systems have been particularly successful in studying the mechanical and dynamic properties of lipid bilayers, membrane protein interactions, and phase separation, as discussed below.
One of the conceptually simplest membrane systems is the supported lipid bilayer (SLB), which consists of a single lipid bilayer spread over an hydrophobic solid support, typically glass, mica or silica [144, 145, 146]. Variations of this configuration include polymer or protein layer between the support and the bilayer [147, 148, 149], porous supports to produce suspended membranes [150], and patterned supports to induce membrane curvature [51]. Supported and suspended lipid bilayers are relatively easy to prepare, and can be used to regulate the chemical and physical environment on both sides of the bilayer. Furthermore, the control on conformation of the membranes greatly facilitates the use of surface based methods to measure and manipulate with high resolution its morphology (thickness, area per lipid, organization), composition (domain, raft, heterogeneities), and dynamics (fluidity, diffusivity) [151, 152, 153]. SLBs are also a system of choice for the study of actin-membrane interactions [154, 155, 156, 157, 158]. In order to get closer to a three-dimensional cell geometry, lipid bilayers can be coated over a solid bead instead of a planar surface [159]. One of the most popular assays of this coated bead system is the supported membrane with excess reservoir (SUPER), which allows the study of protein-induced tubulation and fission of negatively charged lipid mixtures in high-ionic strength conditions [160, 161, 61].
In the absence of a solid support, suspended vesicles of sizes ranging from ten nanometers to micrometers can be produced artificially from defined lipid mixture solutions. Thermodynamically, multilamellar vesicles are more stable that unilamellar vesicles [162], necessitating the addition of mechanical energy to produce unilamellar vesicles. For instance, large unilamellar vesicles (LUVs) – several hundreds nanometers in diameter – can be formed by sonication or extrusion, while further high frequency sonication can break LUVs into small unilamellar vesicles (SUVs) – ten to hundreds nanometers large. SUVs can also be produced by extrusion through filters of the desired pore diameter [163]. Giant unilamellar vesicles (GUVs), which are around ten micrometers in diameter, are most often produced by electroformation [164, 165], although microfluidic techniques have recently emerged, allowing a control on the inner content [166, 167, 168]. Because they have low curvature, and can be micro-manipulated, GUVs offer a convenient platform to probe the physical properties of quasi-flat membranes. Due to their micron size, GUV are particularly well suited for fluorescence microscopy observations, and mechanical micromanipulation techniques. Early mechanical investigations of lipid vesicles were carried with micropipette aspiration techniques by Evans and coworkers[169, 170, 171, 95, 172, 173, 174], allowing the measurement of the viscoelastic properties of membranes of various lipid compositions. The capability to measure or control membrane tension by micropipette aspiration is also useful when combined with other methods. For instance, tube or tether pulling, either with molecular motors [175], micropipiette [176], or an optical trap [177] can be used to study curvature sensitive processes such as lipid sorting or protein binding [178, 179]. However, a drawback of micropipette aspiration experiments is the heterogeneity of applied membrane tension due to contact with the pipette tip. Alternatively, membrane tension can be modulated by varying the osmolarity of the system. In hypotonic conditions (lower solute concentration outside of the vesicle than inside), an osmotic pressure proportional to the concentration differential will be established in the vesicle, homogeneously increasing membrane tension according to Laplace law [180, 181, 59]. Yet, estimations of osmotically induced membrane tension can only be indirectly deduced from the size variation of the GUV, assuming a knowledge of the resting radius of the GUV. On the other hand, decreasing the volume to area ratio, and therefore lowering membrane tension, either by providing a membrane reservoir, applying hypertonic conditions [182], or photo-oxidation [183], produces intriguing shapes driven by a thermodynamic balance between curvature energy and geometrical constraints [184, 185].
Combined with fluorescent probes that have affinities to either liquid ordered or disordered phases [186], LUVs and GUVs are powerful systems to study the phase behavior of lipid mixtures under various thermodynamic conditions [52, 53, 54]. Phase diagrams for several composition of ternary mixtures were determined by Veatch and Keller[55, 56, 57], emphasizing the importance of the miscibility transition temperature between homogeneous and phase separated states [187]. Although it is well established that the miscibility temperature depends on membrane tension, contradictory observations have been reported depending on the experimental method. While tension induced by micropipette aspiration decreases the miscibility temperature over which the membrane is homogeneous [58], osmotically tense GUVs favor the formation of domains [188, 59]. These studies highlight the complexity and often non-intuitive behavior of lipid membranes due to the intricate coupling between their chemical and physical properties.
Unilamellar vesicles are also a system of choice for the study of curvature-inducing proteins on lipid membranes. The main mechanisms by which proteins interact with lipid membranes to create curvature are illustrated in Figure 3, and include scaffolding, asymmetric insertion, clustering, and steric repulsion by crowding [189, 132]. Typically, GUVs at low membrane tension incubated with sufficiently high concentrations of curvature-inducing protein display membrane tubules or invaginations [190, 191, 192]. Curvature sensing by proteins can be quantitatively measured using microtubules pulled out from GUVs, and simultaneously monitoring protein binding as a function of the tube radius (or curvature) and concentration [191, 193, 194].
A step further in complex membrane interaction studies is the incorporation of cytoskeletal components [195]. GUVs are a particularly interesting system for the study of membrane-cytoskeleton interaction because they resemble cell compartments in size and shape, and can allow an interdependent dynamic between network polymerization and membrane shape. Pioneering work on encapsulating actin filaments inside GUVs showed vesicles exhibiting morphological changes due to actin polymerization, leading to various shapes by varying actin activity, concentrations and binding affinities [196, 197, 198, 199, 200]. Yet these early studies where limited by the maximum concentration of actin and ions that could be encapsulated without compromising the integrity of the GUV. A better control on the actin polymerization and binding components can be achieved by generating a network on the outer side of the membrane [201, 202, 203]. Liu and Fletcher produced dendritic actin network growth toward the inside of GUVs by activating the Arp2/3 complex through N-WASP and PI(4,5)P2 binding. Recently, N-WASP and Arp2/3 complex were successfully encapsulated with actin filaments in GUVs, forming an actin shell (or cortex) inside the vesicle [204, 205, 206]. These various membrane-cytskeleton systems offer interesting platforms to study force generation by actin polymerization occurring in cell motility and cytokinesis.
Despite the complexity and variety of biological membrane behavior, several membrane systems have been successfully developed to understand the key design principles that govern membrane processes. In the next section, we review and discuss how mathematical and computational modeling have helped this process.
4 Modeling cellular membrane processes
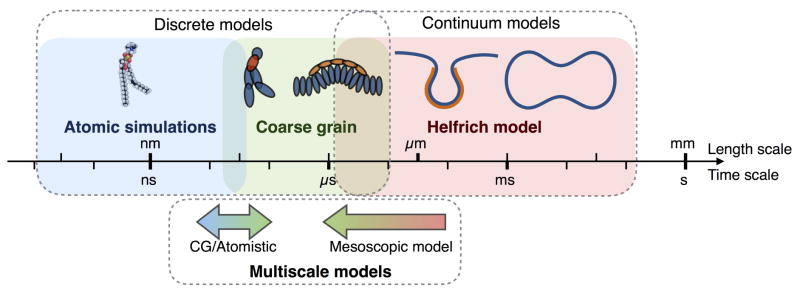
While the building blocks of biological membranes (lipids, proteins) are of the length scale of the membrane thickness (~ 3–6 nm), many biological processes occur simultaneously over several length scales (~100 nm-μm) [207]. When developing membrane models, this length scale separation represents both a challenge and an opportunity. First it is a challenge, because representing all the known biophysical and biochemical interactions in large systems is quite expensive, and often practically impossible from a computational point of view [208, 207]. On the other hand, details at the atomic scale of the lipid are rarely required to study relevant membrane phenomena. Therefore, length scale separation gives us the opportunity to deal with homogenized quantities, allowing for the description of the membrane as a continuum surface with average properties [207]. As shown in Figure 5, these two approaches, namely discrete and continuum approaches, are interdependent, as the quality of the continuum approach relies on the physical relevance of its parameters, which are obtained from small scale analysis. In what follows, we discuss both continuum and discrete modeling approaches of biological membranes, with a particular focus on continuum approaches as they allow for the representation of cellular scale geometries. We conclude by discussing multiscale modeling approaches that attempt to bridge the small and large length scale of cellular membranes.
Figure 5.
Different computational methods developed to study cellular membranes are valid in different length and time scales. Although some methods have attempted to bridge these scales, no unifying multiscale framework currently exists. This provides an opportunity for future model development.
4.1 Discrete membrane models
Atomistic molecular dynamic simulations trace the motion and energy of each atom through an empirical force field [209, 210]. The biggest advantage of these models is that they output information at the molecular level of the lipid lipid and lipid protein interactions that are difficult to obtain experimentally [211, 212, 213, 214]. One of the drawbacks with these approaches, however, is the large number of degrees of freedom that have to be computed, limiting atomistic simulations to represent phenomena on nano to microseconds for domains between ten and hundreds nanometers [208, 215].
To overcome these limitations, coarse grained (CG) methods have been developed. In this approach, the interactions between effective groups of atom are considered instead of the individual atoms [216]. Such methods, including the MARTINI model [217] and the dissipative particle dynamics [218], lose some details at the scale of the atom but allow larger computational domains and longer time scales. CG methods have been successful in modeling domain formation by lipid demixing, and protein inclusion [219, 220]. These models are also well suited to study local topological changes of lipid bilayers. For instance, membrane fusion has been modeled using dissipative particle dynamics [221, 222], showing that when two fusing membranes are in close proximity, the lipids tilt, splay and flip from one monolayer to another [223, 221, 222]. Recently, lipid-protein interactions have been successfully studied using CG simulations to evaluate the bending ability of membrane proteins, as well the interrelations between membrane tension and protein-membrane association [224, 225].
4.2 Continuum membrane models
For biological phenomena spanning over hundreds of nanometers, the discrete modeling approaches discussed above become computationally expensive and impracticable. Continuum models of bilayer membranes have been used to study the deformation of membranes and to explain many biological phenomena. The most widely used model of lipid bilayers is the Helfrich model [104], where the energy per unit area depends only on the mean and Gaussian curvatures of the membrane. This model has been extensively applied to the study the shape of red blood cells [226, 227] and to explain biological phenomena such as endocytic processes [228], the formation of membrane tubes [229] and the shapes of lipid vesicles [230, 231, 232, 233]. The Helfrich model assumes that the lipids are aligned normal to the membrane surface at all times and that curvatures are of the order of the bilayer thermal wavelength (≈20 nm). This approach captures the changes in membrane shape that occur at length scales larger than the thickness of the bilayer (≈ 5 nm). Subsequently, models including the area-difference across the leaflets [234, 235, 236] have been developed, providing further detail for curvature generation.
Incorporation of in-plane transport, including diffusion and lipid flow is an important feature of membrane curvature generation and maintenance. Saffron and Delbruck developed a framework for Brownian motion in biological membranes, which has been the cornerstone of protein diffusion in membranes [237]. Yet, the Saffman-Delbruck model is valid for flat surfaces and breaks down for highly curved membranes. More recently, a few models describing the flow and diffusion of lipids and proteins have been proposed [238, 239]. The role of lipid flow in governing the deformation of membranes was analyzed by modeling the membrane as a viscoelastic material [238, 234]. These efforts demonstrated how the lipid flow velocity is coupled to the changes in membrane shape, and showed that the membrane velocity field satisfies a Killing vector field on minimal surfaces [240]. We subsequently showed that lipid flow plays an important role in governing local membrane tension in response to protein-induced spontaneous curvature [241]. The coupling of membrane diffusion and elasticity results in membrane shapes that are either diffusion-dominated or curvature-dominated [239].
The elastic models used so far to study lipid membranes focus mainly on length scales that are much larger than the thickness of the bilayer. As a result, these models assume that all the lipids are oriented normal to the membrane [104]. There are, however, circumstances under which lipids are not aligned with the surface normal. The lipids are then said to tilt relative to the membrane surface; this in turn induces a change in membrane thickness, which is simply the projection of the lipid length onto the surface normal. For example, the tilt angle of gel-phase dipalmitoylphosphatidylcholine (DPPC) was found to be approximately 32° [242]. Even in the liquid phase, lipids can tilt in regions adjacent to protein inclusions [243]. Experimentally observed ripple phases are characterized by oscillatory thickness variations induced by spatially non-uniform tilt [244]. We developed a model for lipid tilt to study how protein inclusions could change lipid orientation and showed that for small length scales, lipid orientation, and tilt energy dominates the bending energy contributions [245, 246]. Other models accounting for lipid tilt and the attendant thickness variation have been developed by a number of research groups [247, 248, 249, 250, 251, 252, 253]. A continuing challenge with the development of these models is the dearth of experimental data at the level of lipid orientation and tilt angles to validate the models and extract the moduli. However, recently some experimental studies have measured the tilt angle using grazing incidence x-ray diffraction [254, 255] and these will provide valuable inputs for further model development.
4.3 Multiscale models
The modeling approaches described above are all suited to represent phenomena at a specific length scale. Yet the nature of cellular membranes motivate the need for models able to represent phenomena over multiple length scales. Although the development of multiscale membrane models is still an emerging research field, substantial progress has been made in bridging the atomic and CG scales. For instance, the effective properties of CG elements have been derived from atomistic molecular dynamics for lipids [256, 257, 258]. Alternatively, the equilibrium structure of the lipid lipid or lipid protein system can be obtained by CG simulations, and then refined to atomistic resolution to allow the simulation of atom interactions and configuration [259, 260, 261]. Such methodology is particularly useful for computing the dynamics of multicomponent lipid mixtures, where the arrangement of lipids happens on a longer time scale than the specific atom interactions [220]. A step further in multiscale resolution has been proposed by coupling the CG/Atomistic simulations with reaction modeling using hybrid quantum mechanics/molecular mechanics methods, in order to study the interactions of the cytochrome P450 enzyme with the plasma membrane and its environment [262].
Mesoscale model approaches aim to bridge continuum and coarse grain representations. Among them are the dynamically triangulated membrane model [263, 264, 265], and Elastic Membrane 2 (EM2) model [266, 267, 268]. The main idea behind these models is to discretize the Helfrich energy for continuum membranes into pseudo membrane-particles that interact through a bending potential [266]. This takes advantage of the CG simulations methodologies to represent the effect of local phenomena on micrometric scales membrane structures. These mesoscale models are particularly well suited to study the influence of curvature-inducing proteins such as BAR proteins on dynamics of membranes [267, 268, 264, 265, 269, 270]. In an attempt to extend the multiscale capabilities of mesoscale models, an inverse “coarse-graining” method was proposed, to refine the pseudo membrane-particles into CG simulations [271]. In this case the mesoscale EM2 model is run to obtain the equilibrium membrane configuration and the local spontaneous curvature. This information at the pseudo membrane nodes is then converted into CG systems, with one site per lipid and 26 sites per N-BAR protein.
Although the modeling approaches presented here are promising steps towards a true multiscale representation of cellular membranes, we are still quite far from a modeling framework that can encompass the multiscale nature of cellular membranes (see Figure 5). Indeed, the current coupling of scales is sequential and often unidirectional. One first computes an equilibrium configuration at one scale, and then refines this equilibrium to obtain local details at a finer scale. The limitations of this top-down approach are: (i) The effective properties at the larger scale are assumed a priori, instead of arising from an up-scaling approach. (ii) the current methodologies are heavily computational due to the use of particle-based approaches. Therefore, a theory of lipid bilayers that rigorously up-scales the local lipid/protein physics into a continuous mesoscale model with effective properties arising from the local scale would be extremely valuable. Such a model would ease the integration of membrane dynamics into a systemic representation of the cell, enabling its coupling with biological processes occurring away from cellular membranes, such as cytoplasmic transport and extracellular signaling.
5 Some perspectives for the future
Cell biology continues to face challenges that emerge from its complexity, especially bridging the whole with the parts. In particular, in systems biology, these challenges are manifested as the need to bridge the gap between model systems and cells. Multidisciplinary efforts including physics, mathematics, engineering, and computation coupled with experiments at different system scales will continue to lead the way for us to answer fundamental questions associated with cellular phenomena. Additionally, top-down approaches should be complemented by bottom-up approaches in order to relate the fundamental processes to their the biological counterparts. This is more so the case in the study of cellular membranes, where composition, shape, chemistry, and mechanics are all interconnected. An example of a process that is now well-understood through the above mentioned approaches is the formation of micron size lipid heterogeneities at physiological temperature [50, 137]. However, many more such approaches are needed to translate the results observed in synthetic systems to cellular systems.
Approaches such as network analyses [24], Boolean methods [26], compartmental models of signaling [24, 25, 27, 32], and ‘-omics’ approaches [9, 10, 8] have propelled the era of systems biology in the study of cellular membranes. Alongside, over the last few decades, the biophysics of membranes, through theory, computation, and experiment has provided great insight into how mechanics and geometry work in tandem to regulate membrane processes. These two areas of research have followed parallel paths, but recent advances in our understanding of how shape regulates signaling [40, 41], how signaling regulates shape [96, 27], the role of membrane curvature in regulating membrane protein diffusion [80, 39, 81], and how membrane shape and actin flow are controlled [111, 112, 113] provide a glimpse of the rich future at the intersection of these two fields. Therefore, we believe that the future lies in coupling mechanics and geometry of the membrane with biochemistry and temporal dynamics of cellular processes.
To that effect, we identify some current interdisciplinary challenges that serve as opportunities to push the boundaries of systems biology. While by no means exhaustive, this list is a good starting place for the development of theory, computation, and experimental resources.
Although self assembly features of lipids are well understood in synthetic systems, we still lack knowledge on how the dynamic composition affects signaling and cytoskeletal rearrangements in a complex system.
The assumption of symmetry in understanding membrane shapes, often used to generate mathematically tractable shapes, can sometimes be to our detriment since cells do not require symmetry in their shape for fulfilling their function. How can we understand complex geometrical aspects of cellular function including asymmetry?
From a biomedical perspective, the interaction of viral proteins and antimicrobial peptides with the lipid membrane need a further understanding both in terms of biophysics and on their impact on cellular function.
Signaling needs to be understood in the context of its coupling to the dynamics of protein and lipid uptake by endocytosis, and trafficking particularly in endomembranes.
The interaction of the membrane with the fluid in the environment and the associated shape changes are complex mathematical problems that invoke fields such as differential geometry and the need for new computational methods including advanced image analysis and fluid-structure interactions.
While molecular dynamic simulations and experiments have indicated that lipid orientation is an important order parameter governing membrane behavior, there is a need for a truly multiscale theory that can rigorously up-scale lipid scale physics to the curvature-dominant scale model.
Another important aspect of membrane biology is that that biological processes are inherently out-of-equilibrium. Unlike models for mechanical equilibrium that are quite well-developed, substantial work needs to be done to study out-of-equilibrium membrane processes.
The continued success of systems approaches in cell biology relies on our ability to integrate these different features and shape the converging futures of systems biology and biophysics of cellular membranes.
Acknowledgments
This work was supported in part by FISP 3030 award for the year 2015–2016 to M.C., and AFOSR FA9550-15-1-0124 and NSF PHY-1505017 awards to P.R. J.C.S. acknowledges support from the National Institutes of Health through grant GM112065. The authors are thankful to the anonymous reviewers whose comments were crucial for improving this article. The authors also thank Miriam Bell and Mrunal Seshadri for their critical reading of the manuscript, and to their many colleagues for discussions over the years that have shaped these ideas.
Figures 2, 3, and 4 used adapted content from Servier Medical Art powerpoint image bank (http://www.servier.com/Powerpoint-image-bank).
Abbreviations used
- ADF
Actin depolymerizing factor
- Arp2/3
Actin-Related Proteins 2/3 complex
- BAR
Bin amphiphysin rvs domain
- cAMP
Cyclic adenosine monophosphate
- AMP
Antimicrobial peptide
- CG
Coarse grain
- COPII
Coat protein complex associated with anterograde transport
- DAG
Diacylglycerol
- EGF
Epidermal growth factor
- EGFR
Epidermal growth factor receptor
- EM2
Elastic Membrane model 2
- ER
Endoplasmic Reticulum
- GPMV
Giant plasma membrane vesicle
- GUV
Giant unilamellar vesicle
- IP3
Inositol trisphosphate
- IP3R
Inositol trisphosphate receptor
- LUV
Large unilamellar vesicle
- MAP kinase
Mitogen-activated protein kinase
- MEK
Mitogen-activated protein kinase kinase
- NIH
National Institutes of Health
- N-WASP
Neural Wiskott-Aldrich syndrome protein
- PC
Phosphatidylclorine
- PE
Phosphatidyl ethanolamine
- PI(4,5)P2
Phosphatidylinositol 4,5-bisphosphate
- PKC
Protein kinase C
- Raf
Rapidly Accelerated Fibrosarcoma (kinase)
- RBC
Red blood cell
- SEM
Scanning electron microscopy
- SLB
Suspended lipid bilayer
- siRNA
Small interfering ribonucleic acid
- SUPER
Supported membrane with excess reservoir
- SUV
Small unilamellar vesicle
- TEM
Transmission electron microscopy
- TRP3
Tritrpticin
- WASP
Wiskott-Aldrich syndrome protein
- WAVE
WASP-family verprolin-homologous protein
References
- 1.Kitano H. Science. 2002a;295:1662. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 2.Kitano H. Nature. 2002b;420:206. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 3.Ideker T, Galitski T, Hood L. Annual review of genomics and human genetics. 2001;2:343. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 4.Berger SI, Ma’ayan A, Iyengar R. Science signaling. 2010;3:ra30. doi: 10.1126/scisignal.2000723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger SI, Iyengar R. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2011;3:129. doi: 10.1002/wsbm.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma’ayan A, Jenkins SL, Neves S, Hasseldine A, Grace E, Dubin-Thaler B, Eungdamrong NJ, Weng G, Ram PT, Rice JJ, et al. Science. 2005;309:1078. doi: 10.1126/science.1108876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennis E, Alex Brown H, Deems R, Glass C, Merrill A, Jr, Murphy R, Raetz C, Shaw W, Subramaniam S, Russell D, et al. Functional Lipidomics. CRC Press; 2005. pp. 1–16. [Google Scholar]
- 8.Dennis EA. Proceedings of the National Academy of Sciences. 2009;106:2089. doi: 10.1073/pnas.0812636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Meer G. The EMBO Journal. 2005;24:3159. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wenk MR. Nature Reviews Drug Discovery. 2005;4:594. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 11.Schmelzer K, Fahy E, Subramaniam S, Dennis EA. vol. 432 of Lipidomics and Bioactive Lipids: Mass-Spectrometry–Based Lipid Analysis. Academic Press; 2007. pp. 171–183. [Google Scholar]
- 12.Christie WW. Journal of Lipid Research. 1985;26:507. [PubMed] [Google Scholar]
- 13.Yamamoto Y, Brodsky MH, Baker JC, Ames BN. Analytical Biochemistry. 1987;160:7. doi: 10.1016/0003-2697(87)90606-3. [DOI] [PubMed] [Google Scholar]
- 14.Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Methods. 2006;39:82. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Brügger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Proceedings of the National Academy of Sciences. 1997;94:2339. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watson AD, Leitinger N, Navab M, Faull KF, Hörkkö S, Witztum JL, Palinski W, Schwenke D, Salomon RG, Sha W, et al. Journal of Biological Chemistry. 1997;272:13597. doi: 10.1074/jbc.272.21.13597. [DOI] [PubMed] [Google Scholar]
- 17.Schiller J, Arnhold J, Benard S, Müller M, Reichl S, Arnold K. Analytical biochemistry. 1999;267:46. doi: 10.1006/abio.1998.3001. [DOI] [PubMed] [Google Scholar]
- 18.Peterson BL, Cummings BS. Biomedical Chromatography. 2006;20:227. doi: 10.1002/bmc.563. [DOI] [PubMed] [Google Scholar]
- 19.Wymann MP, Schneiter R. Nature Reviews Molecular Cell Biology. 2008;9:162. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 20.Schlessinger J. Cell. 2000;103:211. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 21.Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER. Science. 2005;307:1625. doi: 10.1126/science.1106943. [DOI] [PubMed] [Google Scholar]
- 22.Cho W, Stahelin RV. Annual Review of Biophysics and Biomolecular Structure. 2005;34:119. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 23.Bray D. Annual Review of Biophysics and Biomolecular Structure. 1998;27:59. doi: 10.1146/annurev.biophys.27.1.59. [DOI] [PubMed] [Google Scholar]
- 24.Kholodenko BN. Nature Reviews Molecular Cell Biology. 2006;7:165. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eungdamrong NJ, Iyengar R. Biophysical Journal. 2007;92:808. doi: 10.1529/biophysj.106.093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agnati LF, Guidolin D, Leo G, Fuxe K. Journal of Neural Transmission. 2007;114:77. doi: 10.1007/s00702-006-0567-6. [DOI] [PubMed] [Google Scholar]
- 27.Rangamani P, Fardin MA, Xiong Y, Lipshtat A, Rossier O, Sheetz M, Iyengar R. Biophys J. 2011;100:845. doi: 10.1016/j.bpj.2010.12.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clapham DE. Cell. 2007;131:1047. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 29.Rangamani P, Levy MG, Khan S, Oster G. Proceedings of the National Academy of Sciences. 2016;113:E5298. doi: 10.1073/pnas.1610391113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markevich NI, Moehren G, Demin OV, Kiyatkin A, Hoek JB, Kholodenko BN. Systems Biology. 2004;1:104. doi: 10.1049/sb:20045003. [DOI] [PubMed] [Google Scholar]
- 31.Mayinger P. Cold Spring Harbor Perspectives in Biology. 2011;3:a005314. doi: 10.1101/cshperspect.a005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prior IA, Hancock JF. Seminars in Cell & Developmental Biology. 2012;23:145. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Y, Ren Y, Wu B, Decourt B, Lee AC, Taylor A, Suter DM. Molecular Biology of the Cell. 2015;26:3229. doi: 10.1091/mbc.E15-03-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perry MM, Gilbert AB. Journal of Cell Science. 1979;39:257. doi: 10.1242/jcs.39.1.257. [DOI] [PubMed] [Google Scholar]
- 35.Chenette EJ, Der CJ, Begley TP. Wiley Encyclopedia of Chemical Biology. John Wiley & Sons, Inc; 2007. [Google Scholar]
- 36.Deng Y, Mieczkowski M. Protoplasma. 1998;203:16. [Google Scholar]
- 37.Alberts B, Bray D, Hopkin K, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Essential cell biology. Garland Science; 2013. [Google Scholar]
- 38.Terasaki M, Shemesh T, Kasthuri N, Klemm RW, Schalek R, Hayworth KJ, Hand AR, Yankova M, Huber G, Lichtman JW, et al. Cell. 2013;154:285. doi: 10.1016/j.cell.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rangamani P, Lipshtat A, Azeloglu E, Calizo R, Hu M, Ghassemi S, Hone J, Scarlata S, Neves S, Iyengar R. ell. 2013a;154:1356. doi: 10.1016/j.cell.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neves SR, Tsokas P, Sarkar A, Grace EA, Rangamani P, Taubenfeld SM, Alberini CM, Schaff JC, Blitzer RD, Moraru II, et al. Cell. 2008;133:666. doi: 10.1016/j.cell.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thalmeier D, Halatek J, Frey E. Proceedings of the National Academy of Sciences. 2016;113:548. doi: 10.1073/pnas.1515191113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Meer G, Voelker DR, Feigenson GW. Nature reviews molecular cell biology. 2008;9:112. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicolson GL. Cancer research. 2015;75:1169. doi: 10.1158/0008-5472.CAN-14-3216. [DOI] [PubMed] [Google Scholar]
- 44.Escribá PV, Nicolson GL. Biochimica et biophysica acta. 2014;1838:1449. doi: 10.1016/j.bbamem.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Dallner G, Siekevitz P, Palade GE. The Journal of cell biology. 1966;30:73. doi: 10.1083/jcb.30.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolega J, Manabe M, Sun T-T. Differentiation. 1989;42:54. doi: 10.1111/j.1432-0436.1989.tb00607.x. [DOI] [PubMed] [Google Scholar]
- 47.Singer SJ, Nicolson GL. Science. 1972;175:720. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 48.Simons K, Ikonen E. Nature. 1997;387:569. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 49.Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, Baird BA, Webb WW. Proceedings of the National Academy of Sciences. 2007a;104:3165. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veatch SL, Cicuta P, Sengupta P, Honerkamp-Smith A, Holowka D, Baird B. ACS Chemical Biology. 2008;3:287. doi: 10.1021/cb800012x. [DOI] [PubMed] [Google Scholar]
- 51.Parthasarathy R, Yu C-h, Groves JT. Langmuir. 2006;22:5095. doi: 10.1021/la060390o. [DOI] [PubMed] [Google Scholar]
- 52.Korlach J, Schwille P, Webb WW, Feigenson GW. Proceedings of the National Academy of Sciences. 1999;96:8461. doi: 10.1073/pnas.96.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumgart T, Hess ST, Webb WW. Nature. 2003;425:821. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 54.Oglecka K, Sanborn J, Parikh AN, Kraut RS. Frontiers in Physiology. 2012;3:120. doi: 10.3389/fphys.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veatch SL, Keller SL. Physical Review Letters. 2002;89:268101. doi: 10.1103/PhysRevLett.89.268101. [DOI] [PubMed] [Google Scholar]
- 56.Veatch SL, Keller SL. Biophysical Journal. 2003;85:3074. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veatch SL, Keller SL. Physical Review Letters. 2005;94:148101. doi: 10.1103/PhysRevLett.94.148101. [DOI] [PubMed] [Google Scholar]
- 58.Portet T, Gordon SE, Keller SL. Biophysical Journal. 2012;103:L35. doi: 10.1016/j.bpj.2012.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oglecka K, Rangamani P, Liedberg B, Kraut RS, Parikh AN. eLife. 2014;3:e03695. doi: 10.7554/eLife.03695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loerke D, Mettlen M, Yarar D, Jaqaman K, Jaqaman H, Danuser G, Schmid SL. PLOS Biology. 2009;7:e1000057. doi: 10.1371/journal.pbio.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y-W, Neumann S, Ramachandran R, Ferguson SM, Pucadyil TJ, Schmid SL. Proceedings of the National Academy of Sciences. 2011;108:E234. doi: 10.1073/pnas.1102710108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sarabipour S, Ballmer-Hofer K, Hristova K. eLife. 2016;5:e13876. doi: 10.7554/eLife.13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wood JM. Microbiology and Molecular Biology Reviews. 1999;63:230. doi: 10.1128/mmbr.63.1.230-262.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bass RB, Strop P, Barclay M, Rees DC. Science. 2002;298:1582. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- 65.Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. Nature. 2003;423:33. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 66.Schmidt D, del Mármol J, MacKinnon R. Proceedings of the National Academy of Sciences. 2012;109:10352. doi: 10.1073/pnas.1204700109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anishkin A, Loukin SH, Teng J, Kung C. Proceedings of the National Academy of Sciences. 2014;111:7898. doi: 10.1073/pnas.1313364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ge J, Li W, Zhao Q, Li N, Chen M, Zhi P, Li R, Gao N, Xiao B, Yang M. Nature. 2015;527:64. doi: 10.1038/nature15247. [DOI] [PubMed] [Google Scholar]
- 69.Daub H, Ulrich Weiss F, Wallasch C, Ullrich A. Nature. 1996;379:557. doi: 10.1038/379557a0. [DOI] [PubMed] [Google Scholar]
- 70.Bhola N, Grandis J. Frontiers in bioscience : a journal and virtual library. 2008;13:1857. doi: 10.2741/2805. [DOI] [PubMed] [Google Scholar]
- 71.Rosenbaum DM, Rasmussen SGF, Kobilka BK. Nature. 2009;459:356. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Groves JT, Kuriyan J. Nature structural & molecular biology. 2010;17:659. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blenis J. Proceedings of the National Academy of Sciences. 1993;90:5889. doi: 10.1073/pnas.90.13.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Di Fiore PP, De Camilli P. Cell. 2001;106:1. doi: 10.1016/s0092-8674(01)00428-7. [DOI] [PubMed] [Google Scholar]
- 75.Platta HW, Stenmark H. Current opinion in cell biology. 2011;23:393. doi: 10.1016/j.ceb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 76.Kaksonen M, Sun Y, Drubin DG. Cell. 2003;115:475. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 77.Villaseñor R, Nonaka H, Del Conte-Zerial P, Kalaidzidis Y, Zerial M. Elife. 2015;4:e06156. doi: 10.7554/eLife.06156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Nature cell biology. 2007;9:858. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neves SR. Science signaling. 2011;4:tr8. doi: 10.1126/scisignal.2001988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meyers J, Craig J, Odde DJ. Current Biology. 2006;16:1685. doi: 10.1016/j.cub.2006.07.056. [DOI] [PubMed] [Google Scholar]
- 81.Zieske K, Schwille P. Elife. 2014;3:e03949. doi: 10.7554/eLife.03949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Farhan H, Wendeler MW, Mitrovic S, Fava E, Silberberg Y, Sharan R, Zerial M, Hauri H-P. The Journal of Cell Biology. 2010;189:997. doi: 10.1083/jcb.200912082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Farhan H, Rabouille C. J Cell Sci. 2011;124:171. doi: 10.1242/jcs.076455. [DOI] [PubMed] [Google Scholar]
- 84.Bareford LM, Swaan PW. Advanced Drug Delivery Reviews. 2007;59:748. doi: 10.1016/j.addr.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bronfman FC, Escudero CA, Weis J, Kruttgen A. Developmental Neurobiology. 2007;67:1183. doi: 10.1002/dneu.20513. [DOI] [PubMed] [Google Scholar]
- 86.Saksena S, Emr SD. Biochemical Society Transactions. 2009;37:167. doi: 10.1042/BST0370167. [DOI] [PubMed] [Google Scholar]
- 87.Chia J, Goh G, Racine V, Ng S, Kumar P, Bard F. Molecular Systems Biology. 2012;8:629. doi: 10.1038/msb.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cancino J, Luini A. Traffic. 2013;14:121. doi: 10.1111/tra.12022. [DOI] [PubMed] [Google Scholar]
- 89.Simpson JC, Joggerst B, Laketa V, Verissimo F, Cetin C, Erfle H, Bexiga MG, Singan VR, Hériché J-K, Neumann B, et al. Nature Cell Biology. 2012;14:764. doi: 10.1038/ncb2510. [DOI] [PubMed] [Google Scholar]
- 90.Pulvirenti T, Giannotta M, Capestrano M, Capitani M, Pisanu A, Polishchuk RS, Pietro ES, Beznoussenko GV, Mironov AA, Turacchio G, et al. Nature Cell Biology. 2008;10:912. doi: 10.1038/ncb1751. [DOI] [PubMed] [Google Scholar]
- 91.Sallese M, Giannotta M, Luini A. Seminars in Cell & Developmental Biology. 2009;20:801. doi: 10.1016/j.semcdb.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 92.Luini A, Parashuraman S. Current Opinion in Cell Biology. 2016;39:37. doi: 10.1016/j.ceb.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 93.Weller SG, Capitani M, Cao H, Micaroni M, Luini A, Sallese M, McNiven MA. Proceedings of the National Academy of Sciences. 2010;107:5863. doi: 10.1073/pnas.0915123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA. Cell. 2008;135:1276. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Biophysical Journal. 2000;79:328. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zimmerberg J, Kozlov MM. Nature Reviews: Molecular Cell Biology. 2006;7:9. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 97.Kirchhausen T, Owen D, Harrison SC. Cold Spring Harbor perspectives in biology. 2014;6:a016725. doi: 10.1101/cshperspect.a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McMahon HT, Boucrot E. Nature reviews. Molecular Cell Biology. 2011;12:517. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 99.Dannhauser P, Ungewickell E. Nature Cell Biology. 2012;14:634. doi: 10.1038/ncb2478. [DOI] [PubMed] [Google Scholar]
- 100.Ford M, Mills I, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT. Nature. 2002;419:361. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 101.Stachowiak J, Schmid E, Ryan C, Ann HS, Sasaki D, Sherman M, Geissler P, Fletcher D, Hayden C. Nature Cell Biology. 2012;14:944. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- 102.Stachowiak J, Brodsky F, Miller E. Nature Cell Biology. 2013;15:1019. doi: 10.1038/ncb2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Busch DJ, Houser JR, Hayden CC, Sherman MB, Lafer EM, Stachowiak JC. Nat Communications. 2015;6 doi: 10.1038/ncomms8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Helfrich W. Zeitschrift für Naturforschung C. 1973;28:693. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 105.Lipowsky R. Faraday discussions. 2013;161:305. doi: 10.1039/c2fd20105d. [DOI] [PubMed] [Google Scholar]
- 106.Schmidt N, Mishra A, Lai GH, Wong GC. FEBS Letters. 2010;584:1806. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 107.Schmidt NW, Wong GCL. Current Opinion in Solid State and Materials Science. 2013;17:151. doi: 10.1016/j.cossms.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Iwamoto K, Hayakawa T, Murate M, Makino A, Ito K, Fujisawa T, Kobayashi T. Biophysical Journal. 2007;93:1608. doi: 10.1529/biophysj.106.101584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bozelli JC, Jr, Sasahara ET, Pinto MRS, Nakaie CR, Schreier S. Chemistry and Physics of Lipids. 2012;165:365. doi: 10.1016/j.chemphyslip.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 110.Fang RH, Luk BT, Hu C-MJ, Zhang L. Advanced Drug Delivery Reviews. 2015;90:69. doi: 10.1016/j.addr.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dubin-Thaler B, Giannone G, Dobereiner H, Sheetz M. Biophys J. 2004;86:1794. doi: 10.1016/S0006-3495(04)74246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dubin-Thaler B, Hofman J, Cai Y, Xenias H, Spielman I, Shneidman A, David L, Dobereiner H, Wiggins C, Sheetz M. PLoS ONE. 2008;3:e3735. doi: 10.1371/journal.pone.0003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xiong Y, Rangamani P, Fardin M, Lipshtat A, Dubin-Thaler B, Rossier O, Sheetz M, Iyengar R. Biophys J. 2010;98:2136. doi: 10.1016/j.bpj.2010.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Saarikangas J, Zhao H, Lappalainen P. Physiological reviews. 2010;90:259. doi: 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- 115.Sun Y, Martin AC, Drubin DG. Developmental cell. 2006;11:33. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 116.Kamioka Y, Fukuhara S, Sawa H, Nagashima K, Masuda M, Matsuda M, Mochizuki N. Journal of Biological Chemistry. 2004;279:40091. doi: 10.1074/jbc.M404899200. [DOI] [PubMed] [Google Scholar]
- 117.Krauss M, Haucke V. Reviews of Physiology, Biochemistry and Pharmacology 161. Springer; 2009. pp. 45–66. [DOI] [PubMed] [Google Scholar]
- 118.Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. The Journal of cell biology. 2006;172:269. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Takano K, Toyooka K, Suetsugu S. The EMBO journal. 2008;27:2817. doi: 10.1038/emboj.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Henne WM, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, McMahon HT. Science. 2010;328:1281. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roberts-Galbraith RH, Gould KL. Cell cycle. 2010;9:4091. doi: 10.4161/cc.9.20.13587. [DOI] [PubMed] [Google Scholar]
- 122.Wu M, Huang B, Graham M, Raimondi A, Heuser JE, Zhuang X, De Camilli P. Nature cell biology. 2010;12:902. doi: 10.1038/ncb2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lewellyn EB, Pedersen RT, Hong J, Lu R, Morrison HM, Drubin DG. Developmental cell. 2015;35:281. doi: 10.1016/j.devcel.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu J, Sun Y, Drubin DG, Oster GF. PLoS Biology. 2009;7:e1000204. doi: 10.1371/journal.pbio.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pomorski TG, Nylander T, Cárdenas M. Advances in Colloid and Interface Science. 2014;205:207. doi: 10.1016/j.cis.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 126.Brezesinski G, Möhwald H. Advances in Colloid and Interface Science. 2003;100–102:563. doi: 10.1016/s0001-8686(02)00071-4. [DOI] [PubMed] [Google Scholar]
- 127.Oliveira ON, Jr, Pavinatto FJ, Balogh DT. NATO Science for Peace and Security Series C: Environmental Security. In: Bardosova M, Wagner T, editors. Nanomaterials and Nanoarchitectures. Springer; Netherlands: 2015. pp. 301–343. [Google Scholar]
- 128.Slaughter BD, Unruh JR, Das A, Smith SE, Rubinstein B, Li R. Nature communications. 2013;4:1380. doi: 10.1038/ncomms2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.López-Duarte I, Vu TT, Izquierdo MA, Bull JA, Kuimova MK. Chemical Communications. 2014;50:5282. doi: 10.1039/c3cc47530a. [DOI] [PubMed] [Google Scholar]
- 130.Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. Cell. 2004;118:591. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 131.Aguet F, Antonescu CN, Mettlen M, Schmid SL, Danuser G. Developmental cell. 2013;26:279. doi: 10.1016/j.devcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jarsch IK, Daste F, Gallop JL. The Journal of Cell Biology. 2016;214:375. doi: 10.1083/jcb.201604003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Welsher K, Yang H. Nature nanotechnology. 2014;9:198. doi: 10.1038/nnano.2014.12. [DOI] [PubMed] [Google Scholar]
- 134.Adler J, Shevchuk AI, Novak P, Korchev YE, Parmryd I. Nature Methods. 2010;7:170. doi: 10.1038/nmeth0310-170. [DOI] [PubMed] [Google Scholar]
- 135.Keller H, Rentsch P, Hagmann J. Experimental cell research. 2002;277:161. doi: 10.1006/excr.2002.5552. [DOI] [PubMed] [Google Scholar]
- 136.Levental I, Byfield FJ, Chowdhury P, Gai F, Baumgart T, Janmey PA. Biochemical Journal. 2009;424:163. doi: 10.1042/BJ20091283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sezgin E, Levental I, Grzybek M, Schwarzmann G, Mueller V, Honigmann A, Belov VN, Eggeling C, Coskun Ü, Simons K, et al. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2012;1818:1777. doi: 10.1016/j.bbamem.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 138.Machta BB, Papanikolaou S, Sethna JP, Veatch SL. Biophysical Journal. 2011;100:1668. doi: 10.1016/j.bpj.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Drees F, Reilein A, Nelson W. In: Cell Migration. Guan J-L, editor. Vol. 294. Humana Press; 2005. pp. 303–320. Methods in Molecular Biology. [Google Scholar]
- 140.Perez J-B, Martinez KL, Segura J-M, Vogel H. Advanced Functional Materials. 2006;16:306. [Google Scholar]
- 141.Danelon C, Perez J-B, Santschi C, Brugger J, Vogel H. Langmuir. 2006;22:22. doi: 10.1021/la061321c. [DOI] [PubMed] [Google Scholar]
- 142.Pace H, Simonsson Nyström L, Gunnarsson A, Eck E, Monson C, Geschwindner S, Snijder A, Höök F. Analytical Chemistry. 2015;87:9194. doi: 10.1021/acs.analchem.5b01449. [DOI] [PubMed] [Google Scholar]
- 143.Richards MJ, Hsia C-Y, Singh RR, Haider H, Kumpf J, Kawate T, Daniel S. Langmuir. 2016;32:2963. doi: 10.1021/acs.langmuir.5b03415. [DOI] [PubMed] [Google Scholar]
- 144.Sackmann E. Science. 1996;271:43. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- 145.Richter RP, Bérat R, Brisson AR. Langmuir. 2006;22:3497. doi: 10.1021/la052687c. [DOI] [PubMed] [Google Scholar]
- 146.Reimhult E, Baumann MK, Kaufmann S, Kumar K, Spycher PR. Biotechnology and Genetic Engineering Reviews. 2010;27:185. doi: 10.1080/02648725.2010.10648150. [DOI] [PubMed] [Google Scholar]
- 147.Sackmann E, Tanaka M. Trends in Biotechnology. 2000;18:58. doi: 10.1016/s0167-7799(99)01412-2. [DOI] [PubMed] [Google Scholar]
- 148.Subramaniam AB, Guidotti G, Manoharan VN, Stone HA. Nature Materials. 2013;12:128. doi: 10.1038/nmat3492. [DOI] [PubMed] [Google Scholar]
- 149.Rossi C, Doumiati S, Lazzarelli C, Davi M, Meddar F, Ladant D, Chopineau J. PLOS ONE. 2011;6:e19101. doi: 10.1371/journal.pone.0019101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Simon A, Girard-Egrot A, Sauter F, Pudda C, Picollet D’Hahan N, Blum L, Chatelain F, Fuchs A. ournal of Colloid and Interface Science. 2007;308:337. doi: 10.1016/j.jcis.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 151.Crane J, Tamm L. Methods in Membrane Lipids. In: Dopico A, editor. Methods in Molecular Biology™. Vol. 400. Humana Press; 2007. pp. 481–488. [Google Scholar]
- 152.Ries J, Chiantia S, Schwille P. Biophysical Journal. 2009;96:1999. doi: 10.1016/j.bpj.2008.12.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Elfick APD, Downes AR, Mouras R. Analytical and Bioanalytical Chemistry. 2009;396:45. doi: 10.1007/s00216-009-3223-9. [DOI] [PubMed] [Google Scholar]
- 154.Barfoot RJ, Sheikh KH, Johnson BRG, Colyer J, Miles RE, Jeuken LJC, Bushby RJ, Evans SD. angmuir. 2008;24:6827. doi: 10.1021/la800085n. [DOI] [PubMed] [Google Scholar]
- 155.Lee K, Gallop JL, Rambani K, Kirschner MW. Science. 2010;329:1341. doi: 10.1126/science.1191710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Courtemanche N, Lee JY, Pollard TD, Greene EC. Proceedings of the National Academy of Sciences. 2013;110:9752. doi: 10.1073/pnas.1308257110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vogel SK, Petrasek Z, Heinemann F, Schwille P. eLife. 2013;2:e00116. doi: 10.7554/eLife.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Honigmann A, Sadeghi S, Keller J, Hell SW, Eggeling C, Vink R. eLife. 2014;3:e01671. doi: 10.7554/eLife.01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Baksh MM, Jaros M, Groves JT. Nature. 2004;427:139. doi: 10.1038/nature02209. [DOI] [PubMed] [Google Scholar]
- 160.Pucadyil TJ, Schmid SL. Cell. 2008;135:1263. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pucadyil TJ, Schmid SL. Biophysical Journal. 2010;99:517. doi: 10.1016/j.bpj.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jung HT, Coldren B, Zasadzinski JA, Iampietro DJ, Kaler EW. Proceedings of the National Academy of Sciences. 2001;98:1353. doi: 10.1073/pnas.041420998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kunding AH, Mortensen MW, Christensen SM, Stamou D. Biophysical Journal. 2008;95:1176. doi: 10.1529/biophysj.108.128819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Angelova MI, Soléau S, Méléard P, Faucon F, Bothorel P. Trends in Colloid and Interface Science VI. In: Helm C, Lösche M, Möhwald H, Steinkopff, editors. Progress in Colloid & Polymer Science. Vol. 89. 1992. pp. 127–131. [Google Scholar]
- 165.Walde P, Cosentino K, Engel H, Stano P. ChemBioChem. 2010;11:848. doi: 10.1002/cbic.201000010. [DOI] [PubMed] [Google Scholar]
- 166.Stachowiak JC, Richmond DL, Li TH, Liu AP, Parekh SH, Fletcher DA. Proceedings of the National Academy of Sciences. 2008;105:4697. doi: 10.1073/pnas.0710875105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ota S, Yoshizawa S, Takeuchi S. Angewandte Chemie International Edition. 2009;48:6533. doi: 10.1002/anie.200902182. [DOI] [PubMed] [Google Scholar]
- 168.van Swaay D, deMello A. Lab on a Chip. 2013;13:752. doi: 10.1039/c2lc41121k. [DOI] [PubMed] [Google Scholar]
- 169.Kwok R, Evans E. Biophysical Journal. 1981;35:637. doi: 10.1016/S0006-3495(81)84817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Evans E, Kwok R. Biochemistry. 1982;21:4874. doi: 10.1021/bi00263a007. [DOI] [PubMed] [Google Scholar]
- 171.Evans E, Rawicz W. Physical Review Letters. 1990;64:2094. doi: 10.1103/PhysRevLett.64.2094. [DOI] [PubMed] [Google Scholar]
- 172.Evans E, Heinrich V, Ludwig F, Rawicz W. Biophysical Journal. 2003;85:2342. doi: 10.1016/s0006-3495(03)74658-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Henriksen JR, Ipsen JH. The European Physical Journal E. 2004;14:149. doi: 10.1140/epje/i2003-10146-y. [DOI] [PubMed] [Google Scholar]
- 174.Tian A, Johnson C, Wang W, Baumgart T. Physical Review Letters. 2007;98:208102. doi: 10.1103/PhysRevLett.98.208102. [DOI] [PubMed] [Google Scholar]
- 175.Roux A, Cuvelier D, Nassoy P, Prost J, Bassereau P, Goud B. The EMBO Journal. 2005;24:1537. doi: 10.1038/sj.emboj.7600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tian A, Baumgart T. Biophysical Journal. 2009;96:2676. doi: 10.1016/j.bpj.2008.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sorre B, Callan-Jones A, Manneville J-B, Nassoy P, Joanny J-F, Prost J, Goud B, Bassereau P. Proceedings of the National Academy of Sciences. 2009;106:5622. doi: 10.1073/pnas.0811243106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Callan-Jones A, Sorre B, Bassereau P. Cold Spring Harbor Perspectives in Biology. 2011;3:a004648. doi: 10.1101/cshperspect.a004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Simunovic M, Prévost C, Callan-Jones A, Bassereau P. Phil Trans R Soc A. 2016;374:20160034. doi: 10.1098/rsta.2016.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ohno M, Hamada T, Takiguchi K, Homma M. Langmuir. 2009;25:11680. doi: 10.1021/la900777g. [DOI] [PubMed] [Google Scholar]
- 181.Peterlin P, Arrigler V, Haleva E, Diamant H. Soft Matter. 2012;8:2185. [Google Scholar]
- 182.Boroske E, Elwenspoek M, Helfrich W. Biophysical Journal. 1981;34:95. doi: 10.1016/S0006-3495(81)84839-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sankhagowit S, Wu S-H, Biswas R, Riche CT, Povinelli ML, Malmstadt N. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2014;1838:2615. doi: 10.1016/j.bbamem.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 184.Seifert U. Advances in Physics. 1997;46:13. [Google Scholar]
- 185.Ho JCS, Rangamani P, Liedberg B, Parikh AN. Langmuir. 2016 [Google Scholar]
- 186.Baumgart T, Hunt G, Farkas ER, Webb WW, Feigenson GW. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2007b;1768:2182. doi: 10.1016/j.bbamem.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Honerkamp-Smith AR, Veatch SL, Keller SL. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2009;1788:53. doi: 10.1016/j.bbamem.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hamada T, Kishimoto Y, Nagasaki T, Takagi M. Soft Matter. 2011;7:9061. [Google Scholar]
- 189.Baumgart T, Capraro BR, Zhu C, Das SL. Annual Review of Physical Chemistry. 2011;62:483. doi: 10.1146/annurev.physchem.012809.103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sens P, Johannes L, Bassereau P. Current Opinion in Cell Biology. 2008;20:476. doi: 10.1016/j.ceb.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 191.Sorre B, Callan-Jones A, Manzi J, Goud B, Prost J, Bassereau P, Roux A. Proceedings of the National Academy of Sciences. 2012;109:173. doi: 10.1073/pnas.1103594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Scheve CS, Gonzales PA, Momin N, Stachowiak JC. Journal of the American Chemical Society. 2013;135:1185. doi: 10.1021/ja3099867. [DOI] [PubMed] [Google Scholar]
- 193.Ramesh P, Baroji YF, Reihani SNS, Stamou D, Oddershede LB, Bendix PM. Scientific Reports. 2013;3 doi: 10.1038/srep01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Prévost C, Zhao H, Manzi J, Lemichez E, Lappalainen P, Callan-Jones A, Bassereau P. Nature Communications. 2015;6:8529. doi: 10.1038/ncomms9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Vogel SK, Schwille P. Current Opinion in Biotechnology. 2012;23:758. doi: 10.1016/j.copbio.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 196.Cortese JD, Schwab B, Frieden C, Elson EL. Proceedings of the National Academy of Sciences. 1989;86:5773. doi: 10.1073/pnas.86.15.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Miyata H, Hotani H. Proceedings of the National Academy of Sciences. 1992;89:11547. doi: 10.1073/pnas.89.23.11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Miyata H, Nishiyama S, Akashi K-i, Kinosita K. Proceedings of the National Academy of Sciences. 1999;96:2048. doi: 10.1073/pnas.96.5.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Limozin L, Sackmann E. Physical Review Letters. 2002;89:168103. doi: 10.1103/PhysRevLett.89.168103. [DOI] [PubMed] [Google Scholar]
- 200.Li S, Palmer AF. Langmuir. 2004;20:7917. doi: 10.1021/la049035t. [DOI] [PubMed] [Google Scholar]
- 201.Liu AP, Fletcher DA. Biophysical Journal. 2006;91:4064. doi: 10.1529/biophysj.106.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liu AP, Fletcher DA. Nature Reviews Molecular Cell Biology. 2009;10:644. doi: 10.1038/nrm2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Carvalho K, Tsai F-C, Lees E, Voituriez R, Koenderink GH, Sykes C. Proceedings of the National Academy of Sciences. 2013a;110:16456. doi: 10.1073/pnas.1221524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Pontani L-L, van der Gucht J, Salbreux G, Heuvingh J, Joanny J-F, Sykes C. Biophysical Journal. 2009;96:192. doi: 10.1016/j.bpj.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Murrell M, Pontani L-L, Guevorkian K, Cuvelier D, Nassoy P, Sykes C. Biophysical Journal. 2011;100:1400. doi: 10.1016/j.bpj.2011.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Carvalho K, Lemière J, Faqir F, Manzi J, Blanchoin L, Plastino J, Betz T, Sykes C. Phil Trans R Soc B. 2013b;368:20130005. doi: 10.1098/rstb.2013.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ramakrishnan N, Sunil Kumar PB, Radhakrishnan R. Physics Reports. 2014;543:1. doi: 10.1016/j.physrep.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lindahl E, Sansom MS. Current Opinion in Structural Biology. 2008;18:425. doi: 10.1016/j.sbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 209.Tobias DJ, Tu K, Klein ML. Current opinion in colloid & interface science. 1997;2:15. [Google Scholar]
- 210.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Pastor RW. The Journal of Physical Chemistry B. 2010;114:7830. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tieleman DP, Leontiadou H, Mark AE, Marrink S-J. Journal of the American Chemical Society. 2003;125:6382. doi: 10.1021/ja029504i. [DOI] [PubMed] [Google Scholar]
- 212.de Vries AH, Mark AE, Marrink SJ. Journal of the American Chemical Society. 2004;126:4488. doi: 10.1021/ja0398417. [DOI] [PubMed] [Google Scholar]
- 213.MacCallum JL, Bennett WFD, Tieleman DP. The Journal of General Physiology. 2007;129:371. doi: 10.1085/jgp.200709745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.MacCallum JL, Bennett WFD, Tieleman DP. Biophysical Journal. 2008;94:3393. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hartkamp R, Moore TC, Iacovella CR, Thompson MA, Bulsara PA, Moore DJ, McCabe C. Biophysical Journal. 2016;111:813. doi: 10.1016/j.bpj.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shelley JC, Shelley MY, Reeder RC, Bandyopadhyay S, Klein ML. The Journal of Physical Chemistry B. 2001;105:4464. [Google Scholar]
- 217.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The Journal of Physical Chemistry B. 2007;111:7812. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 218.de Meyer FJ-M, Benjamini A, Rodgers JM, Misteli Y, Smit B. The Journal of Physical Chemistry B. 2010;114:10451. doi: 10.1021/jp103903s. [DOI] [PubMed] [Google Scholar]
- 219.Bennett WFD, Tieleman DP. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2013;1828:1765. doi: 10.1016/j.bbamem.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 220.Ackerman DG, Feigenson GW. The Journal of Physical Chemistry B. 2015;119:4240. doi: 10.1021/jp511083z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Grafmüller A, Shillcock J, Lipowsky R. Physical review letters. 2007;98:218101. doi: 10.1103/PhysRevLett.98.218101. [DOI] [PubMed] [Google Scholar]
- 222.Grafmüller A, Shillcock J, Lipowsky R. Biophysical journal. 2009;96:2658. doi: 10.1016/j.bpj.2008.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Marrink SJ, Mark AE. Journal of the American Chemical Society. 2003;125:11144. doi: 10.1021/ja036138+. [DOI] [PubMed] [Google Scholar]
- 224.Simunovic M, Voth GA. Nature Communications. 2015;6:7219. doi: 10.1038/ncomms8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Simunovic M, Voth GA, Callan-Jones A, Bassereau P. Trends in Cell Biology. 2015;25:780. doi: 10.1016/j.tcb.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Iglic̆ A. Journal of Biomechanics. 1997;30:35. doi: 10.1016/s0021-9290(96)00100-5. [DOI] [PubMed] [Google Scholar]
- 227.Deuling HJ, Helfrich W. Biophysical Journal. 1976;16:861. doi: 10.1016/S0006-3495(76)85736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hassinger JE, Oster G, Drubin DG, Rangamani P. Proceedings of the National Academy of Sciences. 2017:201617705. doi: 10.1073/pnas.1617705114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Derényi I, Jülicher F, Prost J. Physical review letters. 2002;88:238101. doi: 10.1103/PhysRevLett.88.238101. [DOI] [PubMed] [Google Scholar]
- 230.Seifert U, Berndl K, Lipowsky R. Physical Review A. 1991;44:1182. doi: 10.1103/physreva.44.1182. [DOI] [PubMed] [Google Scholar]
- 231.Jaric M, Seifert U, Wintz W, Wortis M. Physical review E, Statistical physics, plasmas, fluids and related interdisciplinary topics. 1995;52:6623. doi: 10.1103/physreve.52.6623. [DOI] [PubMed] [Google Scholar]
- 232.Nelson P, Powers T, Seifert U. Physical review letters. 1995;74:3384. doi: 10.1103/PhysRevLett.74.3384. [DOI] [PubMed] [Google Scholar]
- 233.Seifert U, Shillcock J, Nelson P. Physical review letters. 1996;77:5237. doi: 10.1103/PhysRevLett.77.5237. [DOI] [PubMed] [Google Scholar]
- 234.Rahimi M, Arroyo M. Physical Review E. 2012;86:011932. doi: 10.1103/PhysRevE.86.011932. [DOI] [PubMed] [Google Scholar]
- 235.Mui B, Döbereiner H, Madden T, Cullis P. Biophysical journal. 1995;69:930. doi: 10.1016/S0006-3495(95)79967-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Miao L, Seifert U, Wortis M, Döbereiner H-G. Physical Review E. 1994;49:5389. doi: 10.1103/physreve.49.5389. [DOI] [PubMed] [Google Scholar]
- 237.Saffman P, Delbrück M. Proceedings of the National Academy of Sciences. 1975;72:3111. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rangamani P, Agrawal A, Mandadapu K, Oster G, Steigmann D. Biomech Model Mechanobiol. 2013b;12:833. doi: 10.1007/s10237-012-0447-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Agrawal A, Steigmann DJ. Z Angew Math Phys (ZAMP) 2011;62:549. [Google Scholar]
- 240.Bahmani F, Christenson J, Rangamani P. Continuum Mechanics and Thermodynamics. 2015 [Google Scholar]
- 241.Rangamani P, Mandadapu K, Oster G. Biophysical Journal. 2014a;107:751. doi: 10.1016/j.bpj.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tristram-Nagle S, Zhang R, Suter RM, Worthington CR, Sun WJ, Nagle JF. Biophysical journal. 1993;64:1097. doi: 10.1016/S0006-3495(93)81475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Watkins S, Sontheimer H. The Journal of Neuroscience. 2011;31:17250. doi: 10.1523/JNEUROSCI.3938-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Lenz O, Schmid F. Physical Review Letters. 2007;98:058104. doi: 10.1103/PhysRevLett.98.058104. [DOI] [PubMed] [Google Scholar]
- 245.Rangamani P, Benjamini A, Agrawal A, Smit B, Steigmann D, Oster G. Biomech Model Mechanobiol. 2014b;13:697. doi: 10.1007/s10237-013-0528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Rangamani P, Steigmann D. Proc Royal Soc A. 2014 doi: 10.1098/rspa.2014.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Lubensky TC, MacKintosh FC. Physical Review Letters. 1993;71:1565. doi: 10.1103/PhysRevLett.71.1565. [DOI] [PubMed] [Google Scholar]
- 248.Kuzmin PI, Akimov SA, Chizmadzhev YA, Zimmerberg J, Cohen FS. Biophysical Journal. 2005;88:1120. doi: 10.1529/biophysj.104.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hamm M, Kozlov MM. The European Physical Journal E. 2000;3:323. [Google Scholar]
- 250.Hamm M, Kozlov MM. The European Physical Journal B - Condensed Matter and Complex Systems. 1998;6:519. [Google Scholar]
- 251.May S. European Biophysics Journal. 2000;29:17. doi: 10.1007/s002490050247. [DOI] [PubMed] [Google Scholar]
- 252.Zhong-Can O-Y, Ji-Xing L, Yu-Zhang X. Geometric methods in the elastic theory of membranes in liquid crystal phases. World Scientific; 1999. [Google Scholar]
- 253.Steigmann DJ. International Journal of Non-Linear Mechanics. 2013;56:61. [Google Scholar]
- 254.Watkins EB, Miller CE, Liao W-P, Kuhl TL. ACS nano. 2014;8:3181. doi: 10.1021/nn4052953. [DOI] [PubMed] [Google Scholar]
- 255.Jablin MS, Akabori K, Nagle J. Physical review letters. 2014;113:248102. doi: 10.1103/PhysRevLett.113.248102. [DOI] [PubMed] [Google Scholar]
- 256.Izvekov S, Voth GA. The Journal of Physical Chemistry B. 2005;109:2469. doi: 10.1021/jp044629q. [DOI] [PubMed] [Google Scholar]
- 257.Lyubartsev AP. European Biophysics Journal. 2005;35:53. doi: 10.1007/s00249-005-0005-y. [DOI] [PubMed] [Google Scholar]
- 258.Shi Q, Izvekov S, Voth GA. The Journal of Physical Chemistry B. 2006;110:15045. doi: 10.1021/jp062700h. [DOI] [PubMed] [Google Scholar]
- 259.Gurtovenko AA, Vattulainen I. Journal of the American Chemical Society. 2005;127:17570. doi: 10.1021/ja053129n. [DOI] [PubMed] [Google Scholar]
- 260.Rzepiela AJ, Schäfer LV, Goga N, Risselada HJ, De Vries AH, Marrink SJ. Journal of Computational Chemistry. 2010;31:1333. doi: 10.1002/jcc.21415. [DOI] [PubMed] [Google Scholar]
- 261.Stansfeld PJ, Sansom MS. Journal of Chemical Theory and Computation. 2011;7:1157. doi: 10.1021/ct100569y. [DOI] [PubMed] [Google Scholar]
- 262.Lonsdale R, Rouse SL, Sansom MSP, Mulholland AJ. PLOS Computational Biology. 2014;10:e1003714. doi: 10.1371/journal.pcbi.1003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Noguchi H, Gompper G. Physical Review Letters. 2004;93:258102. doi: 10.1103/PhysRevLett.93.258102. [DOI] [PubMed] [Google Scholar]
- 264.Noguchi H. Scientific Reports. 2016;6:20935. doi: 10.1038/srep20935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ramakrishnan N, Sunil Kumar PB, Ipsen JH. Biophysical Journal. 2013;104:1018. doi: 10.1016/j.bpj.2012.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ayton GS, McWhirter JL, Voth GA. The Journal of Chemical Physics. 2006;124:064906. doi: 10.1063/1.2165194. [DOI] [PubMed] [Google Scholar]
- 267.Ayton GS, Blood PD, Voth GA. Biophysical Journal. 2007;92:3595. doi: 10.1529/biophysj.106.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ayton GS, Lyman E, Krishna V, Swenson RD, Mim C, Unger VM, Voth GA. Biophysical Journal. 2009;97:1616. doi: 10.1016/j.bpj.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Simunovic M, Mim C, Marlovits TC, Resch G, Unger VM, Voth GA. Biophysical Journal. 2013;105:711. doi: 10.1016/j.bpj.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Davtyan A, Simunovic M, Voth GA. Journal of Structural Biology. 2016;196:57. doi: 10.1016/j.jsb.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Lyman E, Cui H, Voth GA. 2011;13:10430. doi: 10.1039/c0cp02978e. [DOI] [PMC free article] [PubMed] [Google Scholar]