Abstract
Objective
To compare colonic microbial composition of systemic sclerosis (SSc) patients and healthy controls and to determine whether certain microbial genera are associated with SSc-gastrointestinal (GIT) symptoms.
Methods
Healthy controls were age- and gender-matched to adult SSc patients (1:1). Cecum and sigmoid mucosal lavage samples were obtained during colonoscopy. The microbiota from these samples were determined by Illumina HiSeq 2000 16S sequencing, and operational taxonomic units were selected using the Greengenes database at 97% identity. Linear discriminant analysis effect size was used to identify the genera that showed differential expression in SSc versus controls. Differential expression analysis for sequence count data was used to identify specific genera associated with GIT symptoms.
Results
Among 17 patients with SSc (88% Female; Median age 52.1 years), the mean (SD) total GIT 2.0 score was 0.7 (0.6). Principal coordinate analysis illustrated significant microbial community differences in SSc versus healthy controls in the cecum (p=0.001) and sigmoid (p=0.001) regions. Similar to inflammatory disease states, SSc patients had decreased commensal bacteria, such as Faecalibacterium and Clostridium, and increased pathobiont bacteria, such as Fusobacterium and γ-Proteobacteria, compared with healthy controls. However, SSc patients had increased Bifidobacterium and Lactobacillus, which are typically reduced in inflammation. SSc patients with moderate/severe GIT symptoms had decreased Bacteroides fragilis and increased Fusobacterium compared with SSc patients with none/mild symptoms.
Conclusions
This study demonstrates a distinct colonic microbial signature in SSc patients compared with healthy controls. This unique ecological change may perpetuate immunological aberrations and contribute to clinical manifestations of SSc.
INTRODUCTION
Gastrointestinal tract (GIT) dysfunction is a leading cause of morbidity and mortality in patients with systemic sclerosis (SSc).[1, 2] Symptoms of lower GIT involvement, such as constipation, abdominal pain, diarrhea, fecal incontinence, and weight loss, [3] are among the most disruptive physical problems for SSc patients and compromise patient emotional well-being and quality of life.[4, 5]
The etiology of SSc-related lower GIT dysfunction is largely unknown and no effective treatment options exist.[2] Research findings exploring upper GIT dysfunction have suggested that changes in the microvasculature, autonomic nervous system and immune system may contribute to smooth muscle atrophy and gut wall fibrosis.[6, 7, 8] However, more recent evidence demonstrates that bacterial overgrowth may lead to malabsorption in these patients.[9] Marie et al. [3] reported that nearly half of all SSc patients (N=51) seen consecutively in a single cohort study had a positive H2/CH4 breath test, and those with a positive breath test had more severe GIT symptoms. Moreover, studies suggest that treatment with antibiotic therapy reduces lower GIT symptoms.[3, 10] Thus, alterations in microbial composition may potentially serve as a pathogenic factor in SSc-GIT dysfunction.
While no studies have examined whether imbalances in colonic microbial composition is a feature of SSc, numerous studies have demonstrated that altered microbiota can induce or intensify inflammation in other autoimmune diseases, such as inflammatory bowel disease (IBD).[11] Substantial evidence suggests there is a decreased prevalence of beneficial human commensal genera known to produce key energy metabolites and anti-inflammatory molecules for mucosal health (e.g. Bacteroides, Bifidobacter, Clostridium type IV and XIV), with a concurrent rise in potential pathobiont genera (e.g., sulfate-reducing delta Proteobacteria, invasive gamma Proteobacteria, Fusobacterium, and Actinobacteria) in patients with IBD.[12] Moreover, specific bacterial phylotypes have been associated with increases in local host inflammatory products through metaproteome analysis at the mucosal-luminal interface in patients with IBD.[13, 14]
The present study aimed to investigate the hypothesis that the SSc disease state is associated with altered colonic microbial composition at the human mucosal-luminal interface. This study also aimed to examine the relationship between microbial composition and self-reported symptoms of GIT dysfunction in SSc patients, as an initial test of the hypothesis that certain microbial genera contribute to the GIT phenotype in SSc. If affirmed, such genera could provide specific targets for intervention to avert or treat this important clinical dimension of SSc.
MATERIALS AND METHODS
Study Participants
Patient participants were consecutively enrolled from the outpatient Rheumatology clinic at the University of California, Los Angeles (UCLA). Eligible participants included adult (age ≥18 years) patients with SSc. Exclusion criteria included co-morbid inflammatory bowel disease, contraindication to undergoing a colonoscopy and inability to withstand from taking an antibiotic and a probiotic for at least 3 weeks prior to the colonoscopy. Patients were allowed to continue taking proton pump inhibitor (PPI) medication because this agent exerts minimal to negligible affects on colonic microbiota.[15]
Healthy control colonic lavage specimens were obtained from the UCLA Pathology Microbiome Repository, which consists of specimens from 150 healthy adult patients. Age- and gender-matched healthy controls were selected by block randomization from this healthy cohort and matched with SSc patients in a 1:1 ratio.
The UCLA institutional review board (IRB) approved the study protocol (IRB #13-0011089), and written informed consent was obtained from each participant.
Specimen procurement and pre-processing
Consented participants underwent a colonoscopy by certified gastroenterologists (BER, JLK, TG). We opted to collect mucosal instead of fecal samples given the distinct and predictable compositional differences between colonic microbiota and stool microbiota demonstrated in prior studies.[16, 17] In addition, by collecting lavage specimens we could ensure optimal preservation of the samples prior to pre-processing. Stool samples collection would have required patients to transport samples from home, and if sample thawing occurred, the integrity of the DNA and RNA would be compromised. We also planned to collect colonic biopsies if there was a clinical indication for biopsy (i.e. ulceration, polyp, etc). Please see Supplementary Methods File for complete details of specimen procurement and pre-processing.
16S rRNA gene sequencing and microbial composition analysis
The microbiota from these samples were profiled by multiplex sequencing for bacterial rRNA genes using an Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA, USA) sequence technique. The exact details of this approach have been outlined in our prior publication, [14] and are summarized in the Supplementary Methods File.
Assessment of GIT symptoms
In addition to collecting demographic and disease-related characteristics, participants completed the GIT 2.0 on the morning of their colonoscopy, a valid measure of GIT symptom severity in SSc patients.[18] The questionnaire consists of seven scales (e.g. reflux, distention/bloating, diarrhea, fecal soilage, constipation, emotional well-being, and social functioning). The GIT 2.0 can furthermore discriminate between self-rated severity (i.e. none/mild versus moderate very severe/very severe disease) of GIT involvement,[18] and correlates with objective measures of GIT dysfunction at least in the upper GIT. [19] Participants also completed the health assessment questionnaire disability index (HAQ-DI), a valid measure of self-reported function that is commonly used in SSc studies.[20]
Statistical and bioinformatics analyses
Statistical approach
Analyses were performed using R version 3.1.2. Mean and standard deviation (SD) were used to describe continuous parametric data, whereas the median and interquartile ranges (IR) were used to describe continuous nonparametric data. All tests were 2-sided with an alpha level of 0.05. We used the false discovery rate (FDR) of Benjamini and Hochberg,[21] and a significant association was defined at the FDR q-value threshold ≤0.1. Cecum and sigmoid data were treated as separate datasets because prior studies have demonstrated biogeographic differences in mucosal samples from these two regions.[22] The Supplementary Methods File summarizes the exact details of the bioinformatics analysis performed for this study.
RESULTS
Participant characteristics
Seventeen patients with SSc (88% female) underwent colonoscopy and completed the questionnaire. Sigmoid and cecum lavage samples were obtained from all patients except for one patient (only sigmoid obtained). None of the patients had a clinical indication for colonic biopsy; therefore, these specimens were not collected. The median age was 52.1 years and the median disease duration was 6.6 years (Table 1). The mean GIT 2.0 scores indicated moderate symptom severity [18] for the total score, as well as for the following domains: distension/bloating, social functioning, emotional well-being, and constipation (Table 1). The mean GIT 2.0 scores for fecal soilage and diarrhea indicated mild symptom severity [18] (Table 1). Six patients (35%) had taken antibiotics in the 3 months preceding the colonoscopy; the mean time between cessation of antibiotics and colonoscopy was 6.5 weeks (Range 4 weeks to 12 weeks). None of the patients used tobacco products, and only 5 consumed alcohol regularly within the past month (mean 2.8 [SD 1.5] servings of alcohol per week).
Table 1.
Study patient characteristics
| SSc participants (N=17) | Healthy controls (N=17) | |
|---|---|---|
| Age (years) | Median 52.1 (IR 46.6, 63.0) | 55.0 (IR 51.0, 62.0) |
| Female | 15 (88.2%) | 15 (88.2%) |
| Race | ||
| White | 9 (52.9%) | 15 (88.2%) |
| Asian | 2 (11.8%) | 0 |
| More than one race | 4 (23.5%) | 0 |
| Other | 2 (11.8%) | 1 (5.9%) |
| Hispanic | 6 (35.3%) | 1 (5.9%) |
| Diffuse cutaneous disease | 6 (35.3%) | NA |
| SSc disease duration (years) | Median 6.6 (IR 2.5, 16.4) | NA |
| ANA positive | 15/16 (93.8%) | NA |
| Scl-70 positive | 3/11 (27.3%) | |
| Anti-centromere positive | 5/11 (45.5%) | |
| HRCT-Defined Interstitial Lung Disease | 12/17 (70.6%) | NA |
| Current prednisone use* | 3 (17.6%) | NA |
| Current other immunosuppressant use † | 3 (17.6%) | NA |
| Current use of probiotic oral supplement ‡ | 3 (17.6%) | NA |
| Current use of proton pump inhibitor | 10 (58.8%) | |
| Health Assessment Questionnaire (HAQ-DI), 0–3 | Mean 1.1 (SD 0.6) | NA |
| Gastrointestinal Tract (GIT) 2.0 Total Score § | Mean 0.7 (0.6) | NA |
| Distension/Bloating Score § | Mean 1.5 (0.9) | NA |
| Diarrhea Score¶ | Mean 0.4 (0.6) | NA |
| Fecal Soilage Score¶ | Mean 0.5 (0.9) | NA |
| Constipation Score § | Mean 0.7 (0.7) | NA |
| Emotional Well-Being Score § | Mean 0.5 (0.7) | NA |
| Social Functioning Score § | Mean 0.5 (0.5) | NA |
Values are n (%), except where otherwise noted.
Dosages of prednisone were ≤10 mg daily.
Immunosuppressant medications included mycophenolate (N=1) and azathioprine (N=2).
Probiotics used included Culturelle (N=1), Florify (N=1), and Align (N=1). Probiotics were not consumed within 3 weeks of the colonoscopy.
Score indicates moderate symptom severity.[18]
Score indicates mild symptom severity.[18]
SSc alters colonic microbial diversity
After the OTU selection process, a total of 5,442 and 5,593 species level OTUs were generated from cecum and sigmoid dataset, respectively. The SSc and control participants revealed similar alpha diversity (i.e. the complexity of microbial composition) by the following metrics (cecum and sigmoid p-values): phylogenetic diversity (p=0.2; p=0.1); observed species (p=0.4; p=0.2); and Shannon index (p=0.7; p=0.6). Compared with controls, SSc participants exhibited a trend for increased alpha diversity using Chao 1 diversity metric (p=0.07; p=0.09, for the cecum and sigmoid regions, respectively). The Chao 1 metric estimates OTU richness for microbial communities, [23] suggesting that the SSc disease state is associated with a potential increase in bacterial diversity (Supplementary Figure 1).
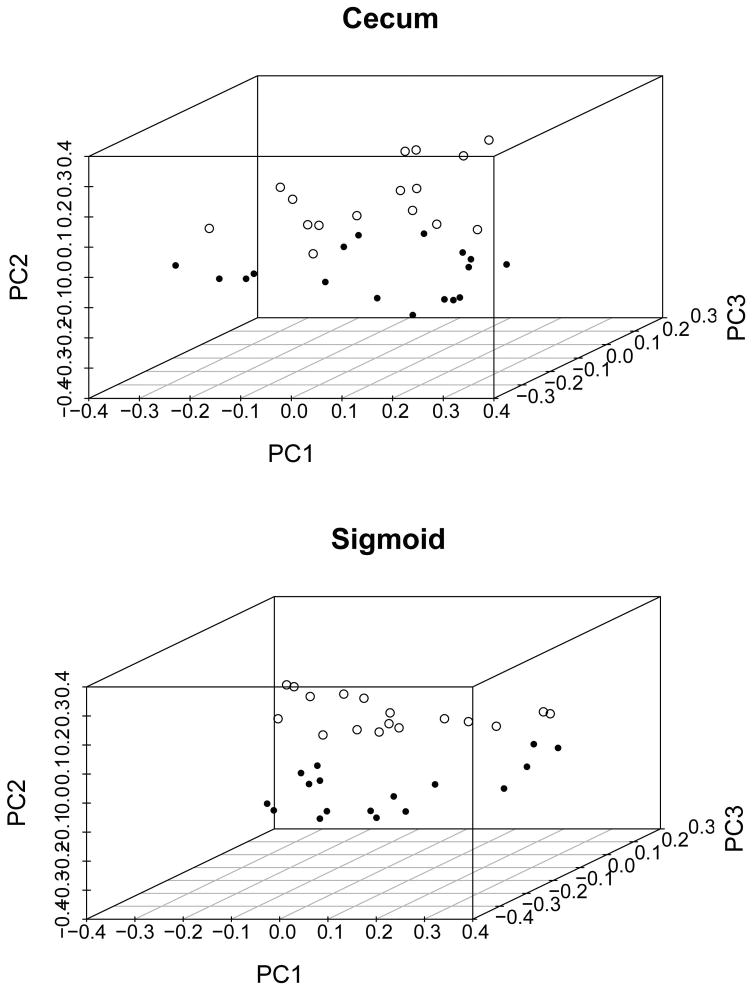
Beta diversity was then computed using unweighted and weighted Unifrac distance metrics to determine whether SSc and control subjects differed in their microbial composition. The PCoA visualization of unweighted Unifrac distances and Adonis analysis of this comparison is shown in Figure 1. In both cecum (p=0.001) and sigmoid (p=0.001) regions, the microbial composition among SSc patients was significantly different from healthy controls. Significant differences between SSc patients and healthy subjects were also observed by weighted Unifrac distances (Adonis values of p=0.03 and p=0.03 for cecum and sigmoid, respectively).
Figure 1. Significant differences in the beta diversity of the SSc and healthy samples as demonstrated by principal coordinate analysis plots of the unweighted UniFrac distance for the cecum (top panel: R2=0.9, P=0.001) and sigmoid (bottom panel: R2=0.9, P=0.001) samples.
Each dot represents a sample from a SSc patient (open circle) or a healthy control (closed circle). The p-values provided were calculated by analysis of variance using distance matrices.
Numerous colonic microbial genera are differentially abundant in SSc patients
To begin to define the compositional differences between SSc and healthy controls predicted by the beta diversity analysis, the relative abundances of microbial composition at different taxonomic levels were computed (Supplementary Figure 2). The three predominant phyla in SSC samples were Bacteroidetes (cecum: 47.2%; sigmoid: 46.7%), Firmicutes (cecum: 23.3%; sigmoid: 25.2%), and Proteobacteria (cecum: 23.1%; sigmoid: 20.5%). Similar to prior microbiome studies in IBD [24], the relative abundances of Verrucomicrobia, Fusobacteria and Actinobacteria were significantly increased in cecum samples from SSc patients compared with controls (q =0.03, 0.02, 0.003, respectively).
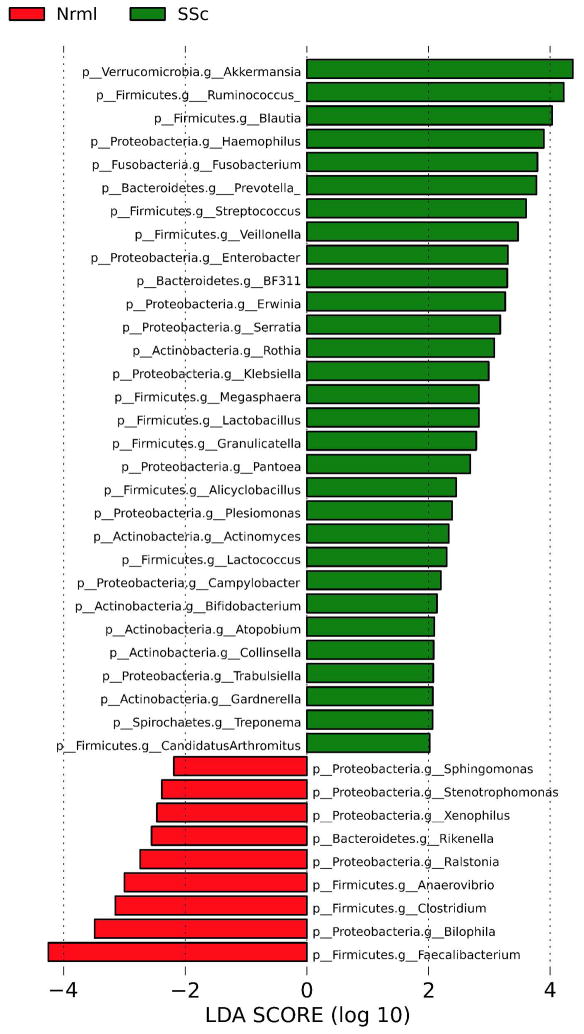
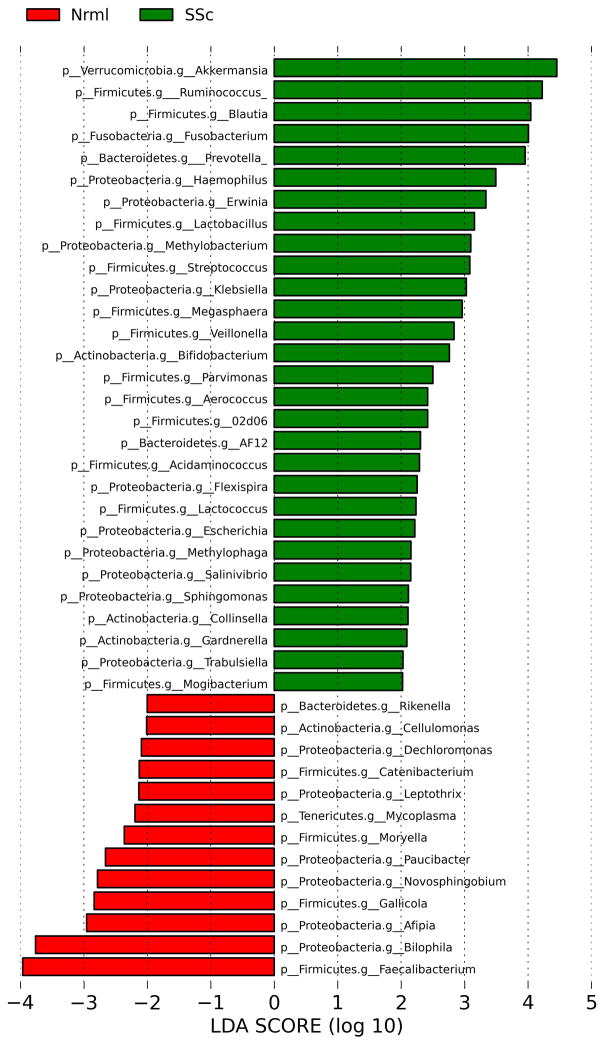
The taxonomic differences between SSc and healthy controls at the genus level were assessed by Linear Discriminant Analysis Effect Size (LefSe) multivariate analysis (to correct for species abundance). The microbial enrichments and depletions reaching significance after correction for multiple testing (q<0.1) are shown for the cecum (Figure 2) and sigmoid (Figure 3).
Figure 2. Genus level taxa associated with SSc patients versus healthy subjects in the cecum.
Linear Discriminant Analysis Effect Size (LefSe) multivariate analysis was used to identify significant associations (q<0.1), and Linear Discriminant Analysis (LDA) was used to calculate the effect size for these associations. Negative and positive effect sizes denote genera decreased (red) or increased (green) in SSc patients, respectively.
Figure 3. Genus level taxa associated with SSc patients versus healthy subjects in the sigmoid.
Linear Discriminant Analysis Effect Size (LefSe) multivariate analysis was used to identify significant associations (q<0.1), and Linear Discriminant Analysis (LDA) was used to calculate the effect size for these associations. Negative and positive effect sizes denote genera decreased (red) or increased (green) in SSc patients, respectively.
In the cecum, commensal genera such as Faecalibacterium, Clostridium, and Rikenella were depleted in SSc patients; whereas, Fusobacterium, Prevotella, and the uncommon γ-Proteobacteria, Erwinia and Trabsulsiella, were enriched in SSc patients. Similarly, in the sigmoid, Faecalibacterium and Rikenella were depleted in SSc patients; whereas, numerous sigmoid genera, such as Fusobacterium and Prevotella were enriched in SSc patients. Surprisingly, two commensal bacterial genera (Lactobacillus and Bifidobacterium) were found in greater abundance in SSc patients compared with controls in both the sigmoid and cecum regions (Figure 2, 3). These two species are typically reduced in abundance in chronic inflammatory states, such as IBD.[25]
Specific microbial genera and species are associated with SSc GIT symptoms
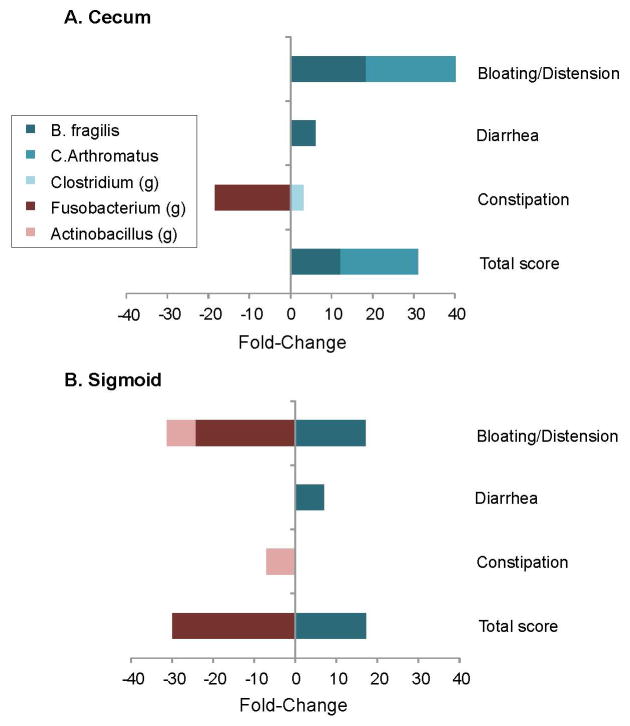
Differential expression analysis for sequence count data (DESeq2) multivariate analysis was used to identify microbial taxa associated with SSc GIT symptoms, including total GIT 2.0 score and individual domains (constipation, diarrhea, or distension/bloating). For the total score or domains, patients were dichotomized into low (none-to-mild) or high (moderate to severe) disease severity groups, and the fold-change and q-values for organisms reaching significance were tabulated (Supplementary Table 1). The findings are visualized in Figure 4.
Figure 4. Bacterial taxa associated with GIT disease score and domains.
Patients were dichotomized into low (none-to-mild) or high (moderate to severe) disease severity groups for the total GIT 2.0 score and its individual domains (constipation, diarrhea, or distension/bloating). Differential expression analysis for sequence count data (DESeq2) multivariate analysis was used to identify microbial taxa significantly associated with low versus high groups (Supplemental Table 1), and calculate the fold-change between groups. Positive and negative factor-change scores denote organisms decreased and increased in high disease severity groups, respectively. Legend provides color code of bacterial taxa; “g” denotes genus-level taxa.
In both the cecum and sigmoid, Bacteroides fragilis was elevated in patients with mild disease (total GIT score, diarrhea domain, and bloating/distension domain). Candidatus Arthromitus had a similar association with mild disease, but only in the cecum. In the cecum, the Clostridium genus also had a small, restricted association with diarrhea. Conversely, genus-level members of Fusobacterium and Actinobacillus were associated with severe disease, but more prominently in the sigmoid region, and more heterogeneously with respect to symptom domains.
Differences in microbial composition by SSc subtype
In an exploratory analysis, we compared beta diversity between patients with limited (N=11) and diffuse (N=6) cutaneous disease. Differences in beta diversity were significant by Adonis in the cecum (R2=0.13, p=0.011), but not in the sigmoid (R2=0.08, p=0.2) using unweighted Unifrac distances. However, only a small amount of the variation (13%, as reflected by the R2) could be explained by this subtype grouping for the cecum. LefSe multivariate analysis was used to examine the taxonomic differences between limited and diffuse patients at the genus level. After correction for multiple testing (q<0.1), Lactobacillus from the Firmicutes phylum was higher in patients with limited disease (LDA 2.7), while Paludibacter from the Bacteroidetes phylum was lower in patients with limited disease (LDA 2.7), compared with patients with diffuse disease.
Altered metabolic proficiencies of microbiota in SSc patients
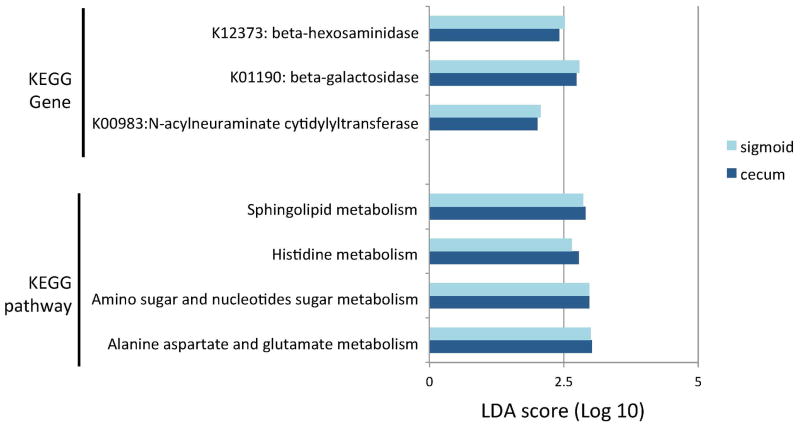
To compare the functional proficiencies of the microbial communities in SSc patients and healthy controls, the imputed metagenomic composition for each patient was determined by PICRUSt, and compared by calculating Bray-Curtis distance matrices and visualizing by PCoA Differences in metagenomic content between SSc patients and healthy controls reached significance by Adonis in both the cecum (R2= 0.9, p=0.016) and sigmoid (R2=0.9, p=0.007) regions. Among all of imputed genes, 66 (cecum) and 89 (sigmoid) genes showed significant differences (q<0.1) in SSc patients compared with healthy controls. Within these significant genes, 41 (cecum) and 47 (sigmoid) genes were mapped to biological pathways through the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. (See Supplementary Table 2 for a complete list of gene differences and their associated pathways). SSc subjects had significantly decreased abundance of genes involved in amino sugar and nucleotide sugar metabolism, such as N-acylneuraminate cytidylyltransferase (K00983; log Linear Discriminant Analysis (LDA) effect size [q-value] in the cecum and sigmoid, respectively: 2.0 [0.001] and 2.1 [0.001]) and beta-hexosaminidase (K12373; log LDA effect size the in cecum and sigmoid, respectively: 2.4 [0.07] and 2.5 [0.04]), as well as genes involved in sphingolipid metabolism, such as beta-galactosidase (K01190; log LDA effect size in the cecum and sigmoid, respectively: 2.7 [0.02] and 2.8 [0.007]) (Figure 5).
Figure 5. Significant differences in metagenomic content between SSc patients and healthy controls.
Linear Discriminant Analysis Effect Size (LefSe) multivariate analysis was used to identify significant associations (q<0.1), and Linear Discriminant Analysis (LDA) was used to calculate the effect size for these associations (Supplemental Table 2). Positive LDA score indicates genes and pathways decreased in SSc patients compared with healthy controls. Legend provides color code for cecum and sigmoid data. KEGG= Kyoto Encyclopedia of Genes and Genomes.
Discussion
To our knowledge, the present study is the first to define colonic microbial composition in adult patients with SSc. We report a unique colonic microbial consortium in SSc patients characterized by significant increases in Fusobacterium, Prevotella and uncommon γ-Proteobacteria (i.e. Erwinia, and Trabsulsiella) genera, and significant decreases in Faecalibacterium and Clostridium genera, compared with age- and gender-matched healthy controls. Interestingly, the SSc microbial consortium was also enriched with Lactobacillus and Bifidobacterium, two commensal genera typically found in lower abundance in chronic inflammatory states.
Fusobacterium species represent a group of gram-negative anaerobes that principally colonize the oral cavity. When present in the colon, these species are considered pathobionts given their invasive nature and ability to translocate into the blood and contribute to systemic processes, including bacteremia, organ abscesses, and possibly coronary artery disease.[26, 27] In patients with Crohn’s disease, Fusobacterium species are increased compared with controls,[28] and human isolates of Fusobacterium varium induce colonic mucosal erosions in mice.[29] Moreover, Fusobacterium isolates recovered from IBD patients demonstrate enhanced invasive and pro-inflammatory properties in cultured epithelial cell assays than those strains isolated from healthy individuals,[30] further implicating these species in the pathogenesis of IBD.
Prevotella species were also enriched in SSc patients compared with healthy controls. These genera are increased in patients with Crohn’s disease (CD), [31] as well as in patients with new onset rheumatoid arthritis. [32]. A recent study found that mice colonized with Prevotella copri, in particular, displayed increased inflammation in dextran sulfate sodium-induced colitis. [32].
The observation that Fusobacterium and Prevotella are increased in SSc patients compared with controls suggests that these species may play a role in the development of the SSc-GIT phenotype. Additional studies are needed to understand their pathogenic potential in SSc. Given the established link between diet and Prevotella, [33] there may be a future role for dietary modifications in averting SSc-GIT symptoms.
The genera depleted in SSc, most notably Faecalibacterium and Clostridium species, are commensal organisms, which may protect the host from mucosal inflammation and against colonization of pathogenic species.[34] For example, Faecalibacterium prausnitzii is decreased in IBD.[35] Low levels of mucosa-associated Faecalibacterium prausnitzii are associated with a higher risk of recurrent CD after surgery;[36] whereas recovery of Faecalibacterium prausnitzii after relapse is associated with maintenance of clinical remission of UC.[37] Moreover, Clostridium species have been found to induce the expansion of regulatory T cells, thereby reducing intestinal inflammation.[38]
A surprising observation in this study was the significant increase in Bifidobacterium and Lactobacillus in SSc patients compared with healthy controls. Lactobacillus was also higher in patients with limited cutaneous sclerosis compared with diffuse cutaneous sclerosis, although this latter finding should be interpreted with caution given the exploratory nature of this subgroup analysis. While Bifidobacterium and Lactobacillus are typically depleted in chronic inflammatory states, a recent study of patients with enthesitis-related arthritis (ERA) also demonstrated an increase in Bifidobacterium in stool samples of children with ERA compared with controls.[39]
Taken together, these findings suggest that our traditional views of “protective,” commensal species need to be considered in the context of the underlying disease state. Given the myriad of ways in which the phenotypic expression of SSc differs from other autoimmune diseases, it is not surprising that the microbial consortium of SSc is rather unique. Moreover, these findings may indicate that therapeutic efforts to increase commensal organisms in SSc (i.e. current commercial probiotics, which commonly include Lactobacillus or Bifidobacter sp.) should be targeted only towards select organisms.
In addition to identifying distinct features of the SSc colonic microbial consortium, the present study elucidated relationships between specific microbial genera/species and the SSc-GIT phenotype. Increased abundance of Fusobacterium and decreased abundance of Bacteroides fragilis and Candidatus Arthromitus were associated with increased GIT symptom severity, as measured by the total GIT 2.0 score, as well as many of the individual GIT 2.0 domains reflecting lower GIT dysfunction. If Bacteroides fragilis and Candidatus Arthromitus are validated in future studies to attenuate mucosal inflammation in SSc, therapy directed at increasing the growth or activity of these species would merit testing to ameliorate SSc-GIT symptoms.
Furthermore, the finding of differences in predicted metagenomic pathways between SSc patients and healthy controls implies that the compositional shifts observed in this study likely correspond to altered functional capacity of the SSc-associated microbiome relative to a healthy microbiome. These pathways may mediate the relationship between microbiota composition and GIT symptoms and warrant further investigation.
The nature and findings of the present study should be placed in the context of certain limitations. First, the present study is cross-sectional. It is unclear whether the relationships observed between specific genera and GIT symptoms persist with time. To address this question, a 12-month longitudinal study of this cohort is currently underway. Second, there may be a small batch effect as the high-throughput sequencing analysis was performed for the control samples prior to the SSc samples. Given that the sequencing protocol was identical for both groups and that fact that the differences observed between SSc and controls were highly significant, a possible batch effect is unlikely to fully explain the observed group differences.
Third, the sample size is small, and there is a need for a validation cohort. Despite the small sample size, however, we observed several significant associations suggesting that the present findings are unlikely to be due to chance alone. Fourth, because of this small sample size and our concern for multiple hypothesis testing (type 1 error), we were unable to perform meaningful subgroup analyses to compare the microbiota of patients with different disease characteristics (i.e. Scl-70 antibody negative versus positive patients). Fifth, in the future, it may be prudent to include a measure of intestinal motility. Since no valid measure of colonic motility presently exists for SSc, this study did not assess colonic motility. However, future studies of this nature may consider using potential surrogate measures of motility, such anal endosonography and/or manometry. This may help us understand the effects (if any) intestinal dysmotility has on microbial composition in SSc, or vice versa. Future studies may also assess dietary intake patterns to ascertain whether certain dietary features (i.e. gluten-free, dairy-free, high animal protein, etc.) affect colonic microbiota.
An additional unanswered question of our study is whether the observed shifts in the colonic microbial consortium in SSc are present prior to the development of SSc-GIT symptoms. A study of patients with very early SSc, such as the VEDOSS cohort (40) may help to discern whether these shifts contribute to the pathogenesis of SSc-GIT.
The present study also has several strengths. First, we obtained our specimens during endoscopy at two colonic regions, which facilitates an in-depth investigation of the microbiota at the mucosal-luminal surface, in contrast with stool collection. Second, we ensured that all patients withheld medications, such as antibiotics and probiotics, at least 3 weeks prior to the colonoscopy, by verifying medications lists three times in the month preceding the colonoscopy. Third, we employed sophisticated statistical analyses to correct for multiple hypothesis testing using a relatively conservative FDR q-value. Fourth, our multivariate method for comparing metagenomic data (LefSe) controlled for relative species abundance and also provided an estimate of the magnitude of the observed difference. Finally, by including a clinical outcome measure, we have endeavored to discern how changes in the microbiome in SSc contribute to GIT symptoms.
To conclude, the present study utilized an innovative experimental strategy to identify bacterial genera associated with SSc. Using an integrative bioinformatics approach, this study also uncovered relationships between specific genera and clinical manifestations of SSc, which merit further investigation. Interestingly, many of the gains and losses appreciated in SSc are similar to those found in Crohn’s disease, a disease that like SSc has both inflammatory and fibrosing pathological features. While future studies are needed to validate and expand upon these findings, the present study is the first to identify a colonic microbial consortium unique to the SSc disease state.
Supplementary Material
Chao1 alpha diversity metric for SSc (dotted line) and healthy controls (solid line) samples. There was a trend for an increase in species richness in SSc patients compared with healthy controls (Nrml) in the cecum (top panel; p=0.07) and sigmoid (lower panel; p=0.09).
Data are plotted to represent cecum (top panel) and sigmoid samples (bottom panel) from the same individual in vertical alignment. One SSc participant had only a sigmoid sample.
Acknowledgments
This study was funded by USPHS grants DK046763 (JB, DPBM), DK062413 (DPBM), AI078885 (JB), AI067068 (DPBM), U54DE023789-01 (DPBM) HS021747 (DPBM), and the NIH/National Center for Advancing Translational Science (NCATS) UL1TR000124; by the Helmsley Charitable Trust and the Crohn’s and Colitis Foundation of America (both to JB, JB, and DPBM). DPBM was supported by the Fineberg Foundation, the Cedars-Sinai F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute, the European Union (305479), and the Joshua L. and Lisa Z. Greer Chair in IBD Genetics (all to DPBM). JPJ was supported by T32DK07180-39. ERV was supported by the Specialty Training and Advanced Research (STAR) Program at the David Geffen School of Medicine at UCLA. None of the authors reports any conflicts of interest.
We thank the patient volunteers for their participation in this study.
References
- 1.Lock G, Holstege A, Lang B, Scholmerich J. Gastrointestinal manifestations of progressive systemic sclerosis. The American journal of gastroenterology. 1997;92:763–71. [PubMed] [Google Scholar]
- 2.Sallam H, McNearney TA, Chen JZ. Systematic review: pathophysiology and management of gastrointestinal dysmotility in systemic sclerosis (scleroderma) Alimentary pharmacology & therapeutics. 2006;23:691–712. doi: 10.1111/j.1365-2036.2006.02804.x. [DOI] [PubMed] [Google Scholar]
- 3.Marie I, Ducrotté P, Denis P, Menard J-F, Levesque H. Small intestinal bacterial overgrowth in systemic sclerosis. Rheumatology. 2009;48:1314–19. doi: 10.1093/rheumatology/kep226. [DOI] [PubMed] [Google Scholar]
- 4.Franck-Larsson K, Graf W, Rönnblom A. Lower gastrointestinal symptoms and quality of life in patients with systemic sclerosis: a population-based study. European journal of gastroenterology & hepatology. 2009;21:176–82. doi: 10.1097/MEG.0b013e32831dac75. [DOI] [PubMed] [Google Scholar]
- 5.Bodukam V, Hays RD, Maranian P, Furst DE, Seibold JR, Impens A, et al. Association of gastrointestinal involvement and depressive symptoms in patients with systemic sclerosis. Rheumatology. 2010:keq296. doi: 10.1093/rheumatology/keq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manetti M, Neumann E, Milia AF, Tarner IH, Bechi P, Matucci-cerinic M, et al. Severe fibrosis and increased expression of fibrogenic cytokines in the gastric wall of systemic sclerosis patients. Arthritis Rheum. 2007;56:3442–7. doi: 10.1002/art.22940. [DOI] [PubMed] [Google Scholar]
- 7.Kawaguchi Y, Nakamura Y, Matsumoto I, Nishimagi E, Satoh T, Kuwana M, et al. Muscarinic-3 acetylcholine receptor autoantibody in patients with systemic sclerosis: contribution to severe gastrointestinal tract dysmotility. Ann Rheum Dis. 2009;68:710–4. doi: 10.1136/ard.2008.096545. [DOI] [PubMed] [Google Scholar]
- 8.Roberts CG, Hummers LK, Ravich WJ, Wigley FM, Hutchins GM. A case-control study of the pathology of oesophageal disease in systemic sclerosis (scleroderma) Gut. 2006;55:1697–703. doi: 10.1136/gut.2005.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaye SA, Lim SG, Taylor M, Patel S, Gillespie S, Black CM. Small bowel bacterial overgrowth in systemic sclerosis: detection using direct and indirect methods and treatment outcome. Rheumatology. 1995;34:265–69. doi: 10.1093/rheumatology/34.3.265. [DOI] [PubMed] [Google Scholar]
- 10.Parodi A, Sessarego M, Greco A, Bazzica M, Filaci G, Setti M, et al. Small intestinal bacterial overgrowth in patients suffering from scleroderma: clinical effectiveness of its eradication. The American journal of gastroenterology. 2008;103:1257–62. doi: 10.1111/j.1572-0241.2007.01758.x. [DOI] [PubMed] [Google Scholar]
- 11.Frank DN, Amand ALS, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences. 2007;104:13780–85. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fava F, Danese S. Intestinal microbiota in inflammatory bowel disease: friend or foe? World journal of gastroenterology. 2011;17:557. doi: 10.3748/wjg.v17.i5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Presley LL, Ye J, Li X, LeBlanc J, Zhang Z, Ruegger PM, et al. Host–microbe relationships in inflammatory bowel disease detected by bacterial and metaproteomic analysis of the mucosal–luminal interface. Inflammatory bowel diseases. 2012;18:409–17. doi: 10.1002/ibd.21793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McHardy IH, Goudarzi M, Tong M, Ruegger PM, Schwager E, Weger JR, et al. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome. 2013;1:17. doi: 10.1186/2049-2618-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuda A, Suda W, Morita H, Takanashi K, Takagi A, Koga Y, Hattori M. Influence of proton-pump inhibitors on luminal microbiota in the gastrointestinal tract. Clin Transl Gastroenterol. 2015;6:e89. doi: 10.1038/ctg.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stearns JC, Lynch MD, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, et al. Bacterial biogeography of the human digestive tract. Scientific reports. 2011:1. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–97. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khanna D, Hays RD, Maranian P, Seibold JR, Impens A, Mayes MD, et al. Reliability and validity of the University of California, Los Angeles scleroderma clinical trial consortium gastrointestinal tract instrument. Arthritis Care & Research. 2009;61:1257–63. doi: 10.1002/art.24730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bae S, Allanor Y, Furst DE, Bodukam V, Coustet B, Morgaceva O, et al. Association between a scleroderma-specific gastrointestinal instrument and objective tests of upper gastrointestinal involvements in systemic sclerosis. Clin Exp Rheumatol. 2013;31:57–63. [PubMed] [Google Scholar]
- 20.Steen VD, Medsger TA. The value of the health assessment questionnaire and special patient-generated scales to demonstrate change in systemic sclerosis patients over time. Arthritis & Rheumatism. 1997;40:1984–91. doi: 10.1002/art.1780401110. [DOI] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- 22.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill TC, Walsh KA, Harris JA, Moffett BF. Using ecological diversity measures with bacterial communities. FEMS Microbiology Ecology. 2003;43:1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 24.Tong M, Li X, Wegener Parfrey L, Roth B, Ippoliti A, Wei B, et al. A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PloS one. 2013;8:e80702. doi: 10.1371/journal.pone.0080702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–99. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts G. Fusobacterial infections: an underestimated threat. British journal of biomedical science. 1999;57:156–62. [PubMed] [Google Scholar]
- 27.Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proceedings of the National Academy of Sciences. 2011;108(Supplement 1):4592–98. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gevers D, Kugathasan S, Denson LA, Vazquiez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naïve microbiome in new-onset Crohn’s disease. Cell Host & Microbe. 2014;15:382–92. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Okayasu I. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 2003;52:79–83. doi: 10.1136/gut.52.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, DeVinney R, et al. Invasive potential of gut mucosa-derived fusobacterium nucleatum positively correlates with IBD status of the host. Inflammatory bowel diseases. 2011;17:1971–78. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 31.Benjamin JL, Hedin CRH, Koutsoumpas A, Ng SC, McCarthy NE, Prescott NJ, et al. Smokers with active Crohn’s disease have a clinical relevant dysbiosis of the gastrointestinal microbiota. Inflammatory Bowel Disease. 2012;18:1092–100. doi: 10.1002/ibd.21864. [DOI] [PubMed] [Google Scholar]
- 32.Scher JU, Sczesnak A, Longman RS, et al. Expansion of intestinal Prevotella copri correlations with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu GD, Chen J, Hoffman C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamada N, Chen G, Núñez G. A complex microworld in the gut: harnessing pathogen-commensal relations. Nature medicine. 2012;18:1190–91. doi: 10.1038/nm.2900. [DOI] [PubMed] [Google Scholar]
- 35.Sokol H, Seksik P, Furet J, Firmesse O, Nion-Larmurier I, Beaugerie L, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflammatory bowel diseases. 2009;15:1183–89. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 36.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences. 2008;105:16731–36. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varela E, Manichanh C, Gallart M, Torrejón A, Borruel N, Casellas F, et al. Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Alimentary pharmacology & therapeutics. 2013;38:151–61. doi: 10.1111/apt.12365. [DOI] [PubMed] [Google Scholar]
- 38.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–36. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 39.Stoll ML, Kumar R, Morrow CD, Lefkowitz EJ, Cui X, Genin A, et al. Altered microbiota associated with abnormal humoral immune responses to commensal organisms in enthesitis-related arthritis. Arthritis research & therapy. 2014;16:486. doi: 10.1186/s13075-014-0486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lepri G, Guiducci S, Bellando-Randone S, Giani I, Bruni C, Blagojevic J, et al. Evidence for oesophageal and anorectal involvement in very early systemic sclerosis (VEDOSS): report from a single VEDOSS/EUSTAR centre. Ann Rheum Dis. 2015;74:124–8. doi: 10.1136/annrheumdis-2013-203889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chao1 alpha diversity metric for SSc (dotted line) and healthy controls (solid line) samples. There was a trend for an increase in species richness in SSc patients compared with healthy controls (Nrml) in the cecum (top panel; p=0.07) and sigmoid (lower panel; p=0.09).
Data are plotted to represent cecum (top panel) and sigmoid samples (bottom panel) from the same individual in vertical alignment. One SSc participant had only a sigmoid sample.