Abstract
Unlike most vertebrates, the shark IgL gene organization precludes secondary rearrangements that delete self-reactive VJ rearranged genes. Nurse sharks express four L chain isotypes, κ, λ, σ, and σ-2, encoded by 35 functional minigenes or clusters. The sequence of gene activation/expression and receptor editing of these isotypes have not been studied. We therefore investigated the extent of isotypic exclusion in separated B cell subpopulations. Surface Ig (sIg)κ–expressing cells, isolated with mAb LK14 that recognizes Cκ, carry predominantly nonproductive rearrangements of other L chain isotypes. Conversely, after depletion with LK14, sIgM+ cells contained largely nonproductive κ and enrichment for in-frame VJ of the others. Because some isotypic inclusion was observed at the mRNA level, expression in the BCR was examined. Functional λ mRNA was obtained, as expected, from the LK14-depleted population, but was also in sIgκ+ splenocytes. Whereas λ somatic mutants from the depleted sample displayed evidence of positive selection, the λ genes in sIgκ+ cells accumulated bystander mutations indicating a failure to express their products at the cell surface in association with the BCR H chain. In conclusion, a shark B cell expresses one L chain isotype at the surface and other isotypes as nonproductive VJ, sterile transcripts, or in-frame VJ whose products may not associate with the H chain. Based on the mRNA content found in the B cell subpopulations, an order of L chain gene activation is suggested as: σ-2 followed by κ, then σ and λ.
Introduction
Lymphocytes have a vast repertoire of Ag receptors generated by V(D)J recombination, but most cells express only one kind on the surface. This is the assumption in Burnet’s clonal selection theory, which explained the specificity of Ab responses. It postulates that individual lymphocytes bearing complementarity to an antigenic determinant can be stimulated by that Ag to proliferation and Ab production (1). Because mammals possess one Ig H chain locus and two kinds of L chain genes, κ and λ, a central question in immunology has been how the B cell is able to restrict expression to one allele of the H chain gene (allelic exclusion) and one allele of one of the L chain genes (allelic and isotypic exclusion). The early observation that κ-expressing cells had the λ locus in germline (GL) configuration but λ-expressing cells often carried deletions at κ genes pointed to a sequential pathway of rearrangement in which silencing of one locus gave way to continued V(D)J recombination at another (see Ref. 2 and references therein). As the numbers of κ and λ gene segments are very different in humans and mice, the order is based not on the recombination substrate numbers but instead a developmental program that activates the loci, one after the other, through temporally accruing transcription factors (3, 4).
L chain rearrangement and expression leading to formation of the BCR mark a crucial point of B cell development. Should the κ-chain pair poorly with H chain such that BCR surface expression is low or unstable, there is no tonic signaling for differentiation, and V(D)J recombination continues (5, 6). Should the BCR react to self components, V(D)J recombination is reinitiated (7, 8). In both instances recombination proceeds with equal probability on either κ allele (9), where continued rearrangement on the original chromosome generates either a replacement of the original VJ or deletion of the Cκ exon (10, 11). Because of the random nature of V(D)J recombination, further Vκ to Jκ rearrangements can be nonproductive, resulting in λ gene activation, and a productive VJ at the λ gene may rescue the B cell. Because primarily single rearrangements were observed despite the presence of four λ genes (12), it is thought that λ gene activation occurs late in the short developmental window that either ends in cell death or progresses to the immature B cell stage. Modification of BCR through secondary rearrangement is called receptor editing (for a recent review, see Ref. 13).
Thus, of the two L chain isotypes in mammals, one apparently has a reserve or salvage function. When three L chain isotypes are present, as in the amphibian Xenopus (14) and some bony fish (15), or four, as in some cartilaginous fish (16), the question arises as to the role of the additional L chains. There is no information on receptor editing and little information on isotype exclusion in cold-blooded vertebrates (17). V(D)J recombination exists in all jawed vertebrates, and the random diversification process produces a vast repertoire of receptors that inevitably includes autoreactive ones (18). The Igκ in Xenopus contains multiple VL and JL in tandem (19), potentially allowing the replacement of a primary VJ. Moreover, in two Xenopus species there is a possibility for gene inactivation in that the 3′-most Jκ is a pseudogene, to which rearrangements have in fact been recovered (20). In bony fish, the IgL are organized as many small loci, each with varying small numbers of V, J, and C exons, some of which share enhancers (15, 21). The tandem positioning of gene segments and the proximity of many loci offer the potential for removing unwanted VJ rearrangements (22).
However, the cluster-type Ig organization of cartilaginous fish is without multiple V or J and does not support secondary recombination of the nested type. Unwanted VJ cannot be edited out at κ miniloci or any others. With this restriction it is anticipated that isotype inclusion at the mRNA level could be extensive.
Our animal model is the nurse shark, a representative of the earliest vertebrate class whose immune system is based on V(D)J recombination. Cartilaginous fish have a unique Ig gene organization for both H and L chains that consists of multiple minigenes, or clusters, each with one V and one to four other rearranging gene segments and exon(s) for a single C region (23). Any species can possess up to >100 clusters. This Ig gene organization existed before the divergence of the two extant cartilaginous fish subclasses 420 million years ago (24). Although these Ig loci appear structurally simple, they have coevolved with TCR organizations that resemble those of tetrapods (25–27) and cannot be considered any less sophisticated a system of Ag receptors.
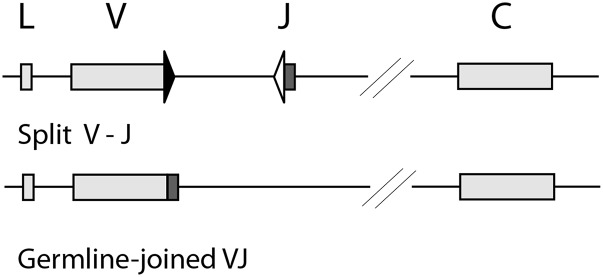
Fig. 1 depicts two kinds of L chain clusters in cartilaginous fish. A special feature is that some clusters carry prerearranged VJ, “GL-joined” (28). These can be in-frame or not, and in skates and rays the only L chains so far defined are all GL-joined, so these are assumed to function as conventional L chains despite their limited diversity (29). The nurse shark is the only model where all the IgL clusters have been characterized (16, 30). Table I shows the four isotypes (κ, λ, σ-2, σ) and their gene organization (split versus GL-joined); only the functional genes are listed by name in column 4. In single nurse shark B cells there are one to three somatically recombined VDJ, of which one is functional, and the other Igμ genes remain in GL configuration (31, 32). However, for L chain genes the GL-joined VJ do not permit analyses of gene activation in genomic B cell DNA. The functional IgL clusters are at least 100 kb apart and assumed to be autonomous, similar to the IgH, but their expression pattern has been difficult to interpret because of the profuse GL-joined L chain mRNA observed in neonates (30). In this study we show that most of this mRNA in pup tissues was not produced in surface Ig (sIg)+ cells. Some of these genes may have arisen for a purpose independent of the adaptive immune system (33, 34).
FIGURE 1.
Cluster organization of shark L chains. Top, Split V-J. IgL cluster with potentially recombining gene segments. Bottom, GL-joined VJ. IgL cluster with fused VJ. Boxes represent signal sequence (L), V gene segment, and C exon, as labeled, and the shaded box is J. Black triangle is RSS with 12-bp spacer, open triangle RSS with 23-bp spacer.
Table I. L chain genes in the nurse shark.
| L Chain | No. | Organization | Subtypes, Functional Genesa | Examined |
|---|---|---|---|---|
| κ (NS4) | ∼39 | GL-joined | 4–6 Clusters, R4, R7, R18, R20, RE18 | mRNA |
| Split | 33 Clusters, ∼21 functional | mRNA | ||
| λ (NS3) | 6 | GL-joined | 5 Clusters, NS3-12, -23, -46 | mRNA |
| 1 Cluster, NS3-8 | mRNA (qPCR only) | |||
| σ-2 (NS5) | 4 | GL-joined | 1 Cluster, NS5-16 | mRNA (qPCR only) |
| Split | 3 Clusters, NS5-2, -48 | mRNA, DNA | ||
| σ | 3 | Split | 2 Clustersc | mRNA, DNA |
| All L chains | ∼52 | GL-joined | 11–13 Clusters, 8–10 in-frame VJ (5 non-κ) | |
| Split | ∼38 Clusters, 25 functional (4 non-κ) |
Functioning genes listed by name. The GL-joined genes carry in-frame VJ and are transcribed with unexceptional C sequences.
Not every κ GL-joined gene is present in individual sharks. Pup2 carries RE18, R20, R18, R4; Pup1 has R7 in addition. Adult shark carries RE18, R20, R18, and pseudogene RE19.
Three C regions were determined by genomic Southern blotting (16). Amplification of the genomic cluster from different animals revealed two sequences in equal portion, differing only by 1 nt in the V gene segment (this study); it is assumed there are two functional clusters.
Thus, the B lymphocyte repertoire from neonates and adults is largely constituted from the rearranging genes, and Ag receptor selection begins with L chain expression. To study the evolution of B cell self-tolerance, information must be acquired as to L chain isotype exclusion and feedback, if any, among the four isotypes. We made an assessment of isotype exclusion by examining the L chain content in sIgκ-expressing and sIgκ-depleted samples from three individuals by quantitative PCR (qPCR) and established mRNA functionality by the nature of the VJ joints. However, in the isotype λ, all the genes bear prejoined VJ, and their mRNA was recovered from both the sIgκ+ population and the depleted sample. In the former there was the possibility for dual BCR. We show an absence of selection on λ mutants from the sIgκ+ sample, which suggests that isotype inclusion at the mRNA level may not mean their inclusion in the BCR.
Materials and Methods
Animals
Three sharks were captured off the coast of Florida (Dynasty Marine). The “adult” shark was a 2.5-foot-long male, estimated to be 2.5–3 y old, an age where its immune system is comparable to immunocompetent adults (33). Pup1 was a male and Pup2 a female, both 2 mo old or less. Their organs were harvested immediately on sacrifice and placed in shark PBS (PBS with 350 mm urea and 200 mm NaCl). RNA samples from other animals (shark-JS, newborn-AQ) have been described previously (32).
mAb specificity
The LK14 mAb detects L chain in IgM and IgW by immunoprecipitation (33). Its specificity was examined by Western blotting of expressed shark polypeptides. Representatives of four L chain isotypes from neonatal shark were cloned into expression vector pGEX and expressed as GST fusion proteins in Escherichia coli BL21: four κ (VJC clone S40, VJ only, C only), two λ (VJC, subtypes NS3-8 and NS3-12), σ (VJC, Sig26B), and three σ-2 (VJC, subtypes NS5-2, NS5-48, and NS5-16). Sonicates were electrophoresed on a 10% SDS-PAGE gel and checked for correct peptide size by Coomassie blue staining. The proteins were electrophoretically transferred onto membranes according to the manufacturer’s instructions for the Mini Trans-Blot cell (Bio-Rad Laboratories). The membranes were incubated with LK14 supernatant, followed by peroxidase sheep anti-mouse IgG (Sigma-Aldrich) as the secondary Ab and chemiluminescent reagents (Western blotting detection reagents; Amersham ECL). The presence of the GST fusion proteins was ascertained using peroxidase goat anti-GST (GE Healthcare).
Isolation of B cell subpopulations
Shark spleen cells were centrifuged through Ficoll, and the buffy coat was resuspended with anti-shark Cκ mAb LK14. Cells were collected after two rounds of column purification on goat anti-mouse IgG MicroBeads (Miltenyi Biotec). The negative population was also collected. In Pup2, after a second round of depletion, the LK14− cells were subjected to additional selection with a mixture of anti-shark IgM-specific mAbs (CB5, CB11, CB16) (35), whichs bind populations of IgM.
RNA and DNA were extracted from the same cell populations according to the manufacturer’s instructions for TRIzol (Invitrogen). Genomic DNA was obtained from nucleated erythrocytes using standard procedures.
PCR and qPCR
The qPCR procedure and Ig primers were described previously in Iacoangeli et al. (see supplemental table IB in Ref. 30) using nurse shark nucleoside diphosphate kinase (NDK) as the standard. All qPCR primer pairs span two exons, and their sequences were adjusted to enable an annealing temperature at 58°C and PCR products 104–136 bp in length. L chain primers targeted the JC, and those detecting unrearranged GL or sterile transcripts (ST) included one primer in the recombination signal sequence (RSS) (30). Each sample was done in triplicate and performed on three occasions. qPCR data were analyzed using the cycle threshold (Ct) method (36). The Ct for each Ig mRNA was normalized to the Ct for NDK mRNA, resulting in a ΔCt reflecting the relative level of the Ig transcript in that sample. The 2−ΔΔCt was calculated as the measure of fold increase of L chain RNA.
For conventional RT-PCR, first-strand cDNA synthesis (SuperScript III; Invitrogen) was primed with oligo(dT) and PCR was performed as described in other publications (32). The primers for the L chains were listed in Iacoangeli et al. (see supplemental table IA in Ref. 30). They are located in the signal sequence or the VL and in the C region. After 25–30 cycles the PCR product was cloned into pGEM-T Easy.
To isolate cDNA sequences encoded by the RE18 κ gene in the adult shark, first-strand cDNA was primed using the cluster-specific RE18R2 (5′-GTCAGTAGGCTGCTGACA-3′) in the C region, followed by PCR primers RE18F (5′-TCACACATCCAGCTGATC-3′) in the signal sequence and RE18R (5′-CTTCTTCCACTGCACCTC-3′) in the C region, also exploiting unique nucleotide positions in V and in C not shared with most other κ genes. However, in experiments that avoided amplifying RE18, an excluding reverse primer was used (nRE18R, 5′-CTTCTTCCACTGCACCTG-3′) together with the universal κ-specific forward primer in leader (NS4L, 5′-GATTTCACACATCCAGCT-3′).
Probes
Probes specific for the V region of κ (ns4v), λ (ns3v), and σ-2 (ns5v) have been described (30); that for the σ V region (sigv) was generated from a GL sequence with the primers in framework region (FR)1 (SIGF, 5′-TTACAGGTGGACAGTGTC-3′) and CDR3 (SIGVR, 5′-GTAAGTACTAGCTGAGGA-3′). Southern blotting and hybridization conditions were described elsewhere (30, 32).
Results
sIg+ splenocytes were obtained from an adult shark and two pups. PCR quantifying L chain content was performed. Although isotype exclusion seems to apply generally, all L chains were found at some level. Cloning of L chain sequences addressed the question of functionality, as determined by the nature of the rearrangement and accumulated mutations. In-frame and nonproductive σ mutants were used to establish standards for assessing selection during somatic hypermutation (SHM). These criteria were applied to λ genes, whose mRNA was recovered from both the sIgκ+ population and the depleted sample.
MACS
The mAb LK14 binds to the C region of shark κ L chains, which are encoded by ∼25 functional clusters in the nurse shark (Table I). LK14 bound a complete κ-chain (S40) or only its C region (S40C) and not the κ VJ or any λ, σ, or σ-2 polypeptides. We cannot say conclusively that LK14 binds every κ-chain, but the C regions are very well conserved, differing 0–2 of 108 aa among expressed clusters, including the GL-joined genes R4 and R7. Exceptions are the C regions of R18 and RE18, which differ from the majority by three and four residues, respectively, and the outlier R20 differs from RE18 by 10 aa and other κ by 11 aa.
Splenocytes from three animals were subjected to MACS with LK14 or anti-IgM mAb. Adult and Pup1 cells that bound these Abs (Kplus or Mplus) and the flowthrough (FT) were collected. In Pup2, the LK14-depleted FT cells were confirmed negative in a second round of exposure to the LK14. These cells were then incubated with anti-IgM mAbs, and a subpopulation of sIgM+ cells was recovered. It was anticipated that these LK14−/IgM+ (KmMplus) cells express sIgM with one of the other, non-κ L chain isotypes.
qPCR
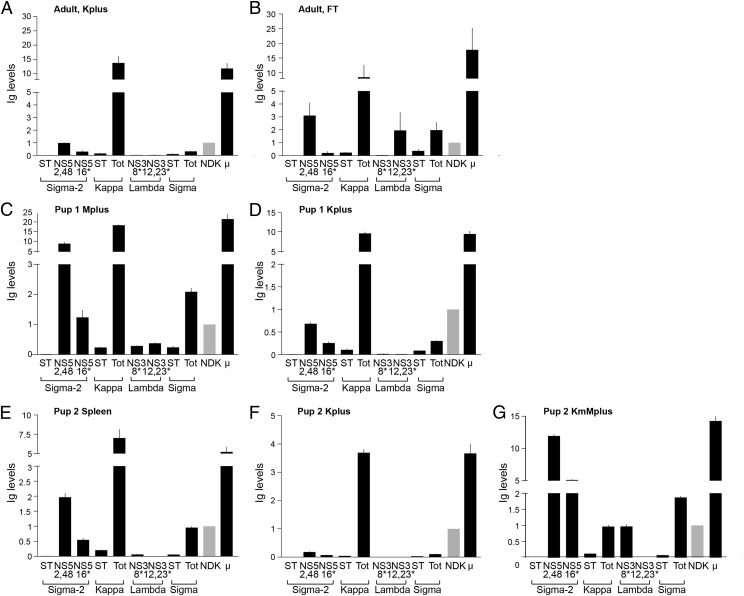
In the shark pups it was anticipated that the Mplus cells express IgM with various L chain isotypes and the Kplus cells IgM or IgW with κ L chain. Expression of every L chain isotype and subtype was examined, including ST from nonrecombined split clusters. In any lymphocyte population at least three groups of transcripts were present: in-frame sequences, nonproductively rearranged sequences, and ST. The presence of ST is regarded as an indication of gene activation and accessibility to the RAG recombinase (37). Because of the proximity of the genetic elements of cluster organization, the V gene segments and the C regions were included in every class of transcripts (30). We had focused on primers spanning JL and CL because the JC splicing event takes place later in RNA processing; the qPCR would generally reflect mature transcripts and preferably in-frame VJ (38). We found that the relative proportions of JC transcripts among the four isotypes and subtypes were similar to their frequency in a cDNA library (30) and therefore the qPCR results were comparable. The μ primers, however, are located in Cμ2–Cμ3, and the array of transcripts detected is not known; hence, it is not expected that μ levels correlate with L chain levels. It was used as a gauge of Ig presence in the mRNA.
In adult Kplus the levels of κ were >13-fold greater than the other L chain types (Fig. 2A). Because Ig can be absorbed unto the surface of adult B cells (M. Flajnik, unpublished results), the LK14-sorted population may be regarded in this instance as enriched (Kenr) in sIgk rather than purified. Nonetheless, the overall levels of σ and λ levels were very low, although σ-2 was discernible. The FT population has not been entirely depleted of κ, but it is enriched in the other isotypes, including λ (Fig. 2B).
FIGURE 2.
Relative L chain transcript levels in splenocyte subpopulations. DNase-treated mRNA samples were analyzed for σ-2, κ, λ, σ, and μ sequences. GL-joined σ-2 and λ genes are named and designated with an asterisk. Graphs show Ig levels normalized to NDK (fold increase to NDK, 2−ΔΔCt); NDK level is shown in gray. (A) Adult, Kplus are the sIgκ+ splenocytes. (B) Adult, FT is FT of the anti-κ column. (C) Pup1, Mplus are the sIgM+ splenocytes. (D) Pup1, Kplus. (E) Pup2, total spleen mRNA. (F) Pup2, Kplus. (G) Pup2, KmMplus. After two depletions of sIgκ+ splenocytes, sIgM+ cells were obtained from the second FT.
In Pup1 the Ig expression profile of sIgM+ (Mplus, Fig. 2C) cells generally was similar to that previously described for its nonsorted splenocytes (30), other than a greater amount of μ detected. In Kplus cells of the pups, σ, σ-2, and λ were considerably depleted (Fig. 2D, 2F). Lastly, after LK14 depletion of Pup2, the sIgM-expressing population (KmMplus) was enriched in σ-2, σ, and the GL-joined λ NS3-8 (Fig. 2G).
In these cell separations the Kplus populations were largely depleted of other L chain isotypes. Because non-κ transcripts were nonetheless present, the nature of these sequences was examined in the next sections.
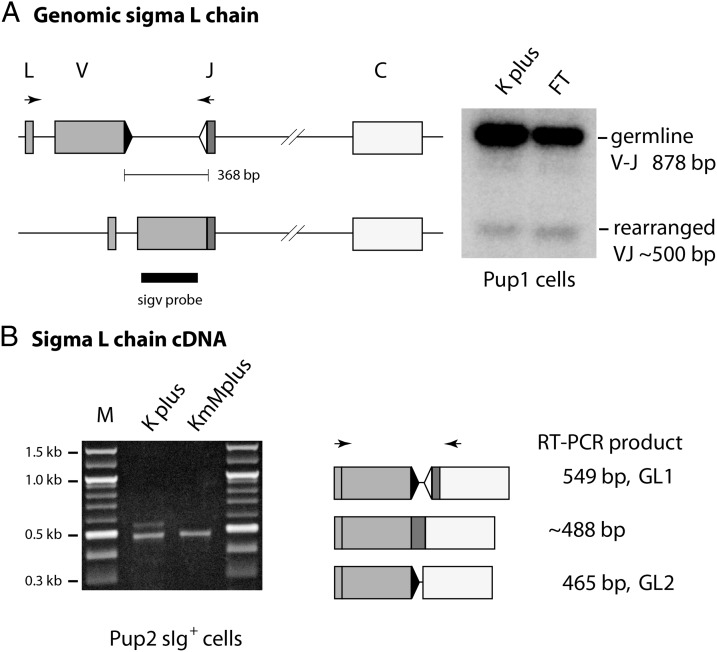
σ L chain
Both σ clusters carry split V-J (Table I), so that their rearrangements can be studied in both genomic and cDNA, as illustrated in Fig. 3. In genomic DNA, primers in leader and JL amplify rearranged σ bands at 500 bp and the GL V-J at 878 bp (Fig. 3A). The rearranged band was gel-purified and cloned. RT-PCR of RNA samples (Fig. 3B) can produce rearranged σ VJC at ∼488 bp and two ST of 549 and 465 bp, respectively, ST1 and ST2, spliced at cryptic sites in the intersegmental region (30). In comparison, qPCR primers target 100 bp in JC so that total σ levels (Tot in Fig. 2) are a combination of VJC and ST1, from which ST can be subtracted to obtain recombined VJ levels.
FIGURE 3.
Isolation of somatically rearranged L chain from genomic DNA and cDNA. (A) σ L chain amplified from genomic DNA. Left, L chain cluster in GL configuration and after deletional rearrangement. PCR primers, as indicated by arrows, amplified GL band at 878 bp and rearranged at ∼500 bp. Right, The PCR products from Pup1 cells were blotted and hybridized with V region probe. (B) σ L chain amplified from cDNA. Left, Agarose gel with PCR products of two sizes, 549 and ∼488 bp, that are ST1 and rearranged VJ, respectively. Right, The ST1 consists of signal sequence spliced to V gene segment, JL spliced to C region, and the intersegmental sequence spliced at cryptic sites. ST2 is intersegmental cryptic site spliced to the C region 5′ acceptor site. Gray boxes represent signal sequence (L) and V gene segment, darker box is JL, open box is C exon. Black triangle is RSS with 12-bp spacer, open triangle RSS with 23-bp spacer.
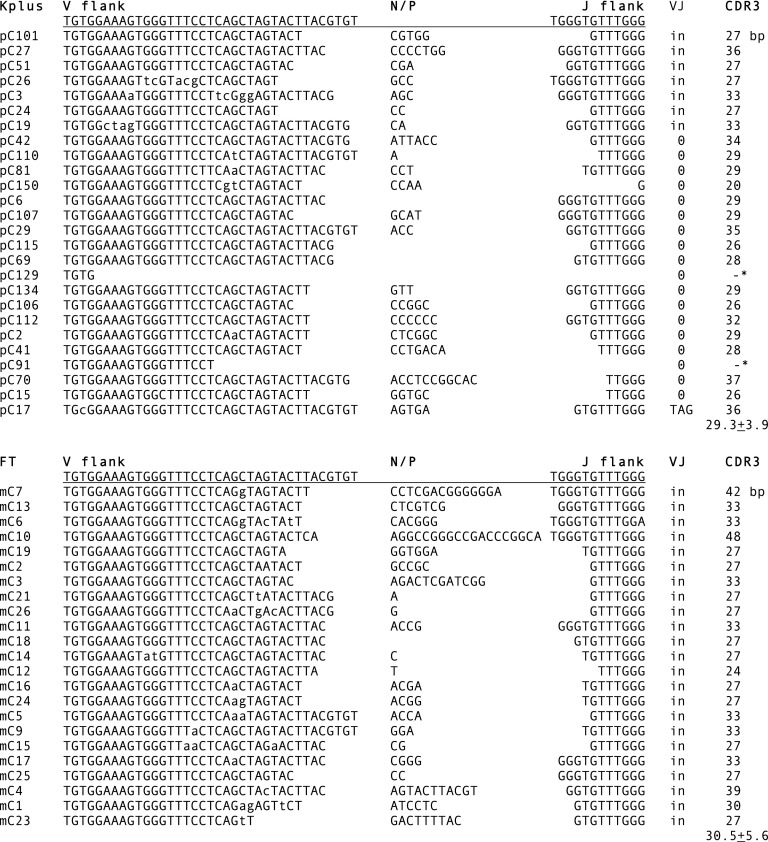
σ VJ raised from adult Kplus genomic DNA produced 31 clones with unique CDR3 (Fig. 4, top). Most (87%) were out of frame (Table II, Adult genomic). The Pup1 Kplus sequences obtained from genomic DNA were also dominant in nonproductive VJ (Table II). The proportion of in-frame VJ was taken as being in fact minor, as there is no known mechanism eliminating unwanted genomic rearrangements. Furthermore, the genomic VJ information allows a corrective assessment of the cDNA results, wherein a higher frequency in-frame VJ in the Kplus groups is likely the effect of RNA surveillance removing the nonproductive sequences (38). The CDR3 of adult σ cDNA is shown in Fig. 4. The lowercase letters indicate mutations, which were plentiful in adult Ig but not the pups.
FIGURE 4.
σ L chain CDR3. RT-PCR was performed on adult Kplus and FT samples. The sequences were compared with the reference GL V and J gene segments, whose coding flank sequences are shown. The sequences were determined for open reading frame (VJ), CDR3 size, and average CDR3 length. Nonproductive rearrangements are indicated by 0 or stop codon (TAG). Somatic mutations are shown in lowercase. An asterisk indicates that much of the J was deleted. N/P, N or P region.
Table II. σ L chain sequences.
| Adult genomic | Kenra | |
| VJ nonproductive (unique CDR3)b | 27 (27) | |
| VJ in-frame (unique CDR3) | 4 (4) | |
| Total | 31 | |
| Adult cDNAc | Kenr | FTd |
| GL | 103 (2)e | 2 (1)e |
| VJ nonproductive (unique CDR3)b | 19 (19) | 0 |
| VJ in-frame (unique CDR3) | 13 (7) | 23 (23) |
| Total | 135 | 25 |
| Pup1 genomic | Kplus | |
| VJ nonproductive (unique CDR3)b | 27 (25) | |
| VJ in-frame (unique CDR3) | 1 | |
| Total | 28 | |
| Pup1 cDNA | Kplus | |
| GL | 12 (2)f | |
| VJ nonproductive (unique CDR3)b | 6 (6) | |
| VJ in-frame (unique CDR3) | 16 (13) | |
| Total | 34 | |
| Pup2 cDNA | Kplus | KmMplus |
| GL | 16 (2)g | 0 |
| VJ nonproductive (unique CDR3)b | 21 (16) | 1 |
| VJ in-frame (unique CDR3) | 6 (6) | 32 (23) |
| Total | 43 | 33 |
Kenr is κ-enriched cells.
VJ out of frame; CDR3 contains stop codon.
CDR3 shown in Fig. 4.
FT is FT after selection for sIgκ+ cells.
Detected as two species, either 549 bp (ST1) or 465 bp (ST2): 82 ST1, 21 ST2 in Kplus; 2 ST1 in Kminus.
Eight ST1, four ST2.
Eleven ST1, five ST2.
Comparing σ cDNA sequences obtained from populations largely depleted of sIgκ show two major differences. As expected, the great majority of rearrangements were in-frame (Fig. 4, bottom, Table II, FT, KmMplus). However, in this case the amount of ST was greatly reduced. Whereas the occurrence of ST1 per total clones in Kplus Pup1 is 8 of 30 (0.27) and in Pup2 11 of 38 (0.29), the Pup1 KmMplus is 0 of 33 (<0.03). This finding was quantitatively confirmed by the qPCR data in Fig. 2D, 2F, and 2G: for Kplus Pup1, ST (ST1) per total σ is 0.09 of 0.3 (0.30), for Kplus Pup2, 0.03 of 0.11 (0.27), and for KmMplus Pup1, 0.08 of 1.87 (0.04).
In summary, in populations enriched for non-sIgκ, the genomic σ VJ from adult FT (Fig. 4) and Pup1 FT (data not shown) were 50% in-frame and the cDNA ≥97% in-frame (Table II, FT, KmMplus). The level of ST was very low (Fig. 2G). In contrast, where the BCR was expected to include κ-chain (Kplus), most σ VJ were nonproductive and a large proportion of the total transcripts were unrecombined. The disproportionate accruing of σ ST transcripts in sIgκ+ cells (27–30%) suggests pre-empted σ rearrangement.
σ-2 L chain
The σ-2 type consists of two rearranging clusters (NS5-2, NS5-48) and one GL-joined (NS5-16). Cloning results obtained for NS5-2 in the various separated populations were overall similar to σ. That is, VJ sequences from genomic DNA samples adult Kplus and Pup1 Kplus were 94 and 87%, respectively, nonproductive (Table III, adult and Pup1 genomic). VJ sequences obtained from adult, Pup1, and Pup2 Kplus cDNA consisted of varyingly portions (35–89%) of nonproductive rearrangements. In contrast, the FT and non–sIgκ-enriched samples (Table III, FT, KmMplus) contained in-frame VJ at 93–94%.
Table III. σ-2 L chain sequences.
| Adult genomic | Kenra | |
| VJ nonproductive (unique CDR3)b | 99 (88) | |
| VJ in-frame (unique CDR3) | 6 (4) | |
| Total | 105 | |
| Adult cDNA | Kenr | FTc |
| GL | 5 (1) | 0 |
| VJ nonproductive (unique CDR3)b | 17 (14) | 2 (2) |
| VJ in-frame (unique CDR3) | 2 (2) | 25 (25) |
| Total | 24 | 27 |
| Pup1 genomic | Kplus | |
| VJ nonproductive (unique CDR3)b | 47 (46) | |
| VJ in-frame (unique CDR3) | 7 (7) | |
| Total | 54 | |
| Pup1 cDNA | Kplus | |
| GL | 5 (1) | |
| VJ nonproductive (unique CDR3)b | 14 (13) | |
| VJ in-frame (unique CDR3) | 35 (25) | |
| Total | 54 | |
| Pup2 cDNA | Kplus | KmMplus |
| GL | 2 (1) | 0 |
| VJ nonproductive (unique CDR3)b | 6 (6) | 2 (2) |
| VJ in-frame (unique CDR3) | 11 (9) | 29 (27) |
| Total | 19 | 31 |
Kenr is κ-enriched cells.
VJ are out of frame or CDR3 contains stop codon.
FT is FT after selection for sIgκ+ cells.
Some σ-2 ST transcripts were cloned from the Kplus cDNA samples, ∼10% of total, but not from the FT or KmMplus.
κ L chain
Because the κ clusters include both split and GL-joined genes, it is only possible to determine κ gene activity through the cDNA. Unlike the σ-2 genes, the κ genes are too well conserved to permit PCR primers in V and J distinguishing somatically recombining clusters in DNA. Only pup κ sequences have been cloned because their paucity of somatic mutation makes it possible to identify individual genes.
In a previous study, 24 of 66 or 36% of κ sequences cloned from Pup1 spleen mRNA were GL-joined, of which RE18 was the prominent one (Table IV). With the same PCR primers, the proportion is strikingly decreased in the Pup1 sIg+ populations. In Mplus 3 of 42 (7%) and in Kplus 3 of 55 (5%) sequences are GL-joined. This suggests that another population of spleen cells is responsible for much of the GL-joined transcript production.
Table IV. κ L chain sequences in pups.
| Total Spleena | Kplus | Mplus | |
|---|---|---|---|
| Pup1 cDNAb | |||
| GL-joined κ | 15 RE18 | 3 R4 | 2 RE18 |
| 6 R4 | 1 R4 | ||
| 1 R20 | |||
| 2 R18 | |||
| VJ nonproductive (unique CDR3)c | 0 | 2 (2) | 0 |
| VJ in-frame (unique CDR3) | 42 (39) | 50 (43) | 39 (37) |
| Total | 66 | 55 | 42 |
| GL κ clones/total | 24/66 (36%) | 3/55 (5%) | 3/42 (7%) |
| Pup2 cDNA, excluding RE18/R20d | |||
| GL-joined κ (R4, R18) | 0 | 0 | |
| VJ nonproductive (unique CDR3)c | 0 | 32 (28) | |
| VJ in-frame (unique CDR3) | 56 (39) | 19 (10) | |
| Total | 56 | 51 |
Sequences published in Ref. 30.
PCR primers in leader (NS4L) and C (NS4C2A).
VJ out of frame or CDR3 contains stop codon.
PCR primers in leader (NS4L) and C (nRE18R); the latter excludes RE18/R20 C regions. Pup2 carries GL-joined RE18, R20, R18, and R4.
As expected, most κ sequences were in-frame, not only because of the sIg selection but also because of preferential decay of nonproductive VJ mRNA. With Pup2 a similar result was seen in the Kplus population (Table IV). Preliminary cloning data showed that Pup2 total spleen mRNA also was abundant in the GL-joined RE18 sequence, and to obtain better split gene representation the new reverse primer (nRE18R) excluded RE18 and R20. Although R4 and R18 could have been amplified along with the rest of known κ genes, only split κ genes were obtained. Pup2 cDNA in Table IV shows that, in contrast to the Kplus sample, the KmMplus mRNA carried a majority of nonproductive VJ. As before, the 30% in-frame VJ is not a reliable indication of its actual frequency in the mRNA population.
The Vκ gene usage was determined for Pup1 and Pup2 samples. In Pup1, Vκ5 was the prominent gene segment in total mRNA (38% of 39 unique sequences) as well as Kplus (42%, 18 of 43) and Mplus (41%, 15 of 37). In contrast, there was no dominant gene found in the Pup2 samples. However, because not all Cκ are defined in Pup2 it cannot be ruled out that the reverse primer nRE18R may bias results. More importantly, in Pup2 there was generally similar Vκ gene usage between the spleen, Kplus, and KmMplus samples. Thirteen individual Vκ genes were tallied, of which there was a sharing of 12 between the latter two groups. Because the 10 in-frame VJ in KmMplus consist of six clusters that were also represented in the Kplus population, this overlap suggests that the 10 VJ were not unrecognized by LK14 but, rather, not expressed as part of the BCR. This conclusion is buttressed by the KmMplus cells having been twice subjected to MACS, ensuring removal of cells bearing L chains that react with the anti-κ LK14 mAb.
Finally, although these results involve small numbers due to the paucity of material from young animals and after processing by MACS, they suggest that probably there is no dissimilarity in κ cluster activation between lymphocytes that express rearranged κ and those that had recombined κ nonproductively and went on to express BCR with a non-κ L chain isotype.
GL-joined genes part of BCR?
We have demonstrated that much of GL-joined κ mRNA was expressed in other than classical sIgM+ or sIgκ+ lymphocytes. However, GL-joined κ sequences were cloned from every sIg+ population, in reduced amounts that correlate better to their gene frequency. One question is whether genes profusely expressed early in development can be found in adult lymphocytes and whether they ever are functionally part of the BCR.
RE18 was selected because it is usually the dominant L chain mRNA in young animals, especially newborns. For instance, in primary neonate spleen and epigonal cDNA libraries it comprised 71 and 73%, respectively, of all κ clones (30). Previously we were unable to find GL-joined sequences among adult RT-PCR products (34), but this time first-strand cDNA priming was specific to the gene, followed by PCR primers designed to target RE18 and R20. Sequences cloned from adult spleen cDNA displayed not only the RE18 CDR3 but also positions throughout unique to RE18 (Fig. 5, underlined and arrows, see legend). We selected the mutants for analysis. SHM in sharks consists not only of point mutations but also changes in adjacent nucleotides (39, 40), which, as it also does in the RE18 mutants, comprises half the substitutions (17 of 36).
FIGURE 5.
GL-joined κ gene RE18 mutants. RT-PCR was performed on adult spleen mRNA. First-strand cDNA and PCR were done with primers targeting RE18/R20. The RE18 reference sequence is shown aligned with four mutants (A–D). They were identified by the fixed CDR3 and nucleotides unique to RE18 (underlined and arrow); the other markers indicated with an arrow are unique to RE18/R20. Mutations are shown in lowercase and highlighted in gray. Changes are indicated by a single-letter amino acid code underneath the codon, with the original/mutated residue. The CDR are underlined.
In Fig. 4 the mutations are highlighted and replacement changes are shown below the affected codon. Among the four cDNA, 4 of 9 mutated codons in FR1–3 resulted in amino acid substitutions in contrast to 16 of 18 in the CDR1–3. These results demonstrate that an active GL-joined κ gene can be transcribed in adult cells and subjected to mutation. The nature and positions of the changes appear to suggest selection acting on the BCR Ag-combining site.
SHM and sequence functionality
The contiguous substitutions that are the hallmark of SHM in sharks present difficulties in evaluating the functional nature of the changes. We decided to compare in-frame and nonproductive VJ mutants to obtain criteria for assessing selection. The adult σ sequences contained many mutations, and those in present in the VJ listed in Fig. 4 were tallied (Table V). The mutations were distributed differently over FR compared with CDR in the two groups. Among nonproductive σ the frequency of mutations was the same between FR and CDR (1.2 and 1.3%). In contrast, there was a greater discrepancy between FR and CDR in the in-frame clones, that is, 0.3 and 2.9%, respectively.
Table V. Analyses of σ mutants over FR and CDR.
| σa | Mutations in |
FR Total |
CDR Total |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FR1 | CDR1 | FR2 | CDR2 | FR3 | CDR3 | FR4 | 249 bp | Average 102 bp | No. cDNA | |||
| FT (in) | 0 | 17 | 6 | 12 | 8 | 36 | 3 | 17 | 0.3% | 65 | 2.9% | 22 |
| Kplus (out) | 3 | 0 | 15 | 13 | 21 | 10 | 16 | 55 | 1.2% | 23 | 1.3% | 18 |
| Amino Acid Substitutions in V |
|||||||
|---|---|---|---|---|---|---|---|
| FR-R | FR-S | RS Ratio | CDR-R | CDR-S | RS Ratio | CDR-R/FR-R | |
| FT (in) | 9, 0.6% | 4, 0.3% | 2.3 | 22, 4.3% | 1, 0.2% | 22 | 4.3/0.6 (7.2) |
| Kplus (out) | 22, 1.6% | 12, 0.9% | 1.8 | 7, 1.5% | 1, 0.2% | 7 | 1.5/1.6 (0.9) |
For σ calculations, 22 in-frame VJ (FT) and 18 nonproductive VJ (Kplus) were used (see Table II); two clones with large deletions were not used. The CDR consisted of CDR1 (33 bp), CDR2 (36 bp), and the average CDR3 (30 bp). The RS ratios were calculated for only the VL because out-of-frame rearrangements rendered CDR3/FR4 not interpretable. RS ratios were calculated for the regions specified. For example, 14 mutations in FR1–3, excluding FR4, in FT cDNA resulted in 9 replacement and 4 synonymous changes; their ratio is 2.3. The 29 mutations in CDR1 and CDR2, excluding CDR3, of FT cDNA resulted in 22 replacement and 1 synonymous changes; their ratio is 22.
To assess the effect on the translated product, amino acid substitutions were tallied excluding CDR3 and FR4 to compare both in-frame and nonproductive sequences. Replacement substitutions (R) and synonymous substitutions (S) in FR and CDR were tallied in Table V. In mouse SHM studies the “RS ratio” is obtained by comparing R versus S in CDR and R versus S in FR. In a functionally active protein, the former ratio is expected to be reasonably higher than the latter because fewer R substitutions are tolerated in the structurally important (FR) parts of the proteins. Because Ig mutants in mammalian systems also undergo affinity maturation, a low CDR-RS/FR-RS is considered indicative of an absence of selection in the CDR, or the Ag-combining regions.
When these methods were applied to the shark σ data, interpretation was difficult. Although the ratios of presumably functional in-frame sequences were higher (22/2.3), nonetheless the score of the nonproductive group would have been considered significant as well (7/1.8). This certainly is the effect of multiple substitutions occurring after a single lesion, and the preferred target motifs of AID tend to occur in the CDR (40). In assessing the overall frequency of changes, with the knowledge that one group was nonfunctional, we observed that the R changes by themselves may provide a better lead. Whereas R changes are irrelevant to the nonfunctional gene’s biological impact, in the selected product they will be distributed nonhomogeneously over FR/CDR. Thus the frequencies of CDR-R (1.5%) and FR-R (1.6%) make 0.9, that is, were nondistinguishing for CDR-FR in the nonproductive σ VJ. In contrast, for the in-frame σ it was 7.2 (4.3%/0.6%), showing that R changes have taken place preferentially in CDR.
λ Mutants in sIgκ-expressing cells
With these new criteria established, the expression of λ genes in sIgκ+ cells was investigated. NS3-23 is a GL-joined VJ with an open reading frame and thus always potentially functional when transcribed. Its presence in Kplus was examined in terms of selection of the mutants. NS3-23 sequences were amplified from adult Kplus, which was enriched for κ-expressing B cells, and from the FT, which appeared to include more λ-expressing cells, as observed in the qPCR (Fig. 2). The FT likely also included plasma cells.
Unique mutant λ sequences were selected from each group and their mutations tallied (Table VI, top). When the R frequencies were calculated, those in the FR (2.5%) versus the CDR (2.9%) in Kplus were on a par. In Kplus the CDR-R/FR-R make 1.2, but in the FT sequences it is 3.8 (7.2%/1.9%), owing to more replacements in their CDR. The λ sequences in Kplus and FT differ in mutational patterns that show those from the latter were subjected to selection pressure. The difference also suggests that sIgκ-expressing B cells that are isotypically included for λ mRNA may exclude them from the BCR.
Table VI. Analyses of λ mutants over FR and CDR.
| λa | Mutations in |
FR Total |
CDR Total |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FR1 | CDR1 | FR2 | CDR2 | FR3 | CDR3 | FR4 | 268 bp | 81 bp | No. cDNA | |||
| FT | 5 | 33 | 3 | 13 | 37 | 23 | 31 | 76 | 1.2% | 69 | 3.7% | 23 |
| Kplus | 3 | 5 | 3 | 2 | 31 | 8 | 11 | 48 | 1.3% | 15 | 1.3% | 14 |
| Amino Acid Substitutions in V |
|||||||
|---|---|---|---|---|---|---|---|
| FR-R | FR-S | RS Ratio | CDR-R | CDR-S | RS Ratio | CDR-R/FR-R | |
| FT | 38, 1.9% | 23, 1.1% | 1.7 | 45, 7.2% | 8, 1.3% | 5.6 | 7.2/1.9 (3.8) |
| Kplus | 31, 2.5% | 8, 0.6% | 3.9 | 11, 2.9% | 0, n.a. | n.a. | 2.9/2.5 (1.2) |
For λ calculations, the entire VJ was used because the NS3-23 gene is GL-joined and in-frame. The CDR consisted of CDR1 (39 bp), CDR2 (21 bp), and the fixed CDR3 (21 bp). The frequencies of FT mutations were calculated as follows: FT, 76/(23 × 268) = 1.2% for FR and 69/(23 × 81) = 3.7% for CDR; Kplus, 48/(14 × 268) = 1.3% for FR and 15/(14 × 81) = 1.3% for CDR.
n.a., not applicable.
Discussion
L chain isotype exclusion in sharks
In this study B cell subpopulations were investigated for the expression of four nurse shark L chain isotypes encoded by 35 functional clusters (Table I). We established that sIgκ-expressing cells (Kplus), isolated through the use of the mAb LK14 that recognizes Cκ, carry mostly nonproductive rearrangements of other L chain isotypes. Conversely, sIgM+ cells not selected by LK14 (KmMplus) contained a large proportion of nonproductive κ and enrichment for in-frame VJ of σ, σ-2, and, by qPCR, λ. These observations suggest that, for the most part, there is L chain isotype exclusion in shark B cells. However, in genomic DNA from Pup1 Kplus cells, 4% of σ VJ and 15% of σ-2 VJ are in-frame (Tables II and III).
To what extent might the Pup1 sIgκ+ cells be isotypically included? To get an estimate, the qPCR levels of functional transcripts of σ and σ-2 are compared that of κ; the percentage of in-frame VJ cDNA was revealed by cloning and applied to the qPCR unit. The rearranged in-frame κ is 90% of κ mRNA in Pup1 (Table IV) and in theory constitutes 6.59 qPCR units (Fig. 2D, qPCR κ total, 7.36). For σ, the qPCR primers detect the rearranged and ST1 forms. The cloned in-frame σ transcripts are 48% of Pup1 Kplus cDNA sequences (13 in-frame versus 8 ST1 and 6 out-of-frame; Table II) and thus form 0.15 units (48% of total 0.3 units) (Fig. 2D, total σ). Thus, in-frame σ VJC are 2% of in-frame κ VJC in the Pup1 Kplus population (0.15/6.59).
Calculated for the split σ-2 genes NS5-2 and -48, in-frame cDNA are 0.39 units (58% of 0.68 units) and thus 6% of in-frame κ VJC (0.39/6.59). Additionally, the third σ-2 gene, NS5-16, is GL-joined in-frame and therefore at 4% of in-frame κ (0.26/6.59). However, NS5-16 has a short fixed CDR3 of 6 codons and is unlikely to compete equally well for H chain when the included L chain is somatically recombined σ-2 or κ, with varying CDR3 of 8–14 codons (30, 41). Examples of this diversity are found in Fig. 4. Although there appears a much greater inclusion of σ-2 in κ-expressing cells (i.e.,10%), a significant portion is unlikely to produce dual receptors of κ/σ-2.
The estimations totaling ∼12% provide some idea of isotype inclusion at the mRNA level. Although we have previously established that the L chain mRNA measurements are comparable in our qPCR assay, promoter strengths may differ, and on a per cell basis κ/σ-2 inclusion may be well above or below 10%. Nonetheless, our results demonstrate that shark B cells generally transcribe one in-frame L chain isotype, and active clusters of other isotypes produce out-of-frame VJ, ST, and in a minority a second in-frame VJ. Until mAb reagents to the non-κ L chain isotypes are generated and the BCR determined, our data show a degree of isotype inclusion at the mRNA level.
We are unfortunately not able to determine the frequency of κ+κ+ but this situation is much the same as for κ/σ-2. Unlike for mammalian pre–B cells, all rearranging shark L chain junctions are extensively modified by TdT (41), so that any two L chains even of the same isotype would have different CDR3 content and lengths and are unlikely to be on a par for H chain pairing. Unequal CDR3 must promote preferential pairing and phenotypic exclusion.
Some single-cell observations
Prior to the discovery of the fourth L chain isotype, σ (16), we attempted single-cell RT-PCR on adult shark lymphocytes (E. Hsu and M. Flajnik, unpublished observations). Most of the cells expressed κ and a minority λ or σ-2; two cells carried all three and cloning revealed one cell with in-frame κ and nonproductive σ-2 and the other cell the reverse. However, one carried the λ pseudogene NS3-56A with its splice defect (40) but the other was a functional λ. Thus, three L chain clusters could be active in a B cell and one cell produced two in-frame L chain mRNA. These data support what we have determined from the separated B cell subpopulations.
The one published study on Ig expression using RT-PCR on single cells was done in PBL of clearnose skate, which possesses σ-2 and λ clusters, called L chain 1 and 2, respectively, all of which exist as fused VJ (17, 42); only the sequence surrounding CDR3 was isolated. One cell carried only λ, but 12 cells contained one to five σ-2 cDNA, four of them with two to four in-frame CDR3. Although it is not unknown whether all the genes are functional, this result shows that multiple L chain genes can be activated in a PBL.
All λ genes in nurse shark and other cartilaginous fish exist as fused VJ (29, 40). Because four of five clusters in nurse shark are functional (Table I), the presence of their mRNA in the sIgκ-expressing population, similar to the σ-2 NS5-16, shows the potential for dual receptors. However, the different patterns of SHM on λ mutants cloned from Kplus and FT populations suggest that there may be phenotypic exclusion of λ in the Kplus population. That λ mutants can consist of two groups, one selected for and one not, also explains previous puzzling results with λ mutants cloned from total spleen or epigonal organ mRNA (40). In that study the replacement changes could not be discerned as following the traditional patterns established for Ig mutants under selection in mice and humans.
The separated populations clearly show enrichment or depletion of L chain mRNA isotypes, indicating that not many L chain clusters were active per cell. We suggest that clusters are stochastically activated and restricted by a developmental time window. If productive VJ arise from rearrangement at split IgL or from functional GL-joined clusters, the L chain polypeptide participates in receptor selection. The presence of σ-2 and in particular σ ST in the Kplus population (Fig. 3B) suggests that, in some cells, the functional κ L chain enabling BCR expression had thwarted recombination at other clusters.
Cluster order
In the clearnose skate, λ is predominant early in life, as embryo and hatchling, but σ-2 became more abundant overall in adult tissues (43). Although both are GL-joined, there seems to be a temporal distinction in L chain isotype utilization, the basis for which is not known. A biased expression of λ in the lymphopoietic organs, the epigonal organ and Leydig gland, compared with spleen is suggestive of functional differences. However, in nurse shark epigonal organ, neonatal or adult, there is no distinction in λ levels from spleen (Ref. 30 and unpublished data).
We have found that κ-expressing Kplus B cells contain largely nonproductive VJ from non-κ isotypes and the KmMplus B cells out-of-frame κ VJ. Assuming that nonproductive VJ were from prior rearrangement events, at first sight there seems to be no activation hierarchy as exists in mammals. Nonetheless, we suggest such results are also compatible with an order of IgL activation among the clusters.
-
1)
σ-2 genes are activated first. σ-2 making innocuous BCR are sIgσ-2+ cells. If σ-2 VJ are nonproductive or form autoreactive BCR, rearrangement continues at other σ-2 genes.
-
2)
With accruing transcription factors (3), κ genes become activated. Rearrangement can occur at σ-2 or κ. Outcomes include:
-
a)
the B cell is sIgσ-2+ and expressing nonproductive or in-frame autoreactive κ (and σ-2) mRNA.
-
b)
the B cell is sIgκ+ with nonproductive or autoreactive σ-2 mRNA.
-
a)
This order explains the high amount of σ-2 in sIgκ+ cells.
-
3)
σ are activated, ST are produced. The large amount of σ ST in the sIgκ+ sample suggests that σ activation occurs after κ. That is, successful κ VJ pre-empted σ rearrangement. Outcomes include:
-
a)
the B cell is sIgσ+ and expressing κ (and σ-2) mRNA.
-
b)
the B cell is sIgκ+ and expressing σ-2 VJ mRNA and σ ST or VJ mRNA.
-
a)
An overlapping activation sequence explains our observations in KmMplus and Kplus clones. Additionally, the relative absence of σ ST in KmMplus cells (Fig. 3B, left) suggests that either the σ genes have yet to be activated (sIgσ-2+ cells) or they had undergone rearrangement upon activation (sIgσ+ cells). Where λ fits in is not clear.
The cluster organization does not support secondary rearrangements at the same gene; if some form of inactivating deletion does occur, relative absences cannot be detected in this study. Perhaps similar to λ genes in the mouse (44), receptor editing in the shark involves additional rearrangements without deleting primary VJ, explaining the isotype inclusion observed in the mRNA. An advantage of four L chain isotypes could be that, by virtue of sequence differences, their L chains are less likely to pair with equal compatibility to a particular H chain. This and the CDR3 diversity would enable stricter surface exclusion. The developing B cell would either continue expressing the autoreactive BCR and be eliminated or undergo receptor selection with the new one. In mammals dual κ receptors are found at 1.4–10% (45, 46) and dual λ in κ-deficient mice at >2% (47), but κ+λ+ are at lower frequencies of 0.2–0.5% (48, 49), perhaps for this reason.
In conclusion, although L chain isotype inclusion has been found in the shark B cell mRNA, a major source of their exclusion in the BCR must lie in competitive pairing with the H chain. The extensive heterogeneity among shark L chains, due to multiple isotypes and CDR3 sequence and length diversity, will make for dissimilar L chains expressed within individual B cells.
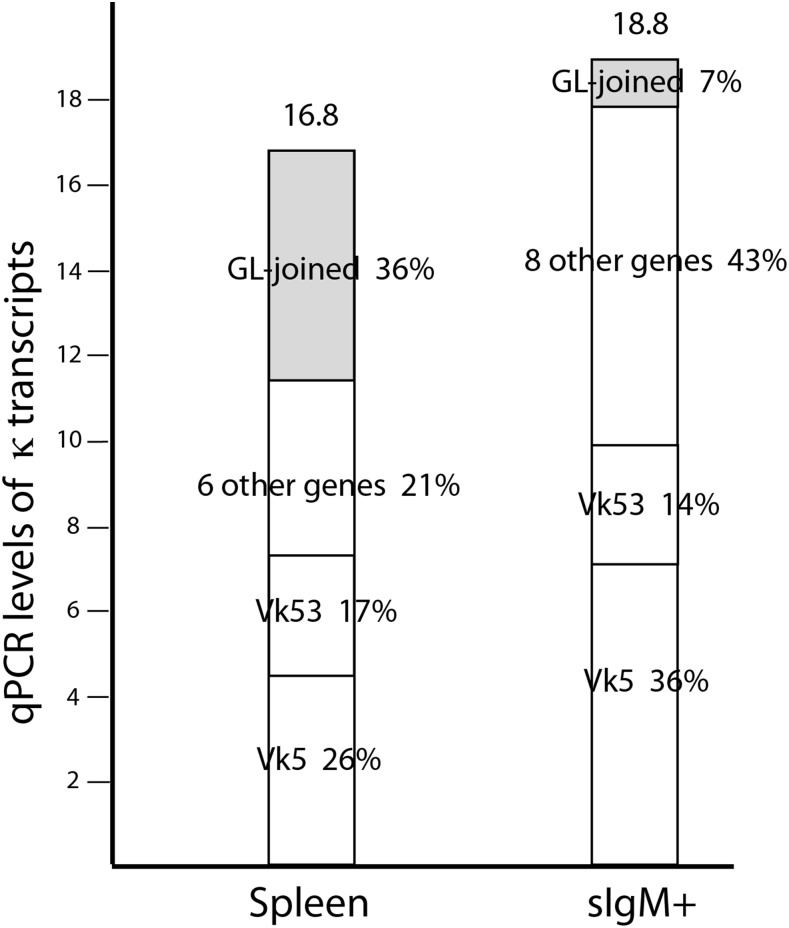
Ig genes that do not require RAG
Fig. 6 compares levels of κ gene expression between total spleen and sIgM+ mRNA, showing that most of the GL-joined κ mRNA is not found in sIgM+ cells (or sIgκ+, Table IV), although usage of the recombining κ genes is similar in the samples.
FIGURE 6.
Reduction of GL-joined κ gene mRNA in sIg+ cells. κ Sequences from the Pup1 Mplus sample were amplified by RT-PCR, cloned, and the gene cluster was identified. The Mplus qPCR levels and expressed genes are compared with Pup1 spleen results (left) from a previous study (30).
The GL-joined κ are largely produced in a cell type that does not express IgM or κ on its surface. The latter result also eliminated IgW+ cells as a source because the mAb LK14 coprecipitates IgW (33). If these GL-joined κ-expressing plasma cells had derived from B cells, their participation in BCR had particularly elicited activation to secretion. This seems unlikely, as the GL-joined κ clusters vary among individual sharks (Table I, footnote b).
Another possibility is that the GL-joined κ combine with H chain encoded by a unique cluster with GL-joined VDJ, called M1gj, particularly as LK14 recognizes the secreted molecule, IgM1gj (33). Secreting cells containing this H chain have been detected in neonates (50). A lymphoid cell type that produces B lineage factors including for Ig gene activation but excluding RAG recombinase could express M1gj with GL-joined L chains. The GL-joined κ are prominent in neonates but decline thereafter (30, 34), similar to IgM1gj expression, which decreases with age inversely to IgM and IgW (33). In fact, C.D. Castro and M. Flajnik (manuscript in preparation) have found association of M1gj H chain with GL-joined L chains, and they cannot find evidence for an M1gj H chain transmembrane form.
In clearnose skate, Litman and coworkers (43) observed a widespread IgH cluster activation, including the GL-joined VDJ that constitute 15% of IgH genes, in embryos and hatchlings. Such Ig would be limited in diversity, but they proposed that Ig cluster firing enabled protective Ab early in ontogeny. In nurse shark early Ig gene expression is specifically dominated by M1gj and at least GL-joined κ. The IgM1gj are limited in diversity and unable to trigger a signal to proliferate, characteristics directly in conflict with Burnet’s conception of Ab-producing cells: it is more like an innate immune component. Its function is unknown but the H and L chain combinations have undoubtedly undergone selection during evolution. This “backward” step from adaptive immune genes demonstrates the flexibility of the Ig gene cluster system for generating de novo immune molecules.
Acknowledgments
We thank Nethania Abraham and Kamala Anumukonda for their help with qPCR.
This work was supported in part by funding from National Institutes of Health Grants R01 GM068095 (to E.H.) and R01 DO0549-25 (to M.F.).
- Ct
- cycle threshold
- FR
- framework region
- FT
- flowthrough
- GL
- germline
- NDK
- nucleoside diphosphate kinase
- qPCR
- quantitative PCR
- R
- replacement substitution
- RSS
- recombination signal sequence
- S
- synonymous substitution
- SHM
- somatic hypermutation
- sIg
- surface Ig
- ST
- sterile transcript.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Burnet M. 1959. The Clonal Selection Theory of Acquired Immunity. Cambridge University Press, London. [Google Scholar]
- 2.Korsmeyer S. J., Hieter P. A., Sharrow S. O., Goldman C. K., Leder P., Waldmann T. A. 1982. Normal human B cells display ordered light chain gene rearrangements and deletions. J. Exp. Med. 156: 975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck K., Peak M. M., Ota T., Nemazee D., Murre C. 2009. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J. Exp. Med. 206: 2271–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derudder E., Cadera E. J., Vahl J. C., Wang J., Fox C. J., Zha S., van Loo G., Pasparakis M., Schlissel M. S., Schmidt-Supprian M., Rajewsky K. 2009. Development of immunoglobulin λ-chain-positive B cells, but not editing of immunoglobulin κ-chain, depends on NF-κB signals. Nat. Immunol. 10: 647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tze L. E., Schram B. R., Lam K. P., Hogquist K. A., Hippen K. L., Liu J., Shinton S. A., Otipoby K. L., Rodine P. R., Vegoe A. L., et al. 2005. Basal immunoglobulin signaling actively maintains developmental stage in immature B cells. PLoS Biol. 3: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannish G., Fuentes-Pananá E. M., Cambier J. C., Pear W. S., Monroe J. G. 2001. Ligand-independent signaling functions for the B lymphocyte antigen receptor and their role in positive selection during B lymphopoiesis. J. Exp. Med. 194: 1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gay D., Saunders T., Camper S., Weigert M. 1993. Receptor editing: an approach by autoreactive B cells to escape tolerance. J. Exp. Med. 177: 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiegs S. L., Russell D. M., Nemazee D. 1993. Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 177: 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X., Linden M., Van Ness B. 2000. Induced κ receptor editing shows no allelic preference in a mouse pre-B cell line. J. Immunol. 165: 7058–7063. [DOI] [PubMed] [Google Scholar]
- 10.Moore M. W., Durdik J., Persiani D. M., Selsing E. 1985. Deletions of kappa chain constant region genes in mouse lambda chain-producing B cells involve intrachromosomal DNA recombinations similar to V-J joining. Proc. Natl. Acad. Sci. USA 82: 6211–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siminovitch K. A., Bakhshi A., Goldman P., Korsmeyer S. J. 1985. A uniform deleting element mediates the loss of kappa genes in human B cells. Nature 316: 260–262. [DOI] [PubMed] [Google Scholar]
- 12.Nadel B., Cazenave P. A., Sanchez P. 1990. Murine lambda gene rearrangements: the stochastic model prevails over the ordered model. EMBO J. 9: 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nemazee D. 2017. Mechanisms of central tolerance for B cells. Nat. Rev. Immunol. 17: 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S. S., Greenberg A., Hsu E. 2000. Evolution and somatic diversification of immunoglobulin light chains. Curr. Top. Microbiol. Immunol. 248: 285–300. [DOI] [PubMed] [Google Scholar]
- 15.Bengtén E., Wilson M. 2015. Antibody repertoires in fish. In Pathogen-Host Interactions: Antigenic Variation v. Somatic Adaptations. Hsu E., Du Pasquier L., eds. Springer International Publishing, Cham, Switzerland, p. 193–234. [Google Scholar]
- 16.Criscitiello M. F., Flajnik M. F. 2007. Four primordial immunoglobulin light chain isotypes, including λ and κ, identified in the most primitive living jawed vertebrates. Eur. J. Immunol. 37: 2683–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eason D. D., Litman R. T., Luer C. A., Kerr W., Litman G. W. 2004. Expression of individual immunoglobulin genes occurs in an unusual system consisting of multiple independent loci. Eur. J. Immunol. 34: 2551–2558. [DOI] [PubMed] [Google Scholar]
- 18.Nemazee D. 1996. Antigen receptor “capacity” and the sensitivity of self-tolerance. Immunol. Today 17: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin T., Ren L., Hu X., Guo Y., Fei J., Zhu Q., Butler J. E., Wu C., Li N., Hammarstrom L., Zhao Y. 2008. Genomic organization of the immunoglobulin light chain gene loci in Xenopus tropicalis: evolutionary implications. Dev. Comp. Immunol. 32: 156–165. [DOI] [PubMed] [Google Scholar]
- 20.Ji Y., Desravines S., Hsu E. 1999. Junctional diversity in Xenopus immunoglobulin light chains. Mol. Immunol. 36: 1159–1168. [DOI] [PubMed] [Google Scholar]
- 21.Bengtén E., Strömberg S., Daggfeldt A., Magor B. G., Pilström L. 2000. Transcriptional enhancers of immunoglobulin light chain genes in Atlantic cod (Gadus morhua). Immunogenetics 51: 647–658. [DOI] [PubMed] [Google Scholar]
- 22.Hsu E., Criscitiello M. F. 2006. Diverse immunoglobulin light chain organizations in fish retain potential to revise B cell receptor specificities. J. Immunol. 177: 2452–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litman G. W., Anderson M. K., Rast J. P. 1999. Evolution of antigen binding receptors. Annu. Rev. Immunol. 17: 109–147. [DOI] [PubMed] [Google Scholar]
- 24.Venkatesh B., Lee A. P., Ravi V., Maurya A. K., Lian M. M., Swann J. B., Ohta Y., Flajnik M. F., Sutoh Y., Kasahara M., et al. 2014. Elephant shark genome provides unique insights into gnathostome evolution. Nature 505: 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rast J. P., Anderson M. K., Strong S. J., Luer C., Litman R. T., Litman G. W. 1997. α, β, γ, and δ T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity 6: 1–11. [DOI] [PubMed] [Google Scholar]
- 26.Chen H., Kshirsagar S., Jensen I., Lau K., Covarrubias R., Schluter S. F., Marchalonis J. J. 2009. Characterization of arrangement and expression of the T cell receptor gamma locus in the sandbar shark. Proc. Natl. Acad. Sci. USA 106: 8591–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Criscitiello M. F., Ohta Y., Saltis M., McKinney E. C., Flajnik M. F. 2010. Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J. Immunol. 184: 6950–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shamblott M. J., Litman G. W. 1989. Genomic organization and sequences of immunoglobulin light chain genes in a primitive vertebrate suggest coevolution of immunoglobulin gene organization. EMBO J. 8: 3733–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Criscitiello M. F. 2015. Shark immunoglobulin light chains. In Immunobiology of the Shark. Smith S. L., Sim R., Flajnik M. F., eds. CRC Press, Boca Raton, FL, p. 221–235. [Google Scholar]
- 30.Iacoangeli A., Lui A., Naik U., Ohta Y., Flajnik M., Hsu E. 2015. Biased immunoglobulin light chain gene expression in the nurse shark. J. Immunol. 195: 3992–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malecek K., Lee V., Feng W., Huang J. L., Flajnik M. F., Ohta Y., Hsu E. 2008. Immunoglobulin heavy chain exclusion in the shark. PLoS Biol. 6: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu C., Feng W., Weedon J., Hua P., Stepanov D., Ohta Y., Flajnik M. F., Hsu E. 2011. The multiple shark immunoglobulin heavy chain genes rearrange and hypermutate autonomously. J. Immunol. 187: 2492–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rumfelt L. L., Avila D., Diaz M., Bartl S., McKinney E. C., Flajnik M. F. 2001. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc. Natl. Acad. Sci. USA 98: 1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S. S., Fitch D., Flajnik M. F., Hsu E. 2000. Rearrangement of immunoglobulin genes in shark germ cells. J. Exp. Med. 191: 1637–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenberg, A. S. 1994. Evolution of the antigen receptor family. Doctoral dissertation, University of Miami, Miami, FL. [Google Scholar]
- 36.Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 37.Abarrategui I., Krangel M. S. 2009. Germline transcription: a key regulator of accessibility and recombination. Adv. Exp. Med. Biol. 650: 93–102. [DOI] [PubMed] [Google Scholar]
- 38.Chemin G., Tinguely A., Sirac C., Lechouane F., Duchez S., Cogné M., Delpy L. 2010. Multiple RNA surveillance mechanisms cooperate to reduce the amount of nonfunctional Igκ transcripts. J. Immunol. 184: 5009–5017. [DOI] [PubMed] [Google Scholar]
- 39.Diaz M., Velez J., Singh M., Cerny J., Flajnik M. F. 1999. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: the translesion synthesis model of somatic hypermutation. Int. Immunol. 11: 825–833. [DOI] [PubMed] [Google Scholar]
- 40.Lee S. S., Tranchina D., Ohta Y., Flajnik M. F., Hsu E. 2002. Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity 16: 571–582. [DOI] [PubMed] [Google Scholar]
- 41.Fleurant M., Changchien L., Chen C.-T., Flajnik M. F., Hsu E. 2004. Shark Ig light chain junctions are as diverse as in heavy chains. J. Immunol. 173: 5574–5582. [DOI] [PubMed] [Google Scholar]
- 42.Rast J. P., Anderson M. K., Ota T., Litman R. T., Margittai M., Shamblott M. J., Litman G. W. 1994. Immunoglobulin light chain class multiplicity and alternative organizational forms in early vertebrate phylogeny. Immunogenetics 40: 83–99. [DOI] [PubMed] [Google Scholar]
- 43.Miracle A. L., Anderson M. K., Litman R. T., Walsh C. J., Luer C. A., Rothenberg E. V., Litman G. W. 2001. Complex expression patterns of lymphocyte-specific genes during the development of cartilaginous fish implicate unique lymphoid tissues in generating an immune repertoire. Int. Immunol. 13: 567–580. [DOI] [PubMed] [Google Scholar]
- 44.Chen J., Trounstine M., Kurahara C., Young F., Kuo C. C., Xu Y., Loring J. F., Alt F. W., Huszar D. 1993. B cell development in mice that lack one or both immunoglobulin kappa light chain genes. EMBO J. 12: 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Velez M. G., Kane M., Liu S., Gauld S. B., Cambier J. C., Torres R. M., Pelanda R. 2007. Ig allotypic inclusion does not prevent B cell development or response. J. Immunol. 179: 1049–1057. [DOI] [PubMed] [Google Scholar]
- 46.Casellas R., Zhang Q., Zheng N. Y., Mathias M. D., Smith K., Wilson P. C. 2007. Igκ allelic inclusion is a consequence of receptor editing. J. Exp. Med. 204: 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalinina O., Doyle-Cooper C. M., Miksanek J., Meng W., Prak E. L., Weigert M. G. 2011. Alternative mechanisms of receptor editing in autoreactive B cells. Proc. Natl. Acad. Sci. USA 108: 7125–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giachino C., Padovan E., Lanzavecchia A. 1995. κ+λ+ Dual receptor B cells are present in the human peripheral repertoire. J. Exp. Med. 181: 1245–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rezanka L. J., Kenny J. J., Longo D. L. 2005. Dual isotype expressing B cells [κ(+)/λ(+)] arise during the ontogeny of B cells in the bone marrow of normal nontransgenic mice. Cell. Immunol. 238: 38–48. [DOI] [PubMed] [Google Scholar]
- 50.Rumfelt L. L., McKinney E. C., Taylor E., Flajnik M. F. 2002. The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand. J. Immunol. 56: 130–148. [DOI] [PubMed] [Google Scholar]