Abstract
After invasion of a susceptible target cell, HIV-1 completes the early phase of its life cycle upon integration of reverse-transcribed viral DNA into host chromatin. The viral capsid, a conical shell encasing the viral ribonucleoprotein complex, along with its constitutive capsid protein plays essential roles at virtually every step in the early phase of the viral life cycle. Recent work has begun to reveal how the viral capsid interacts with specific cellular proteins to promote these processes. At the same time, cellular restriction factors target the viral capsid to thwart infection. Comprehensive understanding of capsid-host interactions that promote or impede HIV-1 infection may provide unique insight to exploit for novel therapeutic interventions.
Keywords: HIV/AIDS, capsid, host proteins, post-entry events, virus-host interactions, virus integration
Capsid-host interactions underlie the unique biology of HIV-1
The capsid core, an outer shell made of ~1300 copies of the viral capsid protein (CA), encloses the viral ribonucleoprotein complex consisting of genomic RNA along with viral proteins nucleocapsid (NC), reverse transcriptase, and integrase (IN) [1]. Throughout this manuscript “capsid” is used to mean the capsid core or shell, whereas “CA” denotes its constitutive capsid protein. CA consists of two alpha-helical domains, the N-terminal domain (NTD) and C-terminal domain (CTD), which connect via a flexible linker [2] (Figure 1a). Individual CA subunits assemble predominantly into hexamers (shown in Figure 1b) as well as a subset of 12 pentamers, which together form the viral capsid [2, 3].
Figure 1. HIV-1 CA structures and common host factor interaction sites.
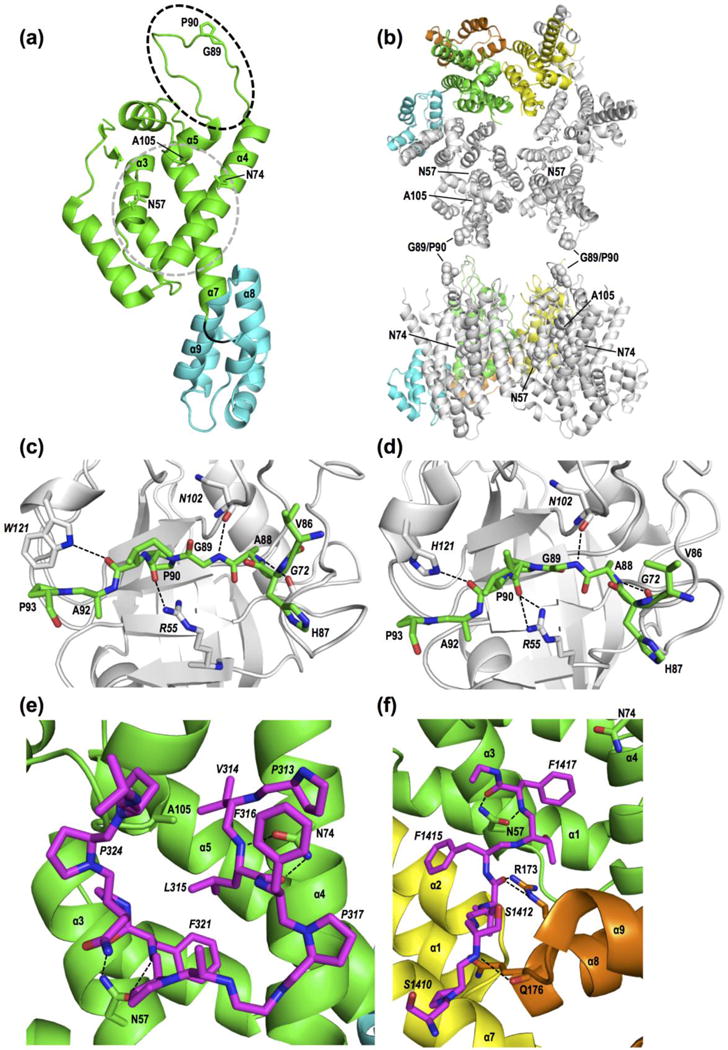
(a) Monomeric CA (NTD, green; CTD, cyan; flexible loop, black) highlighting the CypA-binding loop (dashed oval) as well as residues and secondary structural elements that mediate CPSF6 or NUP153 binding (gray circle). Protein database (PDB) accession code 4XFY [146]. (b) CA hexamer (PDB code 4U0D) [69] views from the cytoplasmic face of the capsid (top) or rotated 90° up into the plane (bottom). Residues highlighted in (a) are shown in space fill in two adjacent gray monomers. Coloring highlights NTD (green and yellow) and CTD (cyan and orange) interactions between adjacent monomers. (c) Close up view of the CypA (light gray)-CA NTD interaction (PDB code 1AK4) [20]. Host factor residues are denoted by italic type; dashed lines, hydrogen bonds. (d) The NUP358 CHD (light gray)-CA NTD interaction (PDB code 4LQW) [81]. (e) Primary interaction site between CPSF6313-327 (magenta) and the CA NTD (PDB code 4U0B) [69]. (f) Co-crystal structure of NUP1531407-1423 (magenta) with hexameric CA (PDB code 4U0D) [69]. In (e) and (f), HIV-1 CA Asn57 engages similarly positioned hotspot Phe residues (F321 in CPSF6; F1417 in NUP153). Yellow and orange in (f) are the NTD and CTD, respectively, of an adjacent CA monomer (see panel b).
Capsid cores must navigate through a hostile environment with both intrinsic and innate antiviral regulators to gain access to the nucleus where the reverse transcript is inserted into host chromatin [4]. HIV-1 additionally hijacks host machinery and manipulates the cellular environment for its survival. The capsid accordingly plays multifactorial roles during the early phase of the HIV-1 life cycle [5, 6], which contributes to shaping the unique biology of lentiviruses. The ability to infect non-dividing cells such as primary macrophages is governed by the capsid [7]. The capsid also contributes to the unique pattern of HIV-1 integration targeting [8–11]. Recent advances in the discovery of capsid-interacting molecules together with imaging and structural studies have provided novel insight into how HIV-1 utilizes its capsid to exploit host machinery and maximize viral fitness. Herein we review the progress in understanding capsid-dependent host factors for HIV-1 replication, focusing on recently published findings (Figure 2). Table 1 lists these factors as well as some other proteins implicated in the early events of HIV-1 replication.
Figure 2. Capsid-dependent host factors for post-entry events of HIV-1 infection.
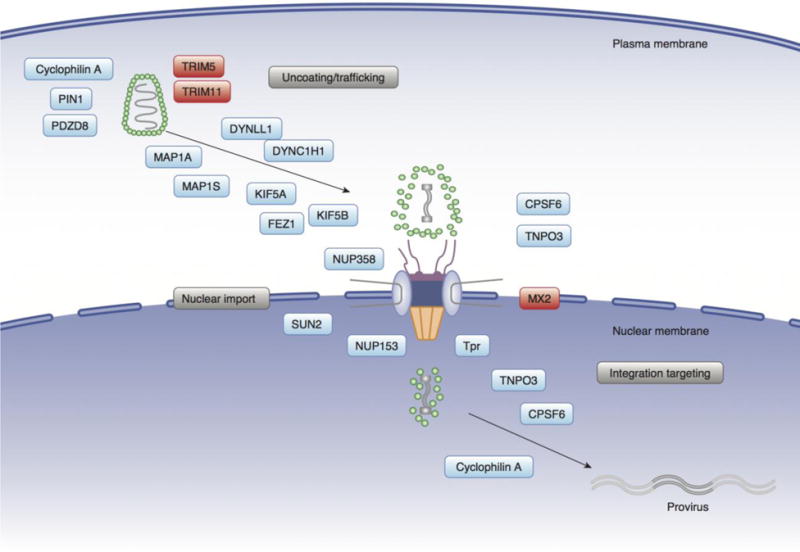
Incoming viral capsids traffic towards the nuclear membrane, and subviral complexes can directly interact with NPC components such as NUP358 and NUP153. HIV-1 nuclear entry is mediated by these NUPs as well as soluble import receptors such as CPSF6/TNPO3. Intranuclear localization of PICs is regulated by CPSF6. Cellular proteins can promote (shown in blue) or block (shown in red) HIV-1 infection. The intact capsid near the cell periphery is depicted partially uncoated at the NPC, and more fully uncoated after nuclear entry. Host factors known to interact with the capsid or modulate uncoating or integration site selection are shown in this illustration. The positioning of the different factors in the figure is not meant to necessarily imply the location or timing of the virus-host interaction.
TABLE 1.
Capsid-dependent host factors for HIV-1
| Gene name | Gene ID | Alias | Gene descriptiona | Binding determinant | Reference |
|---|---|---|---|---|---|
| CPSF6 | 11052 | CFIM68 | Subunit of a cleavage factor for 3′ RNA cleavage and polyadenylation processing | Hexameric CA | [65] |
| Cyclophilin A | 5478 | PPIA | Peptidyl-prolyl cis-trans isomerase | Monomeric CA | [147] |
| Cyclophilin B | 5479 | PPIB | Cyclosporine-binding protein mainly located in the endoplasmic reticulum | Gag | [147] |
| DYNC1H 1 | 1778 | DYNC1H 1 | Member of the cytoplasmic dynein heavy chain family | NDb | [44, 45] |
| DYNLL1 | 8655 | DYNLL1 | Dynein light chain | IN | [48] |
| FEZ1 | 9638 | Fasciculat ion and elongation protein zeta 1 | An ortholog of the Caenorhabditis elegans unc-76 gene that is necessary for normal axonal bundling and elongation within axon bundles | CA-NC | [50] |
| KIF5A | 3798 | KIF5A | Member of the kinesin family protein that functions as a microtubule motor | ND | [50] |
| KIF5B | 3799 | KIF5B | Member of the kinesin family protein that functions as a microtubule motor | ND | [44, 50] |
| MAP1A | 4130 | MAP1A | Microtubule-associated protein family member | Monomeric CA | [43] |
| MAP1S | 55201 | MAP1S | Microtubule-associated protein family member | Monomeric CA | [43] |
| MX2 | 4600 | MXB, MX dynamin like GTPase | Encodes both nuclear and cytoplasmic forms by alternative splicing of the amino terminal end | Capsid lattice | [85–87] |
| PDZD8 | 118987 | PDZ domain containing 8 | Involved in the regulation of cell morphology and cytoskeletal organization | Gag, CA-NC | [37, 148] |
| PIN1 | 5300 | NIMA-interacting 1 | PPIase specifically binding to phosphorylated serine/threonine motifs | Capsid cores | [34] |
| NUP153 | 9972 | Nucleopor in 153 | A nucleoporin that contains characteristic XFXFG pentapeptides | Monomeric/hexameric CA | [57, 58] |
| NUP358 | 5903 | RANBP2 | A very large RAN-binding protein that localizes to the nuclear pore complex | CA NTD | [57, 58] |
| SUN2 | 25777 | UNC84B | An inner nuclear membrane protein | ND | [28, 139] |
| TNPO3 | 23534 | Transporti n (TRN) 3/TRN-SR2 | Nuclear import receptor for serine/arginine-rich proteins | CPSF6 | [56–58] |
| Tpr | 7175 | Translate d promoter region | Forms intranuclear filaments attached to the inner surface of NPCs | No binding | [114, 116] |
| TRIM5α | 85363 | Tripartite motif containing 5α | Member of the tripartite motif (TRIM) family | Capsid lattice | [127] |
| TRIM11 | 81559 | Tripartite motif containing 11 | Member of the TRIM family with no identified function | ND | [36] |
Adapted from National Center for Biotechnology Information
ND, not determined.
Uncoating
Capsid-mediated post-entry events are intertwined with uncoating, the process whereby dynamic structural remodeling leads to shedding of CA subunits [6, 12] (Box 1). Uncoating proceeds concurrently with reverse transcription [13–15] in the confines of the reverse transcription complex (RTC), which yields the integration-competent pre-integration complex (PIC). As described below, a number of host proteins interact with the capsid to modulate uncoating, which may be operationally linked to its intracellular movement towards the nucleus. Capsid is also a target of the intracellular innate immune response (Box 2).
Box 1. HIV-1 capsid uncoating.
There are three popular models for HIV-1 uncoating [6, 12]. The “immediate” model posits that the bulk of HIV-1 CA dissociates from intracellular complexes soon after virus entry. At the other extreme is the “nuclear pore” model whereby the core remains intact until it engages the nuclear pore complex (NPC) at the cytoplasm/nucleus interface where intact capsids may undergo coupled reverse transcription-uncoating [52, 55, 125]. The third “cytoplasmic” model suggests partial CA shedding en route to the nucleus [13, 126]. Modulation of uncoating by depletion of predominantly cytoplasmic host factors supports the cytoplasmic model. Although specific interactions between CA and nucleoporins such as NUP358 and NUP153 may be thought to favor the nuclear pore model, partially uncoated capsids should still engage these factors. Recent research indicates that some CA remains associated with the PIC following nuclear import [60, 72, 74, 78, 108], which is consistent with both the nuclear pore and cytoplasmic models of uncoating. Uncoating is exquisitely fine-tuned through intrinsic core stability and by host factors that can directly interact with CA and either stabilize or destabilize viral capsids [12, 110]. Spatial or temporal deviations from the normal course of uncoating (accelerated or delayed uncoating) severely attenuate infection. For instance, the rhesus macaque TRIM5α restriction factor binds the HIV-1 capsid to accelerate uncoating, which prevents the normal course of reverse transcription [47, 127]. The fate of individual cores (i.e. timing of capsid disassembly) can vary [128, 129], and whether cytoplasmic or nuclear core uncoating, or perhaps their combination, leads to productive infection remains a subject of debate [6]. We tend to favor the cytoplasmic model, though we appreciate that the nuclear pore model may predominate under certain conditions, for example in primary cells that are particularly sensitized to innate immune recognition [27].
Box 2. Innate immunity against HIV-1.
HIV-1 capsid is a target for cell-autonomous antiviral responses [4]. Sensing of viral infection induces an antiviral state, which is initiated by production of interferons (IFNs) and up-regulation of interferon-stimulated genes (ISGs) [130]. ISG products known to target the HIV-1 capsid include TRIM5α and MX2 [130]. However, as yet identified capsid-dependent ISGs likely contribute, because IFNs appear more potent with certain CA mutants to block uncoating and reverse transcription [25, 93, 128]. Innate immune pathways are activated by recognition of pathogen-associated molecular patterns (PAMPs) through pathogen recognition receptors. Various species of nucleic acids as well as other viral structures generated during HIV-1 replication can serve as a PAMP [130]. The hexagonal capsid lattice harbors regularly spaced repeating epitopes that may present a PAMP that is recognized by TRIM5α [131]. De novo synthesized viral DNA in the cytoplasm can be detected by DNA sensors such as cyclic GMP-AMP synthase (cGAS) [132] and gamma-interferon-inducible protein Ifi-16 [133], with work suggesting that the capsid protects viral DNA from the recognition by cGAS through interactions with CA-binding proteins [26, 27]. HIV-1 or HIV-2 CA mutants with altered CypA binding elicit strong innate activation [26, 27, 91]. The CA mutant N74D, which is deficient for CPSF6 binding, induced type I IFN production in macrophages [27, 134], though one study did not reproduce this phenotype [75]. Viral DNA released from disassembled capsid cores is susceptible to degradation by the cytosolic exonuclease TREX1 to avoid innate sensing by DNA sensors [135].
Cyclophilin A
Cyclophilin A (CypA) is a peptidyl-prolyl cis-trans isomerase (PPIase) that facilitates HIV-1 infection through its direct interaction with CA [16–18]. CypA binds a flexible loop in CA between NTD helices 4 and 5 [19, 20] (Figure 1a, black dashed line). The CypA-binding loop protrudes from the outer surface of the capsid (Figure 1b) and thus allows CypA-CA interactions without significant structural remodeling of the conical shell. CA engages a canonical hydrophobic pocket of CypA (Figure 1c) [20], while a non-canonical binding site may enable a single CypA molecule to simultaneously interact with two CA subunits in neighboring hexamers [21].
The requirements of CypA for HIV-1 infection are complex and cell type dependent [22]. For example, CypA knockout impaired viral infectivity in Jurkat cells but not in other cell types [23–25]. Non-immunosuppressive drugs that block the CA-CypA interaction appear to specifically perturb HIV-1 in primary cells [26, 27]. Lahaye et al. showed that Sad1 and UNC (SUN) domain-containing protein 2 (SUN2), which locates to the nuclear envelope, is required to facilitate the role of CypA in HIV-1 infection of primary CD4+ T cells [28], although more recent work failed to reproduce this finding [29]. Notably, the ability of CypA-independent SIV strains to replicate efficiently in primary cells suggest that primate lentiviruses can evolve mechanisms to dispense with CypA.
CypA can regulate nearly every post-entry event of HIV-1 infection. Maximum efficiency of reverse transcription depends on the CypA-CA interaction, though an effect on nuclear entry best correlated with transduction efficiency [22]. CypA regulates the utilization of host factors that either positively (NUP358 and NUP153) or negatively (MX2) impact PIC nuclear entry, as discussed below. Finally, CypA affects integration site selection, as genetic or pharmacological perturbation of its interaction with CA resulted in elevated targeting of gene dense chromosomal regions [9]. Such observations support the notion that both CA and CypA accompany PICs into the nucleus.
CypA additionally affects uncoating. A mechanistic model involves CypA-mediated control of capsid stability to influence reverse transcription and/or interactions with host factors. Although a clear consensus across studies is lacking, it appears that CypA binding to the core promotes its stability [30], and thus delays uncoating [22, 31]. However, CypA can exert an opposing core destabilization activity. One group reported that CypA destabilized the cyclosporine A-dependent A92E CA mutant, though this was not observed in another study [22, 31]. Similarly, CypA destabilized in vitro-assembled HIV-1 CA or CA-NC complexes [21, 32]. Differential CypA activity can be accounted for by a dose-dependent biphasic effect of CypA on core stability; sub-stoichiometric concentrations of CypA stabilize the core as opposed to high amounts of CypA that can destabilize [21, 31]. Liu speculated that steric hindrance by CypA binding to adjacent CA hexamers may weaken the dimer assembly interface to accelerate disassembly [21]. Such core-destabilizing activity may mechanistically mimic premature uncoating by natural and artificial capsid-dependent restriction factors, including rhesus TRIM5α [33] (Box 3). Overall, CypA-mediated regulation of capsid stability affects a range of post-entry steps and is likely to be influenced by capsid interactions with multiple factors, such as PPIases (CypB and PIN1), TRIM11, or PDZ domain-containing protein 8 (PDZD8), each of which was shown to bind to the capsid or modulate core stability [34–37]. It is notable, however, that modulation of core stability by blocking the interaction of CA with some of these factors, such as CypA or PDZD8, did not under certain conditions significantly impact HIV-1 infection [22, 37, 38].
Box 3. Antiviral potential of capsid-binding cofactors.
Although CypA and CPSF6 are generally thought of as positive viral dependency cofactors, each can be a potent HIV-1 antiviral. Indeed, CPSF6 was initially implicated in HIV-1 biology via the finding that the CPSF6-358 truncation variant of CPSF6588, which lacks the RS domain, is a potent restriction factor [65]. Antiviral mechanisms include the trapping of PICs in the cytoplasm (in the case of CPSF6-358) or impairment of reverse transcription (by the CPSF6-375 variant of the CPSF6551 isoform) [65, 136]. TNPO3 depletion induced cytoplasmic localization of CPSF6, which led to inhibition of HIV-1 at a step after reverse transcription [67, 71, 74]. Full-length CPSF6 mis-localized in the cytoplasm or CPSF6-358 stabilizes the viral capsid both in vitro and in vivo, whereas CPSF6-375 accelerates its disassembly [32, 65, 67, 71, 136]. The antiviral potential of endogenous CPSF6 is highlighted by restriction of certain CA mutants [73], including A92E. Such CA mutant viruses are hyper-dependent on cell division for infection, perhaps due to differential activity of CPSF6 in dividing versus non-dividing cells, consistent with the observation that CPSF6-358 restriction is accentuated by cell growth arrest [65]. CPSF6-mediated restriction of these CA mutants depends on CypA-CA interactions. A92E reduces CypA loop dynamics and decreases the efficiency of in vitro CA assembly reactions, thus potentially altering core stability [31, 137]. Thus, changes in core stability may render these mutants sensitive to CPSF6 and CypA in a manner that is not seen for the WT virus. However, according to expression profiling of restrictive vs non-restrictive cell lines, CPSF6 and CypA do not fully account for the restrictive phenotype [22, 138]. SUN2 is also implicated in CypA-mediated restriction [28, 139].
Cytoplasmic trafficking
HIV-1 RTCs/PICs reach nuclear pores by hijacking the host cytoskeleton [39–41]. Cytoplasmic movement depends on both actin microfilaments and microtubules (MTs), including dynamic and stable MTs induced by HIV-1 infection [42, 43], together with host factors involved in cytoskeleton and MT-associated motor proteins such as dynein and kinesin [41]. A functional link between viral trafficking and capsid can be deduced by changes in uncoating kinetics when host factor function is perturbed [44, 45].
Dynein facilitates inward movements of HIV-1 along MTs, as its inhibition caused peripheral accumulation of HIV-1 particles [39, 40, 44]. Dynein also assists HIV-1 uncoating [44–46]. Knockdown of dynein heavy chain (DHC) increased the number of CA foci or the amount of p24 associated with RTCs/PICs as well as pelletable CA in the fate-of-capsid assay [44, 46], a technique that takes advantage of differences in the physical properties of soluble CA versus CA associated with subviral complexes using centrifugation-based separation [47]. A similar effect was observed by disruption of the dynactin complex, which facilitates dynein-mediated intracellular transport [46]. Disruption of dynein-mediated transport by depletion of DHC or overexpression of p50/dynamitin did not significantly impact viral infectivity [45, 46]. Thus, disruption of dynein may delay rather than irreversibly impair uncoating. Contrarily, dynein light chain 1 (DYNLL1) stabilized capsids, and its knockdown accelerated uncoating and blocked reverse transcription [48]. Depletion of dynein axonemal-light chain 1 (DNAL1) also decreased reverse transcription [49]. DYNLL1 was shown to interact with IN [48], but it remains unclear how these dynein molecules interact with the capsid to promote their movement and uncoating.
A yeast two-hybrid screen for CA interacting proteins identified MAP1A and MAP1S, which interact with MTs and bind to HIV-1 cores in vitro and also co-localize with HIV-1 during infection [43]. Depletion of MAP1A/MAP1S blocked infection, with the accumulation of capsid away from the nuclear membrane as a result of impaired retrograde trafficking. It was proposed that MAP1 proteins promote cytoplasmic movement towards the nucleus by tethering viral capsids to MTs [43].
Kinesins, a group of motor proteins, have also been shown to facilitate HIV-1 trafficking and uncoating. KIF5A and KIF5B, which together with KIF5C comprise kinesin-1, are required for maximum infection [44, 50, 51] by promoting a post-reverse transcription step(s), perhaps uncoating. Kinesin-1 is not known to interact with the capsid, but two other cellular factors, FEZ1 and NUP358, can link the capsid with kinesin-1. FEZ1 is a kinesin-1 adaptor that can bind CA-NC complexes in vitro [50]. FEZ1 knockdown impeded HIV-1 trafficking to the nucleus. FEZ1 binding to kinesin-1 is required for its ability to promote HIV-1 infection, suggesting that capsid exploits FEZ1 to utilize kinesin-1 for inward trafficking [50]. This model is seemingly counterintuitive, as kinesins are involved in plus-end-directed motion (toward the cellular periphery). The authors speculated that incoming viral capsids utilize motors of opposing directionality, namely dynein and kinesin, to at steady state achieve MT-dependent inward trafficking of PICs to the nucleus [50].
HIV-1 infection relocates NUP358 in a KIF5B-dependent manner from nuclear pores to the cytoplasm, where NUP358 co-localizes with incoming capsids [51]. NUP358 carries a cyclophilin homology domain (CHD) capable of interacting with CA [9, 52] (Figure 1d) and also interacts with KIF5B, thus NUP358 can link the capsid with kinesin-1. KIF5B depletion impaired WT virus, but not CA mutant N74D or P90A infection, arguing for the direct involvement of kinesins in capsid-mediated post-entry events [51]. KIF5B engagement may provide a similar function as FEZ1, promoting uncoating by a “tug of war” with dynein-like molecules to in this case reduce capsid size for efficient PIC nuclear translocation [51]. These observations reveal complex virus-host interactions that enable the HIV-1 capsid to migrate from the cellular periphery to the nuclear membrane and regulate capsid disassembly.
Nuclear transport
HIV-1 PICs are actively transported through nuclear pore complexes (NPCs), which are composed of ~30 individual components called nucleoporins (NUPs) [53]. PICs could engage NPC components directly, or indirectly through engaging soluble karyopherin transporters that mediate nuclear import of cargo substrates [54]. This is a rich area of HIV-1 research going back decades, with numerous studies supporting both direct and indirect NPC binding scenarios [54, 55]. CA is not known to harbor a functional nuclear localization signal (NLS) but, as mentioned, can bind NUP358 directly [9]. The form(s) of the capsid that engage NUP358 or other nuclear transport factors remain to be determined, though partially uncoated capsids should retain sufficient CA for NUP358 engagement (Box 1).
CPSF6 and TNPO3
Transportin-3 (TNPO3) is a β-karyopherin that has been implicated in HIV-1 nuclear entry. TNPO3 naturally functions to transport serine/arginine-rich splicing factors (SR proteins) into the nucleus. TNPO3 bound HIV-1 IN in a yeast 2-hybrid screen and was identified as an HIV-1 dependency factor through genome-wide RNA interference screens [56–58]. TNPO3 depletion did not prevent PIC formation, but reduced the number of proviruses by blocking replication at or after nuclear entry [56–62]. Requirements for TNPO3 during HIV-1 infection map to CA, not IN [59, 63]. TNPO3 can bind CA-NC complexes [62] and accelerate viral core uncoating in vitro [30]. However, we suspect that the function of TNPO3 in HIV-1 infection is mediated via cleavage and polyadenylation specificity factor 6 (CPSF6) [64]. CPSF6, which functions as a component of the CFIm complex to determine mRNA polyadenylation sites [11], is an SR protein that interacts with CA [65–67]. CPSF6 is composed of an N-terminal RNA-recognition motif, a central proline-rich domain (PRD), and a C-terminal arginine/serine-rich (RS) domain. The PRD (residues 314–322 of the larger CPSF6588 isoform) [68] mediates binding to a hydrophobic pocket created by CA NTD helices 3, 4, and 7 [66, 69] (Figure 1a, gray dashes; shown in detail in Figure 1e). The CPSF6 peptide also contacts CTD helices 8 and 9 from an adjacent monomer [69, 70], and thus binds hexamers with higher affinity than monomeric CA, raising the possibility that CPSF6 engages hexameric CA under physiological conditions [69, 70].
Though CPSF6 is predominantly nuclear [65, 67, 71], it can co-localize with HIV-1 CA in the cytoplasm of infected cells [72]. CPSF6 depletion, which tends to marginally increase virus infection [11, 65, 73] (Box 3), has been reported to reduce nuclear localization of viral DNA and CA [74] and integration [11]. Several CA amino acid substitutions in the CPSF6 binding pocket, including N74D and A77V, reduce the affinity of the virus-host interaction [65, 75]. The N74D mutant virus, though robust in T cell lines, is defective in macrophages due to inefficient reverse transcription [76] and/or innate immune activation [27] (Box 2). Importantly, A77V replicated similarly to the wild type (WT) virus in primary cells, yet lost out to the WT in head-to-head competition and reverted back to WT in the majority of inoculated humanized mice [75]. Thus, although CPSF6 is not an essential viral cofactor, HIV-1 prefers this interaction when push comes to shove. The N74D change delayed uncoating [77] and N74D and N74A mutants displayed fewer CA signals in the nucleus than WT HIV-1 [60, 74, 78]. Interestingly, the N74D mutant as well as other CPSF6-binding deficient mutants render HIV-1 almost completely insensitive to depletion of cofactors implicated in nuclear entry including TNPO3, NUP358, and NUP153 [9, 65, 66, 79]. Thus, one model is that CPSF6 facilitates nuclear entry via acting as a molecular tether between CA and TNPO3 [55, 64, 74].
NUP358
NUP358 is located at the outer surface of NPCs. NUP358 depletion inhibits HIV-1 at a step after reverse transcription, with reduced formation of signals associated with nuclear PICs, including 2-long terminal repeat (LTR) circle DNA and CA [9, 52, 58, 80]. As discussed, the NUP358 CHD can bind the capsid, and CA mutants unable to bind the CHD are insensitive to the effects of NUP358 depletion [9]. The NUP358 CHD can catalyze cis-trans isomerization in vitro [81, 82]. Isomerase activity trends with HIV-1 but not with feline immunodeficiency virus infection, which binds the CHD but does not depend on NUP358, leading to the proposal that isomerization promotes nuclear pore uncoating [81]. However, HIV-1 infection in cells deleted of NUP358 can be rescued by introduction of NUP358 lacking the CHD [83]. CypA may help to guide HIV-1 to the NUP358-dependent nuclear entry pathway [9]. CPSF6-binding deficient mutants also infect cells largely independent of NUP358 [9, 65]. Because NUP358 depletion redistributed TNPO3 to the cytoplasm [84], some CPSF6 could relocate as well, which could account for the insensitivity of CPSF6-binding deficient CA mutants to NUP358 depletion, a possibility raised by Meehan et al. [83].
MX2
MX2 is a capsid-targeting HIV-1 restriction factor [85–87]. MX2 does not block reverse transcription, but inhibits nuclear entry and post entry events such as integration [85–88]. MX2 expression altered the pattern of HIV-1 integration similar to what is observed when host nuclear import cofactors are depleted [8–11, 88], but did not affect viral gene expression or particle production [87].
CA mutations that alter core stability or cofactor binding render HIV-1 insensitive to MX2 [85–91]. Such changes spread across the capsid, including the CTD, which has limited exposure to the core surface [90, 92, 93]. MX2 binds CA or CA-NC assembles, which requires the capsid lattice; MX2 does not exhibit appreciable affinity for monomeric or hexameric CA [89, 94, 95]. Certain CA mutations reduce MX2 binding to assembled CA [89, 93], but binding does not strictly correct with sensitivity to restriction [89, 94]. Binding is therefore necessary but not sufficient for MX2 antiviral activity. MX2 binding is competitively blocked by the CPIPB small molecule, which engages the core surface [89], but it is unclear if CPIPB relieves MX2 antiviral activity during infection. Mutations that impact CypA and CPSF6 binding can alter sensitivity to MX2 restriction [85–91].
MX2 is a dynamin-like GTPase [96], but GTP binding and GTPase activity are dispensable for anti-HIV-1 activity [85, 86, 88, 97]. MX2 is structurally similar to MX1, which is active against a broad range of viruses [96]. One distinct feature of MX2 is its extended NTD [96]. Addition of the human MX2 NTD rendered otherwise inactive MX1 or canine Mx2 active against HIV-1 [92, 97]. Furthermore, artificial chimeric proteins harboring the MX2 NTD appended to otherwise unrelated restriction factors, such as Fv1 or SAMHD1 (SAM domain and HD domain-containing protein 1), potently restricted HIV-1 [98, 99]. The MX2 NTD possesses two critical functions for antiviral activity: subcellular localization and capsid binding. MX2 concentrates at the cytoplasmic face of the nuclear envelope [86, 88, 92, 97]. Its N-terminal 20–25 residues encode a NLS [88, 92, 97, 99], the disruption of which abolished MX2 nuclear localization and antiviral activity [88]. Addition of the heterologous NLS from the simian virus 40 large T antigen conferred anti-HIV activity to MX2 lacking its own NLS [88]. The NTD is critical for MX2 binding to CA [94], with a triple arginine motif playing a particularly important role in binding and restriction, but not subcellular localization [98, 99]. Oligomerization is another MX2 property critical for its antiviral activity, and MX2 forms an extended antiparallel dimer [94, 100]. Dimerization, but not higher-order oligomerization, which is mediated by the MX2 CTD, is essential for anti-HIV-1 activity [88, 89, 94, 98, 100–102]. Oligomerization is required for MX2 binding to capsid [94, 101]. One potential mechanism for MX2 invokes that binding to the capsid blocks uncoating and thereby PIC nuclear entry [86, 87, 94, 97]. Consistent with this model, MX2 was shown to block uncoating and co-localize with CA in virus-infected cells [89].
Human MX2 inhibits a variety of HIV-1 strains as well as other primate lentiviruses, but is minimally active against non-primate lentiviruses [85–87, 90–92]. Certain HIV-1 strains, including transmitted/founder viruses, show natural resistance to MX2 without any obvious fitness cost [90, 91]. Conversely, several primate, but not non-primate MX2 orthologs, are active against HIV-1 [92]. Antiviral specificity locates to MX2 NTD positions 37–39 [92]. Evidence for positive selection in other MX2 regions that are not known to dictate activity against lentiviruses suggests that MX2 has an evolutionary history for interacting with a broad range of pathogens [103].
Nuclear events mediated by NUP153
Nuclear import is apparently a rate-limiting step, as only a fraction of incoming virions is successfully transported [104, 105]. Active nuclear entry and integration targeting are regulated by the capsid and CA-interacting proteins together with IN and its host factors [54, 106]. How the capsid dictated nuclear events was enigmatic because early studies found little PIC-associated CA [107]. However, more recent work detected a noticeable amount of CA in nuclear complexes [60, 72, 74, 108]. Nuclear PICs differ from cytoplasmic PICs in size and amount of CA [108, 109]. This suggests a model where at least some uncoating occurs at the nuclear pore, which is consistent with the cytoplasmic and nuclear pore models of uncoating (Box 1) [6, 110]. We could envision that TNPO3 and/or CPSF6 play a role to release PICs from the confines of the NPC after nuclear entry. Although some studies reported a decrease in the number of nuclear PICs upon TNPO3 depletion [56, 74], this was not reported in another study [105]. The discrepancy may be due to differences in experimental approach, including the strategy for PIC labeling.
NUP153 is required for PIC nuclear entry as shown by decreases in 2-LTR circle DNA [52, 58, 79] and nuclear PICs [74, 105], although its role likely extends beyond transport to include integration and integration targeting [10, 78, 79, 111]. The genetic determinant of NUP153 dependency maps to HIV-1 CA [79]. NUP153 utilization by HIV-1 is regulated by CA binding to CypA and CPSF6 [9, 79, 112]. This is somewhat similar to TNPO3 and NUP358 utilization by HIV-1. However, specific defects of viral nuclear entry via NUP153 depletion were independent from CA-CPSF6 binding [74].
HIV-1 CA directly binds to NUP153 [112]. NUP153 was also reported to bind IN [113], but the physiological relevance of this interaction was subsequently questioned [111, 112]. CA binding is mediated by FG motifs within the CTD of NUP153 in a potentially degenerate way [112] (Figure 1f). These FG motifs are essential for HIV-1 infection, as complementation with CTD-deleted NUP153 failed to rescue virus infectivity [111, 114]. The FG repeat at positions 1415–1418 was critical for binding to multimerized CA [69, 70, 112]. Strikingly, HIV-1 uses the same CA NTD pocket to bind both NUP153 and CPSF6 [66, 69, 70, 112] (Figure 1). FG motifs from both NUP153 and CPSF6 hydrogen bond with a conserved asparagine at position 57 in CA (Figure 1e, f). Coincidentally, mutations at position 57 generate the only CA mutant viruses that are known to lose the ability to infect non-dividing cells regardless of cell type [54]. Equine infectious anemia virus uses an analogous residue (Asp58) to engage a different set of NUP153 FG motifs, suggesting convergent evolution of lentiviruses to utilize NUP153 or other FG motif-carrying NUPs [112].
Similar to CPSF6, NUP153 binds a multi-subunit interface in CA hexamers, with binding extending to the CTD, although there are differences that are specific for each ligand [69] (Figure 1e, f). NUP153 can bind monomeric CA [70, 112] but its binding affinity increased significantly with hexameric CA [69, 70]. One model is that NUP153 binding to CA hexamers alters conformation at key interfaces to regulate interactions that may promote uncoating [69, 70, 112]. The short half-life of NUP153 at the NPC may play a role in mediating irreversible and directional exit from the NPC [52, 112].
Genomic and nuclear architecture in HIV-1 integration targeting
Once inside the nucleus, HIV-1 PICs display slow and diffuse movements [40] and preferentially localized to the nuclear periphery [104, 105]. HIV-1 proviruses likewise accumulate in the nuclear periphery [115, 116]. HIV-1 preferentially associates with decondensed chromatin (euchromatin) and disfavors heterochromatin [11, 104], and integration is biased towards active genes enriched in euchromatin [117]. Cellular genes that repeatedly accommodate HIV-1, so called “recurrent integration genes”, are enriched in the nuclear periphery and contact nuclear pores [116]. The preference to integrate into the nuclear periphery may be dictated by the need to integrate rapidly after nuclear entry [115, 116]. This process is in part regulated by the capsid and CA-interacting proteins [54]. CA changes that specifically affect cofactor binding, as well as depletion of CA-binding host factors implicated in nuclear entry, such as, CypA, CPSF6, NUP358, and NUP153, alter integration site preference [8–11, 75, 111]. Thus, nuclear entry is functionally coupled with integration [8, 58]. A dominant role for targeting transcriptionally active chromatin is dictated by CPSF6 [11], which seemingly acts independently from its CFIm binding partner CPSF5 [118]. Side-by-side comparison of the roles of the IN-binding protein lens epithelium-derived growth factor (LEDGF)/p75 led to a model in which CPSF6 directs HIV-1 to euchromatin where LEDGF/p75 predominantly drives integration within gene bodies [11]. Consistently, LEDGF/p75 is dispensable for PIC localization in euchromatin rich regions [119], although LEDGF/p75 and NUP153 are reportedly important to target the nuclear periphery [116]. CPSF6 is critical for PIC localization in the nucleus [74], though paradoxically it enforces PIC penetration into the nucleus [74], an observation that seemingly is at odds with preferential localization to the nuclear periphery [104, 105, 115, 116]. Thus more research in this area is needed.
The site of HIV-1 integration determines basal transcriptional activity [120, 121]. Depletion of Tpr, which together with NUP153 forms the basket-like structure at the nuclear side of the NPC, affects virus infectivity without altering integration levels [114]. Tpr and NUP153 down-regulation significantly reduced LTR-driven gene expression, although the effects of NUP153 may be caused by its role in stabilizing Tpr [114]. However, both Tpr and NUP153 bind to the LTR downstream of the transcription start site [116]. Thus, coupling nuclear entry and integration [8, 58] may provide proviruses with chromatin environments near the NPC that favor productive expression [114, 116]. Consistent with this model, NPCs harbor “transport”-independent functions that include chromatin organization and regulation of transcription [122]. Redirecting integration by heterologous LEDGF/p75 fusion proteins had little effect on viral gene expression [123, 124], but in vivo passage of an HIV-1 mutant with altered integration targeting selected for virus that restored CPSF6 binding and hence preferential integration into active euchromatin [75]. Thus, although the effects of integration targeting dictated by CPSF6 and other host factors are quantitatively subtle, they provide a critical fitness advantage for HIV-1 propagation under physiological conditions.
Concluding Remarks
The work overviewed herein has created novel paradigms for how the HIV-1 capsid coordinates multiple intertwined processes to ensure integration into host chromatin. Capsid interactions with specific cellular proteins are apparently essential for viral trafficking from the plasma membrane to the nucleus, but also are critical for the unique positioning of the virus within the nucleus. Some of these properties are specific for HIV-1 and may contribute to its unique biology. Structural analyses of CA-host interactions have revealed a remarkable utilization of common capsid interfaces for interacting with multiple binding partners (Figure 1). Yet, many unanswered questions remain (See Outstanding Questions). Work has just begun to offer a glimpse of the diverse host antiviral factors that target the capsid, where additional research should reveal valuable mechanistic information. The viral capsid is underexploited for therapeutic intervention, but may be a vulnerable target owing to its multi-factorial roles and extreme genetic fragility (Box 4).
Outstanding Questions Box.
What kind of molecular interactions allows the viral capsid to utilize the host cytoskeleton for intracellular trafficking?
What are the precise roles of host factors in capsid disassembly? What are the structures and composition of viral complexes that go through nuclear pores, and what type of host-virus interactions allows passage and release of viral complexes from nuclear pores?
How does the unique integration pattern of HIV-1 contribute to viral fitness? Are there roles for CA-dependent integration sites in HIV-1 persistence or latency?
How might the dynamics of CA-host interactions change in the presence of innate immune responses, and how might they modulate core stability to escape from innate immune recognition?
Box 4. Antivirals that block cofactor engagement with the capsid.
Recent efforts led to the discovery of capsid-targeting antiviral compounds, including PF-3450074 (PF74) [140]. PF74 directly binds a composite CA interface made from the NTD and the CTD of an adjacent subunit in the same hexamer [69, 70]. Interestingly, this site overlaps with the binding sites of CPSF6 and NUP153 [69, 70, 112] (see Figure 1), and PF74 can accordingly competitively block CA interactions with CPSF6 and NUP153 [67, 69, 70, 112]. PF74 inhibits HIV-1 infection at multiple steps. Relatively high drug concentrations (~10 μM) block reverse transcription by probably accelerating capsid disassembly, whereas lower drug concentrations (~2 μM) block steps following reverse transcription, most likely nuclear entry [69, 72, 108, 140–142]. In contrast to higher PF74 concentrations that block HIV-1 independent from host molecules, antiviral activity by low doses of PF74 depends on capsid interactions with CypA, NUP153, and CPSF6 [24, 112, 141], host factors implicated in HIV-1 PIC nuclear entry. Drug resistant mutations, which mapped within the binding pocket, altered the requirement for HIV-1 nuclear entry cofactors and conferred fitness costs prominently in primary cells [143, 144]. The limited tolerance to drug resistance by extreme genetic fragility [145] together with the unique multimodal mechanisms of action make the viral capsid a unique target for therapeutic interventions.
Trends Box.
Capsid-binding host factors regulate HIV-1 core disassembly during cytoplasmic trafficking and nuclear entry.
Interactions of the viral capsid with host factors functionally couple nuclear entry and integration targeting.
The assembled viral capsid, a preferred target for cofactor engagement, possesses two distinct interfaces, each of which is capable of interacting with at least two host proteins.
The viral capsid is targeted by multiple antiviral proteins in intrinsic and innate immunity but can recruit host factors to evade innate immune sensing.
Acknowledgments
We apologize to our colleagues whose contributions were not cited due to space constraints. Work in our laboratories is supported by National Institute of Health grants R01AI100720 (MY), R01AI052014 (ANE), and P50GM082251 (MY and ANE).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Perilla JR, Gronenborn AM. Molecular Architecture of the Retroviral Capsid. Trends in biochemical sciences. 2016;41:410–420. doi: 10.1016/j.tibs.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundquist WI, Krausslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harbor perspectives in medicine. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pornillos O, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malim MH, Bieniasz PD. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harbor perspectives in medicine. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fassati A. Multiple roles of the capsid protein in the early steps of HIV-1 infection. Virus research. 2012;170:15–24. doi: 10.1016/j.virusres.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Campbell EM, Hope TJ. HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nature reviews. Microbiology. 2015;13:471–483. doi: 10.1038/nrmicro3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamashita M, Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. Journal of virology. 2004;78:5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ocwieja KE, et al. HIV Integration Targeting: A Pathway Involving Transportin-3 and the Nuclear Pore Protein RanBP2. PLoS pathogens. 2011;7:e1001313. doi: 10.1371/journal.ppat.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaller T, et al. HIV-1 Capsid-Cyclophilin Interactions Determine Nuclear Import Pathway, Integration Targeting and Replication Efficiency. PLoS pathogens. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koh Y, et al. Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. Journal of virology. 2013;87:648–658. doi: 10.1128/JVI.01148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sowd GA, et al. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E1054–1063. doi: 10.1073/pnas.1524213113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ambrose Z, Aiken C. HIV-1 uncoating: connection to nuclear entry and regulation by host proteins. Virology. 2014:454–455. doi: 10.1016/j.virol.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulme AE, et al. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, et al. Inhibition of reverse transcriptase activity increases stability of the HIV-1 core. Journal of virology. 2013;87:683–687. doi: 10.1128/JVI.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosnefroy O, et al. HIV-1 capsid uncoating initiates after the first strand transfer of reverse transcription. Retrovirology. 2016;13:58. doi: 10.1186/s12977-016-0292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sokolskaja E, et al. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. Journal of virology. 2004;78:12800–12808. doi: 10.1128/JVI.78.23.12800-12808.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatziioannou T, et al. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. Journal of virology. 2005;79:176–183. doi: 10.1128/JVI.79.1.176-183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luban J. Cyclophilin A, TRIM5, and resistance to human immunodeficiency virus type 1 infection. Journal of virology. 2007;81:1054–1061. doi: 10.1128/JVI.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franke EK, et al. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 20.Gamble TR, et al. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu C, et al. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nature communications. 2016;7:10714. doi: 10.1038/ncomms10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Iaco A, Luban J. Cyclophilin A promotes HIV-1 reverse transcription but its effect on transduction correlates best with its effect on nuclear entry of viral cDNA. Retrovirology. 2014;11:11. doi: 10.1186/1742-4690-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braaten D, Luban J. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. The EMBO journal. 2001;20:1300–1309. doi: 10.1093/emboj/20.6.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito A, et al. Roles of Capsid-Interacting Host Factors in Multimodal Inhibition of HIV-1 by PF74. Journal of virology. 2016;90:5808–5823. doi: 10.1128/JVI.03116-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulli L, et al. Complex Interplay between HIV-1 Capsid and MX2-Independent Alpha Interferon-Induced Antiviral Factors. Journal of virology. 2016;90:7469–7480. doi: 10.1128/JVI.00458-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lahaye X, et al. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 2013;39:1132–1142. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Rasaiyaah J, et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503:402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lahaye X, et al. Nuclear Envelope Protein SUN2 Promotes Cyclophilin-A-Dependent Steps of HIV Replication. Cell reports. 2016;15:879–892. doi: 10.1016/j.celrep.2016.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donahue DA, et al. SUN2 silencing impairs CD4 T cell proliferation and alters sensitivity to HIV-1 infection independently of Cyclophilin A. Journal of virology. 2017 doi: 10.1128/JVI.02303-02316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shah VB, et al. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. Journal of virology. 2013;87:422–432. doi: 10.1128/JVI.07177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, et al. Target cell type-dependent modulation of human immunodeficiency virus type 1 capsid disassembly by cyclophilin A. Journal of virology. 2009;83:10951–10962. doi: 10.1128/JVI.00682-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fricke T, et al. Human cytosolic extracts stabilize the HIV-1 core. Journal of virology. 2013;87:10587–10597. doi: 10.1128/JVI.01705-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanz-Ramos M, Stoye JP. Capsid-binding retrovirus restriction factors: discovery, restriction specificity and implications for the development of novel therapeutics. The Journal of general virology. 2013;94:2587–2598. doi: 10.1099/vir.0.058180-0. [DOI] [PubMed] [Google Scholar]
- 34.Misumi S, et al. Uncoating of human immunodeficiency virus type 1 requires prolyl isomerase Pin1. The Journal of biological chemistry. 2010;285:25185–25195. doi: 10.1074/jbc.M110.114256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeBoer J, et al. Cyclophilin B enhances HIV-1 infection. Virology. 2016;489:282–291. doi: 10.1016/j.virol.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan T, et al. An HIV-1 capsid binding protein TRIM11 accelerates viral uncoating. Retrovirology. 2016;13:72. doi: 10.1186/s12977-016-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guth CA, Sodroski J. Contribution of PDZD8 to stabilization of the human immunodeficiency virus type 1 capsid. Journal of virology. 2014;88:4612–4623. doi: 10.1128/JVI.02945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S, Sodroski J. Efficient human immunodeficiency virus (HIV-1) infection of cells lacking PDZD8. Virology. 2015;481:73–78. doi: 10.1016/j.virol.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald D, et al. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arhel N, et al. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nature methods. 2006;3:817–824. doi: 10.1038/nmeth928. [DOI] [PubMed] [Google Scholar]
- 41.Gaudin R, et al. HIV trafficking in host cells: motors wanted! Trends in cell biology. 2013;23:652–662. doi: 10.1016/j.tcb.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 42.Sabo Y, et al. HIV-1 induces the formation of stable microtubules to enhance early infection. Cell host & microbe. 2013;14:535–546. doi: 10.1016/j.chom.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez J, et al. Microtubule-associated proteins 1 (MAP1) promote human immunodeficiency virus type I (HIV-1) intracytoplasmic routing to the nucleus. The Journal of biological chemistry. 2015;290:4631–4646. doi: 10.1074/jbc.M114.613133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lukic Z, et al. HIV-1 uncoating is facilitated by dynein and kinesin 1. Journal of virology. 2014;88:13613–13625. doi: 10.1128/JVI.02219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pawlica P, et al. Functional evidence for the involvement of microtubules and dynein motor complexes in TRIM5alpha-mediated restriction of retroviruses. Journal of virology. 2014;88:5661–5676. doi: 10.1128/JVI.03717-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pawlica P, Berthoux L. Cytoplasmic dynein promotes HIV-1 uncoating. Viruses. 2014;6:4195–4211. doi: 10.3390/v6114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jayappa KD, et al. Human immunodeficiency virus type 1 employs the cellular dynein light chain 1 protein for reverse transcription through interaction with its integrase protein. Journal of virology. 2015;89:3497–3511. doi: 10.1128/JVI.03347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gallo DE, Hope TJ. Knockdown of MAP4 and DNAL1 produces a post-fusion and pre-nuclear translocation impairment in HIV-1 replication. Virology. 2012;422:13–21. doi: 10.1016/j.virol.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malikov V, et al. HIV-1 capsids bind and exploit the kinesin-1 adaptor FEZ1 for inward movement to the nucleus. Nature communications. 2015;6:6660. doi: 10.1038/ncomms7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dharan A, et al. KIF5B and Nup358 Cooperatively Mediate the Nuclear Import of HIV-1 during Infection. PLoS pathogens. 2016;12:e1005700. doi: 10.1371/journal.ppat.1005700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Nunzio F, et al. Human Nucleoporins Promote HIV-1 Docking at the Nuclear Pore, Nuclear Import and Integration. PloS one. 2012;7:e46037. doi: 10.1371/journal.pone.0046037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beck M, Hurt E. The nuclear pore complex: understanding its function through structural insight. Nature reviews. Molecular cell biology. 2017;18:73–89. doi: 10.1038/nrm.2016.147. [DOI] [PubMed] [Google Scholar]
- 54.Matreyek KA, Engelman A. Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses. 2013;5:2483–2511. doi: 10.3390/v5102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hilditch L, Towers GJ. A model for cofactor use during HIV-1 reverse transcription and nuclear entry. Current opinion in virology. 2014;4:32–36. doi: 10.1016/j.coviro.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Christ F, et al. Transportin-SR2 imports HIV into the nucleus. Curr Biol. 2008;18:1192–1202. doi: 10.1016/j.cub.2008.07.079. [DOI] [PubMed] [Google Scholar]
- 57.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 58.König R, et al. Global Analysis of Host-Pathogen Interactions that Regulate Early-Stage HIV-1 Replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Iaco A, Luban J. Inhibition of HIV-1 infection by TNPO3 depletion is determined by capsid and detectable after viral cDNA enters the nucleus. Retrovirology. 2011;8:98. doi: 10.1186/1742-4690-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou L, et al. Transportin 3 Promotes a Nuclear Maturation Step Required for Efficient HIV-1 Integration. PLoS pathogens. 2011;7:e1002194. doi: 10.1371/journal.ppat.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Logue EC, et al. The Cargo-Binding Domain of Transportin 3 Is Required for Lentivirus Nuclear Import. Journal of virology. 2011;85:12950–12961. doi: 10.1128/JVI.05384-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valle-Casuso JC, et al. TNPO3 Is Required for HIV-1 Replication after Nuclear Import but prior to Integration and Binds the HIV-1 Core. Journal of virology. 2012;86:5931–5936. doi: 10.1128/JVI.00451-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krishnan L, et al. The Requirement for Cellular Transportin 3 (TNPO3 or TRN-SR2) during Infection Maps to Human Immunodeficiency Virus Type 1 Capsid and Not Integrase. Journal of virology. 2010;84:397–406. doi: 10.1128/JVI.01899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maertens GN, et al. Structural basis for nuclear import of splicing factors by human Transportin 3. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2728–2733. doi: 10.1073/pnas.1320755111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee K, et al. Flexible use of nuclear import pathways by HIV-1. Cell host & microbe. 2010;7:221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price AJ, et al. CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS pathogens. 2012;8:e1002896. doi: 10.1371/journal.ppat.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fricke T, et al. The ability of TNPO3-depleted cells to inhibit HIV-1 infection requires CPSF6. Retrovirology. 2013;10:46. doi: 10.1186/1742-4690-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee K, et al. HIV-1 capsid-targeting domain of cleavage and polyadenylation specificity factor 6. Journal of virology. 2012;86:3851–3860. doi: 10.1128/JVI.06607-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price AJ, et al. Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly. PLoS pathogens. 2014;10:e1004459. doi: 10.1371/journal.ppat.1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhattacharya A, et al. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18625–18630. doi: 10.1073/pnas.1419945112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Iaco A, et al. TNPO3 protects HIV-1 replication from CPSF6-mediated capsid stabilization in the host cell cytoplasm. Retrovirology. 2013;10:20. doi: 10.1186/1742-4690-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng K, et al. Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. Elife. 2014;3:e04114. doi: 10.7554/eLife.04114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Henning MS, et al. In vivo functions of CPSF6 for HIV-1 as revealed by HIV-1 capsid evolution in HLA-B27-positive subjects. PLoS pathogens. 2014;10:e1003868. doi: 10.1371/journal.ppat.1003868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chin CR, et al. Direct Visualization of HIV-1 Replication Intermediates Shows that Capsid and CPSF6 Modulate HIV-1 Intra-nuclear Invasion and Integration. Cell reports. 2015;13:1717–1731. doi: 10.1016/j.celrep.2015.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saito A, et al. Capsid-CPSF6 Interaction Is Dispensable for HIV-1 Replication in Primary Cells but Is Selected during Virus Passage In Vivo. Journal of virology. 2016;90:6918–6935. doi: 10.1128/JVI.00019-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ambrose Z, et al. Human immunodeficiency virus type 1 capsid mutation N74D alters cyclophilin A dependence and impairs macrophage infection. Journal of virology. 2012;86:4708–4714. doi: 10.1128/JVI.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hulme AE, et al. Identification of capsid mutations that alter the rate of HIV-1 uncoating in infected cells. Journal of virology. 2015;89:643–651. doi: 10.1128/JVI.03043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen NY, et al. HIV-1 capsid is involved in post-nuclear entry steps. Retrovirology. 2016;13:28. doi: 10.1186/s12977-016-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matreyek KA, Engelman A. The Requirement for Nucleoporin NUP153 during Human Immunodeficiency Virus Type 1 Infection Is Determined by the Viral Capsid. Journal of virology. 2011;85:7818–7827. doi: 10.1128/JVI.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang R, et al. Perturbation of Host Nuclear Membrane Component RanBP2 Impairs the Nuclear Import of Human Immunodeficiency Virus -1 Preintegration Complex (DNA) PloS one. 2010;5:e15620. doi: 10.1371/journal.pone.0015620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bichel K, et al. HIV-1 capsid undergoes coupled binding and isomerization by the nuclear pore protein NUP358. Retrovirology. 2013;10:81. doi: 10.1186/1742-4690-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin DH, et al. Structural and functional analysis of the C-terminal domain of Nup358/RanBP2. Journal of molecular biology. 2013;425:1318–1329. doi: 10.1016/j.jmb.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meehan AM, et al. A cyclophilin homology domain-independent role for Nup358 in HIV-1 infection. PLoS pathogens. 2014;10:e1003969. doi: 10.1371/journal.ppat.1003969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saitoh N, et al. The distribution of phosphorylated SR proteins and alternative splicing are regulated by RANBP2. Molecular biology of the cell. 2012;23:1115–1128. doi: 10.1091/mbc.E11-09-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goujon C, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kane M, et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013;502:563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Z, et al. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell host & microbe. 2013;14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 88.Matreyek KA, et al. Host and viral determinants for MxB restriction of HIV-1 infection. Retrovirology. 2014;11:90. doi: 10.1186/s12977-014-0090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fricke T, et al. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology. 2014;11:68. doi: 10.1186/s12977-014-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu Z, et al. The highly polymorphic cyclophilin A-binding loop in HIV-1 capsid modulates viral resistance to MxB. Retrovirology. 2015;12:1. doi: 10.1186/s12977-014-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wei W, et al. Accumulation of MxB/Mx2-resistant HIV-1 Capsid Variants During Expansion of the HIV-1 Epidemic in Human Populations. EBioMedicine. 2016;8:230–236. doi: 10.1016/j.ebiom.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Busnadiego I, et al. Host and viral determinants of Mx2 antiretroviral activity. Journal of virology. 2014;88:7738–7752. doi: 10.1128/JVI.00214-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Opp S, et al. MxB Is Not Responsible for the Blocking of HIV-1 Infection Observed in Alpha Interferon-Treated Cells. Journal of virology. 2015;90:3056–3064. doi: 10.1128/JVI.03146-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fribourgh JL, et al. Structural Insight into HIV-1 Restriction by MxB. Cell host & microbe. 2014;16:627–638. doi: 10.1016/j.chom.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kong J, et al. Characterization of the amino-terminal domain of Mx2/MxB-dependent interaction with the HIV-1 capsid. Protein & cell. 2014;5:954–957. doi: 10.1007/s13238-014-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haller O, et al. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends in microbiology. 2015;23:154–163. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 97.Goujon C, et al. Transfer of the amino-terminal nuclear envelope targeting domain of human MX2 converts MX1 into an HIV-1 resistance factor. Journal of virology. 2014;88:9017–9026. doi: 10.1128/JVI.01269-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goujon C, et al. A triple-arginine motif in the amino-terminal domain and oligomerization are required for HIV-1 inhibition by human MX2. Journal of virology. 2015;89:4676–4680. doi: 10.1128/JVI.00169-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schulte B, et al. Restriction of HIV-1 Requires the N-Terminal Region of MxB as a Capsid-Binding Motif but Not as a Nuclear Localization Signal. Journal of virology. 2015;89:8599–8610. doi: 10.1128/JVI.00753-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu B, et al. Structural insight into the assembly of human anti-HIV dynamin-like protein MxB/Mx2. Biochemical and biophysical research communications. 2015;456:197–201. doi: 10.1016/j.bbrc.2014.11.058. [DOI] [PubMed] [Google Scholar]
- 101.Buffone C, et al. Contribution of MxB oligomerization to HIV-1 capsid binding and restriction. Journal of virology. 2015;89:3285–3294. doi: 10.1128/JVI.03730-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dicks MD, et al. Oligomerization Requirements for MX2-Mediated Suppression of HIV-1 Infection. Journal of virology. 2015;90:22–32. doi: 10.1128/JVI.02247-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mitchell PS, et al. Evolutionary Analyses Suggest a Function of MxB Immunity Proteins Beyond Lentivirus Restriction. PLoS pathogens. 2015;11:e1005304. doi: 10.1371/journal.ppat.1005304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Albanese A, et al. HIV-1 pre-integration complexes selectively target decondensed chromatin in the nuclear periphery. PloS one. 2008;3:e2413. doi: 10.1371/journal.pone.0002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Burdick RC, et al. Nuclear import of APOBEC3F-labeled HIV-1 preintegration complexes. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4780–4789. doi: 10.1073/pnas.1315996110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Craigie R, Bushman FD. Host Factors in Retroviral Integration and the Selection of Integration Target Sites. Microbiology spectrum. 2014;2 doi: 10.1128/microbiolspec.MDNA3-0026-2014. 10.1128/microbiolspec.MDNA1123-0026-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Miller MD, et al. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. Journal of virology. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hulme AE, et al. Complementary Assays Reveal a Low Level of CA Associated with Viral Complexes in the Nuclei of HIV-1-Infected Cells. Journal of virology. 2015;89:5350–5361. doi: 10.1128/JVI.00476-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lelek M, et al. Superresolution imaging of HIV in infected cells with FlAsH-PALM. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8564–8569. doi: 10.1073/pnas.1013267109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arhel N. Revisiting HIV-1 uncoating. Retrovirology. 2010;7:96. doi: 10.1186/1742-4690-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Di Nunzio F, et al. Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication. Virology. 2013;440:8–18. doi: 10.1016/j.virol.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Matreyek KA, et al. Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS pathogens. 2013;9:e1003693. doi: 10.1371/journal.ppat.1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Woodward CL, et al. Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1. Journal of virology. 2009;83:6522–6533. doi: 10.1128/JVI.02061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lelek M, et al. Chromatin organization at the nuclear pore favours HIV replication. Nature communications. 2015;6:6483. doi: 10.1038/ncomms7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Di Primio C, et al. Single-cell imaging of HIV-1 provirus (SCIP) Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5636–5641. doi: 10.1073/pnas.1216254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marini B, et al. Nuclear architecture dictates HIV-1 integration site selection. Nature. 2015;521:227–231. doi: 10.1038/nature14226. [DOI] [PubMed] [Google Scholar]
- 117.Schroder AR, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 118.Rasheedi S, et al. The Cleavage and Polyadenylation Specificity Factor 6 (CPSF6) Subunit of the Capsid-recruited Pre-messenger RNA Cleavage Factor I (CFIm) Complex Mediates HIV-1 Integration into Genes. The Journal of biological chemistry. 2016;291:11809–11819. doi: 10.1074/jbc.M116.721647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Quercioli V, et al. Comparative Analysis of HIV-1 and Murine Leukemia Virus Three-Dimensional Nuclear Distributions. Journal of virology. 2016;90:5205–5209. doi: 10.1128/JVI.03188-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Demeulemeester J, et al. Retroviral integration: Site matters: Mechanisms and consequences of retroviral integration site selection. BioEssays : news and reviews in molecular, cellular and developmental biology. 2015;37:1202–1214. doi: 10.1002/bies.201500051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lusic M, Siliciano RF. Nuclear landscape of HIV-1 infection and integration. Nature reviews Microbiology. 2017;15:69–82. doi: 10.1038/nrmicro.2016.162. [DOI] [PubMed] [Google Scholar]
- 122.Raices M, D’Angelo MA. Nuclear pore complexes and regulation of gene expression. Current opinion in cell biology. 2017;46:26–32. doi: 10.1016/j.ceb.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ferris AL, et al. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3135–3140. doi: 10.1073/pnas.0914142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brady T, et al. Quantitation of HIV DNA integration: effects of differential integration site distributions on Alu-PCR assays. J Virol Methods. 2013;189:53–57. doi: 10.1016/j.jviromet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Arhel NJ, et al. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. The EMBO journal. 2007;26:3025–3037. doi: 10.1038/sj.emboj.7601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu H, et al. Evidence for biphasic uncoating during HIV-1 infection from a novel imaging assay. Retrovirology. 2013;10:70. doi: 10.1186/1742-4690-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 128.Da Silva Santos C, et al. A Novel Entry/Uncoating Assay Reveals the Presence of at Least Two Species of Viral Capsids During Synchronized HIV-1 Infection. PLoS pathogens. 2016;12:e1005897. doi: 10.1371/journal.ppat.1005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Francis AC, et al. Time-Resolved Imaging of Single HIV-1 Uncoating In Vitro and in Living Cells. PLoS pathogens. 2016;12:e1005709. doi: 10.1371/journal.ppat.1005709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doyle T, et al. HIV-1 and interferons: who’s interfering with whom? Nature reviews. Microbiology. 2015;13:403–413. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gao D, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jakobsen MR, et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4571–4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Setiawan LC, et al. Mutations in CypA Binding Region of HIV-1 Capsid Affect Capsid Stability and Viral Replication in Primary Macrophages. AIDS research and human retroviruses. 2016;32:390–398. doi: 10.1089/AID.2014.0361. [DOI] [PubMed] [Google Scholar]
- 135.Yan N, et al. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nature immunology. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hori T, et al. A carboxy-terminally truncated human CPSF6 lacking residues encoded by exon 6 inhibits HIV-1 cDNA synthesis and promotes capsid disassembly. Journal of virology. 2013;87:7726–7736. doi: 10.1128/JVI.00124-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lu M, et al. Dynamic allostery governs cyclophilin A-HIV capsid interplay. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:14617–14622. doi: 10.1073/pnas.1516920112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shah VB, Aiken C. Gene expression analysis of a panel of cell lines that differentially restrict HIV-1 CA mutants infection in a cyclophilin a-dependent manner. PloS one. 2014;9:e92724. doi: 10.1371/journal.pone.0092724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Donahue DA, et al. SUN2 Overexpression Deforms Nuclear Shape and Inhibits HIV. Journal of virology. 2016;90:4199–4214. doi: 10.1128/JVI.03202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Blair WS, et al. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS pathogens. 2010;6:e1001220. doi: 10.1371/journal.ppat.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shi J, et al. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. Journal of virology. 2011;85:542–549. doi: 10.1128/JVI.01406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fricke T, et al. BI-2 destabilizes HIV-1 cores during infection and Prevents Binding of CPSF6 to the HIV-1 Capsid. Retrovirology. 2014;11:120. doi: 10.1186/s12977-014-0120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shi J, et al. Compensatory Substitutions in the HIV-1 Capsid Reduce the Fitness Cost Associated with Resistance to a Capsid-Targeting Small-Molecule Inhibitor. Journal of virology. 2015;89:208–219. doi: 10.1128/JVI.01411-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou J, et al. HIV-1 Resistance to the Capsid-Targeting Inhibitor PF74 Results in Altered Dependence on Host Factors Required for Virus Nuclear Entry. Journal of virology. 2015;89:9068–9079. doi: 10.1128/JVI.00340-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rihn S, et al. Extreme genetic fragility of the HIV-1 capsid. PLoS pathogens. 2013;9:e1003461. doi: 10.1371/journal.ppat.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gres AT, et al. STRUCTURAL VIROLOGY. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science. 2015;349:99–103. doi: 10.1126/science.aaa5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Luban J, et al. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 148.Henning MS, et al. PDZD8 is a novel Gag-interacting factor that promotes retroviral infection. Journal of virology. 2010;84:8990–8995. doi: 10.1128/JVI.00843-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


