Abstract
The enteric nervous system (ENS) of the gastrointestinal tract controls many diverse functions, including motility and epithelial permeability. Perturbations in ENS development or function are common, yet there is no human model for studying ENS-intestinal biology and disease. We used a tissue-engineering approach with embryonic and induced pluripotent stem cells (PSCs) to generate human intestinal tissue containing a functional ENS. We recapitulated normal intestinal ENS development by combining human-PSC-derived neural crest cells (NCCs) and developing human intestinal organoids (HIOs). NCCs recombined with HIOs in vitro migrated into the mesenchyme, differentiated into neurons and glial cells and showed neuronal activity, as measured by rhythmic waves of calcium transients. ENS-containing HIOs grown in vivo formed neuroglial structures similar to a myenteric and submucosal plexus, had functional interstitial cells of Cajal and had an electromechanical coupling that regulated waves of propagating contraction. Finally, we used this system to investigate the cellular and molecular basis for Hirschsprung's disease caused by a mutation in the gene PHOX2B. This is, to the best of our knowledge, the first demonstration of human-PSC-derived intestinal tissue with a functional ENS and how this system can be used to study motility disorders of the human gastrointestinal tract.
The ENS is essential for gastrointestinal (GI) motility, secretion, blood flow, epithelial barrier permeability and fluid exchange1. Developmentally, the majority of the ENS arises from vagal NCCs that derive from the dorsal neural tube and migrate ventrally to colonize the foregut around embryonic week 4 in humans. Subsequently, the NCCs proliferate extensively and migrate caudally to colonize the entire GI tract by 7 weeks of human gestation2. When enteric NCCs fail to properly migrate, proliferate, survive and/or differentiate in the GI tract, defects occur in the structure and function of the ENS, and patients present with a spectrum of enteric neuropathies3. In addition, enteric neuropathies are common in digestive diseases, such as inflammatory bowel disease, and often occur secondarily in diseases such as Parkinson's disease, diabetes mellitus and age-related degeneration4. The lack of human ENS model systems for studying the physiopathological processes of enteric neuropathies may account for the surprisingly slow progress in their diagnosis and treatment. For example, the treatment of patients with congenital lack of enteric ganglia, known as Hirschsprung's disease (1 in 5,000 births), involves surgical resection of the aganglionic gut segment, which may result in extensive loss of both the colon and small bowel. Although the remaining bowel contains ganglia, it remains unclear why many of these patients experience recurrent bouts of enterocolitis and dysmotility5. Enteric neurons derived from human PSCs, enteric progenitors or neural stem cells derived from the CNS incorporate into and function in aganglionic chick and murine GI explants (reviewed in ref. 6), which suggests that a similar cell-therapy approach might work in humans.
However, much work remains to be done regarding our understanding of human ENS development and disease. Very little is known about the etiology of enteric neuropathies and the mechanisms that drive various aspects of human ENS development, such as the formation of neuronal diversity, remain unclear7. Although there is currently no way to study human ENS development functionally, recent progress using the directed differentiation of human PSCs has resulted in the formation of complex and physiological three-dimensional human organ cultures, known as organoids8. Organoid models have been developed for intestine, liver, stomach, CNS, thyroid and lung, among other tissues, and have allowed for unprecedented studies of human developmental biology and disease. Despite the remarkable complexity of organoids, they lack the cell and tissue types that are required for full organ function. For example, none of the organoid systems contain an integrated peripheral nervous system. We used principles of embryonic organogenesis to generate three-dimensional HIOs containing a functional ENS.
Results
Generation of human intestinal organoids incorporating ENS
Previously, we developed a method of differentiating human embryonic and induced PSCs into intestinal organoids by following a stepwise differentiation first into definitive endoderm, and then into mid/hindgut tube spheroids that expand into three-dimensional intestinal tissue in vitro9,10 (Supplementary Fig. 1). When HIOs were engrafted into the kidney subcapsular space, they continued to develop and formed highly differentiated intestinal tissue with crypts and villi, functional intestinal stem cells and layers of smooth muscle11 (Supplementary Fig. 1c). Although HIOs contained most of the epithelial and mesenchymal cell types found in the developing gut, they did not contain an ENS. To generate HIOs containing ENS cell types, we first generated human vagal NCCs, the precursors of the ENS, using a modified version of a previously reported method of generating cranial NCCs12 (Supplementary Figs. 1d and 2a–d). This involved manipulating signaling pathways that are known to promote a posterior/vagal fate; treatment with retinoic acid (RA) for 2 d at the neurosphere stage resulted in the robust expression of the vagal-level Hox genes HOXB3, HOXB5 and HOXB7 (Supplementary Fig. 2e), similarly to previous observations13. We confirmed differentiation of NCCs at each stage by analysis of early and late neural crest markers by immunofluorescence (HNK-1, PAX6, p75NTR, SOX2, VIM) and quantitative PCR (AP2, PAX3, PAX7, SOX10, SNAIL2, ZIC1) (Supplementary Fig. 2). We then compared PSC-derived NCCs with isolated cranial NCCs from chick embryos and found that embryonic NCCs also expressed vagal Hox genes in response to RA (Supplementary Fig. 3a–c), consistent with the known role of RA in vagal NCC development14. We further tested whether human-PSC-derived NCCs had neural crest cell behaviors by injecting them into chick embryos, where they followed a migratory path similar to that of endogenous NCCs and differentiated into neurons (Supplementary Fig. 3d–f). We demonstrated multipotency of both cranial and RA-treated NCCs by differentiating them in vitro into Peripherin+ neurons and GFAP+ glial cells, as well as mesodermal lineages, including osteocytes and chondrocytes, as indicated by positive staining for alizarin red and alcian blue, respectively (Supplementary Fig. 4a). These data all support the conclusion that this differentiation method generates functional NCCs.
To incorporate vagal NCCs and ENS precursors into developing HIOs, we mechanically aggregated mid/hindgut spheroids with PSC-derived NCCs by low-speed centrifugation and transferred aggregates to three-dimensional growth conditions for 28 d (Supplementary Fig. 1d). HIOs with and without NCCs were comparable in size (1–2 mm in diameter); however, we detected an abundance of βIII-tubulin (TUBB3)+ neurons and S100β+ glia embedded in the mesenchyme of HIOs combined with NCCs (HIOs with NCC-derived ENS will now be referred to as HIOs+ENS) (Fig. 1a). We rarely detected neurons and never detected glia in HIOs without NCCs in vitro. Moreover, we confirmed the NCC origin of neuroglial cells by using NCCs that were derived from an embryonic stem cell line that constitutively expresses GFP (Supplementary Fig. 4b). The spatial relationship between the epithelium, mesenchyme and neuroglial cells in HIOs+ENS closely resembled that of human fetal intestine15 and embryonic day 11.5 (E11.5) mouse small intestine (Supplementary Fig. 4c), suggesting that intrinsic cues in HIOs help to organize the developing ENS cells. Consistently with this, HIOs expressed both GDNF and EDN3, two crucial chemoattractants that are known to regulate NCC migration into the gut mesenchyme6,16,17 (Supplementary Fig. 4d), suggesting that the incorporation of PSC-derived NCCs into HIOs mesenchyme may utilize normal developmental pathways.
Figure 1.
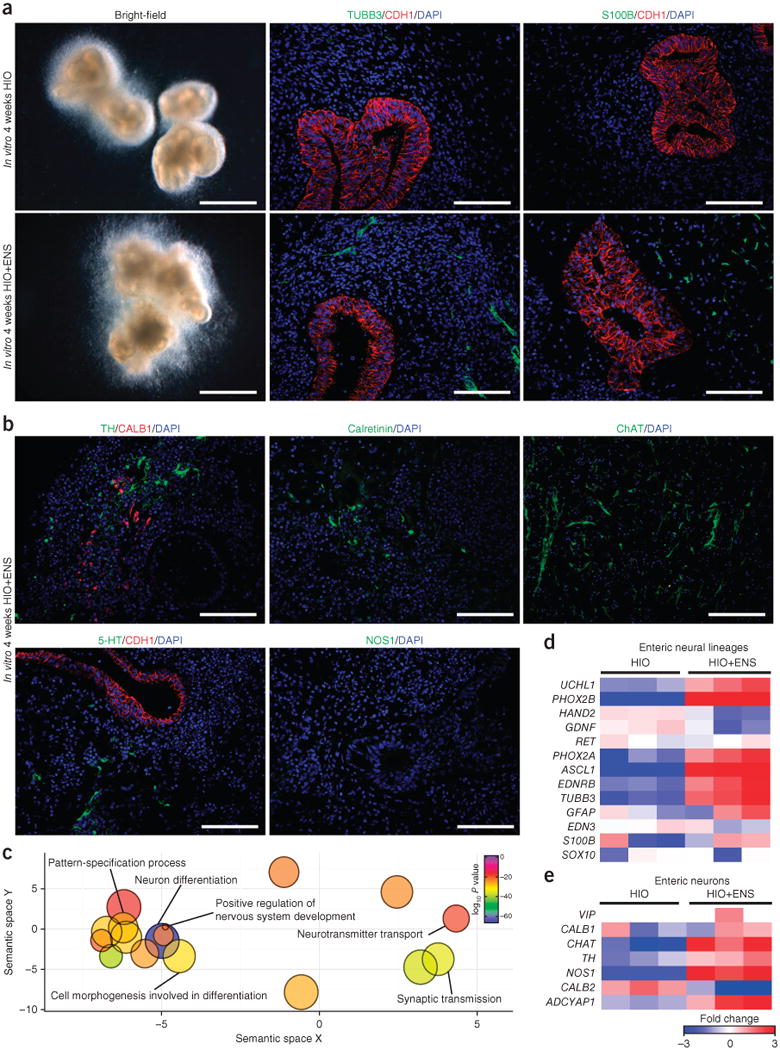
Incorporation of NCCs into developing HIOs in vitro. (a) Immunofluorescence analysis of organoids generated in vitro with (HIO+ENS) and without (HIO) NCC addition. Left, bright-field images. Middle, immunostaining for neurons (TUBB3) and epithelium (CDH1). Right, immunostaining for glial cells (S100β+) and epithelium (CDH1). Scale bars, 1 mm (left) and 100 μm (middle and right). Data are representative of 14 independent experiments combining HIOs with NCCs in vitro. (b) Analysis of different neuronal cell types using neurochemical markers of ENS neurons in HIOs+ENS cultured in vitro. Dopaminergic neurons (TH), interneurons (5-HT), sensory neurons (CALB1), excitatory neurons (calretinin, ChAT) and inhibitory neurons (nNOS) were studied. Scale bars, 100 μm. (c) HIOs and HIOs+ENS generated in vitro were analyzed by RNA-seq, and the gene ontology terms found were visualized using ReVIGO approach, which converts a list of gene-ontology terms into a semantic, similarity-based scatterplot after removing redundant terms. (d,e) HIOs and HIOs+ENS generated in vitro were analyzed by RNA-seq (n = 3 per condition). Expression of a curated list of enteric neural lineage and enteric neuron genes is shown.
The ENS is a complex network of excitatory and inhibitory neuronal subtypes, as well as sensory neurons and interneurons. Histological examination of neuronal markers in HIOs+ENS cultured in vitro for 4 weeks suggested a substantial degree of neuronal diversity. We observed tyrosine hydroxylase (TH), calbindin (CALB1), calretinin (CALB2), choline acetyltransferase (CHAT) and serotonin (5-HT) positive cells, which are expressed by interneurons and dopaminergic and excitatory neurons (Fig. 1b). However, we did not detect neurons expressing neuronal nitric oxide synthase (NOS1) in vitro. To broadly identify the transcriptional changes that occur in whole HIOs following the incorporation of ENS progenitor cells, we performed an RNA-seq analysis. Unsupervised hierarchical clustering of the gene expression data and Spearman rank correlation demonstrated the segregation of the HIOs and HIOs+ENS groups (Supplementary Fig. 5a,b). A differential gene expression analysis revealed that the expression levels of 4,283 genes out of 40,448 were statistically different between HIO and HIO+ENS groups (moderated t test; P < 0.05). With a fold-change greater than 3, 1,240 and 307 transcripts were up- and downregulated in HIOs+ENS, respectively. Using gene ontology analysis (ToppGene Suite) and a reduce and visualize gene ontology (ReViGO), we found that transcripts that were higher in the HIOs+ENS group were largely related to nervous system development and neuron differentiation (Fig. 1c and Supplementary Fig. 5c). Genes related to enteric neuronal development, including PHOX2A, PHOX2B, ASCL1 and EDNRB, were expressed more highly in HIOs+ENS than HIO samples (Fig. 1d). Notably, NOS1 mRNA was present in HIOs+ENS, despite the fact that NOS1 protein was not detected (Fig. 1e).
Maturation of the ENS following in vivo growth
HIOs that were engrafted into mice and allowed to grow for 6–10 weeks in vivo became vascularized, grew to 1–3 cm in diameter and formed highly mature intestinal tissues, with villi and crypts containing functional intestinal stem cells (Fig. 2a and Supplementary Fig. 1c)11. Moreover, the HIO mesenchyme matured into submucosal and myenteric layers of smooth muscle fibers (Fig. 2b). Transplantation of HIOs+ENS resulted in the generation of neurons (TUBB3) and glia (S100β) that were organized into ganglionated structures in close proximity to the submucosal and myenteric layers of smooth muscle fibers (Fig. 2b). We did not detect neurons or glia in transplanted HIOs without NCCs. Although these experiments were performed with vagal/HOX-positive NCC populations, PSC-derived cranial/HOX-negative NCCs were also able to engraft and form ENS neuroglial cells. However, cranial NCCs also formed pigmented epithelial cells in vitro and in vivo (Supplementary Fig. 6) and cartilage (data not shown), consistent with them having a broader multipotency.
Figure 2.

Formation of a three-dimensional neuronal plexus in HIOs+ENS grown in vivo. (a) Gross morphology of HIOs and HIOs+ENS transplanted into the kidney subcapsular space of NOD-SCID IL-2Rγnull NSG mice for 6 weeks. Scale bar, 1 mm. (b) Left, H&E staining of intestinal tissue after transplantation. Middle, the smooth-muscle marker desmin (DES) shows the formation of smooth-muscle fibers located in myenteric and submucosal layers of HIO and HIO+ENS samples. Neurons (TUBB3+) were found only in HIOs+ENS samples. Right, neurons in HIOs+ENS surround the epithelium (CDH1). Scale bar, 100 μm. Data are representative of three independent transplantation experiments (HIOs, n = 9; HIOs+ENS, n = 14). (c) Whole-mount immunostaining and three-dimensional imaging of human intestine and HIOs+ENS. En face view of human adult and infant myenteric plexus and HIOs+ENS, showing the arrangement of neurons (TUBB3) and neuronal bodies (HuC/D) into a neuronal plexus. Scale bars represent 100 μm. (d) En face view of human adult and infant myenteric plexus and HIOs+ENS showing arrangement of glia (S100β) into a plexus. Scale bars, 100 μm (middle) and 200 μm (left and right). Data are representative of two independent experiments (human adult, n = 5; infant, n = 2; HIOs+ENS, n = 5).
We performed whole-mount immunofluorescence imaging of tissues from HIOs+ENS matured in vivo and compared these to adult and infant human small intestine. The ENS of human small bowel contained a network of both neurons (TUBB3) and glia (S100β). The ENS that formed in the developing HIOs similarly contained a neural plexus of TUBB3+ neurons and S100β+ glia that appeared to be closely associated with smooth muscle fibers (Fig. 2c,d, Supplementary Fig. 7a,b and Supplementary Videos 1 and 2). We also observed clusters of HuC/D+ neuronal cell bodies in HIOs+ENS, both in sections and by whole mount, that were reminiscent of ganglia (Supplementary Fig. 7c). However, relative to human intestine, the ganglia in HIOs+ENS were lower in cell body density. Notably, the examination of neural markers in transplanted HIOs+ENS revealed the presence of TH-, CALB1-, calretinin-, CHAT- and 5-HT-positive fibers (Supplementary Fig. 7d). In addition, NOS1 was expressed in the HIOs+ENS after in vivo maturation. This latter is in contrast with in vitro HIOs+ENS, which lack nNOS+ neurons, suggesting that the ENS matured during the period of in vivo growth.
Neuronal calcium oscillations in HIOs+ENS
We next tested the functionality of neurons in HIOs in vitro by using live imaging with the Ca2+ sensor GCaMP6f (ref. 18). To do this, we generated ENS cells from a human iPS cell line expressing GCaMP6f (ref. 19) and incorporated them into HIOs that did not express a reporter. Imaging of ENS cells in HIOs after 8 weeks in vitro revealed broad, spontaneous and rapid calcium transients in ENS cells (Supplementary Fig. 8). When individual cells were monitored, there was an obvious periodicity of calcium efflux in the range of 2–4 min in NCC-derived cells (Fig. 3a and Supplementary Video 3). To determine whether ENS cells are capable of undergoing coordinated activity, we exposed organoids to KCl (30 mM) and observed a large wave of calcium efflux (Fig. 3b and Supplementary Video 4). Using live imaging with the Ca2+ sensor GCaMP6f on explanted HIOs+ENS grown in vivo, we were able to identify bundles of fibers that had calcium transients with spontaneous activity (Fig. 3c and Supplementary Video 5) that were triggered following KCl treatment (30 mM) (Fig. 3c and Supplementary Video 6).
Figure 3.

Live imaging of neural activity in HIOs+ENS. (a) Live imaging of Ca2+ flux in the neural-crest-derived ENS cells revealed periodic activity. Shown are snapshots from a 20-min time-lapse video (see Supplementary Video 3) of neural activity in HIOs+ENS. Colored arrows point to cells whose pixel intensity was measured over time. Numbers in snapshots correspond to numbered peaks in the graph. The graph measures fluorescence values as ΔF/F0, (ΔF = Ft – F0), where Ft is observed fluorescence at time t and F0 is fluorescence at t = 0. The color of the line corresponds to the same colored arrowhead in the snapshots. Scale bars, 50 μm. (b) Live imaging of Ca2+ flux in HIOs+ENS grown in vitro before (left) and after KCl treatment, which induced a rapid calcium efflux in the ENS cells (see Supplementary Video 4). Data are representative of two independent experiments. Scale bars, 100 μm. (c) Live imaging of Ca2+ flux in HIOs+ENS grown in vivo shows calcium transients in nerve bundles before and after KCl treatment, which induces a broad calcium efflux and contraction of the tissue (see Supplementary Videos 5 and 6). In vivo data are representative of two independent experiments with n = 3 organoids engrafted into individual mice per experiment. Scale bars, 100 μm (left) and 500 μm (right).
ENS-mediated contractile activity
Although the organization of epithelium, smooth muscle, neurons and glia in HIOs+ENS tissues is similar to that of human intestine, it is not clear whether any neuromuscular communication exists in HIOs+ENS tissues. To stimulate neurons, we explanted kidney grafts into Tyrode's solution and subjected them to electrical-field stimulation and monitored them for muscular contractions (Fig. 4a and Supplementary Videos 7–9). HIO tissue without an ENS was subjected to a single 1-ms pulse at 100 V, which caused a single contraction, suggesting the direct stimulation of smooth muscle. By contrast, HIOs containing an ENS subjected to a single 1-ms pulse at 50 V (below muscle activation threshold) was sufficient to trigger a sustained wave of contractions. To determine whether the contractions were a result of the activity of the ENS, we blocked neuronal activity with tetrodotoxin (TTX), which binds to voltage-gated Na+ channels on nerves, thereby inhibiting the firing of action potentials. TTX completely inhibited the ability of low-voltage stimulation to trigger contractile activity in HIOs+ENS.
Figure 4.
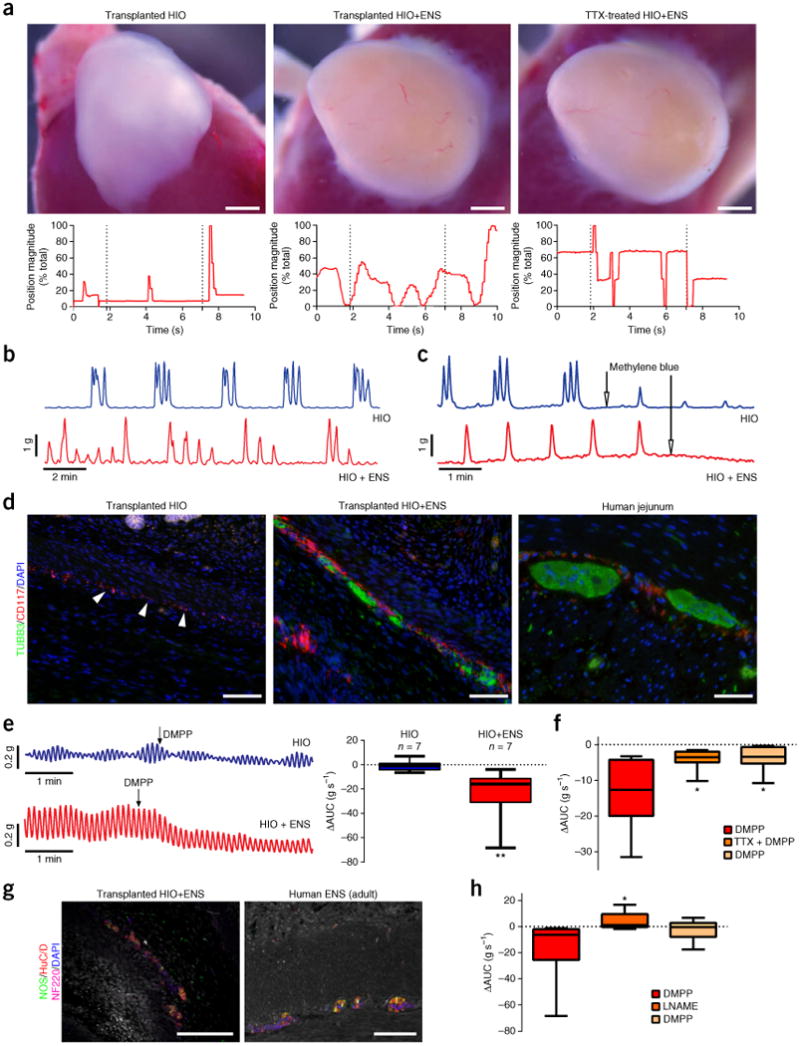
ENS-independent and dependent control of contractile activity. (a) The ENS in HIOs mediates peristaltic-like contractions (see Supplementary Videos 7–9). In vivo grown tissues were explanted and subjected to EFS. HIOs without ENS subjected to high-voltage EFS (1-ms pulse at 100 V) showed one contraction (left, n = 2). HIOs+ENS subjected to low-voltage EFS (1-ms pulse at 50 V) showed a sustained series of wave-like contractions (middle, n = 5) that were lost when tissues were cultured in TTX (TTX-treated HIOs+ENS, right; n = 2). Automated point tracking demonstrated a differential movement in HIOs+ENS, as compared to HIOs, that was lost after TTX treatment. Scale bars, 1 mm. (b) Recordings of spontaneous contractions in transplanted HIO and HIO+ENS tissue strips. Phasic contractions were observed after tissue equilibration (no stimulation), suggestive of ICCs in both HIO (n = 7) and HIO+ENS (n = 7) tissues. (c) Inhibition of ICC activity with methylene blue led to loss of contractile activity (n = 3). (d) Detection of ICCs (CD117, red) in both HIOs and HIOs+ENS in vivo. HIOs without NCCs did not form neurons (TUBB3, green). Scale bars, 100 μm. (e) DMPP stimulation in HIOs and HIOs+ENS. Right, area under the curve (AUC) during DMPP (10 μM) stimulation measured for 2 min before and after stimulation (n = 7). (f) TTX inhibition of ENS activation. DMPP stimulation measured for 2 min, followed by TTX (10 μM) treatment of HIOs+ENS (n = 7). (g) NOS+ neurons were present in HIOs+ENS grown in vivo. Scale bars, 100 μm. (h) ENS-induced relaxation by a NO-dependent mechanism. Shown is the AUC during DMPP stimulation measured for 2 min, followed by L-NAME treatment of HIOs+ENS (n = 7). For box and whisker plots, the black line across the box represents the median, the box represents interquartile range and the whiskers represent the minimum and maximum. *P < 0.05, **P < 0.01, Mann–Whitney test.
GI motility involves the coordination of ENS and pacemaker cells known as ICCs. ICCs mediate rhythmic contraction of smooth muscle independently from the ENS, whereas relaxation of smooth muscle is ENS dependent. To dissect these processes, we isolated tissue strips from HIOs and HIOs+ENS and analyzed them using isometric-force measurement assays in an organ-bath chamber. In the absence of neuronal stimulation, we observed spontaneous phasic contractions in both HIO and HIO+ENS tissues (Fig. 4b and Supplementary Fig. 9a), suggesting the presence of ICCs in both conditions. Methylene blue, which inhibits ICC activity, abolished phasic contractions (Fig. 4c), supporting the conclusion that phasic contractions in HIOs and HIOs+ENS are driven by ICCs. Consistently with this, we observed clusters of cells expressing the ICC marker CD117 in both HIOs and HIOs+ENS (Fig. 4d). This suggests that ICCs develop from the mesenchyme and do not originate from the neural crest. However, ICCs were closely associated with neurons in HIOs+ENS and seemed to form larger clusters in the presence of neural-crest-derived ENS cells.
ENS-mediated neuromuscular coupling
To identify ENS-dependent roles in contraction and relaxation, we activated neurons by using the selective α3-nicotinic receptor agonist dimethylphenylpiperazinium (DMPP) and veratridine, which increases nerve excitability by inhibiting Na+ channel inactivation. Pharmacological activation of the ENS induced a muscle relaxation in HIOs+ENS, but not in HIOs alone (Fig. 4e and Supplementary Fig. 9b). We next confirmed that relaxation was neuron dependent by using TTX, which inhibited the ability of DMPP to induce muscle relaxation (Fig. 4f and Supplementary Fig. 9c). Lastly, we sought to identify a putative neuromediator responsible for the smooth-muscle relaxation that was induced by ENS activation. Given the abundance of nNOS-expressing neurons in transplanted in HIOs+ENS (Fig. 4g), and the fact that nitric oxide (NO) is a well-known enteric muscle inhibitory neuromediator, we inhibited NO synthesis with NG-nitro-l-arginine methyl ester (L-NAME). L-NAME pretreatment significantly decreased the relaxation after DMPP stimulation, indicating that NO was produced by the neurons to induce relaxation (Fig. 4h and Supplementary Fig. 9d). We further found that the addition of a NO-releasing drug, sodium nitroprusside (SNP), induced muscle relaxation in HIOs+ENS, supporting the idea that NO release mediates relaxation (Supplementary Fig. 9e).
Effects of ENS on intestinal organoid development
During embryonic development, the ENS plexus forms functional connections with smooth muscle and the intestinal epithelium, and few studies have investigated whether the ENS influences the development of these tissues (reviewed in ref. 20). Thus, we used this human system to examine the effect of the ENS on the development of other intestinal lineages using RNA-seq data from whole HIOs and HIOs+ENS grown in vitro. We first analyzed curated lists of genes associated with digestive-tract development and digestive functions (Fig. 5a,b) and found that HIOs+ENS differentially expressed genes associated with GI development, as compared to HIOs alone. In one example, the presence of an ENS increased the levels of EGF signaling components and decreased the levels of TGF-β signaling factors (Fig. 5a). The ENS also affected the expression of genes associated with the absorptive and secretory lineages (Fig. 5c). For example, we observed decreased levels of absorptive lineage genes in HIOs+ENS as compared to HIOs alone, including enzyme brush borders genes FABP2, LCT, TREH, SI and MGAM. Secretory lineages were also affected, with a decrease in goblet cell markers, such as MUC2, and a decrease in differentiated Paneth cell markers, but an increase in WNT3. Consistent with this, we saw an increase in some active intestinal stem cell and transit-amplifying cell genes (Fig. 5c). To examine whether the ENS regulates the intestinal stem cell/transit-amplifying compartment, we analyzed the proliferation in the crypts of HIOs and HIOs+ENS grown in vivo. HIOs+ENS exhibited increased proliferation in the crypt compartment to a level that was similar to that seen in the human jejunum (Fig. 5d).
Figure 5.

ENS effects on the epithelium. (a–c) Heat maps of RNA-sequencing data from in vitro HIOs and HIOs+ENS. Curated list of digestive-tract development (a) and digestive function (b) genes revealed a differential regulation in HIOs+ENS, as compared to HIOs. A curated list of epithelial lineage markers, including enterocytes, goblet cells, enteroendocrine cells, Paneth cells and stem cell/transit-amplifying cells, is shown in c. Each lineage seemed to be affected by the presence of an ENS. (d) Double chromogenic staining with CDH1 and KI67 was performed to examine cell proliferation. HIOs+ENS (n = 4) showed increased proliferation, as compared to HIOs (n = 3), that was similar to human intestinal crypts (n = 3). Student's t test, two-tailed, unpaired. Data are presented as means ± s.e.m. (e) Imaging of enteroendocrine (SYN1, synapsin1; CHGA, chromogranin A) and neuronal cells (TUBB3) in HIOs+ENS (n = 6) showed no apparent interaction between neurons and enteroendocrine cells, as compared with human intestinal tissue (n = 4). Left, arrow indicates enteroendocrine cell neuropod in human ENS. Right, arrow indicates association of neurons and enteroendocrine cell in human ENS. Scale bars, 5 μm.
In addition to the regulation of intestinal-barrier homeostasis, the ENS interacts with enteroendocrine cells (EECs) to both transmit and receive information related to luminal nutrients and nutrient homeostasis21. HIOs+ENS contained CHGA+ and SYN1+ EECs in the epithelium, as did the human jejunal epithelial, and in both cases, EECs were associated with ENS cells (Fig. 5e). However, human jejunal EECs contained neuropods associated with neuronal fibers, whereas transplanted HIOs+ENS did not. HIOs+ENS tissues had a less developed neuronal network, reminiscent of fetal intestine, suggesting that EEC and neuronal-fiber interactions had not yet been established (Fig. 5e). The developmental timing of this process is not established in humans and might require additional environmental cues, such as exposure to luminal flow, nutrients and/or microbes.
PHOX2B mutation in HIOs+ENS
Impaired neural crest and ENS development is the underlying cause of several genetic forms of Hirschsprung's disease in humans3. Mutations in the gene encoding the tyrosine kinase receptor RET are among the most common causes of Hirschsprung's disease and have been well studied. However, there are an increasing number of genes associated with Hirschsprung's disease, about which little is known. For example, mutations in the PHOX2B homeobox-containing transcription factor result in complete aganglionosis of the bowel of humans22 and mice23. Despite the obvious phenotype, the molecular pathways that are affected by PHOX2B mutations have not been identified in humans, so we used HIO+ENS as a model system to study this form of Hirschsprung's disease.
We generated induced PSCs containing PHOX2B+/+, PHOX2B+/Y14X and PHOX2BY14X/Y14X alleles and differentiated them into vagal NCCs that expressed posterior HOX genes such as HOXB3, B5 and B7 (refs. 24–26) (Supplementary Fig. 10). NCCs were then recombined with wild-type HIOs generated from H1 embryonic stem cells to generate HIOs+ENS. Transcriptional analysis of organoids in vitro revealed a distinct clustering of transcripts according to genotype, with many genes being altered in both the PHOX2B+/Y14X and PHOX2BY14X/Y14X HIOs+ENS relative to PHOX2B+/+ organoids (Fig. 6a). The expression levels of several neural-crest-related genes were reduced, including PHOX2B, HTR3A, HTR3B, TLX2, EDN1 and EDNRB. However, the most notable increase was in CNS markers. The top biological processes that were upregulated in the PHOX2BY14X/Y14X NCCs were forebrain development, cerebral cortex development, pallium development and telencephalon development. The top biological terms downregulated were all related to muscle development, suggesting that PHOX2BY14X/Y14X NCCs inhibited differentiation of smooth muscle in the HIOs (Fig. 6b and Supplementary Fig. 10d). Consistent with this, HIOs + PHOX2BY14X/Y14X NCCs that were transplanted in mice grew poorly (1 out of 3 grafts grew), as compared with control NCCs (3 out of 3 grafts grew), had few neurons (TUBB3) and showed poorly developed smooth muscle (DES) (Fig. 6c). These data suggest that differentiation of PHOX2B mutant NCCs was impaired and that NCCs had non-cell-autonomous effects on the development of other HIO cell types, such as smooth muscle. Moreover, this human-PSC-derived intestinal model seems well suited for mechanistically studying genetic forms of Hirschsprung's disease in humans.
Figure 6.

Modeling a Hirschsprung's-disease-causing mutation in PHOX2B. (a) Clustering of RNA-seq results from 4-week old in vitro HIOs+ENS with and without the PHOX2BY14X mutation. Genes shown display statistically significant differences of at least twofold change in expression between homozygous and wild-type NCC conditions (ANOVA with Benjamini–Hochberg false discovery at a threshold of P < 0.05). (b) The top gene-ontology categories displaying differences in gene expression between HIOs+ENS generated with PHOX2BY14X/Y14X NCCs, as compared to HIOs+ENS generated with wild-type PHOX2B+/+ NCCs. Categories in red were upregulated in the homozygous condition, whereas categories in blue were downregulated. (c) In vivo growth of HIOs+ENS generated with PHOX2B NCCs. Top row, bright-field images of harvested organoids after 7 weeks of post-transplantation growth under the murine kidney capsule. Fractions indicate the number of organoids that grew and contained intestinal epithelium. Scale bars, 2 mm. Middle row, development of neurons (TUBB3+) in the transplanted organoids. Bottom row, development of glia (S100β+) in the transplanted organoids. Scale bars, 100 μm.
Discussion
We used principles of embryonic intestinal development to engineer human-PSC-derived intestinal tissue containing a functional ENS. Human-PSC-derived vagal-like NCCs that were recombined with PSC-derived intestinal organoids in three-dimensional growth conditions migrated into intestinal mesenchyme, self-organized and differentiated into an array of neuronal and glial cell types of the ENS. After engraftment and growth in vivo, NCCs formed complex ganglionic structures and interganglionic fibers, similar to the embryonic development of the myenteric and submucosal neural plexuses. We further found that the NCC-derived ENS was functionally integrated into intestinal smooth muscle and drove NO-dependent relaxation. The degree of tissue organization seen in transplanted HIOs+ENS suggests that the tissue that we engineered in vitro had the intrinsic information for coordinated cell migration, proliferation, lineage commitment and assembly into a plexus that occurs during embryonic development of the ENS.
We routinely observed the formation of a neuroglial plexus in transplanted HIOs+ENS, in close association with smooth-muscle fibers with organization similar to the neonatal myenteric plexus. However, several lines of evidence suggest that the NCC-derived ENS was less mature and more fetal in nature. For example, development of a submucosal plexus seemed to be delayed in HIO+ENS tissues, and this could be similar to the development in the human fetal gut, where the submucosal plexus develops 2–3 weeks after the myenteric plexus27. Another indicator of developmental immaturity is that the neuroglial plexus in HIOs+ENS contained smaller nerve bundles than the adult human intestine, similar to what is observed in the human fetal gut15. Another possibility is that the smaller nerve bundles were a result of too few starting NCCs in the HIO cultures, as is in the case in Hirschsprung's disease. The immature and fetal nature of HIO+ENS tissues may provide an opportunity to identify specific factors that regulate the maturation of the fetal gut and ENS. For example, it was recently demonstrated that the microbiota of the lumen influences the colonization of the mucosa by glial cells28, and the HIOs ± ENS model could allow for mechanistic dissection of this process. Factors that promote intestinal maturation could be used clinically with premature infants, who are at heightened risk for intestinal infections resulting from an immature mucosa.
We observed a substantial degree of neuronal diversity in HIOs+ENS in vitro, including excitatory and interneurons that had intrinsic and inducible waves of calcium efflux, suggestive of neuronal activity. HIOs+ENS that were grown in vivo acquired additional neuronal diversity in the form of nNOS+ inhibitory neurons, which are known to form after cholinergic neurons in the developing fetal mouse gut29. To our surprise, CHAT-positive neurons, which appear at early stages during ENS development30, were found in HIOs+ENS in vitro, but were not detected in HIOs+ENS in vivo. There is evidence for transient expression of neurochemical markers, such as CHAT-positive neurons20, suggesting that embryonic expression may not mark cells that ultimately form cholinergic neurons. Moreover, neuroglial cells assembled into a myenteric and submucosal plexus that were functionally associated with highly differentiated layers of smooth muscle. Electrical-field stimulation (EFS) of these engineered tissues triggered ENS-dependent waves of contractile activity that resemble human intestinal motility. However, we also observed contractile activity that was dependent on resident ICCs that were present in both HIOs and HIO+ENS. Taken together, our data suggest that ICCs largely drive contraction, whereas relaxation of muscle is ENS dependent and mediated by nNOS+ inhibitory neurons that are present in HIO+ENS tissue. The fact that nNOS+ inhibitory neurons became functional earlier than cholinergic excitatory neurons further supports the conclusion that the ENS in HIOs is fetal in nature.
There are several pronounced differences between rodents and humans regarding development, physiology and diseases of the intestine. For example, the development of crypts occurs in utero in humans, whereas it happens postnatally in mice. During ENS development there are differences in the formation of TH+ neurons between mouse and humans31–33. Given that HIOs+ENS contain TH+ neurons in vitro and in vivo, this system may be the only means by which to study the unique developmental properties of these cell types. Moreover, an experimental system to study human TH+ dopaminergic ENS neurons may provide insight into GI dysmotility symptoms found in patients with Parkinson's34. Because of its highly tractable nature, this system should be particularly useful for studying developmental deficits of the ENS, such as Hirschsprung's disease. Defects in NCC formation, migration, incorporation into the mesenchyme and proliferation can all result in aganglionic sections of the intestine. As we have demonstrated using PSCs with PHOX2B mutations, this in vitro system allows for mechanistic dissection of known and unknown forms of Hirschsprung's disease in humans. There is high enthusiasm for emerging stem-cell-based therapies for the treatment of enteric neuropathies35. Previous therapeutic efforts have focused on engrafting ENS precursors into diseased tissue36–38. Our approach focuses on developmental biology to engineer PSC-derived tissue with functional ENS39, which is suited for generating functional intestinal tissue that could be transplanted into patients with short bowel syndrome. Both approaches highlight the promise of stem-cell-based therapies for the treatment of a variety of GI diseases and how this field has advanced in recent years.
Online Methods
Animals and human tissue
Immune-deficient NOD-SCID IL-2Rγnull (NSG) male and female mice, 8-16 weeks old, were used in all experiments (obtained from the Comprehensive Mouse and Cancer Core Facility). All mice were housed in the animal facility at the Cincinnati Children's Hospital Medical Center (CCHMC). All experiments were performed with the approval of the Institutional Animal Care and Use Committee of CCHMC (IACUC2016-0014).
The study using human tissues was approved by an Institutional Review Board (IRB2014-0427) at CCHMC. Informed consent for the collection and use of tissues was obtained from all donors, parents or legal guardians. Healthy human tissues were obtained from jejunum.
Cell lines and culture conditions
Human ES cells, H1 (WA-01) and H9 (WA-09) were from WiCell, and all human iPS cells WTC11 AAVS1-CAG-GCaMP6f WTC10 PHOX2B WT (+/+), WTC10 PHOX2B het (+/Y14X) and WTC10 PHOX2B null (Y14X/Y14X) were from B.R.C.'s lab. All PSCs were maintained in an undifferentiated state on Matrigel (BD Biosciences) without feeders. The PHOX2B mutant lines used to generate the mutant NCCs were generated from a single iPS line (WTC10) derived from a healthy patient. The mutation was generated using a pair of transcription activator-like effector nucleases (TALENs) targeted at the PHOX2B mutation site and a 60-bp oligonucleotide donor containing the C→A that results in the Y14X mutation. The mutant cells were selected for using droplet digital PCR designed to detect and distinguish a mutant from a wild-type allele. For more complete details about the modification of the PHOX2B allele, refer to ref. 40. H9-GAPDH-GFP hESCs were generated by targeting sequences encoding GFP to the 3′ UTR of the GAPDH gene by using standard procedures41 that incorporated TALEN-facilitated homologous recombination42. GFP is expressed as T2A (ref. 43) pseudofusion protein immediately adjoining the C-terminus of GAPDH. H1 and H9 ESC lines were analyzed for pluripotency karyotype and the absence of mycoplasma contamination by the CCHMC Pluripotent Stem Cell Facility. WTC iPSC lines were analyzed for karyotype by Cell Line Genetics, and the top ten potential off-target sites were sequenced.
Cells were fed mTeSR1 media and routinely passaged using Dispase II (Gibco). We slightly modified previously published protocols to generate NCCs12 and HIOs9,10, and combined them at an early stage of intestinal differentiation to generate HIOs containing ENS (Supplementary Fig. 1). Briefly, for NCC generation human PSCs were treated with collagenase IV (500 U/ml) in mTeSR1 for 60 min to detach colonies. Cells were washed to remove collagenase, then gently triturated and resuspended in neural induction media12 on non-TC-treated petri dishes. Neural induction media was changed daily, and retinoic acid (2 μM) was added on days 4 and 5 for posteriorization. Day-6 free-floating neurospheres were plated on fibronectin (3 μg/cm2) and fed neural induction media without RA for 4 d. Migrated cells were collected using a brief Accutase treatment (2-3 min) and passaged onto fibronectin or used immediately for combining with HIOs. NCCs were differentiated essentially as described for neuroglial12 and mesenchymal lineages44. We generated HIOs in a similar manner to our previous protocols9,10, but most notably, used small molecule CHIR99021 in place of WNT3A. After the formation of gut tube spheroids, we centrifuged spheroids ± 20,000–50,000 NCCs for 3 minutes at 300g and embedded them in Matrigel. Cultures were fed a basic gut media (advanced DMEM/F12, 1× B27 supplement, 1× N2 supplement, 10 μM HEPES, 2 mM l-glutamine, 1× Pen-Strep) supplemented with 100 ng EGF ml−1 and maintained in vitro for up to 8 weeks.
In vivo transplantation and electrical-field stimulation of HIOs
HIOs and HIOs+ENS were ectopically transplanted into the kidney capsule of NSG mice following a previously developed protocol11. Briefly, 4-6-week-old HIOs were embedded in collagen and transplanted into the kidney subcapsular space. Engrafted HIOs were harvested 6-10 weeks after transplantation and analyzed for neural and glial markers, or used for electrical-field stimulation (EFS) experiments and isometric-force organ-bath chambers. For EFS, HIOs and HIOs+ENS were explanted into Tyrode's solution and equilibrated for approximately 5 min before stimulation was begun. Electric stimulation was applied using a Grass S88 Stimulator (Grass Technologies) with single pulse, 1-ms duration and 50- or 100-V settings. HIO and HIOs+ENS were then incubated for 5 min in 10 μM TTX diluted in Tyrode's, rinsed and placed back in fresh Tyrode's. EFS was then repeated. Videos were recorded on a Leica dissection microscope using Leica Application Suite software and processed with VideoLAN and Handbrake to achieve 16× play speed. Videos were analyzed using the video-analysis software Tracker version 4.91 (Douglas Brown). Automated point tracking with position was performed to measure the movement within explanted HIOs and HIOs+ENS during EFS.
Chick-embryo manipulation
Chicken eggs were purchased from Charles River and incubated at 39 °C until they reached the desired Hamburger and Hamilton stage. For chicken NCC cultures, dorsal neural tubes from cranial or trunk regions of HH8 embryos were dissected using a Gastromaster and cultured on matrigel-coated tissue culture dishes with NCC culture medium12 for 24 h. The neural ectoderm was scrapped off the plates with a tungsten needle after NCCs migrated out from the neural tube explants. The NCCs that remained on the plates were then treated with 4 ng/ml FGF4 or 2 μM RA for 48 h. For NCCs injection, GFP-labeled human PSC-derived NCCs were collected and injected intersomitically into HH10-12 chicken embryos. The embryos were harvested around HH23 for analysis.
Directed differentiation of human neural crest cells
For neuroglial differentiation, NCCs were grown to confluency in neural induction media and then switched to growth factor reduced media (1:1 ratio of DMEM/F12-GlutaMAX and Neurobasal media, 0.5× B27 supplement, 0.5× N2 supplement, 1× pen-strep). Media was changed every other day and cells were cultured for 2-3 weeks before assaying for neuroglial markers. For osteogenic and chondrogenic differentiations, NCCs were first differentiated into neural crest–mesenchymal precursor cells (NCMP cells) by plating NCCs on uncoated tissue culture plates at 60,000 cells cm-2. Cells were fed a mesenchymal stem cell (MSC) media every 2–3 d (DMEM/F12, 10% FBS, 2 mM l-glutamine, 1× pen-strep) and passaged using TrypLE Select (Gibco) every 4–6 d before cells reached confluency. After 10-14 d in MSC media, cells were harvested and used for mesenchymal differentiations. For osteogenic differentiation, NCMPs were plated on uncoated tissue culture dishes at 5,000 cells cm-2 and cultured 3-4 weeks with osteogenic media (DMEM/F12, 10% FBS, 0.1 μM dexamethasone, 10 mM β-glycerol phosphate, 200 μM ascorbic acid). Cells were fixed with 4% PFA for 15 min, washed with dH2O, and stained with 2% (w/v) Alizarin red solution, pH 4.2 for 2–3 min at 23 °C. Cultures were washed with dH2O and imaged on a Leica DM IL LED Inverted Microscope (Type 11090137001). For chondrogenic differentiation, micromass cultures were generated by harvesting NCMPs and spotting 200,000 cells in 10 μl chondrogenic media (DMEM/F12, 10% FBS, 200 μM ascorbic acid, 10 ng ml−1 TGFβ-III) onto uncoated tissue culture dishes. Spots were incubated for 2 h at 37 °C and 5% CO2 before gently adding additional chondrogenic media to fill the well. Micromass cultures were fed fresh chondrogenic media every 2 d and maintained in culture for 4 weeks before assaying for chondrogenic differentiation. Cultures were fixed with 4% PFA for 15 min, washed with PBS, rinsed once with 0.1 N HCl, and stained with a 1% Alcian blue solution, prepared in 0.1 N HCl for 30 min at 23 °C. Cultures were imaged on a Leica S8APO stereomicroscope with Leica TL BFDF
Immunohistochemistry and microscopy
NCCs, cell monolayers, and day 0 spheroids were fixed with 4% paraformaldehyde (PFA) at 23 °C for 15 min, washed and then stained directly. Four-week-old in vitro HIOs and in vivo transplants were fixed in 4% PFA at 4 °C for 1 h to overnight. Tissues were processed and embedded in paraffin or OCT, sectioned at 5 and 10 μm, respectively, and affixed to Superfrost Plus slides (Fisherbrand). Parraffin sections were hydrated and underwent heat-induced epitope retrieval. Sections and cells were permeabilized with 0.25% Triton-X100 for 10 min and then blocked with 5% normal donkey serum (NS, Jackson ImmunoResearch) in PBS for 30 min at 23 °C. Primary antibodies diluted in PBS were applied to slides and cells overnight at 4 °C, followed by washes and incubation with secondary antibodies at 23 °C for 2 h or overnight at 4 °C (see Supplementary Table 1 for antibodies and dilutions). For double colorimetric staining, the ABC vectastain system (Vector Laboratories) was used for signal amplification. For signal detection, Vector Red and diaminobenzidine substrate kits were used (Vector Laboratories). Lillie-Mayer's modified hematoxylin was used as nuclear counterstain (Dako). Slides were mounted using Fluoromount-G or Cytoseal 60 and images were obtained using a Zeiss ApoTome Imager Z1 or Zeiss LSM510 confocal microscope. Whole-mount 3D images were obtained by fixing tissues in 4% PFA overnight at 4 °C then equilibrating in 100% methanol for 1 h on ice. Tissues were permeabilized with Dent's bleach (4:1:1 MeOH: DMSO: 30% H2O2) for 2 h at 21–23 °C before rehydrating with PBS. Tissues were then blocked with 5% NDS in PBS for 2 h at 21–23 °C, incubated with primary antibodies in PBS overnight at 4 °C, washed and incubated with secondary antibodies in PBS overnight at 4 °C. Secondary antibodies are routinely tested for background using a minus primary antibody control. Following staining, tissues were dehydrated through a methanol series and cleared with Murray's Clear (2:1 Benzyl benzoate: Benzyl alcohol) before imaging on a Nikon A1 inverted confocal microscope. Images were processed using NIS Elements, Bitplane Imaris, Zeiss Axiovision, and Adobe Photoshop CS7 software.
RNA isolation, quantitative real-time PCR and RNA-Seq
Total RNA from cells and organoids was isolated using NucleoSpin RNA isolation Kit (Macherey-Nagel). Complementary DNA was generated immediately following RNA isolation using SuperScript VILO cDNA Synthesis Kit (Invitrogen). Both isolation and synthesis were carried out according to the manufacturers' protocols. We performed qPCR using QuantiTect SYBR Green PCR Kit (Qiagen) on a BioRad CFX96 Real-Time PCR Detection System. Primer sequences were generally obtained from qPrimerDepot (http://primerdepot.nci.nih.gov) and can be found in Supplementary Table 2.
Whole-transcriptome (RNA) sequencing was performed at the CCHMC DNA Sequencing and Genotyping Core. RNA-seq libraries were prepared using the Illumina TruSeq RNA preparation kit and sequenced on the Illumina Hi-Seq 2000 with single-end 75-bp, 30 million reads (Illumina). Following removal of primers and barcodes, RNA-seq analysis was performed entirely in Strand NGS software (Agilent Technologies). The samples were aligned against the GRCh37 genome model. Multiply-mapping reads were removed, and aligned reads were filtered on base quality, with a quality threshold ≥30 and zero ‘Ns’ allowed in each read. The aligned gene read counts were quantified and used to compute reads per kilobase per million reads (RPKMs) for each transcript in each sample. Raw counts were normalized using the DESeq algorithm, thresholded to 1 and baselined to the median of all samples.
Expression values were compared between samples and assessed for statistical significance using an ANOVA with Benjamini–Hochberg false discovery at a threshold of P < 0.05 and a 2.0-fold cut-off filter, requiring a threefold differential. Heat maps of the relative expression of differentially regulated genes were generated, clustering on both gene and condition, using the Euclidean distance metric and average linkage rule. A Spearman correlation matrix was performed on differentially regulated genes to determine the sample cluster patterns on the basis of gene expression. Ontology analysis and biological network were performed using ToppGene Suite45 and ReVIGO46.
Data-availability statement
The RNA sequencing data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE84666.
GCamP6f calcium imaging
Detection of calcium transients was performed using WTC11 AAVS1-CAG-GCaMP6f reporter line. HIOs+ENS grown in vivo or in vitro were cultured on 8-well Ibidi micro-slide (ibiTreat plates, Ibidi, Klopferspitz) 24 h prior imaging. They were then imaged every minute for 20 min using 10× objective on a Nikon Eclipse Ti with NIS elements software to obtain background fluorescence level. HIOs+ENS were then treated with 30 mM KCl. Fluorescence values were reported as ΔF/F0, (ΔF = Ft − F0) where Ft is observed fluorescence at time t and F0 is fluorescence at t = 0. Experiments were carried out at 21–23 °C.
Ex vivo intestinal motility
Engrafted HIOs and HIOs+ENS were harvested and placed in ice-cold HBSS. Muscle strips (4-6 mm in length and 1-2 mm in width) were cut from the engrafted HIOs and HIOs+ENS. Preparations were suspended vertically in an organ bath filled with Krebs solution (NaCl, 117 mM; KCl, 4.7 mM; MgCl2, 1.2mM; NaH2PO4, 1.2 mM; NaHCO3, 25 mM; CaCl2, 2.5 mM and glucose, 11 mM), warmed at 37 °C and gassed with 95% O2 + 5% CO2. After an equilibration period of 60 min at initial tension of 0.5g, the contractile response of the muscle was continuously recorded, using eight-chamber tissue-organ bath with isometric-force transducers (0-25g; AD Instruments) coupled to a computer equipped with LabChart Pro software (AD Instruments). Muscle preparations were stimulated with dimethylphenylpiperazinium (DMPP; 10 μM; Sigma-Aldrich) and veratridine (3 μM; Sigma-Aldrich). Chemical stimulations were applied at 15-min intervals followed by three washes. Tetrodotoxin (TTX; 10 μM; Tocris) or NG-nitro-l-arginine methyl ester (L-NAME; 50 μM; Sigma) was applied 5 min before DMPP stimulation. NOS was inhibited with sodium nitroprusside (SNP; 100 μM; Sigma-Aldrich). Methylene blue (50 μM) was used to inhibit ICC activity. The effects of chemical stimulation on tension were evaluated by measuring the area under the curve (AUC). Data are expressed in ΔAUC i.e., “stimulated” AUC measured 120 s after stimulation minus “control” AUC measured 120 s before stimulation.
Statistical analyses
All data are represented as mean ± s.e.m. t tests and Mann-Whitney tests were completed using Prism (GraphPad). The determined significance cutoff was P < 0.05. No statistical method was used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessment except for the proliferation counting. No randomization was made.
Supplementary Material
Acknowledgments
We thank A. Zorn, N. Shroyer and members of the Wells and Zorn laboratories for reagents and feedback. We thank M. Kofron for assistance with confocal imaging. We thank S. Danzer, R. Pun, J. Piero, M. Marotta and M. Oria for help with the equipment for the electrical field stimulation experiments. We thank K. Campbell and J. Kuerbitz for providing antibodies for the neurochemical analysis. This work was supported by US National Institutes of Health grants U18TR000546 (J.M.W.), U18EB021780 (J.M.W. and M.A.H.), U01DK103117 (J.M.W. and M.A.H.), R01DK098350 (J.M.W.) and R01DK092456 (J.M.W.), and an Athena Blackburn Research Scholar Award in Neuroenteric Diseases (M.M.M.). We also acknowledge core support from the Cincinnati Digestive Disease Center Award (P30 DK0789392; Pilot and Feasibility Award), Clinical Translational Science Award (U54 RR025216) and technical support from Cincinnati Children's Hospital Medical Center (CCHMC) Confocal Imaging Core and the CCHMC human Pluripotent Stem Cell Facility.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHoR CoNTRIBUTIoNS: M.J.W., M.M.M. and J.M.W. conceived the study and experimental design, performed and analyzed experiments and wrote the manuscript. M.M.M., H.M.P., C.L.W., N.S. and M.A.H. helped to design and execute the mouse engraftment experiments, and M.M.M. performed the functional ENS assays. S.T. performed the experiments using the PHOX2B lines. P.A. and M.N. helped to design and execute the ex vivo organ-bath studies. S.A.B and C.-F.C. designed and performed the chick experiments. B.R.C., M.A.M. and Y.M. suggested the use of and provided the GCaMP6f and PHOX2B induced PSC lines. A.G.E., J.S. and E.G.S. provided the GAPDH-GFP HESC line. All of the authors contributed to the writing or editing of the manuscript.
CoMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/ reprints/index.html.
References
- 1.Furness JB. The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol. 2012;9:286–294. doi: 10.1038/nrgastro.2012.32. [DOI] [PubMed] [Google Scholar]
- 2.Sasselli V, Pachnis V, Burns AJ. The enteric nervous system. Dev Biol. 2012;366:64–73. doi: 10.1016/j.ydbio.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Obermayr F, Hotta R, Enomoto H, Young HM. Development and developmental disorders of the enteric nervous system. Nat Rev Gastroenterol Hepatol. 2013;10:43–57. doi: 10.1038/nrgastro.2012.234. [DOI] [PubMed] [Google Scholar]
- 4.Saffrey MJ. Cellular changes in the enteric nervous system during ageing. Dev Biol. 2013;382:344–355. doi: 10.1016/j.ydbio.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 5.McKeown SJ, Stamp L, Hao MM, Young HM. Hirschsprung disease: a developmental disorder of the enteric nervous system. Wiley Interdiscip Rev Dev Biol. 2013;2:113–129. doi: 10.1002/wdev.57. [DOI] [PubMed] [Google Scholar]
- 6.Burns AJ, Thapar N. Neural stem cell therapies for enteric nervous system disorders. Nat Rev Gastroenterol Hepatol. 2014;11:317–328. doi: 10.1038/nrgastro.2013.226. [DOI] [PubMed] [Google Scholar]
- 7.Hao MM, Young HM. Development of enteric neuron diversity. J Cell Mol Med. 2009;13:1193–1210. doi: 10.1111/j.1582-4934.2009.00813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 9.McCracken KW, Howell JC, Wells JM, Spence JR. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc. 2011;6:1920–1928. doi: 10.1038/nprot.2011.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson CL, et al. An in vivo model of human small intestine using pluripotent stem cells. Nat Med. 2014;20:1310–1314. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajpai R, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mica Y, Lee G, Chambers SM, Tomishima MJ, Studer L. Modeling neural crest induction, melanocyte specification, and disease-related pigmentation defects in hESCs and patient-specific iPSCs. Cell Rep. 2013;3:1140–1152. doi: 10.1016/j.celrep.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kudoh T, Wilson SW, Dawid IB. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129:4335–4346. doi: 10.1242/dev.129.18.4335. [DOI] [PubMed] [Google Scholar]
- 15.Fu M, Tam PK, Sham MH, Lui VC. Embryonic development of the ganglion plexuses and the concentric layer structure of human gut: a topographical study. Anat Embryol (Berl) 2004;208:33–41. doi: 10.1007/s00429-003-0371-0. [DOI] [PubMed] [Google Scholar]
- 16.Young HM, Ciampoli D, Hsuan J, Canty AJ. Expression of Ret-, p75(NTR)-, Phox2a-, Phox2b-, and tyrosine hydroxylase-immunoreactivity by undifferentiated neural crest-derived cells and different classes of enteric neurons in the embryonic mouse gut. Dev Dyn. 1999;216:137–152. doi: 10.1002/(SICI)1097-0177(199910)216:2<137::AID-DVDY5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 17.Young HM, et al. GDNF is a chemoattractant for enteric neural cells. Dev Biol. 2001;229:503–516. doi: 10.1006/dbio.2000.0100. [DOI] [PubMed] [Google Scholar]
- 18.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huebsch N, et al. Automated video-based analysis of contractility and calcium flux in human-induced pluripotent stem cell-derived cardiomyocytes cultured over different spatial scales. Tissue Eng Part C Methods. 2015;21:467–479. doi: 10.1089/ten.tec.2014.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao MM, et al. Enteric nervous system assembly: functional integration within the developing gut. Dev Biol. 2016;417:168–181. doi: 10.1016/j.ydbio.2016.05.030. [DOI] [PubMed] [Google Scholar]
- 21.Bohórquez DV, et al. Neuroepithelial circuit formed by innervation of sensory enteroendocrine cells. J Clin Invest. 2015;125:782–786. doi: 10.1172/JCI78361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajaj R, et al. Congenital central hypoventilation syndrome and Hirschsprung's disease in an extremely preterm infant. Pediatrics. 2005;115:e737–e738. doi: 10.1542/peds.2004-1910. [DOI] [PubMed] [Google Scholar]
- 23.Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- 24.Fu M, Lui VC, Sham MH, Cheung AN, Tam PK. HOXB5 expression is spatially and temporarily regulated in human embryonic gut during neural crest cell colonization and differentiation of enteric neuroblasts. Dev Dyn. 2003;228:1–10. doi: 10.1002/dvdy.10350. [DOI] [PubMed] [Google Scholar]
- 25.Lui VC, et al. Perturbation of hoxb5 signaling in vagal neural crests down-regulates ret leading to intestinal hypoganglionosis in mice. Gastroenterology. 2008;134:1104–1115. doi: 10.1053/j.gastro.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 26.Denham M, et al. Multipotent caudal neural progenitors derived from human pluripotent stem cells that give rise to lineages of the central and peripheral nervous system. Stem Cells. 2015;33:1759–1770. doi: 10.1002/stem.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace AS, Burns AJ. Development of the enteric nervous system, smooth muscle and interstitial cells of Cajal in the human gastrointestinal tract. Cell Tissue Res. 2005;319:367–382. doi: 10.1007/s00441-004-1023-2. [DOI] [PubMed] [Google Scholar]
- 28.Kabouridis PS, et al. Microbiota controls the homeostasis of glial cells in the gut lamina propria. Neuron. 2015;85:289–295. doi: 10.1016/j.neuron.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergner AJ, et al. Birthdating of myenteric neuron subtypes in the small intestine of the mouse. J Comp Neurol. 2014;522:514–527. doi: 10.1002/cne.23423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erickson CS, et al. Appearance of cholinergic myenteric neurons during enteric nervous system development: comparison of different ChAT fluorescent mouse reporter lines. Neurogastroenterol Motil. 2014;26:874–884. doi: 10.1111/nmo.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baetge G, Gershon MD. Transient catecholaminergic (TC) cells in the vagus nerves and bowel of fetal mice: relationship to the development of enteric neurons. Dev Biol. 1989;132:189–211. doi: 10.1016/0012-1606(89)90217-0. [DOI] [PubMed] [Google Scholar]
- 32.Blaugrund E, et al. Distinct subpopulations of enteric neuronal progenitors defined by time of development, sympathoadrenal lineage markers and Mash-1-dependence. Development. 1996;122:309–320. doi: 10.1242/dev.122.1.309. [DOI] [PubMed] [Google Scholar]
- 33.Anlauf M, Schäfer MK, Eiden L, Weihe E. Chemical coding of the human gastrointestinal nervous system: cholinergic, VIPergic, and catecholaminergic phenotypes. J Comp Neurol. 2003;459:90–111. doi: 10.1002/cne.10599. [DOI] [PubMed] [Google Scholar]
- 34.Anderson G, et al. Loss of enteric dopaminergic neurons and associated changes in colon motility in an MPTP mouse model of Parkinson's disease. Exp Neurol. 2007;207:4–12. doi: 10.1016/j.expneurol.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burns AJ, et al. White paper on guidelines concerning enteric nervous system stem cell therapy for enteric neuropathies. Dev Biol. 2016;417:229–251. doi: 10.1016/j.ydbio.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fattahi F, et al. Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature. 2016;531:105–109. doi: 10.1038/nature16951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hotta R, et al. Transplanted progenitors generate functional enteric neurons in the postnatal colon. J Clin Invest. 2013;123:1182–1191. doi: 10.1172/JCI65963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindley RM, et al. Human and mouse enteric nervous system neurosphere transplants regulate the function of aganglionic embryonic distal colon. Gastroenterology. 2008;135:205–216. doi: 10.1053/j.gastro.2008.03.035. [DOI] [PubMed] [Google Scholar]
- 39.Burns AJ, Roberts RR, Bornstein JC, Young HM. Development of the enteric nervous system and its role in intestinal motility during fetal and early postnatal stages. Semin Pediatr Surg. 2009;18:196–205. doi: 10.1053/j.sempedsurg.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Miyaoka Y, et al. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat Methods. 2014;11:291–293. doi: 10.1038/nmeth.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costa M, et al. A method for genetic modification of human embryonic stem cells using electroporation. Nat Protoc. 2007;2:792–796. doi: 10.1038/nprot.2007.105. [DOI] [PubMed] [Google Scholar]
- 42.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang W, et al. Faithful expression of multiple proteins via 2A-peptide self-processing: a versatile and reliable method for manipulating brain circuits. J Neurosci. 2009;29:8621–8629. doi: 10.1523/JNEUROSCI.0359-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee G, et al. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat Biotechnol. 2007;25:1468–1475. doi: 10.1038/nbt1365. [DOI] [PubMed] [Google Scholar]
- 45.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


