Abstract
KCNQ1 is the pore forming subunit of cardiac slow-delayed rectifier potassium (IKs) channels. Mutations in the kcnq1 gene are the leading cause of congenital long QT syndrome (LQTS). Here we present the cryo-EM structure of a KCNQ1/CaM complex. The conformation corresponds to an “uncoupled”, PIP2-free state of KCNQ1, with activated voltage sensors and a closed pore. Unique structural features within the S4-S5 linker permit uncoupling of the voltage sensor from the pore in the absence of PIP2. CaM contacts the KCNQ1 voltage sensor through a specific interface involving a residue on CaM that is mutated in a form of inherited LQTS. Using an electrophysiological assay we find that this mutation on CaM shifts the KCNQ1 voltage-activation curve. This study describes one physiological form of KCNQ1, depolarized voltage sensor with closed pore in the absence of PIP2, and reveals a regulatory interaction between CaM and KCNQ1 that may explain CaM-mediated LQTS.
Graphical abstract
Cryo-EM structure of the potassium channel KCNQ1 in complex with calmodulin provides insights into molecular underpinnings of congenital long QT syndrome.

Introduction
Cardiac rhythm is triggered and maintained by synchronized electrical impulses throughout the heart. The slow delayed rectifier current, IKs, plays a vital role in shaping the repolarization phase of human cardiac action potentials, which are about 250-350 ms long (Abbott, 2014; Jespersen et al., 2005; Robbins, 2001). The molecular constituent of IKs is a channel complex formed by KCNQ1 (Kv7.1 or KvLQT1) in association with KCNE1 (minK) (Barhanin et al., 1996; Sanguinetti et al., 1996). KCNQ1 belongs to the voltage-gated potassium channel superfamily and is the pore forming subunit of IKs channels. Mutations in the kcnq1 gene are associated with several congenital cardiac diseases including long QT syndromes, familial atrial fibrillation, and short QT syndromes (Hedley et al., 2009). In the United States 3,000 to 4,000 cases of sudden death in children and young adults occur each year associated with inherited long QT syndrome; about 40% of these are due to loss of function mutations in the kcnq1 gene (Pagon et al., 1993a). In addition to their role in the human heart, KCNQ1 channels are also essential for the function of epithelial cells in various organs, including colon, kidney, inner ear and small intestine (Abbott, 2014; Jespersen et al., 2005). Consequently, patients with Jervell and Lange-Nielsen syndrome, a subtype of long QT syndrome, also experience hearing loss (Pagon et al., 1993b). For these reasons KCNQ1 is a potential drug target for treatment of multiple diseases (Wulff et al., 2009).
KCNQ1 channel activity is regulated by the signaling lipid phosphatidylinositol 4,5-bisphosphate (PIP2) (Ford et al., 2004; Loussouarn et al., 2003; Suh and Hille, 2002; Zhang et al., 2003). PIP2 serves as a cofactor for many ion channels and transporters (Suh and Hille, 2008). KCNQ channels are completely dependent on PIP2: their activity is thoroughly suppressed by stimulation of Gq-coupled receptors, which deplete PIP2 through activation of PLC (Delmas and Brown, 2005). PIP2 acts as if to stabilize the open state of KCNQ1 (Loussouarn et al., 2003; Zhang et al., 2003). Open state stabilization is thought to stem from the ability of PIP2 to maintain competent coupling between the voltage sensor and pore (Zaydman and Cui, 2014; Zaydman et al., 2013). This mechanism is supported by the fact that the KCNQ1 voltage sensors undergo voltage-dependent conformational changes but fail to open the pore in the absence of PIP2 (Zaydman et al., 2013).
CaM is likely an obligate subunit of KCNQ1, interacting with cytosolic IQ motifs and contributing to channel assembly and surface localization (Ghosh et al., 2006; Shamgar et al., 2006). CaM also functions as a regulator of channel gating, relieving inactivation in a Ca2+-dependent manner, which is consistent with the observation that increased intracellular Ca2+ concentration augments the delayed rectifier potassium current in ventricular myocytes of guinea pig (Tohse, 1990). Of note, human genetic studies have also identified multiple CaM mutations associated with cardiac arrhythmia, including long QT syndromes (Boczek et al., 2016; Jiménez-Jáimez et al., 2016). However, the molecular mechanisms by which CaM mutations lead to disease are uncertain due to the large number of CaM binding partners.
For a more complete understanding of KCNQ1 structure and function we have determined the molecular structure of KCNQ1 in complex with CaM.
Results
Architecture of KCNQ1/CaM
A construct (KCNQ1EM) encompassing amino acids 67-610 of frog (Xenopus laevis) KCNQ1, which shares 78% sequence identity with human KCNQ1 (Figure S1), was used for single-particle electron cryo-microscopy (cryo-EM) analysis. The construct exhibits good biochemical stability and displays similar channel kinetics to full-length KCNQ1 (Figure 1A-1C). Half-activation voltages (V1/2) of full-length frog KCNQ1 and KCNQ1EM are -15.5 ± 0.6 mV and -10.3 ± 0.3 mV, respectively. KCNQ1EM was expressed using a mammalian cell expression system and was initially co-purified with endogenous CaM (Figure 1C), which was confirmed by mass spectrometry. Further sample preparations were carried out by co-expressing KCNQ1EM and CaM.
Figure 1. Functional validation, overall architecture and domain organization of KCNQ1EM.
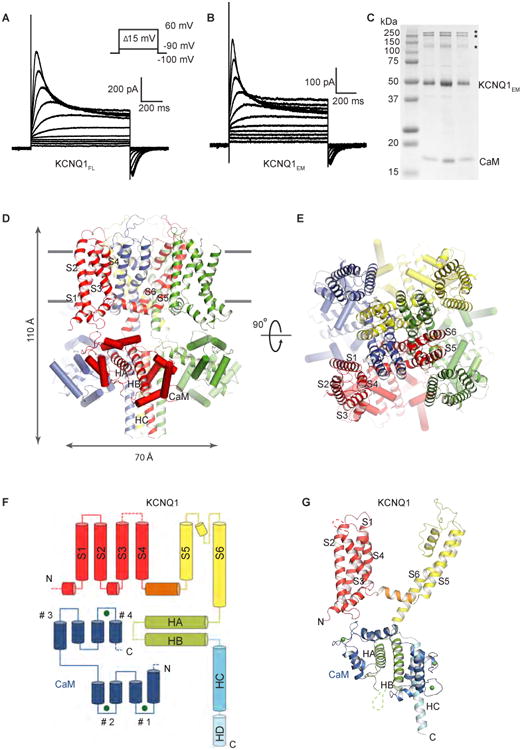
(A-B) Representative voltage family current traces of the full-length frog KCNQ1 (KCNQ1FL) and the cryo-EM construct (KCNQ1EM). Voltage steps are applied from -90 mV to 60 mV at 15 mV increments.
(C) SDS-PAGE gel of purified KCNQ1EM/CaM complex from peak fractions of size exclusion chromatography with protein markers on the left. The bands indicated by asterisks are potential KCNQ1EM dimers, trimers and tetramers.
(D-E) Side view and top view of the KCNQ1EM/CaM complex. Each protomer is shown in a different color, and CaM is represented as cylinders. The S1-S6 and HA-HC are labeled.
(F) Domain organization of one subunit. The HD, which is masked out in 3D reconstruction, is indicated by cylinder with dashed outlines. The EF hands of CaM are labeled as #1-#4 from the N-terminal to C-terminal end. The first two EF-hand regions form the N-lobe, and the other two form the C-lobe. Green spheres represent calcium ions.
(G) Model of one subunit with domains colored as in (F).
The KCNQ1EM/CaM complex structure was determined to an overall resolution of 3.7 Å with C4 symmetry imposed (Figure S2A and S2B), and the final cryo-EM map allows de novo model building (Figure S3A and S3B). As indicated by the local-resolution calculation, the central region of the map is better resolved than the periphery (Figure S2C). Model refinement was carried out in both real and reciprocal space against one of the independently calculated half maps (working set), and validated against the other half map (free set) and the full map (Figure S3C) (Hite et al., 2015). The final model correlates well with the cryo-EM density and contains good stereochemistry (Figure S3D).
The four-fold symmetrical structure of the KCNQ1EM/CaM complex has dimensions of ∼ 70 Å × 70 Å × 110 Å (Figure 1D and 1E). Each protomer contains one KCNQ1EM subunit and one CaM. CaM has two approximately symmetrical N- and C-lobes, each containing a pair of EF-hand motifs connected by a flexible central loop (Figure 1F, 1G and S5A). The KCNQ1EM monomer contains six transmembrane helices (S1-S6), and 4 intracellular helices (HA-HD) (Figure 1F and 1G). Like Kv1.2 (Long et al., 2005), the transmembrane domain of KCNQ1EM has a classical voltage-gated potassium channel topology, featuring a domain-swapped homotetramer with the voltage sensor (S1-S4) interacting with helix S5 from the neighboring subunit (Figure 1D and 1E). The cytosolic HA and HB helices, which are connected to the S6 transmembrane helix, are sandwiched between the N- and C-lobes of CaM (Figure 1F and 1G). HC and HD form two long helical bundles that facilitate tetramerization of the channel. In contrast to a previous SAXS study carried out in the absence of the transmembrane domain (Sachyani et al., 2014), which identified flexibility in a location proximal to HC, we observe flexibility distal to the HC helix. This flexibility produced disorder in the position of the HD helix and therefore a higher resolution map could be achieved by masking out the HD bundle during single-particle reconstruction (Figure S2A).
KCNQ1EM and Kv1.2-2.1 show very interesting structural differences within their transmembrane regions. First, a nine amino acid insertion in the S2-S3 ‘foot’ that connects S2 and S3 helices is unique to KCNQ1 among the entire set of Kv1 to Kv9 channels (Figure 2B). The functional significance of this insertion, which extends into the cytoplasm and interacts with CaM, will be addressed later. Second, the S3 helix in KCNQ1EM is straight throughout its length: this contrasts the S3 helix in Kv1.2-2.1 (PDB code: 2R9R), which is bent by ∼25° owing to a proline residue that is conserved in most Kv channels but is not present in KCNQ1 (Long et al., 2007) (Figure 2B, 2C, S4A and S4B). Third, the extracellular loop between S5 and the pore helix forms a negatively charged cap element surrounding the channel entrance (Figure S4C and S4E). Finally, the S6 helix of KCNQ1EM lacks the conserved glycine and PXP motif that serve as potential gating hinges (Hackos et al., 2002; Jiang et al., 2002; Long et al., 2005) and instead has a functionally important PAG motif (Seebohm et al., 2006) (Figure S4D and S4E). KCNQ1EM also possesses a signature GSG motif forming the narrowest restriction along the ion conduction pathway (Figure S4D, S4E and 2D).
Figure 2. The activated voltage senor and closed pore.
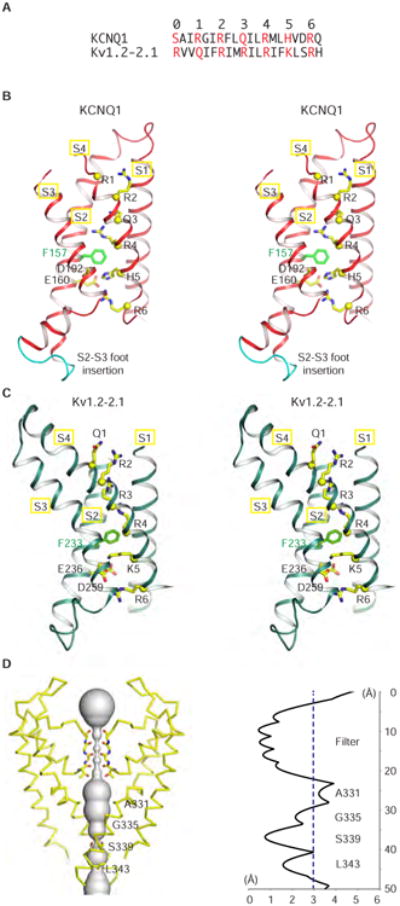
(A) Sequence alignment of S4 for KCNQ1EM and Kv1.2-2.1.
(B-C) Stereoview of the voltage sensors of KCNQ1EM and Kv1.2-2.1 (PDB: 2R9R). Only the helical regions of S1-S4 are shown for clarity. The c-alphas of the positive charged (or polar) resides on S4 are shown in yellow spheres with stick side chains, and the gating charge transfer center residues F157/F233 (green), E160/E236 (yellow) and D192/D259 (yellow) are shown as sticks. The nine amino acid insertion in KCNQ1 S2-S3 ‘foot’ is colored in cyan in (B).
(D) Left, view of the KCNQ1EM pore with front and back subunits excluded for clarity. Right, radius of the pore calculated using HOLE program. The amino acids facing the pore are labeled.
A “decoupled” conformation
The voltage sensors in KCNQ1EM in many respects resemble those in the Kv1.2-2.1 chimera (Long et al., 2007), with four positive charged (or polar) S4 amino acids above (extracellular to) the phenylalanine (F157 in KCNQ1 and F233 in Kv1.2-2.1) that caps the gating charge transfer center and two positive charged S4 amino acids below the phenylalanine (Figure 2A-2C) (Tao et al., 2010). This similarity leads us to conclude that the voltage sensor in KCNQ1 adopts a depolarized conformation, like that in Kv1.2-2.1. Such a conformation of the voltage sensor is consistent with the absence of an applied field on the KCNQ1 channels in solution. The pore in KCNQ1 is quite obviously closed, with a radius at its narrowest position (S339) of ∼0.8 Å, much less than the radius of K+ ion (Figure 2D).
In a depolarization-activated K+ channel one should generally expect a depolarized voltage sensor to be associated with an open pore, as is the case for the Kv1.2-2.1 channel (Long et al., 2007). But KCNQ1 is both depolarization-activated and PIP2-dependent: membrane depolarization opens the pore only in the presence of PIP2. But gating charge displacement currents still occur in the absence of PIP2. This fact would suggest that in the absence of PIP2 the voltage sensors depolarize even though the pore does not open (Zaydman et al., 2013). Such a condition has been referred to as an uncoupling of the pore and voltage sensor (Zaydman et al., 2013). The structure presented here is consistent with such an uncoupled state, which occurs in cells when Gq-coupled GPCRs are stimulated, leading to PIP2 depletion from the membrane.
The KCNQ1EM structure reveals an apparently disengaged connection between the S4 helix and S4-S5 linker (Figure 3). Structure comparison was carried out between transmembrane domains of KCNQ1EM and the open Kv1.2-2.1 channel, which exhibits activated voltage sensors and an open pore (Figure 3). We have superimposed the two structures by aligning their S2 helices and selectivity filters. These two reference points are selected because they are similar and because they fix the structures at central and peripheral locations. Aside from the expected differences within the pore domains (because Kv1.2-2.1 is open and KCNQ is closed, Figure S4E), superimposition reveals a large displacement of the hinge region (residues 234-236) that connects S4 to the S4-S5 linker (Figure 3). Instead of a helix, KCNQ1EM contains a loop that is shifted by ∼10.5 Å and an S4-S5 linker that is angled outward (away from S6) by ∼25° relative to that in Kv1.2-2.1. The diminished interaction surface area between the S4-S5 linker and S6 in KCNQ1 relative to Kv1.2-2.1 seems consistent with an uncoupled pore and voltage sensor.
Figure 3.

Conformational differences between open Kv1.2-2.1 and decoupled KCNQ1EM. Only the helical regions of S1-S6 are shown for clarity. The center of the hinge region, shown as yellow spheres, is displaced ∼10.5 Å in KCNQ1EM compared to its position in Kv1.2-2.1. The S4-S5 linker tilts by ∼25 degrees (indicated by black dashed lines) away from S6 in KCNQ1EM compared to its position in Kv1.2-2.1.
The region in close proximity to the loop connecting S4 to the S4-5 linker might also contribute to the binding of PIP2, which is known to facilitate the coupling between the voltage sensor and pore domain. As a signaling lipid that interacts directly with KCNQ1, PIP2 is proposed to involve the HB helix (Tobelaim et al., 2017) and the interface between the voltage sensor and pore domain (Zaydman et al., 2013). Coloring of the surface of KCNQ1EM/CaM according to electrostatic potential shows two distinct positively charged pockets (Figure 4A) that could serve as potential binding sites for negatively charged PIP2 (Kasimova et al., 2015). One of the pockets faces the inner leaflet of the membrane and consists mainly of HB and the C-terminus of S6. The other pocket faces away from the inner leaflet of the lipid bilayer and is located between voltage sensor and pore domain. This second pocket consists of residues surrounding the loop connecting S4 and S4-S5 linker. Strengthening the idea that PIP2 binds within this region, mutations have shown that these same amino acids influence PIP2 modulation of channel activity (Figure 4B) (Zaydman and Cui, 2014). It thus seems likely that PIP2 binds in this region to strengthen the coupling between the voltage sensor and the pore. This concept is conveyed in a cartoon, which illustrates a requirement of PIP2 to enable the voltage sensor to engage the pore and impart work on it (Figure 4D).
Figure 4. The hinge connecting S4 and the S4-S5 linker.

(A) Surface charge representation of KCNQ1EM. Two positive charged pockets, which are next to the hinge region (loop connecting S4 to the S4-S5 linker) and HB helix, are indicated by yellow arrows.
(B) Zoomed-in view of the hinge region and map of the key residues that are important for PIP2 regulation of the KCNQ1EM channel. The hinge region is colored in red, and c-alphas of key residues are shown as yellow spheres with side chains in stick representation.
(C) Sequence alignment of the hinge region connecting S4 and the S4-S5 linker.
(D) Cartoon depicting the idea that PIP2 couples the voltage senor domain and the pore domain, in contrast with Kv1.2-2.1. The grey box and orange cylinder represents the voltage sensor domain containing positive charged “+” amino acids on the S4 helix. The pore domain is colored yellow and the S4-S5 linker green. PIP2 is shown as a purple hexagon. The red loop represents the hinge region connecting S4 to the S4-S5 linker. The black arrow signifies the process of membrane depolarization.
CaM and long QT mutations
The cryo-EM structure shows that CaM contacts KCNQ1EM through two separate interfaces. One of these was identified previously in crystal structures of cytoplasmic domains (Sachyani et al., 2014). In this contact the HA-HB helices of KCNQ1EM insert into the middle of a clamshell-like structure formed by the N- and C-lobes of CaM (Figure 1 F and 1G). The structure of the CaM/HA-HB complex is very similar in the cryo-EM and crystal structures, with an RMSD of 0.82 Å (Figure S5A). One difference, however, is that we observe density in the 4th EF hand that was not observed in the crystal structure (PDB code: 4V0C) (Figure S3B). This density is consistent with (but not proof of, because Ca2+, Na+ and water are indistinguishable at this resolution) the presence of a Ca2+ ion. The other contact region between CaM and KCNQ1EM was not observed in crystal structures because the crystal structures did not contain the transmembrane channel. This second contact is formed between the S2-S3 loop of the voltage sensor (part of the transmembrane channel) and the 3rd EF hand of CaM (Figure 5A and S5B). Sequence alignment of the voltage-gated potassium channel (Kv1-9) families shows that the nine-amino acid S2-S3 loop in KCNQ1EM (and other Kv7s) is conserved in length and sequence among both KCNQ1 orthologs and paralogs, but is absent in the other voltage-gated potassium channels (Figure 5B-5C). This loop, unique to the Kv7s, forms the second contact surface, specifically with the 3rd EF hand of CaM. No density for a Ca2+ ion is observed here, even at the high Ca2+ concentration (0.5 mM) under which the structure was determined (Figure S3B). By interacting with both the voltage sensors and the HA-HB helices, CaM could provide an alternative functional linkage between voltage sensors and the pore of KCNQ1EM separate from the S4-S5 linker.
Figure 5. Interaction between the KCNQ1EM S2-S3 loop and CaM.
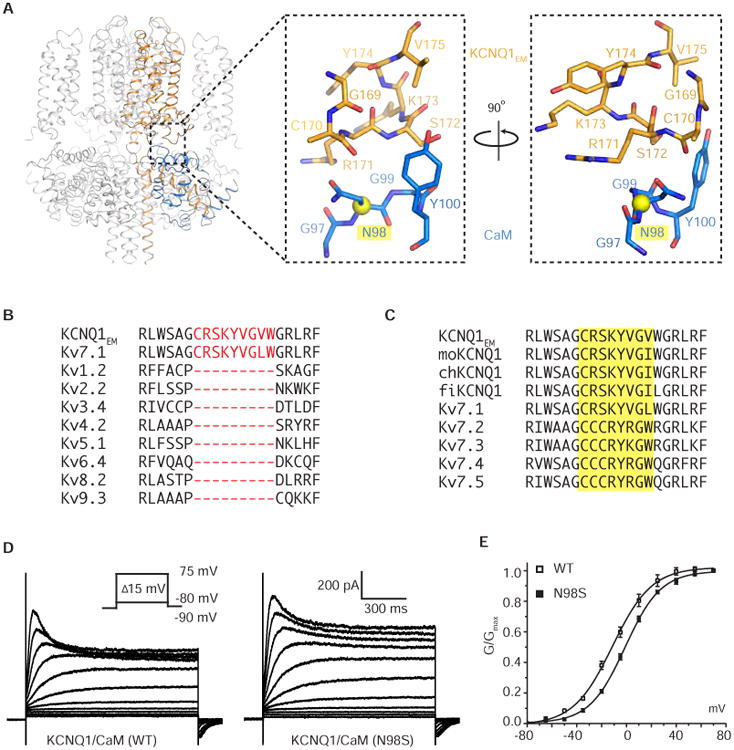
(A) Magnified view of the newly discovered interface between KCNQ1EM and CaM. The long QT disease mutant site, Asn98, is highlighted and its C-alpha is shown as a sphere. The side chain of C170 was truncated during modeling due to the absence of density.
(B-C) Sequence alignment of the S2-S3 loop region among Kv1-9 family and KCNQ1 orthologs and paralogs. Protein accession codes are as follows, Kv1.2: NP_004965.1, Kv2.2: NP_004761.2, Kv3.4: NP_004969.2, Kv4.2: NP_036413.1, Kv5.1: NP_002227.2, Kv6.4: NP_758857.1, Kv8.2: NP_598004.1, Kv9.3: NP_002243.3, Kv7.1: NP_000209.2, Kv7.2: NP_742105.1, Kv7.3: NP_004510.1, Kv7.4: NP_004691.2 and Kv7.5: NP_062816.2. All the above sequences are from Homo sapiens. moKCNQ1: NP_032460.2 (Mus musculus), chKCNQ1: XP_421022.3 (Gallus gallus) and fiKCNQ1: NP_001116714.1 (Danio rerio).
(D) Current traces recorded at different voltages for KCNQ1EM/CaM_WT and KCNQ1EM/CaM_N98S.
(E) Tail current (G/Gmax) versus voltage (G-V) curve was plotted from (D) and fitted with the Boltzmann function with V1/2 for KCNQ1EM/CaM_WT and KCNQ1EM/CaM_N98S equal to -11.3 ± 0.7 mV (SEM, n=4) and -1.1 ± 0.5 mV (SEM, n=7), respectively.
The potential relevance of interactions between KCNQ1 and CaM is underscored by the important role that CaM plays in channel modulation as well as its connection to human disease mutations. Mutations in kcnq1 are the leading cause of congenital long QT syndromes, more than 100 of which we have mapped onto the cryo-EM structure (Figure S1 and S5C) (Hedley et al., 2009). These mutations are thought to alter protein folding/trafficking or channel function, thus affecting the IKs current and the QT interval on the electrocardiogram. By classifying these mutations based on their locations within the 3D structure (Figure S5C) we hope to advance further study of their mechanisms of pathogenesis.
Several long QT mutations also map to CaM genes, however, mechanistic interpretation has been difficult owing to the fact that CaM interacts with so many different binding partners, including other long QT related proteins such as Nav1.5 and Cav1.2 (Limpitikul et al., 2014; Sarhan et al., 2012). We call to attention one particular CaM mutation, CaM_N98S, which is associated with prolonged QT intervals and sudden death in young patients (Jiménez-Jáimez et al., 2016; Makita et al., 2014). This mutation maps directly onto the interface between CaM and the voltage sensor (Figure 5A and S5B). When studied in CHO cells the CaM_N98S did not affect channel assembly or complex formation (Figure S5D) but it did affect gating (Figure 5D and 5E). Co-expression of KCNQ1 and CaM_N98S was accomplished by inserting both genes into the same transfected vector. Even though some channel assembly with endogenous CaM might occur, the half activation curve shifted by ∼10 mV (V1/2 of KCNQ1EM/CaM: -11.3 ± 0.7 mV, V1/2 of KCNQ1EM/CaM_N98S: -1.1 ± 0.5 mV). Thus, it is possible that CaM_N98S could produce long QT syndrome by affecting channel gating through the CaM-voltage sensor contact region. We also attempted to determine whether the CaM_N98S mutation affects gating in KCNQ1 when co-expressed with KCNE1. Unfortunately a shift in voltage-dependent activation could not be assessed (in either frog or human species) because the channel in complex with KCNE1 does not reach full activation even during 5 sec depolarizing steps to +80 mV. Thus it is impossible to determine a properly normalized voltage-dependent activation curve in the presence of KCNE1.
Summary
The cryo-EM structure of the KCNQ1EM/CaM complex shows us the architecture of a channel from the Kv7 branch of the K+ channel family tree. This channel exhibits a domain-swapped transmembrane topology similar to Shaker-like voltage-dependent K+ channels (Kv1.2) and dissimilar to Eag1 and hERG voltage-dependent K+ channels (Kv10 and Kv11) (Whicher and MacKinnon, 2016) to HCN channels (Lee and MacKinnon, 2017) and to Slo1 and Slo2 channels (Hite and MacKinnon, 2017; Hite et al., 2017; Tao et al., 2017). Among the domain-swapped voltage-dependent potassium channel structures so far observed (Long et al., 2005; 2007), KCNQ1 is unique in the following respect. Its S4-S5 linker, which connects the voltage sensor to the pore (and “domain-swaps” the voltage sensor to a neighboring pore subunit), is not entirely helical, but instead is partly an extended chain, or loop. This feature appears to result in a disengagement of the S4-S5 linker from the pore domain. The loop also coincides with a potential PIP2 binding site. These findings support the hypothesis that PIP2 facilitates voltage-dependent pore opening by altering the structure of the S4-S5 linker and enabling it to engage and open the pore when the membrane is electrically depolarized (Figure 4D). The cryo-EM structure also provides an explanation for long QT syndrome mediated by a mutation in CaM, which coincides with a newly discovered interface through which CaM is in direct contact with the voltage sensor.
To our knowledge KCNQ1 in situ always functions in association with a second ancillary subunit called KCNE1 (mink) (Barhanin et al., 1996; Sanguinetti et al., 1996) to conduct the slow delayed rectifier current, IKs. The KCNQ1/KCNE1 complex activates much more slowly than KCNQ1 alone, and therefore a structure of the complex will be needed to better understand this channel's role in electrical signaling. In the present study of KCNQ1 in the absence of KCNE1, we think that the main conclusions reached regarding a structural basis for voltage sensor-pore uncoupling in the absence of PIP2 and the identification of an interface through which a CaM mutation mediates long QT syndrome will likely still hold.
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Roderick MacKinnon (mackinn@rockefeller.edu)
Experimental Model and Subject Details
Cell lines
Chinese hamster ovary (CHO)-K1 cells were cultured in DMEM/F-12 medium (GIBCO) supplemented with 10% fetal bovine serum at 37°C. S f9 cells were cultured in Sf-900 II SFM medium (GIBCO) at 27°C. HEK293S GnTI - cells were cultured in Freestyle 293 medium supplemented with 2% fetal bovine serum at 37°C.
Method Details
Cloning, expression and purification of KCNQ1EM/CaM
DNA encoding frog KCNQ1 (Xenopus laevis, NP_001116347.1) was synthesized by Genewiz Inc. To improve the biochemical and thermal stability of KCNQ1, the N-terminal and C-terminal regions were truncated, leaving a construct (KCNQ1EM) with residues 67-610. EcoR I and Xho I sites were used for cloning the KCNQ1EM construct into a BacMam expression vector (Goehring et al., 2014) with a C-terminal green fluorescent protein (GFP)-6×His tag linked by a preScission protease site. DNA encoding CaM, which is absolutely conserved from frog to human, was also synthesized by Genewiz Inc. and cloned into a BacMam expression vector.
Recombinant baculoviruses of KCNQ1EM and CaM were generated separately using the Bac-to-Bac system (Invitrogen) as previous described (Goehring et al., 2014). Then P3 viruses of KCNQ1EM and CaM were mixed at a 5:1 ratio and used for transfection of HEK293S GnTI- cells for protein expression. For 1L cultures of HEK293S GnTI- cells (1.5-3×106 cells/mL) in Freestyle 293 media (Gibco) supplemented with 2% FBS (Gibco), 100 ml P3 virus mixture was used. Infected cells were incubated at 37°C overnight, and protein expression was induced by adding 10 mM sodium butyrate. Cells were cultured at 30°C for another 48-60 hrs before harvest (Goehring et al., 2014).
4L of cell pellet was resuspended in 150 ml lysis buffer (40 mM HEPES pH7.2, 150 mM KCl, 0.5 mM CaCl2, 2 μM leupepetin, 1 μM pepstatin, 100 μM AEBSF, 1 mM benzamidine and 1 μg/mL aprotinin), and then solubilized by adding a detergent mixture DDM/CHS (5:1 ratio, 6% stock) to a final concentration of 1.5% while stirring at 4°C for 2 hrs. Solubilized KCNQ1EM/CaM was separated from the insoluble fraction by high-speed centrifugation (38,000g for 1 hr), and incubated with 4 ml CNBR-activated sepharose beads (GE healthcare) coupled with 4 mg high-affinity GFP nanobodies (GFP-NB) (Kirchhofer et al., 2010). The GFP tag was cleaved by preScission protease overnight at 4°C, and KCNQ1EM/CaM complex was further purified by size-exclusion chromatography with a Superose 6, 10/300 GL column (GE Healthcare) equilibrated with 20mM HEPES pH7.2, 150mM KCl, 0.5mM CaCl2, 2mM Dithiothreitol (DTT), 0.05% DDM, 0.005% CHS. The peak fractions were pooled, concentrated to 4-5 mg/ml using a 100-kDa MWCO centrifugal device (Ambion) and immediately used for cryo-EM grid preparation.
Electrophysiology of KCNQ1
Chinese hamster ovary (CHO) cells, cultured in DMEM-F12 (Gibco) with 10% FBS and L-glutamine, were transfected with the KCNQ1EM expression plasmid with lipofectamine 2000 reagent (Thermo Fisher Scientific). A medium exchange was carried out after 4hr incubation with lipofectamine reagent. 48 hrs following transfection, the media was replaced with bath solution and experiments were performed at room temperature using whole-cell patch clamp techniques with polished borosilicate glass pipettes with resistance between 2-4 MΩ. All recordings were carried out using pClamp10.5 software (Molecular Devices), an Axopatch 200B amplifier (Molecular Devices), and an Axon digidata 1550 digitizer (Molecular Devices). Data were filtered at 1 kHz and digitized at 10 kHz.
The bath solution contains 10 mM HEPES pH 7.2, 10 mM KCl, 140 mM NaCl, 2 mM MgCl2, and 1 mM CaCl2, and the pipette solution was 10 mM HEPES pH 7.2, 50 mM KF, 100 mM KCl, 5mM EGTA. Normalized tail current vs. voltage was plotted and fit with a Boltzmann function using Origin 9 (OriginLab Corp.).
Electrophysiology of CaM long QT mutations
For measurement of effects of the CaM_N98S mutant, KCNQ1EM and CaM_N98S (or CaM_WT as control) were constructed into one BacMam vector with each expression cassette containing a CMV promoter. CHO cell transfection and recording were carried out as described above except for slight differences in the recording solutions. The bath solution contained 10 mM HEPES pH 7.2, 15 mM KCl, 136 mM NaCl, 2 mM MgCl2, and 1 mM CaCl2.
Cryo-EM analysis
Cryo-EM grids were prepared with a Vitrobot Mark IV (FEI). Quantifoil R1.2/1.3 holey carbon grids (Quantifoil) were glow-discharged for 15s. Then 3.5 μL of 5 mg/ml KCNQ1EM/CaM was pipetted onto the grids, which were blotted for 4s under blot force 1 at ∼90% humidity and frozen in liquid nitrogen cooled liquid ethane. The grids were loaded onto a 300 keV Titan Krios (FEI) with a K2 direct electron detector (Gatan).
Images were recorded with SerialEM (Mastronarde, 2005) in super-resolution mode with a super resolution pixel size of 0.65 Å and a defocus range of -1.2 to -2.8 μm. Data were collected with a dose rate of 10 electrons per physical pixel per second, and images were recorded with a 15 s exposure and 300 ms subframes (50 total frames) corresponding to a total dose of 89 electrons per Å2.
Dose-fractionated images (movies) were first gain-corrected and 2×binned (resulting in a pixel size of 1.3 Å). Frame-based motion-corrected, summed micrographs were generated by the Unblur program (Grant and Grigorieff, 2015) using all 50 frames and Unblur's exposure filter (radiation damage filter) option. Defocus values were estimated using CTFFIND4 (Rohou and Grigorieff, 2015). All subsequent image processing except noted, was carried out with RELION 1.4 (Scheres, 2012a; 2012b) using a particle box size of 256 pixels and a mask diameter of 195 pixels (Figure S2). Initially, ∼5000 particles were manually picked and subjected to reference-free 2D classification. 6 representative 2D class averages were selected as templates for automated particle picking. Auto-picked particles from ∼3,500 micrographs were cleaned up by visual inspection to remove non-channel particles and aggregates, resulting in 382,245 particles, which were further divided into 3 subsets and subjected to 3 rounds of reference-free 2D classification to remove outlier particles. An initial 3D reference was obtained using EMAN2 (Tang et al., 2007) from a subset of particles. To obtain ‘polished’ particle images, corrected for individual particle movements, frame-based motion-corrected movies, generated by Unblur without using its exposure filter option, were 2× frame-binned (resulting in a total of 25 frames per movie), and then applied to the alignparts-lmbfgs program (Rubinstein and Brubaker, 2015) together with coordinates of ‘cleaned’ particles. Alignparts-lmbfgs's exposure filter option was used to filter radiation damage. All ‘polished’ particle images were then subjected to one more round of 2D classification for further removal of outlier particles. All particles were used for 3D refinement in Relion, followed by masked 3D classification without angular refinement, sorting into 9 classes. By visual inspection in UCSF Chimera (Pettersen et al., 2004), the 2 classes with continuous, unbroken features (147,749 particles) and highest resolutions were combined and subjected to 3D refinement with C4 symmetry restraints and a mask applied (Figure S2). At the same time, electron dose was reduced to 30 e-/ Å2 by selecting 8 frames (frames 2-9 out of the total 25 frames) in the alignparts-lmbfgs program. The refined particles were further subjected to another round of 3D classification without particle alignment, and 2 out of the 6 classes were combined for further 3D refinement in RELION 1.4 or in Frealign (Lyumkis et al., 2013). Both programs yielded maps with similar quality, but with slightly higher resolution in Frealign. Both maps from Frealign and Relion were used for model building. Structure refinement was carried out using the Frealign map.
Model building
For map sharpening, BFACTOR.EXE (written by Nikolaus Grigorieff) was used to sharpen the maps from FREALIGN at a b-factor value between -100 and -150 Å2.
The KCNQ1EM/CaM model was built in COOT (Schmitt et al., 2014). First, a model for the transmembrane domain was generated using the I-TASSER server (Zhang, 2008). This, together with the crystal structure of CaM/HA-HB (PDB code: 4V0C) (Sachyani et al., 2014) was docked into the cryo-EM map. From this starting point manual rebuilding and refinement were carried out. Protein structure quality was monitored using the Molprobity server (Chen et al., 2010). The resulting PDB model includes the majority of KCNQ1EM construct (domain boundary 67-610). Several disordered loops (N-terminal loop: 67-93, S3-S4 linker: 207-218, HA-HB loop: 385-496) and C-terminal region including the HD helix (557-610) are not modeled due to their disorder, and they are indicated as dash lines in Figure S1. We did not model the K+ ions in selectivity filter, as their densities are weak. We observed similarly weak ion density in two other potassium channels solved by cryo-EM at comparable resolution (Hite and MacKinnon, 2017; Whicher and MacKinnon, 2016). We do not think this represents the absence of ions in the selectivity filter.
Model refinement and validation
Real and reciprocal space refinement of the model in P4 crystallographic symmetry, with one monomer per “asymmetric unit”, was carried out using phases and amplitudes extracted from one of the independently calculated half maps generated by FREALIGN. At the end of refinement Fourier shell correlations (FSC) were calculated between the refined model and the half map used for refinement (work), the other half map (free), and the full map to assess over fitting (Figure S3C).
To minimize computation during refinement of the model, both the model and the half map were translated into a box that extended 5Å beyond the model and map in each direction (Winn et al., 2011). Then the model was refined against the working half map in real space using PHENIX real space refine (Adams et al., 2010) with secondary structure restraints. Following real space refinement, the working half map was solvent flattened by creating a mask extending ∼3Å beyond the model and setting regions of the map outside the mask to 0. Structure factors were calculated for the solvent flattened working half map and the model was refined against the working half map in reciprocal space using Refmac (Brown et al., 2015). ProSMART (Nicholls et al., 2014) was used to generate secondary structure restraints used during reciprocal space refinement. Figures were generated with Chimera (Pettersen et al., 2004), Pymol (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.), HOLE (Smart et al., 1996), APBS (Dolinsky et al., 2007; 2004), and the Bsoft package (Cardone et al., 2013) and structure calculations were performed with the SBGrid suite of programs (Morin et al., 2013).
Quantification and Statistical Analysis
See Methods Details for details on cryo-EM analysis
Data and Software Availability
Data Resources
Cryo-EM density map of KCNQ1/CaM complex has been deposited in the electron microscopy data bank under accession code EMD-8712. Atomic coordinates of KCNQ1/CaM complex have been deposited in the protein data bank under accession code 5VMS.
Supplementary Material
Figure S1. Sequence Information of KCNQ1, Related to Figure 1.
Alignment and secondary structure assignments of KCNQ1 orthologs from Xenopus laevis (froKCNQ1, NP_001116347.1), Homo sapiens (humKCNQ1, NP_000209.2), Mus musculus (mouKCNQ1, NP_032460.2), Gallus gallus (chiKCNQ1, XP_421022.3), Danio rerio (fisKCNQ1, NP_001116714.1). Secondary structure of unobserved regions is represented as dash lines. Sequence conservation is indicated using red/blue spectrum. Red triangles and cyan circles indicate long QT missense mutations mapped onto the 3D model in Figure S5C and CaM-interacting residues identified by the Protein Interaction Calculator (Tina et al., 2007), respectively. Two potential ATP binding residues in the ordered region are highlighted (Li et al., 2013).
Figure S2. Cyo-EM structure determination, Related to Figure 1.
(A) Flow chart of the cryo-EM data processing procedure.
(B) Fourier shell correlation (FSC) of the final reconstruction. The resolution is 3.7 Å at the cutoff of 0.143.
(C) Cut-through view of KCNQ1EM local resolution map estimated by blocres program (Cardone et al., 2013).
(D) Orientation distribution of the particles used in final reconstruction of Relion. Only a quarter of a sphere is shown because the structure is 4-fold symmetric. The height of the bar is proportional to the orientation density distribution.
Figure S3. Model building, Related to Figure 1 and Figure 5.
(A-B) Representative local densities with refined models for KCNQ1EM and CaM. The green spheres represent calcium ions. A magnified view of the density map of the 4th EF hand is shown with higher threshold.
(C) Model validation. FSC between model and half map 1 (working set), model and half map 2 (free set), and model and full map are plotted in green, red and blue traces, respectively.
(D) Refinement statistics for the KCNQ1EM/CaM model. The Molprobility scores were calculated using the KCNQ1EM/CaM tetramer.
Figure S4. Important structural features of KCNQ1EM, Related to Figure 1 and Figure 2.
(A) Sequence alignment of S3 segments among Kv1-9 family members. The sequences used are the same as in Figure 5.
(B) Comparison of S3 between KCNQ1EM and Kv1.2-2.1. The Kv1.2-2.1 S3 contains a 25-degree bend compared to S3 in KCNQ1EM, highlighted by dashed lines.
(C) Two views of KCNQ1EM colored according to surface potential (-5.0 KT/e to 5.0 KT/e)(Dolinsky et al., 2007; Holst and Saied, 1993). The loops between S5 and the pore form negative charged caps (indicated by the arrow) surrounding the channel entrance.
(D) Sequence alignment of the S6 segment among KCNQ (Kv7) family members (frKCNQ1:NP_001116347.1, Kv7.1: NP_000209.2, Kv7.2: NP_742105.1, Kv7.3: NP_004510.1, Kv7.4:NP_004691.2, Kv7.5: NP_062816.2) and Kv1-4 (sequences identifiers are the same as in Figure 5). Key residues important for gating are highlighted.
(E) Structure alignment of the pore domain between KCNQ1EM (yellow) and Kv1.2-2.1 (grey). A magnified view of the narrowest region in KCNQ1EM is shown in the dash box. Key residues lining the pore are labeled in black with yellow highlight. Residues forming the negatively charged ion entrance are labeled in red with yellow highlight.
Figure S5. Interactions between KCNQ1EM and CaM, Related to Figure 5.
(A) Superposition of CaM/HA-HB cryo-EM and crystal structures (PDB code: 4V0C).
(B) Density of the interface between the S2-S3 loop of KCNQ1EM and EF hand #3 of CaM.
(C) Stereo view of KCNQ1EM/CaM with disease mutations mapped. Mutation sites in KCNQ1EM voltage senor, S4-S5 linker, pore domain, HA-HB helices, HC helix and CaM are shown as red, blue, yellow, purple, brown and orange spheres, respectively.
(D) Complex formation between KCNQ1EM and CaM (wild type or N98S mutation) by fluorescence-detection size-exclusion chromatography (FSEC) (Kawate and Gouaux, 2006) using a Superose 6, 10/300 GL column (GE healthcare). The GFP tag is labeled on either CaM or CaM-N98S. Green: KCNQ1EM/CaM(WT)-GFP, brown: CaM-GFP alone, blue: KCNQ1EM/CaM(N98S)-GFP, dark green: CaM(N98S)-GFP, magenta: KCNQ1EM alone.
Highlights.
Cryo-EM structure of KCNQ1/CaM complex at 3.7 Å resolution
KCNQ1 is captured in an ‘uncoupled’, PIP2-free state
Insights into the molecular basis for calmodulin-mediated long QT syndrome
Acknowledgments
We thank M. Ebrahim at the Rockefeller University cryo-EM resource center for help with microscope operation, Y. C. Hsiung for help with large-scale cell culture, members of the MacKinnon lab for helpful discussions, and C. H. Lee and J. Whicher for critical reading of the manuscript. J. Sun is an American Heart Association Fellow (Award ID: 17POST32260003). R. MacKinnon is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Author Contributions: J.S performed the experiments. J.S. and R.M. designed the experiments, analyzed the results, and prepared the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott GW. Biology of the KCNQ1 Potassium Channel. New Journal of Science. 2014;2014:1–26. [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. KvLQT1 and IsK (minK) proteins associate to form the IKs cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- Boczek NJ, Gomez-Hurtado N, Ye D, Calvert ML, Tester DJ, Kryshtal DO, Hwang HS, Johnson CN, Chazin WJ, Loporcaro CG, et al. Spectrum and Prevalence of CALM1-, CALM2-, and CALM3-Encoded Calmodulin Variants in Long QT Syndrome and Functional Characterization of a Novel Long QT Syndrome-Associated Calmodulin Missense Variant, E141G. Circ Cardiovasc Genet. 2016;9:136–146. doi: 10.1161/CIRCGENETICS.115.001323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Long F, Nicholls RA, Toots J, Emsley P, Murshudov G. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone G, Heymann JB, Steven AC. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J Struct Biol. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas P, Brown DA. Pathways modulating neural KCNQ/M (Kv7) potassium channels. Nat Rev Neurosci. 2005;6:850–862. doi: 10.1038/nrn1785. [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford CP, Stemkowski PL, Smith PA. Possible role of phosphatidylinositol 4,5 bisphosphate in luteinizing hormone releasing hormone-mediated M-current inhibition in bullfrog sympathetic neurons. Eur J Neurosci. 2004;20:2990–2998. doi: 10.1111/j.1460-9568.2004.03786.x. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Nunziato DA, Pitt GS. KCNQ1 assembly and function is blocked by long-QT syndrome mutations that disrupt interaction with calmodulin. Circ Res. 2006;98:1048–1054. doi: 10.1161/01.RES.0000218863.44140.f2. [DOI] [PubMed] [Google Scholar]
- Goehring A, Lee CH, Wang KH, Michel JC, Claxton DP, Baconguis I, Althoff T, Fischer S, Garcia KC, Gouaux E. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. Elife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackos DH, Chang TH, Swartz KJ. Scanning the intracellular S6 activation gate in the shaker K+ channel. J Gen Physiol. 2002;119:521–532. doi: 10.1085/jgp.20028569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley PL, Jørgensen P, Schlamowitz S, Wangari R, Moolman-Smook J, Brink PA, Kanters JK, Corfield VA, Christiansen M. The genetic basis of long QT and short QT syndromes: a mutation update. Hum Mutat. 2009;30:1486–1511. doi: 10.1002/humu.21106. [DOI] [PubMed] [Google Scholar]
- Hite RK, MacKinnon R. Structural Titration of Slo2.2, a Na(+)-Dependent K(+) Channel Cell. 2017;168:390–399.e11. doi: 10.1016/j.cell.2016.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite RK, Tao X, MacKinnon R. Structural basis for gating the high-conductance Ca2+-activated K+ channel. Nature. 2017;541:52–57. doi: 10.1038/nature20775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite RK, Yuan P, Li Z, Hsuing Y, Walz T, MacKinnon R. Cryo-electron microscopy structure of the Slo2.2 Na(+)-activated K(+) channel. Nature. 2015;527:198–203. doi: 10.1038/nature14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst M, Saied F. Multigrid solution of the Poisson—Boltzmann equation. J Comput Chem. 1993;14:105–113. [Google Scholar]
- Jespersen T, Grunnet M, Olesen SP. The KCNQ1 potassium channel: from gene to physiological function. Physiology (Bethesda) 2005;20:408–416. doi: 10.1152/physiol.00031.2005. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. The open pore conformation of potassium channels. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- Jiménez-Jáimez J, Palomino Doza J, Ortega Á, Macías-Ruiz R, Perin F, Rodríguez-Vázquez del Rey MM, Ortiz-Genga M, Monserrat L, Barriales-Villa R, Blanca E, et al. Calmodulin 2 Mutation N98S Is Associated with Unexplained Cardiac Arrest in Infants Due to Low Clinical Penetrance Electrical Disorders. PLoS ONE. 2016;11:e0153851. doi: 10.1371/journal.pone.0153851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimova MA, Zaydman MA, Cui J, Tarek M. PIP2-dependent coupling is prominent in Kv7.1 due to weakened interactions between S4-S5 and S6. Sci Rep. 2015;5:7474. doi: 10.1038/srep07474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, et al. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- Lee CH, MacKinnon R. Structures of the Human HCN1 Hyperpolarization-Activated Channel. Cell. 2017;168:111–120.e111. doi: 10.1016/j.cell.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Gao J, Lu Z, McFarland K, Shi J, Bock K, Cohen IS, Cui J. Intracellular ATP binding is required to activate the slowly activating K+ channel IKs. Pnas. 2013;110:18922–18927. doi: 10.1073/pnas.1315649110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpitikul WB, Dick IE, Joshi-Mukherjee R, Overgaard MT, George AL, Jr, Yue DT. Calmodulin mutations associated with long QT syndrome prevent inactivation of cardiac L-type Ca2+ currents and promote proarrhythmic behavior in ventricular myocytes. Journal of Molecular and Cellular Cardiology. 2014;74:115–124. doi: 10.1016/j.yjmcc.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SB, Campbell EB, MacKinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- Loussouarn G, Park KH, Bellocq C, Baró I, Charpentier F, Escande D. Phosphatidylinositol-4,5-bisphosphate, PIP2, controls KCNQ1/KCNE1 voltage-gated potassium channels: a functional homology between voltage-gated and inward rectifier K+ channels. Embo J. 2003;22:5412–5421. doi: 10.1093/emboj/cdg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita N, Yagihara N, Crotti L, Johnson CN, Beckmann BM, Roh MS, Shigemizu D, Lichtner P, Ishikawa T, Aiba T, et al. Novel calmodulin mutations associated with congenital arrhythmia susceptibility. Circ Cardiovasc Genet. 2014;7:466–474. doi: 10.1161/CIRCGENETICS.113.000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P. Collaboration gets the most out of software. Elife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RA, Fischer M, McNicholas S, Murshudov GN. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr D Biol Crystallogr. 2014;70:2487–2499. doi: 10.1107/S1399004714016241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJ, Bird TD, Fong CT, Mefford HC, Smith RJ, et al. Long QT Syndrome 1993a [Google Scholar]
- Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJ, Bird TD, Fong CT, Mefford HC, Smith RJ, et al. Jervell and Lange-Nielsen Syndrome 1993b [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Robbins J. KCNQ potassium channels: physiology, pathophysiology, and pharmacology. Pharmacol Ther. 2001;90:1–19. doi: 10.1016/s0163-7258(01)00116-4. [DOI] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein JL, Brubaker MA. Alignment of cryo-EM movies of individual particles by optimization of image translations. J Struct Biol. 2015;192:188–195. doi: 10.1016/j.jsb.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Sachyani D, Dvir M, Strulovich R, Tria G, Tobelaim W, Peretz A, Pongs O, Svergun D, Attali B, Hirsch JA. Structural basis of a Kv7.1 potassium channel gating module: studies of the intracellular c-terminal domain in complex with calmodulin. Structure. 2014;22:1582–1594. doi: 10.1016/j.str.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. Coassembly of KvLQT1 and minK (IsK) proteins to form cardiac IKs potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- Sarhan MF, Tung CC, Van Petegem F, Ahern CA. Crystallographic basis for calcium regulation of sodium channels. Proc Natl Acad Sci U S a. 2012;109:3558–3563. doi: 10.1073/pnas.1114748109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012a;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012b;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N, Grunnet M, Olesen SP. Cardiac Potassium Channel Subtypes: New Roles in Repolarization and Arrhythmia. Physiological Reviews. 2014;94:609–653. doi: 10.1152/physrev.00022.2013. [DOI] [PubMed] [Google Scholar]
- Seebohm G, Strutz-Seebohm N, Ureche ON, Baltaev R, Lampert A, Kornichuk G, Kamiya K, Wuttke TV, Lerche H, Sanguinetti MC, et al. Differential roles of S6 domain hinges in the gating of KCNQ potassium channels. Biophys J. 2006;90:2235–2244. doi: 10.1529/biophysj.105.067165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamgar L, Ma L, Schmitt N, Haitin Y, Peretz A, Wiener R, Hirsch J, Pongs O, Attali B. Calmodulin is essential for cardiac IKS channel gating and assembly: impaired function in long-QT mutations. Circ Res. 2006;98:1055–1063. doi: 10.1161/01.RES.0000218979.40770.69. [DOI] [PubMed] [Google Scholar]
- Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–60. 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- Suh BC, Hille B. Recovery from muscarinic modulation of M current channels requires phosphatidylinositol 4,5-bisphosphate synthesis. Neuron. 2002;35:507–520. doi: 10.1016/s0896-6273(02)00790-0. [DOI] [PubMed] [Google Scholar]
- Suh BC, Hille B. PIP2 is a necessary cofactor for ion channel function: how and why? Annu Rev Biophys. 2008;37:175–195. doi: 10.1146/annurev.biophys.37.032807.125859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tao X, Hite RK, MacKinnon R. Cryo-EM structure of the open high-conductance Ca2+-activated K+ channel. Nature. 2017;541:46–51. doi: 10.1038/nature20608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tina KG, Bhadra R, Srinivasan N. PIC: Protein Interactions Calculator. Nucleic Acids Res. 2007;35:W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobelaim WS, Dvir M, Lebel G, Cui M, Buki T, Peretz A, Marom M, Haitin Y, Logothetis DE, Hirsch JA, et al. Competition of calcified calmodulin N lobe and PIP2 to an LQT mutation site in Kv7.1 channel. Proc Natl Acad Sci U S a. 2017;114:E869–E878. doi: 10.1073/pnas.1612622114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohse N. Calcium-sensitive delayed rectifier potassium current in guinea pig ventricular cells. Am J Physiol. 1990;258:H1200–H1207. doi: 10.1152/ajpheart.1990.258.4.H1200. [DOI] [PubMed] [Google Scholar]
- Whicher JR, MacKinnon R. Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science. 2016;353:664–669. doi: 10.1126/science.aaf8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff H, Castle NA, Pardo LA. Voltage-gated potassium channels as therapeutic targets. Nat Rev Drug Discov. 2009;8:982–1001. doi: 10.1038/nrd2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaydman MA, Cui J. PIP2 regulation of KCNQ channels: biophysical and molecular mechanisms for lipid modulation of voltage-dependent gating. Front Physiol. 2014;5:195. doi: 10.3389/fphys.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaydman MA, Silva JR, Delaloye K, Li Y, Liang H, Larsson HP, Shi J, Cui J. Kv7.1 ion channels require a lipid to couple voltage sensing to pore opening. Proc Natl Acad Sci U S a. 2013;110:13180–13185. doi: 10.1073/pnas.1305167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Craciun LC, Mirshahi T, Rohács T, Lopes CMB, Jin T, Logothetis DE. PIP(2) activates KCNQ channels, and its hydrolysis underlies receptor-mediated inhibition of M currents. Neuron. 2003;37:963–975. doi: 10.1016/s0896-6273(03)00125-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence Information of KCNQ1, Related to Figure 1.
Alignment and secondary structure assignments of KCNQ1 orthologs from Xenopus laevis (froKCNQ1, NP_001116347.1), Homo sapiens (humKCNQ1, NP_000209.2), Mus musculus (mouKCNQ1, NP_032460.2), Gallus gallus (chiKCNQ1, XP_421022.3), Danio rerio (fisKCNQ1, NP_001116714.1). Secondary structure of unobserved regions is represented as dash lines. Sequence conservation is indicated using red/blue spectrum. Red triangles and cyan circles indicate long QT missense mutations mapped onto the 3D model in Figure S5C and CaM-interacting residues identified by the Protein Interaction Calculator (Tina et al., 2007), respectively. Two potential ATP binding residues in the ordered region are highlighted (Li et al., 2013).
Figure S2. Cyo-EM structure determination, Related to Figure 1.
(A) Flow chart of the cryo-EM data processing procedure.
(B) Fourier shell correlation (FSC) of the final reconstruction. The resolution is 3.7 Å at the cutoff of 0.143.
(C) Cut-through view of KCNQ1EM local resolution map estimated by blocres program (Cardone et al., 2013).
(D) Orientation distribution of the particles used in final reconstruction of Relion. Only a quarter of a sphere is shown because the structure is 4-fold symmetric. The height of the bar is proportional to the orientation density distribution.
Figure S3. Model building, Related to Figure 1 and Figure 5.
(A-B) Representative local densities with refined models for KCNQ1EM and CaM. The green spheres represent calcium ions. A magnified view of the density map of the 4th EF hand is shown with higher threshold.
(C) Model validation. FSC between model and half map 1 (working set), model and half map 2 (free set), and model and full map are plotted in green, red and blue traces, respectively.
(D) Refinement statistics for the KCNQ1EM/CaM model. The Molprobility scores were calculated using the KCNQ1EM/CaM tetramer.
Figure S4. Important structural features of KCNQ1EM, Related to Figure 1 and Figure 2.
(A) Sequence alignment of S3 segments among Kv1-9 family members. The sequences used are the same as in Figure 5.
(B) Comparison of S3 between KCNQ1EM and Kv1.2-2.1. The Kv1.2-2.1 S3 contains a 25-degree bend compared to S3 in KCNQ1EM, highlighted by dashed lines.
(C) Two views of KCNQ1EM colored according to surface potential (-5.0 KT/e to 5.0 KT/e)(Dolinsky et al., 2007; Holst and Saied, 1993). The loops between S5 and the pore form negative charged caps (indicated by the arrow) surrounding the channel entrance.
(D) Sequence alignment of the S6 segment among KCNQ (Kv7) family members (frKCNQ1:NP_001116347.1, Kv7.1: NP_000209.2, Kv7.2: NP_742105.1, Kv7.3: NP_004510.1, Kv7.4:NP_004691.2, Kv7.5: NP_062816.2) and Kv1-4 (sequences identifiers are the same as in Figure 5). Key residues important for gating are highlighted.
(E) Structure alignment of the pore domain between KCNQ1EM (yellow) and Kv1.2-2.1 (grey). A magnified view of the narrowest region in KCNQ1EM is shown in the dash box. Key residues lining the pore are labeled in black with yellow highlight. Residues forming the negatively charged ion entrance are labeled in red with yellow highlight.
Figure S5. Interactions between KCNQ1EM and CaM, Related to Figure 5.
(A) Superposition of CaM/HA-HB cryo-EM and crystal structures (PDB code: 4V0C).
(B) Density of the interface between the S2-S3 loop of KCNQ1EM and EF hand #3 of CaM.
(C) Stereo view of KCNQ1EM/CaM with disease mutations mapped. Mutation sites in KCNQ1EM voltage senor, S4-S5 linker, pore domain, HA-HB helices, HC helix and CaM are shown as red, blue, yellow, purple, brown and orange spheres, respectively.
(D) Complex formation between KCNQ1EM and CaM (wild type or N98S mutation) by fluorescence-detection size-exclusion chromatography (FSEC) (Kawate and Gouaux, 2006) using a Superose 6, 10/300 GL column (GE healthcare). The GFP tag is labeled on either CaM or CaM-N98S. Green: KCNQ1EM/CaM(WT)-GFP, brown: CaM-GFP alone, blue: KCNQ1EM/CaM(N98S)-GFP, dark green: CaM(N98S)-GFP, magenta: KCNQ1EM alone.


