Abstract
Agrobacterium-mediated genetic transformation of plants, a unique example of transkingdom DNA transfer, requires the presence of several proteins encoded by the host cell. One such cellular factor is VIP1, an Arabidopsis protein proposed to interact with and facilitate import of the bacterial DNA–protein transport (T) complexes into the plant cell nucleus. Thus, VIP1 is required for transient expression of the bacterial DNA, an early step in the transformation process. However, the role of VIP1 in subsequent transformation events leading to the stable expression of bacterial DNA was unexplored. Here, we used reverse genetics to dissect VIP1 functionally and demonstrate its involvement in the stable genetic transformation of Arabidopsis plants by Agrobacterium. Our data indicate that the ability of VIP1 to interact with the VirE2 protein component of the T-complex and localize to the cell nucleus is sufficient for transient genetic transformation, whereas its ability to form homomultimers and interact with the host cell H2A histone in planta is required for tumorigenesis and, by implication, stable genetic transformation.
Keywords: T-complex, VirE2, nuclear import, histones, chromatin targeting
Agrobacterium genetically transforms plants and, in nature, elicits neoplastic growths in many host species. Moreover, although plants represent the natural hosts for Agrobacterium, it can also transform a wide range of other eukaryotes, from fungi (1) to human cells (2). This genetic transformation is achieved by transporting a single-stranded (ss) copy (T-strand) of the transferred DNA (T-DNA) from the bacterial tumor-inducing (Ti) plasmid into the host cell nucleus and integrating the T-strand into the host cell genome (3, 4).
Nuclear import of T-strands is mediated by two bacterial virulence (Vir) proteins, VirD2 and VirE2, which are thought to associate directly with the T-strand, forming a transport (T) complex (5). In the T-complex, one molecule of VirD2 is covalently attached to the 5′ end of the T-strand whereas VirE2 is presumed to coat the rest of the T-strand molecule (reviewed in refs. 3 and 5). Interestingly, VirE2 alone is sufficient to transport ssDNA into the plant cell nucleus (6).
In plant cells, the T-complex likely interacts with cellular factors during its nuclear import and T-DNA integration (reviewed in ref. 3). One such plant cell factor is the Arabidopsis VirE2-interacting protein (VIP1) that binds VirE2 and acts as a molecular adaptor between VirE2 and the nuclear import machinery of the host cell (7, 8). The full extent of VIP1 participation in the transformation process, however, remains unknown because the VIP1 requirement for nuclear import, an early stage of infection, precludes studies of its potential involvement in the later infection steps, such as T-DNA integration and tumorigenesis. Here, we use reverse genetics to dissect VIP1 functionally and demonstrate that the capacity of VIP1 to interact with VirE2 and localize to the cell nucleus is sufficient for transient genetic transformation, whereas its ability to form homomultimers and interact with the plant H2A histone is required for the subsequent step of stable genetic transformation and tumor formation.
Materials and Methods
Identification of vip1-1 Homozygous Lines. T-DNA insertion line SALK_001014 of Arabidopsis (Col-0 strain) (9) was received from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). Plants homozygous for T-DNA insertion in VIP1 were identified by PCR by using T-DNA left border-specific (9) (http://signal.salk.edu/cgi-bin/tdnaexpress) and gene-specific primers and genomic DNA was extracted by using the DNeasy Plant Kit (Qiagen, Valencia, CA). For Southern blotting, 10 μg of purified DNA was digested with HindIII or BamHI and analyzed by using standard techniques (10) and a digoxigenin-labeled probe, which did not contain HindIII or BamHI sites and spanned nucleotides 1–781 of the T-DNA kanamycin-resistance gene.
RT-PCR. Total RNA was extracted from 200 mg of plant tissues, treated with RQ1 RNase-free DNase (Promega), and reverse-transcribed with Moloney murine leukemia virus reverse transcriptase (New England Biolabs), and the resulting cDNAs were PCR-amplified (11) and detected by ethidium bromide staining of agarose gels. Forward 5′-TCTCGGTTGATTCCGATTTC-3′ and reverse 5′-CCGCAGATTGTCTATTCGCT-3′ primers generated a 280-bp product from the vip1-1 transcript, and forward 5′-CGAACGGTGTTGTTCCTCCTAATTCTCTT-3′ and reverse 5′-GACACA A ACTCAGCCTCTCT TGGTGAAAT-3′ primers generated a 780-bp product from the VIP1 transcript. All PCRs were performed by using a high-fidelity Pfu DNA polymerase (Stratagene) and verified by DNA sequencing.
Bombardment and Nuclear Import in Plant Tissues. PCR-amplified green fluorescent protein (GFP) ORF was ligated into the BglII–HindIII sites of pEGFP-C1 (Clontech), producing GFP dimer, (GFP)2, and transferred into the NcoI–BamHI sites of pRTL2-GUS (12), replacing β-glucuronidase (GUS) and resulting in pRTL2-(GFP)2. PCR-amplified VIP1-N164 (corresponding to amino acids 1–164 of VIP1) was cloned into the SalI–BamHI sites of pRTL2-(GFP)2, and the expression cassette was transferred into the SphI site of pGDR, which also expresses free DsRed2 (13), resulting in pGDR-(GFP)2-VIP1-N164 that was bombarded into the leaf epidermis of Nicotiana tabacum (7). After 16 h, the leaves were examined under a Zeiss LSM 5 Pascal confocal laser scanning microscope. Each experiment examined 20–100 GFP/DsRed2-expressing cells and was repeated at least four times. Note that DsRed2 represents a useful marker for confocal microscopy analysis of nuclear import because it does not require excitation by UV light and, thus, can be visualized by using a He/Ne laser. In addition to identifying the cell nucleus, it allows one to see the cell outline (13, 14) otherwise invisible on a confocal image.
GUS-VirE2 expression plasmid (15) was bombarded into leaves of the wild-type or vip1-1 Arabidopsis and, 16 h later, the tissues were stained histochemically for GUS activity and with DAPI (7, 16) and were observed under a Zeiss Axiophot microscope. Each experiment examined 10–20 expressing cells and was repeated at least four times.
For detection of nuclear import by using immunofluorescence, 0.25- to 0.5-cm leaf samples were fixed, embedded, sectioned into 8-μm slices, reacted with anti-VIP1 antibodies (17) as described (18), stained with Cy5-conjugated secondary antibody (Jackson ImmunoResearch), and examined by confocal microscopy.
Bimolecular Fluorescence Complementation (BiFC) Assay for Protein Interactions in Planta. Yellow fluorescent protein (YFP) spectral variant of GFP was dissected into two parts: the N-terminal part (nYFP) that terminated at amino acid residue 174, and the C-terminal part (cYFP) that began with a methionine residue preceding the residue 175 of YFP. pRTL2-nYFP and pRTL2-cYFP have been described (19). For cYFP fusions, VirE2 and VIP1 were first cloned into the SalI–BamHI sites of pEYFP-C1, from which cYFP-VIP1 and cYFP-VirE2 were PCR-amplified and cloned into the NcoI–BamHI sites of pRTL2-GUS (12), replacing GUS and producing pRTL2-cYFP-VIP1 and pRTL2-cYFP-VirE2. PCR-amplified VIP1-N164 was cloned into the SalI–BamHI sites of pRTL2-cYFP-VIP1, replacing VIP1. For nYFP fusions, PCR-amplified nYFP was cloned into the NcoI–SalI sites of pRTL2-GFP (20) and pRTL2-(GFP)2-VIP1-N164, replacing GFP and (GFP)2, and the entire expression cassettes were transferred into the SphI site of pGDR, which expresses free DsRed2 (13). PCR-amplified Arabidopsis cDNA encoding histone H2A-1 (GenBank accession no. AF204968.1) was cloned into the SalI–BamHI sites of pRTL2-cYFP-VIP1, replacing VIP1. The tested pairs of constructs were bombarded into tobacco leaves and examined by confocal microscopy. The transformed cells were identified by the presence of DsRed2; each experiment examined 50–100 DsRed2-expressing cells and was repeated at least three times, with 30–40% of the transformed cells exhibiting BiFC.
Agrobacterium-Mediated Transient and Stable Genetic Transformation of Arabidopsis. For transient T-DNA expression, 50–80 root segments from 10- to 14-day-old Arabidopsis seedlings grown in baby food jars (16) were cultivated for 48 h at 25°C on a hormone-free MS medium (HFMS) (21) with Agrobacterium tumefaciens EHA105 (OD600 = 0.1) harboring the GUS-expressing binary vector pKIWI105 (22). For histochemical GUS staining, root segments were transferred to fresh HFMS for 4 days (16), and stained with 5-bromo-4-chloro-3-indolyl β-d-glucuronide (BioVectra, Canada) (16). For quantification of GUS activity, root segments were ground and assayed by using 4-methylumbelliferyl β-d-galactoside (Sigma-Aldrich) (16). All experiments were performed in triplicate. For tumor assays (16), 30–50 root segments per tested plant were submerged in an OD600 = 0.1 culture of A. tumefaciens A208, incubated for 10 min at 25°C, cultivated for 48 h at 25°C in HFMS, washed, and cultured for 4 weeks in HFMS with 300 μg/ml carbenicillin. Tumors were counted, and their phenotype was determined. Each experiment was repeated three times.
Genetic Complementation of the vip1-1 Mutant. To produce the full-length VIP1 transgene, we used a fluorescent tagging of full-length proteins technique (23). We constructed a transgene containing the native VIP1 promoter, coding region with introns, and the 3′ UTR sequences; because most regulatory sequences in Arabidopsis are contained in relatively small (2-kb) regions (24), we included 1,757 bp upstream of the VIP1 translation initiation codon and 517 bp downstream of the STOP codon in our constructs. First, VIP1 was amplified by using the forward 5′-GCTCGATCCACCTACGCTGTTGTATTATGAGAAAGCTTGAGCG-3′ and reverse 5′-CGTAGCGAGACCACAGGAGACGGCACCATTTCTCATTT-3′ primers that contained gene-specific sequences tailed with sequences complementary to the Gateway (Invitrogen) primers. The attB1 and attB2 recombination sites (25) were then added by using the standard Gateway forward 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGCTCGATCCACCTAGGCT-3′ and reverse 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGTAGCGAGACCACAGGA-3′ primers (23). All PCR conditions were as described (23). The resulting VIP1 gene was verified by DNA sequencing and inserted into pSAT6-DEST [constructed by inserting the Gateway conversion cassette C.1 (Invitrogen) into the AgeI–BspHI sites of pAUX3133 (26)] by Gateway recombination as described (23). Then, the VIP1 gene was transferred into the PI-PspI site of pPZP-RCS2 (26), which contained, within its AscI site, a hygromycin-resistance gene expression cassette. This binary construct was introduced into A. tumefaciens GV3101 and used to transform the vip1-1 plants by flower dipping (27); seven hygromycin-resistant T1 transformants were then selected and tested for transient T-DNA expression and tumorigenesis.
Results
vip1-1, an Arabidopsis T-DNA Insertional Mutant in the VIP1 gene. An Arabidopsis mutant from the SALK collection (9) with a T-DNA insertion(s) in the third exon of the VIP1 gene (Fig. 1A) was self-pollinated, and the homozygous line was identified by PCR analysis. The location of the 5′ end of the T-DNA insert in this line was confirmed by DNA sequencing (Fig. 1D).
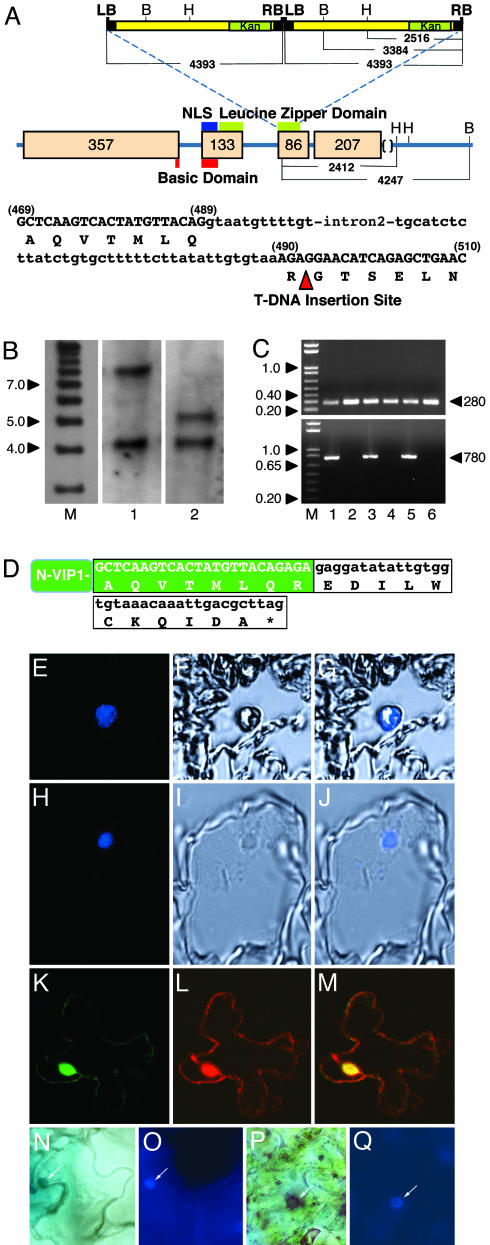
Fig. 1.
vip1-1 mutant and its protein product. (A) Schematic structure of the VIP1 gene and its flanking sequences on chromosome 1 (GenBank accession no. AC009526.3), pattern of integration of the mutagenic T-DNA, and VIP1 sequence flanking the T-DNA insertion site. LB and RB, T-DNA left and right borders; B and H, BamHI and HindIII restriction sites. The arrowhead indicates the location of T-DNA insertion. The kanamycin-resistance gene (Kan) within T-DNA is indicated by a green box. Positions of nuclear localization signal (NLS domain), basic domain, and leucine zipper domain of VIP1 are indicated by blue, red, and green bars, respectively. VIP1 exons and their sizes in base pairs are indicated by beige boxes with the corresponding numbers. The distances between LB, RB, and various restriction sites are indicated in base pairs. In the VIP1 sequence, amino acids are shown in one-letter code, nucleotide sequences of exons and introns are indicated by upper and lowercase, respectively, and exon nucleotide positions are shown in parentheses. (B) Southern blot analysis of the homozygous vip1-1 line. Lane M, molecular size markers; lanes 1 and 2, BamHI- or HindIII-digested vip1-1 DNA, respectively. Numbers on the left indicate marker sizes in thousands of base pairs. (C) RT-PCR detection of VIP1 transcripts. RT-PCR products specific for the 5′-terminal half (Upper) and full-length VIP1 (Lower) are 280 and 780 bp, respectively, as indicated by arrowheads on the right. Lanes 1 and 2, total seedlings; lanes 3 and 4, roots; lanes 5 and 6, leaves; lanes 1, 3, and 5, wild-type plants; lanes 2, 4, and 6, vip1-1 plants. Numbers on the left indicate marker sizes in thousands of base pairs. (D) Nucleotide and amino acid sequences of the N-terminal portion of VIP1 (N-VIP1, green box) terminating with 11 aa encoded by the mutagenic T-DNA (white box) and a stop codon (asterisk). (E–G) Leaf sections of wild-type plants probed with anti-VIP1 antibody. (H–J) Leaf sections of vip1-1 plants probed with anti-VIP1 antibody. E and H, F and I, and G and J are Cy5 confocal fluorescence images, phase-contrast images, and merged images, respectively. (K) Nuclear import of (GFP)2-VIP1-N164 in tobacco cells. (L) Free DsRed2 produced in the (GFP)2-VIP1-N164-expressing cell. (M) Merged (GFP)2-VIP1-N164/DsRed2 image. GFP is in green, DsRed2 is in red, and overlapping GFP and DsRed2 are in yellow. All images are single confocal sections. (N and O) GUS-VirE2 in wild-type plants. (P and Q) GUS-VirE2 in vip1-1 plants. N and P show GUS staining, and O and Q show DAPI staining. Arrows indicate the cell nucleus.
We then used Southern blot analysis to determine the number of T-DNA insertion sites and the number of individual T-DNA inserts in vip1-1 plants. vip1-1 genomic DNA was digested either with BamHI or with HindIII, each of which has a single recognition site in the T-DNA (Fig. 1A). Based on the sequences of the T-DNA insert (http://signal.salk.edu) and the VIP1 gene, the resulting bands of 7.8 and 4.3 kb (Fig. 1B, lane 1) and 5.2 and 4.3 kb (Fig. 1B, lane 2) suggested insertion of two T-DNA molecules in a head-to-tail orientation into a single genomic site (Fig. 1A), which was indeed located within VIP1. Subsequent PCR amplification of the junction between left and right borders of the integrated T-DNA molecules confirmed that their integration had indeed occurred in a head-to-tail orientation (not shown).
vip1-1 Produces a Truncated Transcript Corresponding to the N-Terminal Portion of VIP1 and a Protein Product That Localizes to the Cell Nucleus. The T-DNA integration site was located within the second half of the 780-bp VIP1 ORF, specifically at 492 bp downstream of the translation initiation codon (Fig. 1 A). Thus, it is possible that the first half of the VIP1 ORF still produces a truncated transcript. To test this possibility, we used RT-PCR to detect transcripts specific for the 5′-terminal half of the VIP1 gene and for the full-length VIP1 in the wild-type and vip1-1 plants. Fig. 1C Upper shows that both types of plants accumulated RT-PCR products corresponding to transcripts of the 5′-terminal half of the VIP1 ORF in their various tissues, such as total seedlings (lanes 1 and 2), roots (lanes 3 and 4), and leaves (lanes 5 and 6). These 280-bp RT-PCR products were derived from the VIP1 mRNA rather than from the larger, 380-bp, intron-containing VIP1 genomic sequence. As expected, no full-length VIP1 transcripts were detected in the vip1-1 plants, whereas the wild-type plants accumulated RT-PCR products corresponding to the complete, 780-bp VIP1 mRNA (Fig. 1C Lower). In control experiments, actin-specific transcripts generated similar amounts of RT-PCR products in all samples, indicating similar efficiencies of the RT-PCRs (not shown).
Next, we directly confirmed that the vip1-1 plants indeed generated a VIP1-specific protein product and determined the subcellular location of this VIP1 mutant. To this end, we used immunostaining with anti-VIP1 antibodies followed by an indodicarbocyanine (Cy5)-conjugated secondary antibody and confocal fluorescence microscopy. Cy5 is excited near 650 nm and fluoresces near 670 nm, thus substantially circumventing the autofluorescence usually associated with the plant cell cytoplasm. In confocal sections of leaf tissues of young wild-type Arabidopsis plants, VIP1 was found within the cell nucleus (Fig. 1 E–G). The vip1-1 plants also produced a VIP1-specific protein that exhibited a nuclear accumulation pattern virtually identical to that of the wild-type VIP1 protein (compare Fig. 1 H–J to E–G). No signal was observed when preimmune antiserum was used instead of anti-VIP1 antibodies (not shown).
Biological Activities of the N-Terminal Portion of VIP1. That the vip1-1 mutation resulted in production of a truncated transcript allowed us to examine whether different parts of the VIP1 protein may play different roles in the process of Agrobacterium-mediated genetic transformation. First, we produced a recombinant cDNA corresponding to the residual 5′-terminal VIP1 sequence in the vip1-1 mutant. The 164-aa protein product encoded by this cDNA, designated VIP1-N164, was assayed for the two known biological activities of VIP1, import into the plant cell nucleus and interaction with the Agrobacterium VirE2 protein.
To determine the subcellular localization of VIP1-N164, its coding sequence was fused to a GFP reporter and expressed in plant cells after biolistic delivery of the fusion construct into intact tobacco leaves. Because VIP1-N164 is a small protein, ≈18 kDa, we fused it to (GFP)2, to produce an ≈71-kDa fusion product, which exceeds the 40- to 60-kDa size exclusion limit of the nuclear pore (reviewed in ref. 28). Fig. 1K shows that (GFP)2-VIP1-N164 was predominantly intranuclear as determined by using optical sections through the cell nucleus. The location of the cell nucleus was verified by accumulation of DsRed2 expressed from the same vector as (GFP)2-VIP1-N164 (Fig. 1L). The combined image of GFP and DsRed2 fluorescence showed overlapping signal (Fig. 1M) within the cell nucleus, confirming nuclear targeting of (GFP)2-VIP1-N164. Thus, the subcellular localization of VIP1-N164 was identical to that of the wild-type VIP1 and of the VIP1-specific protein produced in the vip1-1 mutant plants (Fig. 1 E–J).
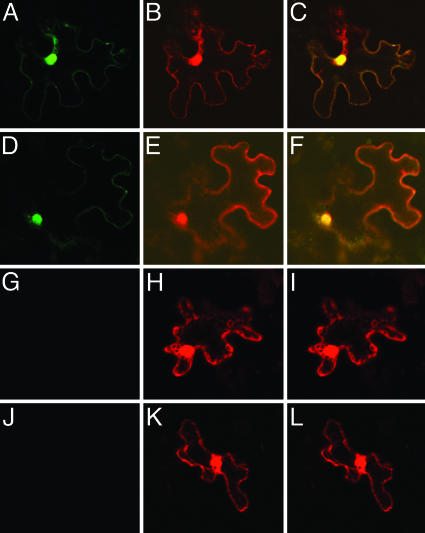
Next, we examined whether VIP1-N164 can interact with VirE2 in planta. To this end, we used BiFC assay, in which a molecule of YFP is separated into two parts, N-terminal (nYFP) and C-terminal (cYFP). Neither of these YFP fragments fluoresces when expressed alone, but the fluorescence is restored when nYFP and cYFP are brought together as fusions with interacting proteins (29), allowing detection of protein interactions and determination of the subcellular localization of the interacting proteins (29). Fig. 2 A–C shows that nYFP-tagged VIP1-N164 interacted with cYFP-VirE2 in the nuclei of tobacco leaf cells, resulting in reconstruction of the YFP fluorescence, which colocalized with the nuclear DsRed2 signal. In positive control experiments, nYFP-VIP1 interacted with cYFP-VirE2 (Fig. 2 D–F), whereas in negative control experiments no YFP signal was detected after coexpression of nYFP-VIP1 or cYFP-VIP1 with unfused cYFP or nYFP, respectively (Fig. 2 G–I and J–L).
Fig. 2.
VIP1-VirE2 interaction and VirE2 nuclear import in tobacco cells. (A–C) nYFP-VIP1-N164, cYFP-VirE2, and free DsRed2. (D–F) nYFP-VIP1, cYFP-VirE2, and free DsRed2. (G–I) nYFP-VIP1, cYFP, and free DsRed2. (J–L) nYFP, cYFP-VIP1, and free DsRed2. (A, D, G, and J) YFP signal. (B, E, H, and K) DsRed2 signal. (C, F, I, and L) Merged signals. All images are single confocal sections.
Finally, we directly demonstrated the ability of the vip1-1 plants to support nuclear import of VirE2. As expected, VirE2 tagged with GUS and transiently expressed in leaf epidermal cells accumulated in the cell nucleus of the wild-type Arabidopsis plants (Fig. 1N); the location of the cell nucleus was verified by DAPI staining (Fig. 1O). Similarly, vip1-1 plants exhibited efficient nuclear import of GUS-VirE2 (Fig. 1 P and Q), indistinguishable from that in the wild-type plants. In control experiments, free GUS expressed in all wild-type and vip1-1 plants remained cytoplasmic (not shown). These results further support the idea that the N-terminal portion of VIP1 generated by the vip1-1 mutation most likely retains its ability to facilitate nuclear import of VirE2 in plant cells.
Effects of the N-Terminal Portion of VIP1 on Transient and Stable Plant Genetic Transformation by Agrobacterium. That VIP1-N164 retained the ability of the full-length protein to bind VirE2 and promote its nuclear import allowed us to examine whether the role of VIP1 during genetic transformation by Agrobacterium is limited to the step of nuclear import, or whether this protein is also involved in other, later stages of the infection. Specifically, we focused on the steps of transient and stable genetic transformation. Transient transformation, which closely follows nuclear import of the T-DNA, occurs from the T-DNA molecules that have not yet integrated into the plant genome and, in Arabidopsis plants, can be detected 4–6 days after infection (16). Stable transformation and tumorigenesis, on the other hand, require integration of the T-DNA into the host genome and are usually assayed 4–5 weeks after infection (16). Thus, transient T-DNA expression and tumorigenesis have become standard criteria for the ability of plants to support early stages of T-DNA transfer and the final stage of T-DNA integration, respectively (16).
For transient transformation, Arabidopsis root segments were inoculated with an Agrobacterium strain carrying in its T-DNA a uidA gene for the GUS reporter, and GUS activity was detected histochemically 6 days after inoculation. Fig. 3 A and B shows that wild-type and vip1-1 plants exhibited comparable degrees of transient T-DNA expression as estimated from the size and number of indigo-blue-stained areas of root segments. When GUS expression was further quantified by using a sensitive fluorometric assay, we detected no quantitative differences in GUS activity between wild-type Arabidopsis and vip1-1 mutant plants (Fig. 3C), indicating equal efficiency of their transient transformation. Because the uidA reporter gene contained an intron which prevents its expression in bacteria (30), our measurements represented the GUS activity directed by the T-DNA after its transfer to plant cells. Thus, the N-terminal portion of VIP1 in the vip1-1 mutant most likely contained protein activities sufficient to promote transient expression of the Agrobacterium T-DNA.
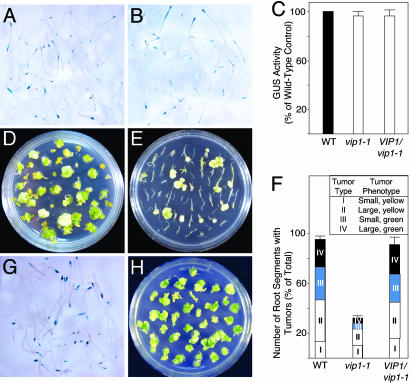
Fig. 3.
vip1-1 phenotype: effects on transient and stable genetic transformation by Agrobacterium and complementation by the wild-type VIP1 gene. (A) Transient T-DNA expression in wild-type plants. (B) Transient T-DNA expression in vip1-1 plants. (C) Quantification of GUS activity. Black, white, and gray bars indicate transient GUS expression in Agrobacterium-infected wild-type (WT), vip1-1, and VIP1-expressing vip1-1 (VIP1/vip1-1) plants, respectively. GUS activity in control wild-type plants was defined as 100%. All data represent average values of three independent experiments with indicated standard deviations. (D) Tumor formation in wild-type plants. (E) Reduced tumor formation in vip1-1 plants. (F) Quantification of tumorigenicity in wild-type (WT), vip1-1, and VIP1-expressing vip1-1 (VIP1/vip1-1) plants. White, light gray, dark gray, and black bars indicate numbers of root segments that developed tumors classified as type I, II, III, and IV, respectively. Inset describes tumor phenotypes used for tumor classification. Total number of root segments per experimental condition was defined as 100%. (G) Transient T-DNA expression in VIP1/vip1-1 plants. (H) Restored tumor formation in VIP1/vip1-1 plants.
Stable genetic transformation was assayed by the formation of tumors after inoculation with wild-type oncogenic Agrobacterium. Fig. 3D shows that Agrobacterium elicited numerous tumors on root segments derived from the wild-type Arabidopsis. In contrast, the vip1-1 mutant plants exhibited a decrease in their susceptibility to Agrobacterium tumorigenicity, developing relatively few tumors (Fig. 3E). Overall, the tumor-inducing activity of Agrobacterium in vip1-1 plants was reduced to ≈40% of that observed in the wild-type plants (Fig. 3F). To facilitate description of the effect of the vip1-1 mutation on Agrobacterium tumorigenicity, the tumors were scored according to their severity. Four tumor types were distinguished, with type IV being the most severe. Individual types were assigned according to tumor phenotypes, such as size and color (Inset of Fig. 3F), which correlate with the susceptibility of Arabidopsis to Agrobacterium transformation (16). Indeed, the vip1-1 plants developed a significantly lower percentage of type II tumors than the wild-type plants; type III and IV tumors in vip1-1 were even more rare, amounting to only 15–20% of the similar type tumors developed in the wild-type Arabidopsis (Fig. 3F). These data suggest that VIP1 is involved in stable genetic transformation by Agrobacterium and that its C-terminal portion is required for this function, although a chance that the vip1-1 mutation specifically affects tumorigenesis, rather than stable transformation per se, cannot be ruled out.
Complementation of the vip1-1 Mutant. Although no T-DNA insertions were detected outside the VIP1 locus in the vip1-1 mutant line (Fig. 1), another, unrelated, mutation may have contributed to the vip1-1 phenotype. To exclude this possibility, we performed a genetic complementation analysis. The vip1-1 mutant was transformed with a transgene corresponding to the full-length genomic sequence of VIP1 with its native regulatory elements. Fig. 3 G and H shows that the resulting transgenic plants were susceptible to both transient and stable transformation by Agrobacterium. Quantification of transient gene expression and tumor formation (Fig. 3 C and F) demonstrated that these transformation parameters in the vip1-1 plants expressing the VIP1 transgene were virtually identical to those in the wild-type plants, indicating genetic complementation of the vip1-1 mutation. Similar results were observed with other independent vip1-1 lines transgenic for VIP1 (not shown). Thus, the vip1-1 phenotype, in regard to Agrobacterium infection, is most likely because of disruption of the native VIP1 function by the mutation in this gene.
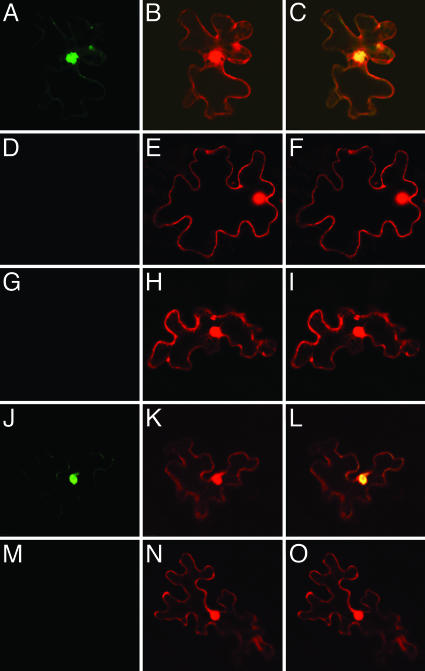
The C-Terminal Portion of VIP1 Is Required for Protein Multimerization and Binding to the Host H2A Histone in Planta. Many bZIP proteins dimerize in vivo by means of their leucine zipper domains (reviewed in ref. 31). We used BiFC to examine whether VIP1, a bZIP protein (7), also dimerizes within plant cells, and whether this functionality is affected by the vip1-1 mutation. Fig. 4 A–C shows that nYFP-tagged VIP1 interacted with cYFP-tagged VIP1, producing a strong signal of the reconstructed YFP and demonstrating the predominantly nuclear location of the interacting VIP1 molecules. However, nYFP-VIP1-N164 was unable to interact with cYFP-VIP1-N164 (Fig. 4 D–F) or even with the cYFP-tagged full-length VIP1 (Fig. 4 G–I), indicating that the C-terminal parts of both interacting VIP1 molecules are required for protein multimerization.
Fig. 4.
BiFC assay for VIP1-VIP1 dimerization and VIP1–H2A interaction in tobacco cells. (A–C) nYFP-VIP1, cYFP-VIP1, and free DsRed2. (D–F) nYFP-VIP1-N164, cYFP-VIP1-N164, and free DsRed2. (G–I) nYFP-VIP1, cYFP-VIP1-N164, and free DsRed2. (J–L) nYFP-VIP1, cYFP-H2A, and free DsRed2. (M–O) nYFP-VIP1-N164, cYFP-H2A, and free DsRed2. (A, D, G, J, and M) YFP signal. (B, E, H, K, and N) DsRed2 signal. (C, F, I, L, and O) Merged signals. All images are single confocal sections.
Recently, VIP1 has been suggested to interact with the host cell chromatin (3). Fig. 4 J–L shows that nYFP-tagged VIP1 interacted with the cYFP-tagged Arabidopsis H2A histone, a chromatin component required for stable genetic transformation by Agrobacterium (32), within the plant cell nucleus. On the other hand, nYFP-VIP1-N164 exhibited virtually no interaction with cYFP-H2A (Fig. 4 M–O).
Discussion
Agrobacterium is unique in its ability to genetically transform eukaryotic cells (33). The molecular mechanism of the Agrobacterium-mediated transformation is complex, comprising a wide spectrum of diverse biological reactions. To understand this process better, it is important to characterize its molecular players, i.e., bacterial and host cell factors required for transformation. Whereas our knowledge about the Agrobacterium proteins that take part in the infection is relatively advanced, the information about the cellular participants is just emerging (reviewed in refs. 3 and 4). One recently identified host cell factor is VIP1, an Arabidopsis protein proposed to facilitate import of the invading T-complex into the plant cell nucleus (7, 8). Because nuclear import is an early event in the transformation process, antisense knockdown of the VIP1 gene expression blocked all subsequent steps, from transient T-DNA expression to integration and stable transformation (7). Thus, although indicating an important role for VIP1 during the Agrobacterium–plant cell interaction and identifying at least one transformation step, i.e., nuclear import that requires VIP1, the knockdown experiments were inherently unable to detect potential involvement of VIP1 in transformation events downstream of nuclear import. Our use of reverse genetics helped circumvent this difficulty and provided an indication for VIP1 activity beyond nuclear import.
Because in the vip1-1 line the mutagenic T-DNA had inserted into the second half of the VIP1 ORF, the N-terminal half of VIP1 was still produced and localized to the cell nucleus. Importantly, the N-terminal portion of VIP1 was biologically active in vivo, within plant tissues. Specifically, it retained two biological activities of the full-length protein: binding to VirE2 and nuclear targeting. These activities were sufficient to promote nuclear import of VirE2, suggesting that the N-terminal part of VIP1 is still capable of directing the entire T-complex into the cell nucleus. Indeed, the vip1-1 mutant plants supported efficient nuclear import of VirE2 and remained highly susceptible to transient genetic transformation by Agrobacterium, for which nuclear import of the T-DNA is an absolute prerequisite. In contrast, vip1-1 plants exhibited a significant recalcitrance to Agrobacterium tumorigenicity as compared with wild-type Arabidopsis. Transgenic expression of the full-length VIP1 gene restored the susceptibility of the vip1-1 mutant to Agrobacterium tumorigenesis, complementing the mutation and demonstrating that alteration of the VIP1 function indeed underlies the vip1-1 phenotype.
Tumor formation represents a major hallmark and indicator of stable genetic transformation, an ultimate step of the Agrobacterium infection process that occurs after transient transformation has waned (16, 22). The C-terminal portion of VIP1 therefore carries protein determinants likely required for its function within the cell nucleus, during tumorigenesis. An insight into this function was obtained from our in planta experiments demonstrating that VIP1 forms homomultimers in the plant cell nucleus, and that this VIP1 multimerization requires that each of the two interacting VIP1 molecules contains their C-terminal domains. The VIP1 sequence between the amino acid residue 165 and the C terminus includes three of seven leucine repeats (leucine zipper) of VIP1 (7). Consistent with the known involvement of leucine zippers in dimerization of bZIP proteins (reviewed in ref. 31), the loss of the integrity of the VIP1 leucine zipper in the vip1-1 mutant is likely responsible for the inability of the mutant protein to form homomultimers.
As with many other plant bZIP proteins (34), VIP1 may be involved in transcription, suggesting that, after its nuclear import, VIP1 targets to and associates with the chromosomal DNA. Because during Agrobacterium infection VIP1 also likely associates with the T-complex by means of its interaction with VirE2 (7), it may transport the T-complex to chromosomal regions where T-DNA integration will occur. We hypothesize that this potential intranuclear targeting of VIP1 requires protein multimerization that is inhibited by the C-terminal truncation. This inhibition blocks the ability of VIP1 to transport T-complexes to the plant cell chromatin, thereby reducing the efficiency of tumorigenesis, most likely by interfering with T-DNA integration. This notion is consistent with the role that formation of bZIP protein dimers is thought to play in their interactions with the host chromatin (31). That the ability of VIP1 to interact with the H2A histone is required for tumorigenesis further supports this hypothesis and sheds new light on the well known role of H2A in T-DNA integration (32). However, whereas biological evidence indicates that VIP1 can form ternary complexes with VirE2 and VirF, a bacterial F-box protein likely involved in proteasomal uncoating of the T-complex (35), further experiments are required to determine whether VIP1 can bind both VirE2 and host histones.
In summary, we used the vip1-1 mutation to dissect endogenous VIP1 in vivo, functionally and structurally, by uncoupling its two activities: one located within the N-terminal portion of the protein and required to bind VirE2, target to the cell nucleus, and facilitate transient genetic transformation by Agrobacterium, and the other located, at least partly, within the C-terminal portion of the protein and required to form homomultimers, interact with the host H2A histone, and facilitate the Agrobacterium-induced tumorigenesis and, by implication, stable genetic transformation.
Acknowledgments
We thank Dr. S. Gelvin for the gift of H2A. Our laboratory is supported by grants (to V.C.) from the National Institutes of Health, the National Science Foundation, the U.S. Department of Agriculture, the U.S.–Israel Binational Agricultural Research and Development Fund, and the U.S.–Israel Binational Science Foundation, and by grants (to T.T.) from the U.S.–Israel Binational Agricultural Research and Development Fund and the Human Frontier Science Program.
Author contributions: J.L., T.T., and V.C. designed research; J.L., A.K., and M.V. performed research; A.K. contributed new reagents/analytic tools; J.L., A.K., and T.T. analyzed data; and J.L. and V.C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: T-complex, DNA–protein transport complex; T-DNA, transferred DNA; VIP1, Arabidopsis VirE2-interacting protein; BiFC, bimolecular fluorescence complementation; YFP, yellow fluorescent protein; bZIP, basic leucine zipper; GUS, β-glucuronidase.
Note Added in Proof. In addition to H2A, VIP1 interaction with other core histones has been reported by Loyter et al. (36).
References
- 1.Piers, K. L., Heath, J. D., Liang, X., Stephens, K. M. & Nester, E. W. (1996) Proc. Natl. Acad. Sci. USA 93, 1613–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunik, T., Tzfira, T., Kapulnik, Y., Gafni, Y., Dingwall, C. & Citovsky, V. (2001) Proc. Natl. Acad. Sci. USA 98, 1871–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzfira, T. & Citovsky, V. (2002) Trends Cell Biol. 12, 121–129. [DOI] [PubMed] [Google Scholar]
- 4.Gelvin, S. B. (2003) Microbiol. Mol. Biol. Rev. 67, 16–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zupan, J. & Zambryski, P. C. (1997) Crit. Rev. Plant Sci. 16, 279–295. [Google Scholar]
- 6.Zupan, J., Citovsky, V. & Zambryski, P. C. (1996) Proc. Natl. Acad. Sci. USA 93, 2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzfira, T., Vaidya, M. & Citovsky, V. (2001) EMBO J. 20, 3596–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzfira, T., Vaidya, M. & Citovsky, V. (2002) Proc. Natl. Acad. Sci. USA 99, 10435–10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R., et al. (2003) Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- 10.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Smith, J. A., Seidman, J. G. & Struhl, K. (1987) Current Protocols in Molecular Biology (Greene & Wiley, New York).
- 11.Kang, J., Kuhn, J. E., Schafer, P., Immelmann, A. & Henco, K. (1995) in PCR 2, A Practical Approach, eds. McPherson, M. J., Hames, B. D. & Taylor, G. R. (IRL, Oxford).
- 12.Restrepo, M. A., Freed, D. D. & Carrington, J. C. (1990) Plant Cell 2, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodin, M. M., Dietzgen, R. G., Schichnes, D., Ruzin, S. & Jackson, A. O. (2002) Plant J. 31, 375–383. [DOI] [PubMed] [Google Scholar]
- 14.Dietrich, C. & Maiss, E. (2002) BioTechniques 32, 286–293. [DOI] [PubMed] [Google Scholar]
- 15.Citovsky, V., Zupan, J., Warnick, D. & Zambryski, P. C. (1992) Science 256, 1802–1805. [DOI] [PubMed] [Google Scholar]
- 16.Nam, J., Mysore, K. S., Zheng, C., Knue, M. K., Matthysse, A. G. & Gelvin, S. B. (1999) Mol. Gen. Genet. 261, 429–438. [DOI] [PubMed] [Google Scholar]
- 17.Citovsky, V., Kapelnikov, A., Oliel, S., Zakai, N., Rojas, M. R., Gilbertson, R. L., Tzfira, T. & Loyter, A. (2004) J. Biol. Chem. 279, 29528–29533. [DOI] [PubMed] [Google Scholar]
- 18.Ueki, S. & Citovsky, V. (2002) Nat. Cell Biol. 4, 478–485. [DOI] [PubMed] [Google Scholar]
- 19.Lacroix, B., Vaidya, M., Tzfira, T. & Citovsky, V. (2005) EMBO J. 24, 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avivi, Y., Morad, V., Ben-Meir, H., Zhao, J., Kashkush, K., Tzfira, T., Citovsky, V. & Grafi, G. (2004) Dev. Dyn. 230, 12–22. [DOI] [PubMed] [Google Scholar]
- 21.Murashige, T. & Skoog, F. (1962) Physiol. Plant. 15, 473–497. [Google Scholar]
- 22.Janssen, B. J. & Gardner, R. C. (1990) Plant Mol. Biol. 14, 61–72. [DOI] [PubMed] [Google Scholar]
- 23.Tian, G. W., Mohanty, A., Chary, S. N., Li, S., Paap, B., Drakakaki, G., Kopec, C. D., Li, J., Ehrhardt, D., Jackson, D., et al. (2004) Plant Physiol. 135, 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Arabidopsis Genome Initiative (2000) Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 25.Walhout, A. J., Temple, G. F., Brasch, M. A., Hartley, J. L., Lorson, M. A., van den Heuvel, S. & Vidal, M. (2000) Methods Enzymol. 328, 575–592. [DOI] [PubMed] [Google Scholar]
- 26.Goderis, I. J., De Bolle, M. F., Francois, I. E., Wouters, P. F., Broekaert, W. F. & Cammue, B. P. (2002) Plant Mol. Biol. 50, 17–27. [DOI] [PubMed] [Google Scholar]
- 27.Kim, J. Y., Yuan, Z. & Jackson, D. (2003) Development (Cambridge, U.K.) 130, 4351–4362. [DOI] [PubMed] [Google Scholar]
- 28.Dingwall, C. & Laskey, R. A. (1991) Trends Biochem. Sci. 16, 478–481. [DOI] [PubMed] [Google Scholar]
- 29.Hu, C. D., Chinenov, Y. & Kerppola, T. K. (2002) Mol. Cell 9, 789–798. [DOI] [PubMed] [Google Scholar]
- 30.Liu, C. N., Li, X. Q. & Gelvin, S. B. (1992) Plant Mol. Biol. 20, 1071–1087. [DOI] [PubMed] [Google Scholar]
- 31.Baxevanis, A. D. & Vinson, C. R. (1993) Curr. Opin. Genet. Dev. 3, 278–285. [DOI] [PubMed] [Google Scholar]
- 32.Mysore, K. S., Nam, J. & Gelvin, S. B. (2000) Proc. Natl. Acad. Sci. USA 97, 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stachel, S. E. & Zambryski, P. C. (1989) Nature 340, 190–191. [DOI] [PubMed] [Google Scholar]
- 34.van der Krol, A. R. & Chua, N.-H. (1991) Plant Cell 3, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tzfira, T., Vaidya, M. & Citovsky, V. (2004) Nature 431, 87–92. [DOI] [PubMed] [Google Scholar]
- 36.Loyter, A., Rosenbluh, J., Zakai, N., Li, J., Kozlovsky, S. V., Tzfira, T. & Citovsky, V. (2005) Plant Physiol. 138, in press. [DOI] [PMC free article] [PubMed]