Abstract
Objective
IBS is a common gut disorder of uncertain pathogenesis. Among other factors, genetics and certain foods are proposed to contribute. Congenital sucrase–isomaltase deficiency (CSID) is a rare genetic form of disaccharide malabsorption characterised by diarrhoea, abdominal pain and bloating, which are features common to IBS. We tested sucrase–isomaltase (SI) gene variants for their potential relevance in IBS.
Design
We sequenced SI exons in seven familial cases, and screened four CSID mutations (p.Val557Gly, p.Gly1073Asp, p.Arg1124Ter and p.Phe1745Cys) and a common SI coding polymorphism (p.Val15Phe) in a multicentre cohort of 1887 cases and controls. We studied the effect of the 15Val to 15Phe substitution on SI function in vitro. We analysed p.Val15Phe genotype in relation to IBS status, stool frequency and faecal microbiota composition in 250 individuals from the general population.
Results
CSID mutations were more common in patients than asymptomatic controls (p=0.074; OR=1.84) and Exome Aggregation Consortium reference sequenced individuals (p=0.020; OR=1.57). 15Phe was detected in 6/7 sequenced familial cases, and increased IBS risk in case–control and population-based cohorts, with best evidence for diarrhoea phenotypes (combined p=0.00012; OR=1.36). In the population-based sample, 15Phe allele dosage correlated with stool frequency (p=0.026) and Parabacteroides faecal microbiota abundance (p=0.0024). The SI protein with 15Phe exhibited 35% reduced enzymatic activity in vitro compared with 15Val (p<0.05).
Conclusions
SI gene variants coding for disaccharidases with defective or reduced enzymatic activity predispose to IBS. This may help the identification of individuals at risk, and contribute to personalising treatment options in a subset of patients.
Keywords: IRRITABLE BOWEL SYNDROME, GENETICS, POLYMORPHIC VARIATION, DIARRHOEA
Significance of this study.
What is already known on this subject?
IBS shows genetic predisposition, but specific causative genes have not been unequivocally identified.
Certain foods, particularly carbohydrates, are among the proposed triggers of IBS symptoms, at least in some patients.
The sucrase–isomaltase (SI) gene, which is mutated in hereditary recessive forms of sucrose intolerance (congenital sucrase–isomaltase deficiency (CSID)) characterised by diarrhoea, represents an excellent candidate to play a role in IBS predisposition.
What are the new findings?
Although rare, CSID mutations with known defective disaccharidase (SI) properties are found more often in patients with IBS than controls.
A common SI variant (15Phe), which shows reduced enzymatic activity in vitro, is strongly associated with increased risk of IBS.
How might it impact on clinical practice in the foreseeable future?
Screening for functional SI genetic variants may help the identification of subsets of patients with suboptimal carbohydrate (disaccharide) digestion rates.
This holds potential for stratifying patients with IBS and personalising treatment options in those with SI genetic defects.
Introduction
IBS is the most common gut disorder, affecting more than 10% of the general population in Westernised countries; it is associated with significant healthcare expenditure and considerably affects patients' quality of life.1 2 IBS is a functional GI disorder (FGID), diagnosed and classified according to expert consensus guidelines, the Rome criteria, based on recurrent symptoms including abdominal discomfort or pain associated with diarrhoea (IBS-D), constipation (IBS-C) or mixed symptoms (IBS-M).3 The aetiology of IBS is unknown, although psychological stressors, prior infections, dietary irritants, gut dysbiosis, epithelial barrier dysfunction and mucosal immune activation are among the recognised risk factors.4 Genetic predisposition has been demonstrated in classical family/twin studies and epidemiological surveys, but unequivocal susceptibility genes have yet to be identified.5 Because of incomplete understanding of the mechanisms underpinning IBS pathophysiology, effective treatment options are limited and primarily aimed at targeting symptoms, resulting in suboptimal efficacy.
The role of nutrition and dietary factors is increasingly recognised in IBS. Patients (particularly IBS-D) often report postprandial symptoms,6 and many IBS sufferers claim that certain foods are the triggering factors.7 Avoidance of carbohydrates due to perceived maldigestion is common, and a diet low in fermentable oligosaccharides, disaccharides, monosaccharides and polyols (FODMAPs, which are poorly absorbed in the small intestine) has been proposed as effective in reducing IBS symptoms.8 9 At least in some patients, the food–symptom relation may involve malabsorption of carbohydrates due to inefficient enzymatic breakdown of polysaccharides, which may find indirect support in the observed symptom improvement in patients with postprandial IBS-D treated with pancrealipase,10 and the detection of disaccharidase deficiency in children with FGID.11
Sucrase–isomaltase (SI) deficiency (also called sucrose intolerance) is a form of carbohydrate malabsorption characterised by diarrhoea, abdominal pain and bloating, which are features common to IBS-D. The symptoms result from defective glucosidase (disaccharidase) activity of the SI enzyme in the small intestine.12 This enzyme is key to the degradation of starch and sugars digested daily,13 and its functional impairment leads to colonic accumulation of unabsorbed carbohydrates causing osmotic diarrhoea. At the same time, this induces a shift in gut microbiota-associated activities of carbohydrate metabolism and fermentation, with increased production of short-chain fatty acids and gases, which contribute to symptom generation. In the congenital form of SI deficiency (congenital sucrase–isomaltase deficiency (CSID)), patients harbour two defective copies of the SI gene due to recessive homozygous or compound heterozygous mutations that abolish or dramatically reduce enzymatic activity.14 15 CSID usually manifests early in life, but the phenotype and severity of symptoms can vary depending on the specific nature and position (sucrase or isomaltase domain) of different SI mutations and their homozygous or heterozygous combinations.16 In addition, adult patients, previously misdiagnosed with IBS, have been described.17 18 Overall, this speaks for a potentially higher clinical impact of SI genetic variation than that based solely on the detection of rare homozygous mutations in patients with CSID, evoking the hypothesis that SI functional polymorphisms may contribute also to IBS predisposition and symptom generation. If confirmed, this may have important implications in the management of IBS because of the potential for dietary intervention or enzyme supplementation in a subset of genetically exposed patients. To test this hypothesis, we performed a series of independent experiments: (1) sequencing of the entire SI coding region in seven familial cases of severe postprandial IBS-D; (2) detailed in vitro functional characterisation of a common coding variant leading to the amino acid change p.Val15Phe in the SI protein; (3) genotyping and association testing of the most common known CSID mutations,19 and the p.Val15Phe variant in 1887 individuals from four independent cohorts of IBS cases and controls; (4) in a small Swedish general population sample, pilot analyses of correlation between p.Val15Phe genotype and (i) IBS status (ii) stool frequency and (iii) faecal microbiota composition. The results obtained from these experiments support a role for SI genetic variation in IBS.
Materials and methods
Study subjects
IBS probands
Eight Caucasian individuals, namely seven postprandial IBS-D cases (Rome criteria) and one asymptomatic relative, from four unrelated families were selected for sequencing of the SI gene (see online supplementary figure S1). Two of the symptomatic patients were parents of two of the probands. Patients experienced meal-related symptoms for more than 2 years and were currently using digestive enzyme supplements to reduce postprandial symptoms (described more in detail in the online supplementary methods).
IBS case–control cohorts
We studied a total of 1031 IBS cases and 856 controls, all non-Hispanic/Latino whites from four independent cohorts from Sweden, Italy and USA, who have been described in detail elsewhere and already included in previous genetic studies.20–25 Their demographics and clinical characteristics are reported in table 1, and detailed information is provided in the online supplementary methods and table S1.
Table 1.
Demographics and clinical characteristics of study subjects
| Case–control cohorts | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sweden |
Italy |
US (Mayo) |
US (UCLA) |
Total |
||||||
| IBS | CTRL | IBS | CTRL | IBS | CTRL | IBS | CTRL | IBS | CTRL | |
| N | 387 | 355 | 319 | 255 | 187 | 114 | 138 | 132 | 1031 | 856 |
| Mean age, year | 42 | 42.5 | 39.6 | 34.8 | 47.9 | 46.6 | 35.0 | 33.5 | 41.4 | 39.4 |
| % F:M | 82:18 | 43:57 | 73:27 | 68:32 | 91:9 | 67:33 | 75:25 | 58:42 | 80:20 | 56:44 |
| IBS-D | 127 | 159 | 89 | 42 | 417 | |||||
| IBS-C | 95 | 128 | 56 | 45 | 324 | |||||
| IBS-M | 162 | 32 | 41 | 34 | 269 | |||||
| IBS-U | 3 | 0 | 1 | 17 | 321 | |||||
| PopCol | Total | IBS* | CTRL* | Microbiota | Diary | |||||
| N | 250 | 30 | 163 | 136 | 133 | |||||
| Mean age, year | 53.6 | 51.6 | 54.7 | 54.8 | 54.7 | |||||
| % F:M | 64:36 | 60:40 | 59:41 | 62:38 | 65:35 | |||||
| IBS-D | 11 | |||||||||
| IBS-C | 5 | |||||||||
| IBS-M | 9 | |||||||||
| IBS-U | 5 | |||||||||
*IBS cases (IBS) and asymptomatic controls (CTRL) were identified based on questionnaire data according to Rome criteria.
Diary, subjects with available daily recordings of defaecation patterns used for correlation analysis with the Val15Phe variant and stool frequency; Microbiota, subjects with available faecal 16S sequencing data; IBS-C, constipation-predominant IBS; IBS-D, diarrhoea-predominant IBS; IBS-M, IBS mixed phenotype; IBS-U, unclassified IBS; PopCol, Population-based Colonoscopy Study; UCLA, University of California, Los Angeles.
PopCol participants
The Population-based Colonoscopy study (PopCol) is a cohort representative of the general population from Stockholm, Sweden, which includes a data-rich set of individuals with available information from bowel symptom questionnaires, 1-week and 2-week bowel pattern diaries, clinical records from gastroenterology visits, blood and stool samples for genetic and microbiota analyses, and histology from biopsies obtained at ileocolonoscopy (table 1, online supplementary table S1 and online supplementary methods).26 27
gutjnl-2016-312456supp001.pdf (784.4KB, pdf)
All study participants provided informed consent, and the study protocols were approved by local ethics committees.
Sequencing (SI and 16S) and genotyping
Sequencing of the SI coding region was performed as described previously28 and in the online supplementary methods. For the microbiota studies, faecal bacterial DNA was extracted from each sample and 16S rRNA gene amplicon (V1–V2 region) sequencing was performed on the MiSeq platform (Illumina, USA). Read coverage was normalised to 10 000 read per sample, and taxonomic classification was performed as described in the online supplementary methods. Targeted genotyping of known CSID mutations and the p.Val15Phe rs9290264 single nucleotide polymorphism (SNP) in patients with IBS and controls was carried out using the iPLEX chemistry on the MassARRAY platform (Sequenom, USA), while p.Val15Phe PopCol genotypes were extracted from available HumanOmniExpressExome data (Illumina, USA).
Measurement of SI enzymatic activity in vitro
Functional characterisation of the polymorphism p.Val15Phe was obtained in a well-established SI model system,29 by transiently transfecting monkey fibroblast COS-1 cells with cDNA vectors encoding the 15Val and 15Phe variants. Sucrase activity was measured using sucrose as a substrate as previously described.30 Briefly, immunoprecipitates from 15Val or 15Phe transfected cells were incubated with sucrose (28 mmol/L) for 1 hour at 37°C, and the amount of glucose generated from sucrose hydrolysis was detected using the glucose oxidation-phenol aminopyrine (GOD-PAP) monoreagent method (Axiom GmbH, Germany). To control for transfection efficiency, 15Val and 15Phe activities were normalised relative to the specific SI protein amounts, as measured by immunoblot analysis of the corresponding immunoprecipitates. A detailed description of the protocols adopted for the full in vitro functional characterisation of the p.Val15Phe polymorphism is reported in the online supplementary methods.
In silico and statistical analyses
PHRED-like scores from the Combined Annotation-Dependent Depletion (CADD) database V.1.3 were used for predicting functional effects of known CSID mutations and SI common coding polymorphisms (see online supplementary methods).31 One-sided (testing predisposing risk effects) association analysis of CSID mutations carriage was performed using 2×2 contingency table statistics (Fisher's exact test and χ2 as appropriate) in IBS cases versus (i) controls and (ii) reference genotypes from publicly available large-scale sequence data for individuals of European descent (http://exac.broadinstitute.org/).19 Association with the p.Val15Phe variant was tested on pooled data using logistic regression under a (best powered) additive genetic model adjusting for sex and batch (recruitment centre). A meta-analysis of results from individual cohorts was also carried out to verify the absence of statistical heterogeneity (see online supplementary methods). Spearman's correlation was used to assess the relationship between 15Phe copy number, stool frequency and gut microbiota composition in the PopCol cohort. Functional differences in 15Val and 15Phe SI protein properties in in vitro experiments were evaluated using Student's t-tests. More detailed descriptions of the statistical procedures are reported in the online supplementary methods.
Results
Sequencing of the SI gene in IBS families
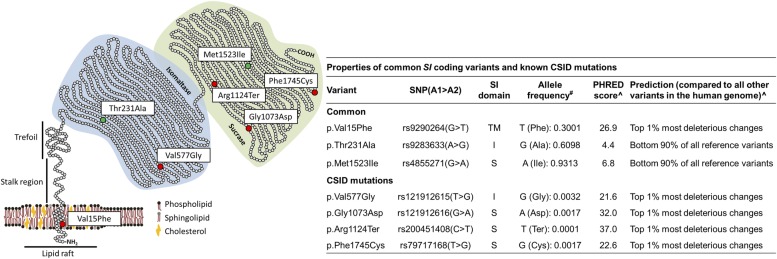
Seven familial cases and one asymptomatic relative (control) from four IBS-D families were included in the SI sequencing effort (see online supplementary figure S1 and online supplementary methods), aiming to identify SI mutations or variants of potential relevance to IBS. No new variants were detected in this experiment (data not shown), and all sequenced family members were homozygous for SI reference coding alleles, except at two sites, namely missense SNPs rs9290264 (p.Val15Phe) and rs9283633 (p.Thr231Ala) (see online supplementary figure S1). Together with rs4855271 (p.Met1523Ile), rs9290264 and rs9283633 are the only common (minor allele frequency >0.01) missense SNPs in the SI gene (http://exac.broadinstitute.org/gene/ENSG00000090402 and figure 1). No conclusive experimental data exist on the potential functional effects of these amino acid changes on SI properties. However, opposite to p.Thr231Ala and p.Met1523Ile that result in benign changes, computational predictions at the CADD database (see online supplementary methods) rank p.Val15Phe among the top 1% most deleterious amino acid substitutions in the human genome,31 similar to known CSID mutations (figure 1). Although also common in the general population (allele frequency=0.3), the 15Phe was present in 6/7 (five heterozygous and one homozygous) sequenced affected family members, and cosegregated with IBS in the two families where parental SI sequence data were also available (see online supplementary figure S1). Based on the above information, we set to undertake a functional in vitro characterisation of the 15Phe coding allele.
Figure 1.
Properties of sucrase–isomaltase (SI) mutants and common coding polymorphisms. Left: schematic representation of SI protein structure and functional domains; the position of congenital sucrase–isomaltase deficiency (CSID) mutations and common coding variants is reported and colour-coded according to their functional effects (red=damaging, green=benign). Right: the variants, corresponding dbSNP IDs (http://www.ncbi.nlm.nih.gov/SNP), SI protein domain location (TM, transmembrane; I, isomaltase; S, sucrase), allele frequency, PHRED-like score (range 1–99, ranking a variant relative to all possible substitutions in the human genome) and predicted functional consequences are reported. #Exome Aggregation Consortium browser (http://exac.broadinstitute.org); ^Combined Annotation-Dependent Depletion database (http://cadd.gs.washington.edu/info). SNP, single nucleotide polymorphism.
Functional characterisation of the Phe15Val coding polymorphism
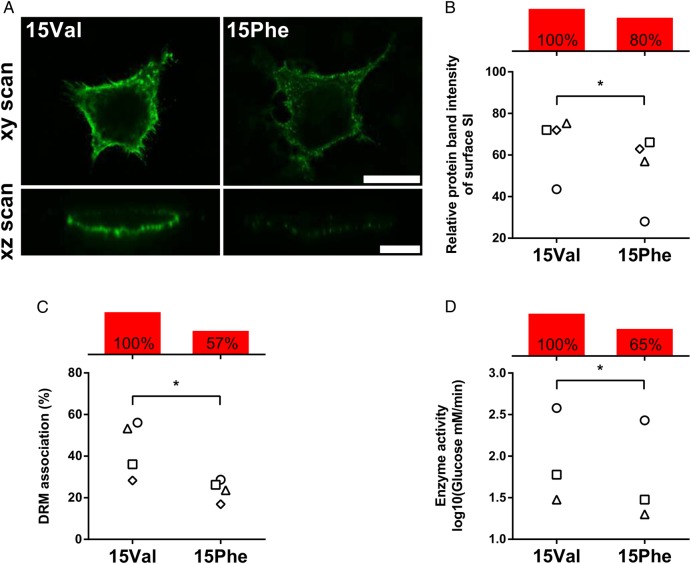
Adopting an established in vitro model previously used to study known CSID mutants, SI 15Val and 15Phe protein variants were individually expressed in COS-1 cells and tested for potential differences in their functional properties. The patterns of glycosylation and trypsin digestion, as well as the intracellular distribution and trafficking kinetics, appeared to be similar for the two variants (not shown). However, when tested for their relative cell surface expression, which requires association with lipid rafts through the stalk region adjacent to residue 15 where the 15Phe substitution occurs, the two variants' behaviour was consistently different under a series of experimental conditions, and the 15Phe variant showed a 20% reduction of cell surface localisation compared with 15Val (figure 2A–C). Of note, the 15Phe SI variant had an overall 35% reduction of enzyme activity compared with 15Val, after normalisation and quantification of immunoprecipitated proteins from COS-1 transfected cells (p<0.05; figure 2D).
Figure 2.
Functional characterisation of the p.Val15Phe coding polymorphism. COS-1 cells were transiently transfected with either 15Val or 15Phe cDNAs and studied 48 hours after transfection. Individual values for 15Val and 15Phe cells from the same experiment are indicated with identical symbols. Net differences are reported with red bars as per cent average relative to 15Val arbitrarily set as 100% reference. (A) Cell surface localisation via immunofluorescence. Non-permeabilised cells were immunostained with a mixture of anti-sucrase–isomaltase (SI) antibodies and Alexa 488 secondary antibody, and analysed by confocal laser scanning microscopy on the xy (scale bar 25 µm) and xz planes (scale bar 10 µm). (B) Quantification of cell surface expression. SI surface proteins were labelled with biotin and immunoprecipitated using anti-Si antibodies after cell lysis. Immunoprecipitates were divided into two equal aliquots and analysed by immunoblotting with either anti-SI or anti-streptavidin antibodies. Relative quantification of surface-bound SI versus total cell SI was performed, and results are expressed in relation to values obtained for 15Val, which is set to 100%. (C) Quantification of association with sphingolipid/cholesterol-rich microdomains (lipid rafts) via detergent-resistant membrane (DRM) analysis. Following non-ionic detergent cell lysis, SI proteins were immunoprecipitated, fractionated by ultracentrifugation into insoluble (pellet, raft) and soluble (supernatant, non-raft) fractions, and DRM association (raft) quantified by immunoblotting with anti-SI antibodies. (D) Quantification of enzymatic activity. Sucrase activity was determined on immunoprecipitated SI proteins by measuring glucose release with the GOD–PAP method, upon normalisation for total protein amount by immunoblotting. *p<0.05.
Association analysis of CSID mutations and the 15Phe variant in IBS cases and controls
A total of 1887 individuals from four independent IBS case–control cohorts were included in the analysis of the SI gene in IBS (table 1). The four most common known CSID mutations (p.Val557Gly, p.Gly1073Asp, p.Arg1124Ter and p.Phe1745Cys) were genotyped in these cohorts and tested for their potential to confer IBS risk. Twenty-two heterozygous IBS cases and ten heterozygous controls were identified, consistent with a trend for patients with IBS to have nearly two times higher odds of carrying a CSID mutation than asymptomatic controls (p=0.074, OR=1.84; table 2). This finding (statistically weak due to the control sample size likely inadequate to test rare variants) could be consolidated by inspecting public data from a reference panel of >30 000 sequenced European individuals from the Exome Aggregation Consortium. As shown in table 2, significant results were obtained both for p.Val577Gly and a cumulative analysis of CSID mutations (p=0.0029, OR=2.11 and p=0.020, OR=1.57, respectively). The distribution of mutations into different IBS subtypes was even for p.Val557Gly, which maps to the isomaltase domain of the SI protein, though the other mutations affecting the sucrase domain (p.Gly1073Asp, p.Arg1124Ter and p.Phe1745Cys; figure 1) were found only in patients with IBS-D and IBS-M (table 2). The IBS predisposing potential of the 15Phe variant was also tested by genotyping the entire sample for the rs9290264 (Val15Phe) SNP. As shown in table 3, 15Phe was significantly associated with increased risk of IBS in a logistic regression analysis of the combined cohorts under an additive genetic model adjusted for sex and recruitment centre (p=0.0030, OR=1.26). Similar findings (associations and effect sizes) were obtained using a dominant model, while recessive association testing yielded negative results (data not shown). Of note, the strength of association appeared to vary in different IBS subtypes, as 15Phe significantly affected disease risk only in the subtypes characterised by diarrhoea, and the D and M subtypes combined provided the best evidence of association and stronger genetic risk effects (p=0.00051, OR=1.34 for the IBS-D/IBS-M; table 3). Similar IBS genetic risk effects were observed across all four datasets, and a meta-analysis of associations from individual cohorts yielded results identical to the pooled analyses and devoid of statistical heterogeneity (Cochran's Q range 0.55–0.97 and heterogeneity index I2=0 for all IBS traits; online supplementary figure S2).
Table 2.
Association of known CSID mutations with IBS
| IBS | Subtypes | CTRLS | p Value* | OR | ExAC | p Value* | OR | |
|---|---|---|---|---|---|---|---|---|
| N=1031 | N=856 | N=31 402 | ||||||
| p.Val577Gly | 14 | 4 IBS-D, 5 IBS-M, 5 IBS-C | 6 | 0.12 | 1.95 | 203 | 0.0029 | 2.11 |
| p.Gly1073Asp | 3 | 3 IBS-D | 3 | 0.57 | 0.83 | 109 | 0.42 | 0.89 |
| p.Arg1124Ter | 1 | 1 IBS-D | 0 | – | – | 8 | 0.077 | 4.04 |
| p.Phe1745Cys | 4 | 1 IBS-D, 2 IBS-M, 1 IBS-U | 1 | 0.25 | 3.33 | 110 | 0.42 | 1.11 |
| Any mutation | 22 | 9 IBS-D, 7 IBS-M, 5 IBS-C, 1 IBS-U | 10 | 0.074 | 1.84 | 430 | 0.020 | 1.57 |
*p Value for carriage of SI mutations in IBS cases (IBS) versus controls (CTRLS) and versus ExAC-sequenced individuals of European descent (ExAC).
CSID, Congenital sucrase–isomaltase deficiency; ExAC, Exome Aggregation Consortium; SI, sucrase–isomaltase; IBS-C, constipation-predominant IBS; IBS-D, diarrhoea-predominant IBS; IBS-M, IBS mixed phenotype; IBS-U, unclassified IBS. Significant p Values (<0.05) highlighted in bold italics.
Table 3.
Association of the 15Phe variant with IBS
| CTRL | IBS |
IBS-C |
IBS-D |
IBS-M |
IBS-D/M |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AF | AF | p Value | OR | AF | p Value | OR | AF | p Value | OR | AF | p Value | OR | AF | p Value | OR | |
| Case–control | 0.264 | 0.306 | 0.0030* | 1.26 | 0.279 | 0.42 | 1.09 | 0.312 | 0.0082 | 1.30 | 0.329 | 0.0026* | 1.41 | 0.319 | 0.00051* | 1.34 |
| PopCol | 0.29 | 0.417 | 0.045 | 1.89 | 0.400 | 0.45 | 1.72 | 0.409 | 0.20 | 1.88 | 0.556 | 0.017 | 3.81 | 0.475 | 0.013 | 2.50 |
| Combined | 0.268 | 0.309 | 0.0013* | 1.27 | 0.281 | 0.39 | 1.10 | 0.314 | 0.0052 | 1.30 | 0.336 | 0.00057* | 1.46 | 0.323 | 0.00012* | 1.36 |
*p Value significant after Bonferroni correction for multiple comparisons (N=15), considering five traits (IBS, IBS-C, IBD-D, IBS-M and IBS-D/M) in three datasets (case–control cohorts, PopCol and both combined). Significant p Values (<0.05) highlighted in bold italics.
AF, 15Phe allele frequency; CTRL, controls; IBS-C, constipation-predominant IBS; IBS-D, diarrhoea-predominant IBS; IBS-D/M, IBS-D+IBS-M; IBS-M, IBS mixed phenotype; p Value, p Value from logistic regression adjusted for sex and batch (centre).
Analysis of the 15Phe variant in relation to bowel function and microbiota in the general population
A data-rich subset of 250 individuals from the PopCol cohort was studied to run pilot tests of the relevance of 15Phe in the general population, in relation to bowel function, symptoms and microbiota composition (table 1). Using Rome III criteria extracted from questionnaire data, 30 IBS and 163 symptom-free individuals were identified, and, despite small sample size, a significant association was detected between 15Phe and increased IBS risk, particularly IBS-M (p=0.017, OR=3.81; table 3). Combining these data with the case–control results further strengthened the evidence of association with IBS, which was strongest in patients with diarrhoea from the IBS-M and IBS-D subtypes (combined p=0.00012, OR=1.36; table 3). In 133 PopCol individuals who had daily recordings of defaecation patterns (see online supplementary methods), a significant correlation (p=0.026) was observed between the number of 15Phe copies and mean stool frequency, with homozygous carriers showing the highest number of bowel movements per day (figure 3). However, no significant findings were obtained from a similar analysis assessing the relationship with stool consistency (measured on the Bristol Stool Form Scale, not shown). Faecal microbiota data (16S rRNA gene sequencing) were available for 136 PopCol-genotyped individuals and used to test the potential relation between host p.Val15Phe genotype and gut bacterial community composition (see online supplementary methods). By testing the 20 most abundant genera (see online supplementary table S2), we detected Bonferroni-corrected significant inverse correlation between the number of 15Phe copies and the abundance of Parabacteroides (p=0.0024; online supplementary figure S3). This was independent of IBS status and bowel complaints, as the correlation was still significant when tested in the subset of 90 PopCol symptom-free individuals (data not shown).
Figure 3.
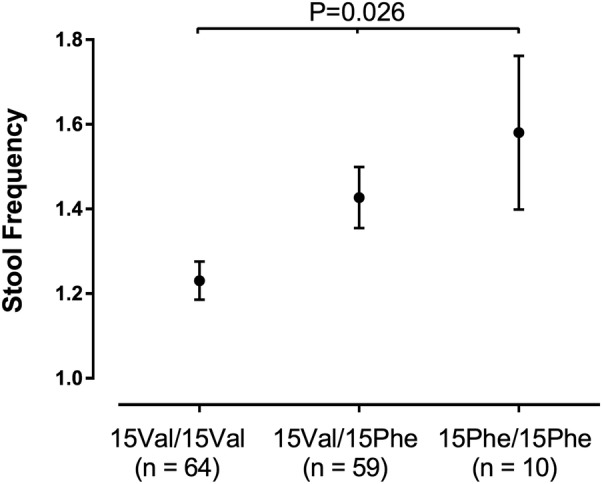
Correlation between p.Val15Phe genotype and stool frequency. Mean (±SD) number of bowel movements per day (stool frequency, y-axis) is reported for Population-based Colonoscopy Study individuals (with available diary data) stratified according to the genotype at the p.Val15Phe single nucleotide polymorphism site (x-axis). The Spearman's p value for the correlation test is also reported.
Discussion
Our studies show that genetic variation in the SI gene is associated with predisposition to IBS. We detected a twofold increased risk of IBS in heterozygous carriers of known rare CSID mutations. In addition, we detected genetic risk effects attributable to a common coding variant, 15Phe at SNP rs9290264, which cosegregated with IBS in some affected families, and was associated with increased disease odds in large multinational IBS case–control cohorts and a pilot general population sample. Although the current study only targeted a few selected functional variants, the SI gene harbours >700 other (mostly rare) coding SNPs. Hence, SI polymorphisms may influence IBS risk across different homozygous, heterozygous and compound heterozygous allelic combinations, and future resequencing and large-scale SI genotyping efforts may lead to the identification of additional IBS risk variants.
As previously demonstrated for CSID mutants with established SI defects, we also discovered that the 15Phe variant imparts deficient enzymatic properties, resulting in 35% reduced disaccharidase activity in vitro. Reduced carbohydrate degradation rates may result in altered concentrations of starch and sugar (sucrose) breakdown products across the intestine, possibly including colonic increase of undigested disaccharides not absorbed through the small intestine. In the large bowel, osmotic luminal water release, microbiota composition and bacterial fermentation with gas production are all potentially affected by these changes, with repercussions across diverse bowel functions and, ultimately, symptoms.12 32 Of interest, in the PopCol cohort, we observed a positive correlation between the number of 15Phe copies and defaecation frequency, which might be due to the effects on transit time mediated by similar mechanisms. Hence, genotype-dependent reduction of SI disaccharidase function appears to affect disease risk across a clinical spectrum ranging from severe monogenic CSID forms to milder complex IBS manifestations. The latter is particularly true for IBS phenotypes characterised by diarrhoea, since, similar to the effect of rare mutations in CSID, 15Phe shows strongest association with the IBS-M and IBS-D subtypes while no significant results were obtained for IBS-C. This notion is important because it may help identify a subgroup of patients with IBS with inherited predisposition to disaccharide maldigestion, which holds potential for personalised approaches to their clinical management.
Dietary carbohydrates are recognised triggers in many IBS sufferers, who often implement food avoidance regimes in the attempt to control or reduce their symptoms; thus, 52% of 1242 US patients with IBS who completed an IBS-Patient Education Questionnaire in 2007 also believed IBS is caused by lack of digestive enzymes.33 The results from our study provide a biological basis for these beliefs, by demonstrating potential mechanisms mediating the interplay between carbohydrate consumption and enzymatic (SI) defects in their digestion. At the same time, they also provide a rationale for future studies of bowel symptoms in patients with IBS stratified according to SI genotype in eventual trials of dietary exclusion of sucrose or enzyme supplementation. With increasing recognition of the importance of dietary factors in IBS, our results may contribute to inform FODMAP-based strategies for the treatment of IBS. For instance, it may be of high interest to further evaluate low-FODMAP diets or similarly effective traditional dietary intervention strategies34 for their efficacy in reducing IBS symptoms by also taking into account patients' SI genotype.
Finally, the results from our pilot study of PopCol microbiota composition in relation to Val15Phe genotype are interesting, because faecal microbiota concentration of Parabacteroides has been shown to decrease with higher dietary carbohydrate intake,35 and this genus was under-represented in patients with IBS, at least in some studies.36 37 Although unlikely of causative nature, the negative correlation between 15Phe allele dosage and Parabacteroides abundance may thus contribute to the identification and further stratification of patients with IBS with SI disturbances.
In conclusion, we report the first experimental evidence pointing to nutrigenetic mechanisms associated with IBS predisposition and symptom generation. This holds potential for stratifying patients with IBS and personalising treatment options in those with SI genetic defects.
Acknowledgments
The authors thank Professor Dr Hans-Peter Hauri (Biocenter, Basel, Switzerland) and Professor Dr Erwin Sterchi (University of Bern, Bern, Switzerland) for providing monoclonal anti-sucrase–isomaltase antibodies.
Footnotes
Contributors: HYN and MDA: study concept, design and supervision; MvK-B, CD, MHei, UP, OD, MEM, GN, RC, PU-S, FG, MN, SW, MS, PK, BO, PTS, GL, AD, LA, AA, EM, LE, PP, MB, VS, GB, LC and MC: characterisation of study individuals and data acquisition; LD, E-MK and HYN: functional in vitro experiments; FHa, LBT, MB, F-AH, JFB, AF and MDA: microbiota analyses; GA and MDA: genotyping; MHe, LD, FH, FB, TZ, MB, F-AH, JR, JFB, AF, HYN and MDA: data integration, analysis and interpretation; MHe, LD, HYN and MDA: drafting of the manuscript with input and critical revision from all other authors.
Funding: This work was supported by grants from the Swedish Research Council (Vetenskapsrådet), the Olle Engkvist Byggmästare Foundation and an unrestricted research grant from Medical Need Europe AB to MDA; the European Union Seventh Framework Programme (FP7/2007–2013, ESGI) to MDA and AF; the German Research Foundation (DFG) Research Training Grant 1743 and Excellence Cluster 306 to AF; the German Research Foundation DFG to HYN; the Soderbergs Foundation to LE; NIH grants P50 DK64539, P01 DK33506 and DK047343 to EM and LC.
Competing interests: The work was partially financed by an unrestricted grant from Medical Need Europe AB to MDA. MDA and HYN have received unrestricted research grants and lecturing honoraria from QOL Medical, and LC has served on a scientific advisory board for QOL Medical.
Ethics approval: Local ethics committees at Karolinska Institutet, Mayo Clinic, University of California, Los Angeles (UCLA), Bologna University, University of Veterinary Medicine Hannover.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Sperber AD, Dumitrascu D, Fukudo S, et al. . The global prevalence of IBS in adults remains elusive due to the heterogeneity of studies: a Rome Foundation working team literature review. Gut Published Online First: 27 Jan 2016. doi: 10.1136/gutjnl-2015-311240 10.1136/gutjnl-2015-311240 [DOI] [PubMed] [Google Scholar]
- 2. Chey WD, Kurlander J, Eswaran S. Irritable bowel syndrome: a clinical review. JAMA 2015;313:949–58. 10.1001/jama.2015.0954 [DOI] [PubMed] [Google Scholar]
- 3. Drossman DA. The functional gastrointestinal disorders and the Rome III process. Gastroenterology 2006;130:1377–90. 10.1053/j.gastro.2006.03.008 [DOI] [PubMed] [Google Scholar]
- 4. Camilleri M. Peripheral mechanisms in irritable bowel syndrome. N Engl J Med 2012;367:1626–35. 10.1056/NEJMra1207068 [DOI] [PubMed] [Google Scholar]
- 5. Gazouli M, Wouters MM, Kapur-Pojskić L, et al. . Lessons learned—resolving the enigma of genetic factors in IBS. Nat Rev Gastroenterol Hepatol 2016;13:77–87. 10.1038/nrgastro.2015.206 [DOI] [PubMed] [Google Scholar]
- 6. Ragnarsson G, Bodemar G. Pain is temporally related to eating but not to defaecation in the irritable bowel syndrome (IBS). Patients’ description of diarrhea, constipation and symptom variation during a prospective 6-week study. Eur J Gastroenterol Hepatol 1998;10:415–21. [DOI] [PubMed] [Google Scholar]
- 7. Böhn L, Störsrud S, Törnblom H, et al. . Self-reported food-related gastrointestinal symptoms in IBS are common and associated with more severe symptoms and reduced quality of life. Am J Gastroenterol 2013;108:634–41. 10.1038/ajg.2013.105 [DOI] [PubMed] [Google Scholar]
- 8. Gibson PR, Varney J, Malakar S, et al. . Food components and irritable bowel syndrome. Gastroenterology 2015;148:1158–74. 10.1053/j.gastro.2015.02.005 [DOI] [PubMed] [Google Scholar]
- 9. De Giorgio R, Volta U, Gibson PR. Sensitivity to wheat, gluten and FODMAPs in IBS: facts or fiction? Gut 2016;65:169–78. 10.1136/gutjnl-2015-309757 [DOI] [PubMed] [Google Scholar]
- 10. Money ME, Walkowiak J, Virgilio C, et al. . Pilot study: a randomised, double blind, placebo controlled trial of pancrealipase for the treatment of postprandial irritable bowel syndrome-diarrhoea. Frontline Gastroenterol 2011;2:48–56. 10.1136/fg.2010.002253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karnsakul W, Luginbuehl U, Hahn D, et al. . Disaccharidase activities in dyspeptic children: biochemical and molecular investigations of maltase-glucoamylase activity. J Pediatr Gastroenterol Nutr 2002;35:551–6. 10.1097/00005176-200210000-00017 [DOI] [PubMed] [Google Scholar]
- 12. Treem WR. Congenital sucrase-isomaltase deficiency. J Pediatr Gastroenterol Nutr 1995;21:1–14. 10.1097/00005176-199507000-00001 [DOI] [PubMed] [Google Scholar]
- 13. Diaz-Sotomayor M, Quezada-Calvillo R, Avery SE, et al. . Maltase-glucoamylase modulates gluconeogenesis and sucrase-isomaltase dominates starch digestion glucogenesis. J Pediatr Gastroenterol Nutr 2013;57:704–12. 10.1097/MPG.0b013e3182a27438 [DOI] [PubMed] [Google Scholar]
- 14. Alfalah M, Keiser M, Leeb T, et al. . Compound heterozygous mutations affect protein folding and function in patients with congenital sucrase-isomaltase deficiency. Gastroenterology 2009;136:883–92. 10.1053/j.gastro.2008.11.038 [DOI] [PubMed] [Google Scholar]
- 15. Naim HY, Heine M, Zimmer K-P. Congenital sucrase-isomaltase deficiency: heterogeneity of inheritance, trafficking, and function of an intestinal enzyme complex. J Pediatr Gastroenterol Nutr 2012;55(Suppl 2):S13–20. 10.1097/01.mpg.0000421402.57633.4b [DOI] [PubMed] [Google Scholar]
- 16. Ouwendijk J, Moolenaar CE, Peters WJ, et al. . Congenital sucrase-isomaltase deficiency. Identification of a glutamine to proline substitution that leads to a transport block of sucrase-isomaltase in a pre-Golgi compartment. J Clin Invest 1996;97:633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muldoon C, Maguire P, Gleeson F. Onset of sucrase-isomaltase deficiency in late adulthood. Am J Gastroenterol 1999;94:2298–9. 10.1111/j.1572-0241.1999.01320.x [DOI] [PubMed] [Google Scholar]
- 18. Ringrose RE, Preiser H, Welsh JD. Sucrase-isomaltase (palatinase) deficiency diagnosed during adulthood. Dig Dis Sci 1980;25:384–7. 10.1007/BF01308064 [DOI] [PubMed] [Google Scholar]
- 19. Uhrich S, Wu Z, Huang J-Y, et al. . Four mutations in the SI gene are responsible for the majority of clinical symptoms of CSID. J Pediatr Gastroenterol Nutr 2012;55(Suppl 2):S34–5. 10.1097/01.mpg.0000421408.65257.b5 [DOI] [PubMed] [Google Scholar]
- 20. Wouters MM, Lambrechts D, Knapp M, et al. . Genetic variants in CDC42 and NXPH1 as susceptibility factors for constipation and diarrhoea predominant irritable bowel syndrome. Gut 2014;63:1103–11. 10.1136/gutjnl-2013-304570 [DOI] [PubMed] [Google Scholar]
- 21. Zucchelli M, Camilleri M, Andreasson AN, et al. . Association of TNFSF15 polymorphism with irritable bowel syndrome. Gut 2011;60:1671–7. 10.1136/gut.2011.241877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ek WE, Reznichenko A, Ripke S, et al. . Exploring the genetics of irritable bowel syndrome: a GWA study in the general population and replication in multinational case-control cohorts. Gut 2015;64:1774–82. 10.1136/gutjnl-2014-307997 [DOI] [PubMed] [Google Scholar]
- 23. Beyder A, Mazzone A, Strege PR, et al. . Loss-of-function of the voltage-gated sodium channel NaV1.5 (channelopathies) in patients with irritable bowel syndrome. Gastroenterology 2014;146:1659–68. 10.1053/j.gastro.2014.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grasberger H, Chang L, Shih W, et al. . Identification of a functional TPH1 polymorphism associated with irritable bowel syndrome bowel habit subtypes. Am J Gastroenterol 2013;108:1766–74. 10.1038/ajg.2013.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Orand A, Gupta A, Shih W, et al. . Catecholaminergic gene polymorphisms are associated with gi symptoms and morphological brain changes in irritable bowel syndrome. PLoS ONE 2015;10:e0135910 10.1371/journal.pone.0135910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walter SA, Kjellström L, Nyhlin H, et al. . Assessment of normal bowel habits in the general adult population: the Popcol study. Scand J Gastroenterol 2010;45:556–66. 10.3109/00365520903551332 [DOI] [PubMed] [Google Scholar]
- 27. Kjellström L, Molinder H, Agréus L, et al. . A randomly selected population sample undergoing colonoscopy: prevalence of the irritable bowel syndrome and the impact of selection factors. Eur J Gastroenterol Hepatol 2014;26:268–75. 10.1097/MEG.0000000000000024 [DOI] [PubMed] [Google Scholar]
- 28. Sander P, Alfalah M, Keiser M, et al. . Novel mutations in the human sucrase-isomaltase gene (SI) that cause congenital carbohydrate malabsorption. Hum Mutat 2006;27:119 10.1002/humu.9392 [DOI] [PubMed] [Google Scholar]
- 29. Moolenaar CE, Naim HY. Cloning and expression of human intestinal sucrase-isomaltase. Biochem Soc Trans 1995;23:304S 10.1042/bst023304s [DOI] [PubMed] [Google Scholar]
- 30. Dahlqvist A. Assay of intestinal disaccharidases. Anal Biochem 1968;22:99–107. 10.1016/0003-2697(68)90263-7 [DOI] [PubMed] [Google Scholar]
- 31. Kircher M, Witten DM, Jain P, et al. . A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310–15. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chassard C, Lacroix C. Carbohydrates and the human gut microbiota. Curr Opin Clin Nutr Metab Care 2013;16:453–60. 10.1097/MCO.0b013e3283619e63 [DOI] [PubMed] [Google Scholar]
- 33. Halpert A, Dalton CB, Palsson O, et al. . What patients know about irritable bowel syndrome (IBS) and what they would like to know. National Survey on Patient Educational Needs in IBS and development and validation of the Patient Educational Needs Questionnaire (PEQ). Am J Gastroenterol 2007;102:1972–82. [DOI] [PubMed] [Google Scholar]
- 34. Bohn L, Storsrud S, Liljebo T, et al. . Diet low in FODMAPs reduces symptoms of irritable bowel syndrome as well as traditional dietary advice: a randomized controlled trial. Gastroenterology 2015;149:1399–407. 10.1053/j.gastro.2015.07.054 [DOI] [PubMed] [Google Scholar]
- 35. Wu GD, Chen J, Hoffmann C, et al. . Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–8. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Noor SO, Ridgway K, Scovell L, et al. . Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol 2010;10:134 10.1186/1471-230X-10-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rangel I, Sundin J, Fuentes S, et al. . The relationship between faecal-associated and mucosal-associated microbiota in irritable bowel syndrome patients and healthy subjects. Aliment Pharmacol Ther 2015;42:1211–21. 10.1111/apt.13399 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2016-312456supp001.pdf (784.4KB, pdf)