ABSTRACT
Polymicrobial interactions are complex and can influence the course of an infection, as is the case when two or more species exhibit a synergism that produces a disease state not seen with any of the individual species alone. Cell-to-cell signaling is key to many of these interactions, but little is understood about how the host environment influences polymicrobial interactions or signaling between bacteria. Chronic wounds are typically polymicrobial, with Staphylococcus aureus and Pseudomonas aeruginosa being the two most commonly isolated species. While P. aeruginosa readily kills S. aureus in vitro, the two species can coexist for long periods together in chronic wound infections. In this study, we investigated the ability of components of the wound environment to modulate interactions between P. aeruginosa and S. aureus. We demonstrate that P. aeruginosa quorum sensing is inhibited by physiological levels of serum albumin, which appears to bind and sequester some homoserine lactone quorum signals, resulting in the inability of P. aeruginosa to produce virulence factors that kill S. aureus. These data could provide important clues regarding the virulence of P. aeruginosa in albumin-depleted versus albumin-rich infection sites and an understanding of the nature of friendly versus antagonistic interactions between P. aeruginosa and S. aureus.
KEYWORDS: quorum sensing, Pseudomonas aeruginosa, Staphylococcus aureus, polymicrobial infection, wound, albumin
INTRODUCTION
Interactions between microbes are complex and range from fierce competition for nutrients and niches, manifested by antagonistic behavior, to highly evolved cooperative mechanisms between different species that support their mutual growth in specific environments (1). These interactions influence not only which species live or die but also the course of an infection. A growing appreciation of the importance of polymicrobial interactions during infection has resulted in increased efforts to develop representative in vitro and in vivo infection models with which to elucidate these mechanisms of interaction.
Our group has focused on understanding the interactions between microbial species in wound infections (2–5), which are typically polymicrobial, associated with the presence of biofilms, and exhibit increased tolerance to antimicrobials (6, 7). Staphylococcus aureus and Pseudomonas aeruginosa are the two most commonly isolated species from chronic wounds, many of which are coinfected with both species (8–10). There is also evidence that patients with P. aeruginosa-S. aureus coinfected wounds have worse outcomes than those with P. aeruginosa or S. aureus monospecies infections (11–13). Despite clinical evidence that S. aureus and P. aeruginosa coinfect chronic wounds, there have been few studies investigating their interspecies interactions, which may be partially explained by the technical difficulty of growing them together in vitro.
Under standard microbiological culturing conditions (planktonic growth in complex liquid medium), S. aureus is typically eradicated from the coculture within 8 h, presumably due to killing by P. aeruginosa (5). P. aeruginosa produces multiple exoproducts that inhibit the growth of, or kill, S. aureus, such as the LasA protease (14, 15), 4-hydroxy-2-heptylquinoline-N-oxide (HQNO) (16), pel and psl products (17), and phenazines such as pyocyanin (18). While we have a good understanding of the mechanisms controlling the regulation and expression of these P. aeruginosa virulence factors, little is known about how the composition of the bacterial environment influences their production.
We previously described an in vitro wound-like model that prevented P. aeruginosa from killing S. aureus and allowed the two species to persist in coculture for up to 7 days, similar to coinfections seen in mouse wounds (5). This model was used to demonstrate synergism in regard to antibiotic tolerance when S. aureus and P. aeruginosa were grown together in a wound-like environment (5). However, one intriguing facet of the study that was not elucidated was how the wound environment modulates the interactions between S. aureus and P. aeruginosa to promote coinfection. Therefore, in this study we sought to understand how components of the wound environment alter the ability of P. aeruginosa to kill S. aureus in coculture.
RESULTS AND DISCUSSION
Albumin prevents P. aeruginosa from killing S. aureus in the in vitro wound environment.
Our group (5), in addition to others (11, 19, 20), have noted the ability of Pseudomonas aeruginosa to rapidly kill Staphylococcus aureus in standard laboratory cocultures. This is an active killing process that occurs as P. aeruginosa approaches stationary phase and is dependent on quorum sensing (QS). Killing is conserved across a wide range of isolates and growth conditions (see Fig. S1 in the supplemental material); however, our group recently reported that P. aeruginosa and S. aureus can be stably cocultured in vitro using a “wound-like” medium (WLM) that contains many of the same components found in the wound environment (5). In order to determine the specific medium components required for stable coculture, we systematically tested the components of WLM.
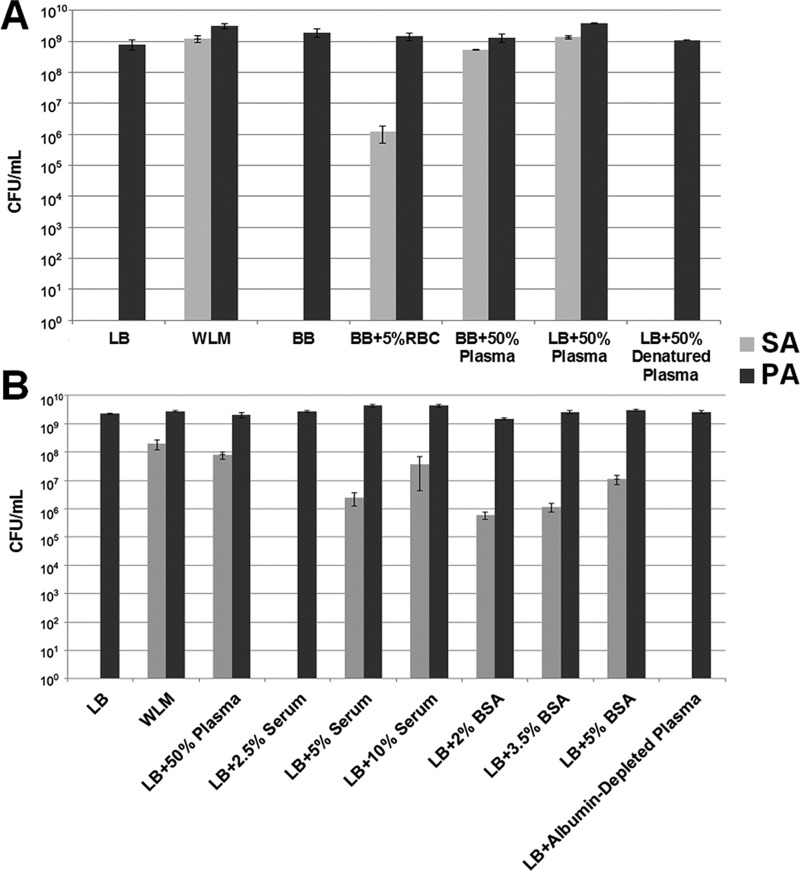
WLM components were tested in the same concentrations used in WLM, which includes 45% Bolton broth, 50% adult bovine plasma, and 5% laked horse red blood cells (RBC). Bolton broth alone was not able to rescue S. aureus in coculture (Fig. 1A). This result is not surprising since Bolton broth is a rich, undefined complex medium, similar to Luria-Bertani (LB) broth, which also cannot rescue S. aureus. A marginal rescue effect was observed when Bolton broth was supplemented with RBC (Fig. 1A), but it was not nearly as robust as that seen with whole WLM. Coculturing S. aureus and P. aeruginosa in Bolton broth supplemented with adult bovine plasma dramatically increased the numbers of S. aureus bacteria that survived coculture with P. aeruginosa, similar to the results seen with whole WLM (Fig. 1A). This effect was also observed when LB broth was supplemented with plasma. These data demonstrate that the plasma fraction of the WLM is largely responsible for preventing P. aeruginosa from killing S. aureus in WLM coculture.
FIG 1.
The albumin component of WLM rescues S. aureus (SA) from eradication when it is in coculture with P. aeruginosa (PA). (A) PAO1-SA31 cocultures were grown overnight in Luria-Bertani (LB) broth, full WLM (wound-like medium), or specific components of WLM, including Bolton broth (BB), Bolton broth plus red blood cells (BB+RBC) or plasma, or Bolton broth plus heat-denatured plasma. (B) PAO1-SA31 cocultures were grown overnight in LB broth, full WLM, or LB broth supplemented with the indicated components. Overnight cultures were then serially diluted and plated on Pseudomonas and Staphylococcus isolation agar to determine the number of CFU per milliliter. Plasma was heat denatured by heating whole plasma to 80°C for 15 min. Albumin was depleted from plasma using DEAE Affi-Gel Blue gel columns (Bio-Rad) according to the manufacturer's recommendations. Experiments were repeated at least three times, with at least three technical replicates.
We next investigated which component of plasma allowed S. aureus to coexist with P. aeruginosa in coculture. When plasma was heat denatured (21), the rescue effect was lost (Fig. 1A), suggesting a role for plasma proteins. Serum is the component of plasma generated when clotting factors and immune cells are removed while proteins and other molecules present in whole blood are retained (22). At physiologically relevant concentrations of serum, there was a stepwise increase in the amount of S. aureus rescued in coculture with P. aeruginosa (Fig. 1B), indicating that the protective factor was present in serum. As bovine serum albumin (BSA) is the most abundant protein found in serum, at about 65%, and constitutes 5% of the total blood volume (23, 24), we reasoned that this protein may be responsible for the protective effect. Addition of BSA at physiological concentrations caused a stepwise increase in the amount of S. aureus that remained viable in cocultures (Fig. 1B). In addition, depleting albumin from plasma resulted in a loss of the rescue effect, and P. aeruginosa regained the ability to kill S. aureus (Fig. 1B). Importantly, significant inhibition of S. aureus killing was not seen when S. aureus and P. aeruginosa were cocultured in LB broth supplemented with other serum proteins, such as gamma globulins and fibrinogen (Fig. S2), suggesting that the inhibitory effect observed was not simply due to the addition of excess protein and implying that the mechanism of action is albumin specific. Human serum albumin (HSA) also rescued S. aureus from killing by P. aeruginosa at a level similar to that of BSA, as did Albuminar (CSL Behring), which contains 25% human albumin in normal saline and is a common commercial albumin replacement therapy administered to patients suffering from hypoalbuminemia (Fig. S3). Taken together, these results support a role for albumin in preventing loss of S. aureus from in vitro WLM cocultures.
The ability of P. aeruginosa to kill S. aureus is quorum sensing controlled and does not require cell-to-cell contact.
In order to elucidate how albumin prevents P. aeruginosa from killing S. aureus in the presence of albumin, we sought to determine the mechanism of S. aureus killing by P. aeruginosa in coculture. P. aeruginosa produces many virulent exoproducts, many of which have been shown to kill or inhibit the growth of S. aureus (14–18). P. aeruginosa exoproducts can be either secreted into the extracellular environment or delivered in a contact-dependent manner (25–27). Thus, we next sought to determine whether cell-to-cell contact was required for S. aureus eradication in S. aureus-P. aeruginosa cocultures.
To test whether P. aeruginosa requires cell-to-cell contact to kill S. aureus, cocultures were grown in transwells, with the two species separated by a 0.4-μm-pore-size filter. This experimental setup allows the bacterial species to grow separately, without the ability to physically interact, while still allowing secreted exoproducts to move freely through the membrane. We observed that S. aureus was similarly eradicated from coculture when the two species were grown mixed in both compartments or when they were separated in different compartments by the membrane (Fig. S4). These data indicate that P. aeruginosa is able to kill S. aureus in coculture independent of direct cell-to-cell contact, suggesting that some P. aeruginosa-secreted exoproduct(s) must be responsible for the eradication of S. aureus from coculture.
The P. aeruginosa quorum sensing systems affect expression of over 300 genes either directly or indirectly, controlling up to 6% of the total P. aeruginosa genome (28). P. aeruginosa possesses two prototypical acyl-homoserine lactone (AHL) QS systems, termed Las and Rhl. These systems function through production of diffusible signal molecules by the enzymes LasI and RhlI, which are subsequently sensed by the cognate transcriptional regulator proteins LasR and RhlR. These systems do not function independently but are arranged hierarchically, with Las controlling expression of Rhl (29). The production of many P. aeruginosa-secreted exoproducts known to inhibit or kill S. aureus is controlled by QS (14–18), including the Las system-controlled staphylolytic enzyme LasA and the Rhl system-controlled rhamnolipid biosurfactant, hydrogen cyanide, and pyocyanin (29). P. aeruginosa also possesses a third QS system termed PQS (Pseudomonas quinolone signal). The PQS system directs the synthesis of a quinolone-based autoinducer, 2-heptyl-3-hydroxy-4-quinolone (HHQ), which is sensed by the transcriptional regulator PqsR (MvfR) (30). PQS production and sensing require activation by Las and Rhl systems. As with the Las and Rhl systems, the PQS system controls production of several antimicrobials, including pyocyanin and 4-hydroxy-2-heptylquinoline N-oxide (HQNO) (31).
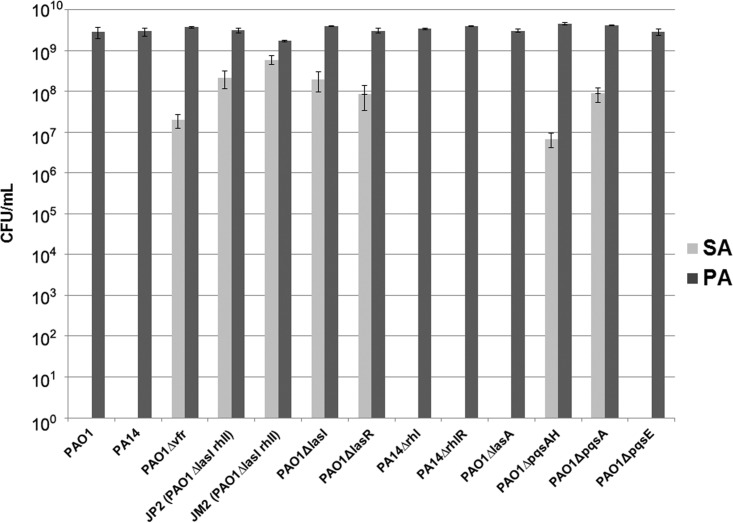
To evaluate the contribution of P. aeruginosa QS to S. aureus eradication, we cocultured S. aureus with a panel of P. aeruginosa strains that have mutations in different genes in the QS regulatory hierarchy. We observed that P. aeruginosa strains with mutations that control the Las and Rhl QS systems (PAO1 Δvfr), with mutations in the Las system (PAO1 ΔlasI rhlI, PAO1 ΔlasI, and PAO1 ΔlasR), or with mutations in the PQS system were unable to kill S. aureus in coculture (Fig. 2). However, mutations in the Rhl system (PAO1 ΔrhlI and PAO1Δ rhlR) did not affect the ability of P. aeruginosa to kill S. aureus in coculture. As the Las system is known to regulate the production of the staphylolytic LasA protease (14, 15), we tested whether a PAO1 strain unable to produce LasA could kill S. aureus. Surprisingly, we found that the ability of PAO1 ΔlasA to kill S. aureus was equivalent to that of PAO1. This indicates that in this coculture system LasA is not a significant contributor to S. aureus killing.
FIG 2.
P. aeruginosa mutants with defects high in the QS regulatory system and in the PQS system are unable to kill S. aureus. A total of 104 CFU of the indicated strains was inoculated into LB medium and grown overnight with SA31. Cocultures were initiated as a 1:1 mix of P. aeruginosa and S. aureus. Samples were then taken for serial dilution and plating on Pseudomonas and Staphylococcus isolation agar to determine the number of CFU per milliliter. Experiments were repeated at least three times, with at least three technical replicates.
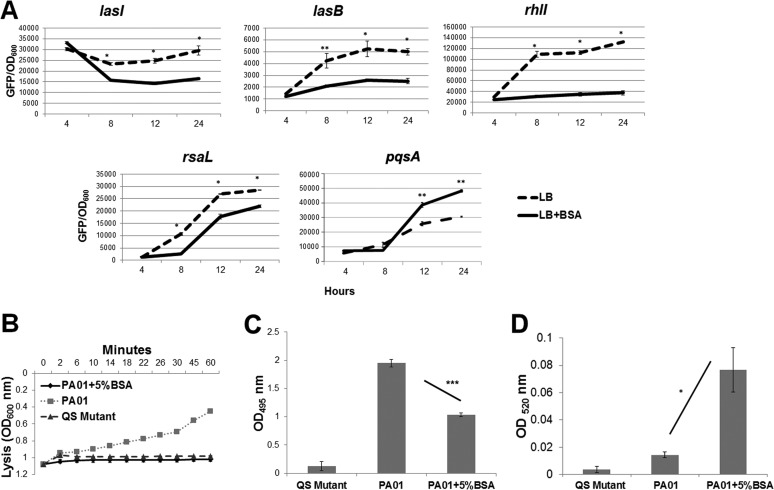
Based on these results, we hypothesized that albumin was inhibiting P. aeruginosa killing of S. aureus via inhibition of QS. To test this hypothesis, we first measured the impact of albumin addition on the expression of several QS-controlled genes. While albumin did not affect the growth of P. aeruginosa (Fig. S5), the expression levels of the Las-controlled genes lasI, lasB, rhlI, and rsaL were all significantly decreased by the addition of albumin (Fig. 3A). As expected, albumin also decreased production of the Las-controlled extracellular products LasA and LasB (Fig. 3B). Surprisingly, the production of pyocyanin, a redox-active small molecule primarily controlled by the PQS QS system, was increased (Fig. 3D). Taken together, these data demonstrated that albumin impacts P. aeruginosa QS; however, while the Las and Rhl systems are repressed by albumin, the PQS system was stimulated. Our results are consistent with those of Kruzcek et al. (32), who reported that pyocyanin and PQS were increased in the presence of serum at late stages of P. aeruginosa growth. Thus, while pqsA mutants are attenuated in their ability to kill S. aureus, this effect does not appear to be due to a reduction in pyocyanin or the expression of pqsA.
FIG 3.
Albumin inhibits P. aeruginosa QS. (A) P. aeruginosa GFP reporters were grown for 24 h in LB broth with and without 5% BSA, monitored for their level of GFP fluorescence, and normalized to growth (OD600). In the presence of BSA, the expression of most P. aeruginosa QS-controlled genes was significantly reduced in comparison to growth in LB alone. Activities of the Las system-regulated exoproducts LasA (B) and LasB (C) and the PQS system-regulated pyocyanin (D) were measured from the supernatants of overnight cultures of PAO1 grown in LB medium with or without 5% BSA. The QS mutant JP2 (PAO1 ΔlasI rhlI) supernatant was utilized as a negative control. *, P < 0.01; **, P < 0.001; ***, P < 0.0001 (one-way analysis of variance, Tukey posttest). Experiments were repeated at least three times, with at least three technical replicates.
It should also be noted that we also tested P. aeruginosa strains with mutations in other genes, including lasB, pilA, pel and psl, narGH, phzA, phzE, pcrV, pchE, algD, and others, and they were not attenuated in their ability to kill S. aureus in planktonic coculture (data not shown). In addition we tested several purified exoproducts and signals for their ability to kill S. aureus (without P. aeruginosa), including 2-heptyl-4-quinolone (HHQ), HQNO, PQS, and the Las- and Rhl-associated QS signals, and we saw no effect on S. aureus growth at the concentrations tested. So, despite great effort, we were not able to pinpoint the specific exoproduct(s) responsible for S. aureus killing in our system. We think that it is likely to be a combination of exoproducts, possibly at specific concentrations or produced at specific times during growth. Importantly, while the specific S. aureus-killing products were not identified, it is clear from our data that they are QS controlled.
Albumin binds and sequesters P. aeruginosa QS molecules.
Albumin is a common serum transport protein that carries numerous metabolites, hormones, and fatty acids throughout the body (23). The protein consists of three homologous domains, with nine binding sites that can bind a variety of molecules with different affinities (33). Examples of known albumin ligands include cholesterol, ibuprofen, and fatty acids (34–36). When we compared the structure of the P. aeruginosa QS signaling molecules to that of known BSA ligands, we noticed a great deal of structural similarity. Since albumin is known to nonspecifically bind to an array of molecules present in the physiological environment, we hypothesized that serum albumin can inhibit P. aeruginosa QS by binding and sequestering QS signaling molecules. This would effectively remove them from the environment, and therefore P. aeruginosa QS gene expression would be decreased.
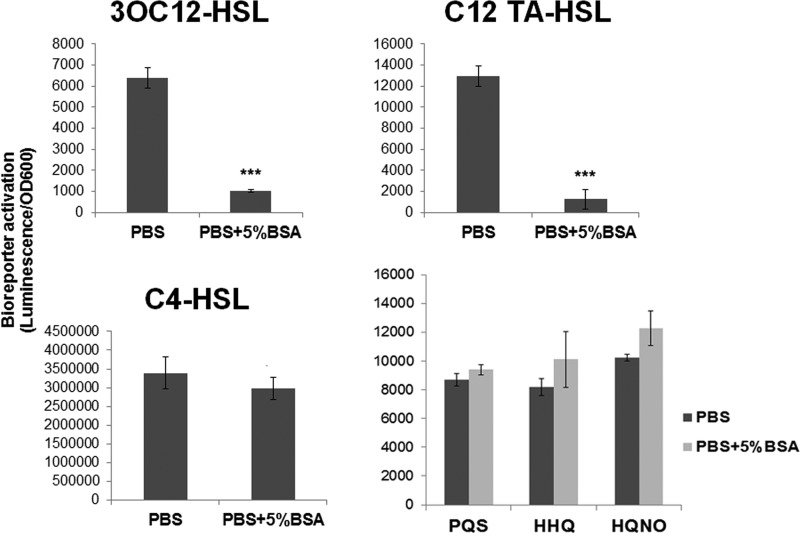
To test the hypothesis that albumin inhibits P. aeruginosa QS by binding and sequestering autoinducers, we incubated purified N-3-oxo-dodecanoyl homoserine lactone (3OC12-HSL) and N-butanoyl homoserine lactone (C4-HSL) (the cognate signaling molecules of the LasI/R and RhlI/R QS systems, respectively) with phosphate-buffered saline (PBS) alone or PBS supplemented with 5% BSA for 3 h. The solutions were then passed through a 30-kDa-molecular-mass-cutoff filter to remove the BSA and anything bound to it. Any QS molecules remaining in the eluate were then extracted with ethyl acetate and detected using luminescent bioreporters as described in Materials and Methods. We observed a significant, 6.25-fold, decrease in the amount of 3OC12-HSL, but not C4-HSL, in the eluates of solutions that were incubated with BSA prior to column separation (Fig. 4A).
FIG 4.
BSA binds 3OC12-HSL and its degradation product C12-TA-HSL. 3OC12-HSL, C12-TA-HSL, C4-HSL, PQS, HHQ, or HQNO was added to PBS or PBS–5% BSA at a concentration of 5 μM. After a 3-h incubation the solutions were passed through a 30-kDa column to remove the BSA and any of its ligands. Autoinducers were extracted from the eluates, and their levels were measured using LasR, RhlR, or PQS-based luciferase bioreporter strains as described in Materials and Methods. ***, P < 0.0001 (one-way analysis of variance, Tukey posttest). Experiments were repeated at least three times, with at least three technical replicates.
We also examined the ability of BSA to bind C12-TA-HSL (where TA is tetramic acid), which is a degradation product of 3OC12-HSL. C12-TA-HSL is generated by nonenzymatic deprotonation and has been detected in P. aeruginosa cultures and in sputum from patients infected with P. aeruginosa (37, 38). C12-TA-HSL has bactericidal activity and can also act as a siderophore (37). In our experiments, C12-TA-HSL activated our LasR-based bioreporter just as well, if not better than, 3OC12-HSL. This suggests that C12-TA-HSL could physiologically activate the Las QS system in addition to 3OC12-HSL. We detected a significant, 10.4-fold decrease in the amount of C12-TA-HSL, similar to level of 3OC12-HSL, in eluates from solutions incubated with BSA. We saw no evidence of albumin binding to the quinolone-type autoinducers PQS, HHQ, and HQNO (Fig. 4). Taken together, these results indicate that albumin decreases expression of Las QS genes through sequestration of 3OC12-HSL.
To determine the affinity of QS molecules to albumin, we used isothermal titration calorimetry (ITC), which has been used extensively to study albumin and fatty acid binding (39–42). With ITC, affinities are determined by directly measuring the binding of a ligand to a protein by recording the heat absorbed or released by the binding process. ITC with 3OC12-HSL and BSA resulted in an oscillating thermogram (Fig. 5), which is indicative of multiple complex binding events. This implies that one ligand binds more strongly, and as the particular binding site involved becomes saturated or the ligand becomes exhausted, a second, less strongly bound ligand begins to bind. While it is clear from our ITC measurements that 3OC12-HSL binds BSA, computing accurate binding affinity (Kd) values for multiple binding events of many different affinities would not be possible.
FIG 5.
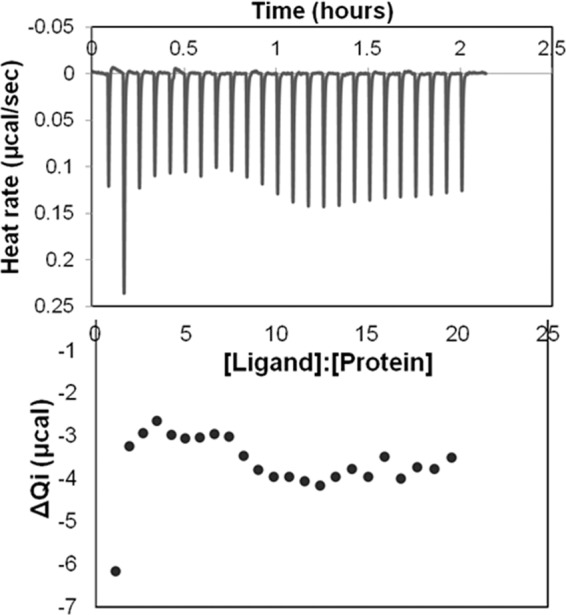
ITC thermograph demonstrates the binding of 3OC12-HSL to BSA. 3OC12-HSL (2 mM) in 5% ethyl acetate was utilized as a ligand for measuring the binding affinity to 30 μM fatty-acid-free BSA in 5% ethyl acetate protein by ITC as described in Materials and Methods. Qi, heat effect.
Our observations correlate with the findings of Davis et al. (43), who also proposed that albumin may act as “a sink or perhaps a reservoir for AHLs.” This group demonstrated that, in the presence of HSA, the critical micelle concentration (the amount of AHL or HSL needed to self-aggregate into a micelle) for 3OC14-HSL increased from 17 μM to over 500 μM, suggesting that albumin effectively reduced the concentration of free AHLs in the medium. A similar mechanism has also been described in S. aureus, where human apolipoprotein B binds to the S. aureus autoinducing pheromone and prevents its attachment to bacteria and subsequent signaling through its receptor, AgrC (44). Mice deficient in plasma apolipoprotein B were significantly more susceptible to invasive agr+ strain infection but not to infection with an agr deletion mutant. Therefore, the authors proposed that apolipoprotein B at homeostatic levels in blood is an essential innate defense effector against invasive S. aureus infection.
Potential clinical significance of our findings and future studies.
If albumin is capable of binding and sequestering P. aeruginosa HSLs, this could have important clinical implications during P. aeruginosa infections. Albumin concentrations differ depending on anatomical location. For example, body sites rich in blood, such as wounds, are rich in albumin. Based on our data, it is possible that when P. aeruginosa is in an albumin-rich environment, such as in chronic wounds, QS will be downregulated, or even nonfunctional, due to the binding of QS molecules by serum albumin. Conversely, there are body sites that have reduced levels of albumin, such as the urinary tract, lung, and the peripheral blood during burn trauma (45, 46). In these types of infections, P. aeruginosa is more likely to become the predominant microbe (47, 48). We propose that the reduced levels of albumin in these environments allow the P. aeruginosa QS to function, thus enhancing P. aeruginosa fitness through production of Las-controlled antimicrobials that eliminate coinfecting bacteria. This may be particularly relevant to burn patients who have extremely low levels of albumin in the infection site (46, 49, 50), and P. aeruginosa is generally the predominant pathogen. P. aeruginosa is highly virulent in burns, quickly disseminating and causing sepsis and septic shock, which can be lethal (51), and the contribution of QS has been shown to be paramount for virulence and dissemination in animal burn models (52, 53).
While our study represents a first step in understanding both how specific components present in the infection site can modulate cell-to-cell signaling and impact polymicrobial interactions, there are still many questions to answer. One major limitation of this study was that our observations were largely based on planktonic growth, which may not correlate with the biofilm growth occurring in wounds. Thus, important future studies will be to investigate the effect of albumin on QS in P. aeruginosa biofilms, both in vitro and in vivo. We were also unable to determine a specific binding affinity (Kd) for HSL-albumin. This is critical to understanding the in vivo relevance of our findings as albumin binds many small molecules, such as free fatty acids or steroid-based ligands, that will likely compete with AHLs for binding. Finally, while P. aeruginosa QS has been demonstrated to be an important factor controlling virulence in burn wounds (52, 53), the same is not true for chronic wounds. Based on our in vitro findings, we would predict that P. aeruginosa QS gene expression is higher in the albumin-depleted burn wound than in the albumin-rich chronic wound. If this is true, it is possible that P. aeruginosa virulence can be attenuated by the administration of exogenous albumin given locally at the burn wound. This finding would also be extremely relevant to investigators who are developing P. aeruginosa QS inhibitors for the treatment of chronic wound infections, which may ultimately prove ineffective. Thus, our future studies will focus on experiments conducted in vivo to characterize the interaction of albumin (and other host proteins) with P. aeruginosa quorum signals.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. P. aeruginosa and S. aureus clinical isolates were provided by Abdul Hamood (54) and obtained from patients at the Texas Tech University Health Sciences Center's affiliated hospital, University Medical Center, under an approved IRB protocol. S. aureus and P. aeruginosa planktonic cultures were grown in Luria-Bertani (LB) broth or LB broth supplemented as specified in the figure legends. The in vitro wound-like medium (WLM) used was previously described by DeLeon et al. (5). WLM consists of 45% Bolton broth (complex meat-based broth), 50% bovine plasma, and 5% laked horse red blood cells (RBC). A 5-ml volume of medium was inoculated with 104 to 105 cells from overnight cultures of S. aureus, P. aeruginosa, or a 1:1 mixture of both and grown, shaking at 220 rpm, at 37°C overnight. Samples were then serially diluted, and plated on Pseudomonas isolation agar to determine the number of CFU of P. aeruginosa per milliliter and/or on mannitol salt agar (MSA) or Staphylococcus isolation agar to determine the number of CFU of S. aureus per milliliter (Difco, Sparks, MD); (Remel, Thermo Fisher Scientific, Lenexa, KS).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Description | Reference or source |
|---|---|---|
| P. aeruginosa PAO1 | Wild type | 58 |
| P. aeruginosa PA14 | Wild-type P. aeruginosa | 59 |
| S. aureus SA31 | S. aureus sp. aureus strain ATCC 29213 | Remel Microbiology Products |
| P. aeruginosa mutants | ||
| PAO Δvfr | vfr deletion of PAO-SW | 60 |
| PAO-JP2 | PAO1 ΔlasI ΔrhlI mutant, Tcr Hgr | 61 |
| PAO-JM2 | PAO1 ΔrhlI mutant, Tcr | 62 |
| PAO1 lasI::Gmr | PAO1 insertion mutant (Gmr) in the lasI gene | 63 |
| PAO1 lasR::Gmr | PAO1insertion mutant (Gmr) in the lasR gene | 64 |
| PA14 rhlI::Tcr | PA14 insertion mutant (Tcr) in the rhlI gene | 65 |
| PA14 rhlR::Tcr | PA14 insertion mutant (Tcr) in the rhlR gene | 65 |
| PAO1 lasA::Tcr | PAO1 insertion mutant (Tcr) in the lasA gene | 66 |
| PAO1 ΔpqsA | pqsA deletion mutant; PQS negative | 67 |
| PAO1 ΔpqsA pqsH | Double deletion mutant in PAO1 | 30 |
| PAO1 ΔpqsE | pqsE in-frame deletion mutant in PAO1 | 68 |
| Bioreporters | ||
| PAO1(prhlI-LVAgfp) | P. aeruginosa PAO1 carrying the rhlI-gfp transcriptional reporter on plasmid prhlI-LVAgfp | 69 |
| PAO1(plasI-LVAgfp) | P. aeruginosa PAO1 carrying the lasI-gfp transcriptional reporter on plasmid plasI-LVAgfp | 69 |
| PA14(pGJB5) | P. aeruginosa PA14 carrying the transcriptional rsaL-gfp reporter on plasmid pGJB5 | 70 |
| PAO1 lasB::GFP | MH451 lac::lasR lasB::gfp(ASV), GFP reporter of the PAO1 lasB gene | 71 |
| PA14(pAW1) | P. aeruginosa PA14 carrying the pqsA-gfp transcriptional reporter on plasmid pAW1 | This study |
| E. coli(pSB1075)a | E. coli 3OC12-HSL luminescent reporter strain | 72 |
| E. coli(p536) | E. coli C4-HSL luminescent reporter strain | 73 |
| PAO1 pqsA CTX-lux::pqsA | pqsA mutant containing a chromosomal pqsA-luxCDABE transcriptional reporter | 74 |
E. coli, Escherichia coli.
DNA manipulations.
PCR was performed using an Expand Long Template PCR system (Roche). Qiagen kits were used for purification of plasmids (QIAprep spin miniprep kit), gel purifications (QIAquick gel extraction kit), and PCR purifications (MinElute PCR purification kit). Restriction endonucleases and buffers were purchased from New England BioLabs or Fermentas Life Sciences. DNA sequencing was performed at the DNA Core Facility at the University of Texas at Austin Institute for Cell and Molecular Biology.
Plasmid construction.
To construct the pqsA transcriptional reporter plasmid pAW1, the region upstream of pqsA was amplified from P. aeruginosa chromosomal DNA using primers CGGAATTCGTAGGTGTCCTCTTCGGCAG and GCTCTAGATCAATCAATCAGCGATATGCATCCGGATCAG. The resulting amplicon was digested with EcoRI/XbaI and ligated into EcoRI/XbaI-digested pGJB5 (Table 1). The resulting plasmid, pAW1, was confirmed by sequencing and transformed into P. aeruginosa by electroporation.
Staphylolytic assay.
A staphylolytic assay was performed as previously described (55). LasA protease activity was measured by determining the ability of P. aeruginosa culture supernatants to lyse S. aureus cells. Briefly, an overnight culture of S. aureus was boiled for 10 min, followed by centrifugation for 10 min at 10,000 × g. The resulting pellet was resuspended in 10 mM Na2PO4 (pH 7.5) to an optical density at 600 nm (OD600) of approximately 0.8. An overnight culture of P. aeruginosa was centrifuged at 10,000 × g for 10 min to pellet the cells, and then the supernatant was passed through a 0.2-μm-pore-size filter for sterilization. A 100-μl aliquot of supernatant was then added to 1 ml of S. aureus suspension. LasA activity results in clearance of the suspension (reduced turbidity), indicative of S. aureus cell lysis, as measured by the OD600 at 2, 6, 10, 14, 18, 22, 26, 30, 45, and 60 min.
Pyocyanin assay.
A pyocyanin assay was performed as previously described (56). Briefly, overnight cultures were centrifuged at 6,000 × g for 10 min at room temperature, and sterile supernatant was collected after filtration through a 0.2-μm-pore-size filter. A 5-ml portion of the supernatant was mixed with 3 ml of chloroform. Following centrifugation for 3 min at 1,200 × g at room temperature, 2.5 ml of the phase under the chloroform was collected and mixed with 1 ml of 0.2 M HCl. A sample of 200 μl was collected, and the absorbance was measured at 530 nm in a spectrophotometer.
Detection of QS gene expression.
Reporter strains carrying green fluorescent protein (GFP) transcriptional fusions to QS promoters (Table 1) were utilized to monitor QS gene expression in environments with and without 5% BSA supplementation. A total of 104 to 105 cells from overnight cultures were inoculated into LB broth with or without 5% BSA supplementation and grown at 37°C, with shaking at 220 rpm, for 24 h. Samples from these cultures were diluted 1:100 in 1× phosphate-buffered saline (PBS), and the level of GFP fluorescence (excitation wavelength, 480 nm; emission wavelength, 535 nm) was detected using a Synergy H1 hybrid microtiter plate reader (BioTek, Winooski, VT).
Detection of autoinducers.
Solutions (5 μM) of purified 3OC12-HSL, C4-HSL (Fluka Chemie, Sigma-Aldrich, Saint Louis, MO), and the quinolones (PQS, HHQ, and HQNO; obtained from Steve Diggle, Nottingham University) were prepared in 1× PBS and incubated for 3 h, with shaking at 130 rpm at 37°C, either with or without 5% BSA. Solutions were then passed through a 30-kDa-molecular-mass-cutoff Vivaspin column (Corning, Lowell, MA) via centrifugation at 8 × g for 10 min. QS molecules were separated from the eluate via three successive rounds of acidified ethyl acetate (high-performance liquid chromatography [HPLC]-grade) extraction as previously described (57). Final extracts were dried in an Eppendorf 5301 concentrator (Eppendorf, Hauppauge, NY) and resuspended in 30 μl of methanol (HPLC grade). Overnight cultures of the 3OC12-HSL, C4-HSL, or PQS luciferase bioreporter strains (Table 1) were diluted to an OD600 of 0.5 in LB broth. Five-microliter aliquots of methanol-dissolved extracts were combined with 100-μl aliquots of diluted reporter strain cultures in the wells of a 96-well, clear, flat-bottom microtiter plate (Costar, Corning, NY) and incubated statically at 37°C for 3 h. Luminescence was detected using a luminometer (Modulus microplate reader; Turner Biosystems, Promega, Madison, WI), and values were normalized to the value for the reporter biomass (OD600).
ITC.
Isothermal titration calorimetry (ITC) measurements were characterized using a TA Instruments small-volume ITC instrument at 20°C. Titrations were performed using a stir rate of 250 rpm and either 1.5- or 2-μl injection volume. Titrations of 2 mM 3OC12-HSL into 30 μM defatted BSA in 5% ethyl acetate and 1× PBS buffer showed drifting of the thermogram binding profile upon repeated measurements of the same sample over time due to the degradation of the 3OC12-HSL. Measurement of the C12-TA-HSL degradation product of 3OC12-HSL was measured by allowing a reconstituted 3OC12-HSL to degrade completely (for 72 h) before the titration was performed. All titrations were corrected for any heat of dilution of the ligand through separate control experiments in which the ligand was injected into sample buffer lacking BSA in the cell.
Statistics.
Statistical analyses were performed using GraphPad Prism, version 6, or InStat, version 3. Specific tests for significance are indicated in figure legends.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by grants AI105763 (K.P.R.) and GM116547 (M.W.) from the National Institute of Allergy and Infectious Diseases and the National Institute of General Medical Sciences, respectively, and grant 62507-LS from the U.S. Army Research Office (K.P.R. and M.W.). M.W. is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease.
We thank Steve Diggle (University of Nottingham) for the QS bioreporters, mutant strains, and purified PQS, HHQ, and HQNO and Gunnar Kaufmann (The Scripps Research Institute) for the C12-TA-HSL.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00116-17.
REFERENCES
- 1.Murray JL, Connell JL, Stacy A, Turner KH, Whiteley M. 2014. Mechanisms of synergy in polymicrobial infections. J Microbiol 52:188–199. doi: 10.1007/s12275-014-4067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. 2013. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A 110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. 2014. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet 10:e1004518. doi: 10.1371/journal.pgen.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, Rumbaugh KP. 2011. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 6:e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR, Rumbaugh KP. 2014. Synergistic Interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun 82:4718–4728. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James GA, Swogger E, Wolcott R, Pulcini E, Secor P, Sestrich J, Costerton JW, Stewart PS. 2008. Biofilms in chronic wounds. Wound Repair Regen 16:37–44. doi: 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 7.Bjarnsholt T, Kirketerp-Moller K, Jensen PO, Madsen KG, Phipps R, Krogfelt K, Hoiby N, Givskov M. 2008. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen 16:2–10. doi: 10.1111/j.1524-475X.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 8.Fazli M, Bjarnsholt T, Kirketerp-Moller K, Jorgensen B, Andersen AS, Krogfelt KA, Givskov M, Tolker-Nielsen T. 2009. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J Clin Microbiol 47:4084–4089. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trivedi U, Parameswaran S, Armstrong A, Burgueno-Vega D, Griswold J, Dissanaike S, Rumbaugh KP. 2014. Prevalence of multiple antibiotic resistant infections in diabetic versus nondiabetic wounds. J Pathog 2014:173053. doi: 10.1155/2014/173053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gjodsbol K, Christensen JJ, Karlsmark T, Jorgensen B, Klein BM, Krogfelt KA. 2006. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J 3:225–231. doi: 10.1111/j.1742-481X.2006.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pastar I, Nusbaum AG, Gil J, Patel SB, Chen J, Valdes J, Stojadinovic O, Plano LR, Tomic-Canic M, Davis SC. 2013. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 8:e56846. doi: 10.1371/journal.pone.0056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendricks KJ, Burd TA, Anglen JO, Simpson AW, Christensen GD, Gainor BJ. 2001. Synergy between Staphylococcus aureus and Pseudomonas aeruginosa in a rat model of complex orthopaedic wounds. J Bone Joint Surg Am 83–A:855–861. [DOI] [PubMed] [Google Scholar]
- 13.Rosenbluth DB, Wilson K, Ferkol T, Schuster DP. 2004. Lung function decline in cystic fibrosis patients and timing for lung transplantation referral. Chest 126:412–419. doi: 10.1378/chest.126.2.412. [DOI] [PubMed] [Google Scholar]
- 14.Mansito TB, Falcon MA, Moreno J, Carnicero A, Gutierrez-Navarro AM. 1987. Effects of staphylolytic enzymes from Pseudomonas aeruginosa on the growth and ultrastructure of Staphylococcus aureus. Microbios 49:55–64. [PubMed] [Google Scholar]
- 15.Kessler E, Safrin M, Olson JC, Ohman DE. 1993. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem 268:7503–7508. [PubMed] [Google Scholar]
- 16.Hoffman LR, Deziel E, D'Argenio DA, Lepine F, Emerson J, McNamara S, Gibson RL, Ramsey BW, Miller SI. 2006. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 103:19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Z, Yang L, Qu D, Molin S, Tolker-Nielsen T. 2009. Pseudomonas aeruginosa extracellular products inhibit staphylococcal growth, and disrupt established biofilms produced by Staphylococcus epidermidis. Microbiology 155:2148–2156. doi: 10.1099/mic.0.028001-0. [DOI] [PubMed] [Google Scholar]
- 18.Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol 61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 19.Palmer KL, Aye LM, Whiteley M. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol 189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer KL, Mashburn LM, Singh PK, Whiteley M. 2005. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol 187:5267–5277. doi: 10.1128/JB.187.15.5267-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pena I, Dominguez JM. 2010. Thermally denatured BSA, a surrogate additive to replace BSA in buffers for high-throughput screening. J Biomol Screen 15:1281–1286. doi: 10.1177/1087057110379768. [DOI] [PubMed] [Google Scholar]
- 22.Tuck MK, Chan DW, Chia D, Godwin AK, Grizzle WE, Krueger KE, Rom W, Sanda M, Sorbara L, Stass S, Wang W, Brenner DE. 2009. Standard operating procedures for serum and plasma collection: early detection research network consensus statement standard operating procedure integration working group. J Proteome Res 8:113–117. doi: 10.1021/pr800545q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennis MS, Zhang M, Meng YG, Kadkhodayan M, Kirchhofer D, Combs D, Damico LA. 2002. Albumin binding as a general strategy for improving the pharmacokinetics of proteins. J Biol Chem 277:35035–35043. doi: 10.1074/jbc.M205854200. [DOI] [PubMed] [Google Scholar]
- 24.Fasano M, Curry S, Terreno E, Galliano M, Fanali G, Narciso P, Notari S, Ascenzi P. 2005. The extraordinary ligand binding properties of human serum albumin. IUBMB Life 57:787–796. doi: 10.1080/15216540500404093. [DOI] [PubMed] [Google Scholar]
- 25.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, Low DA. 2010. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 30.Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Camara M, Williams P. 2007. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol 14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 31.Machan ZA, Taylor GW, Pitt TL, Cole PJ, Wilson R. 1992. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J Antimicrob Chemother 30:615–623. doi: 10.1093/jac/30.5.615. [DOI] [PubMed] [Google Scholar]
- 32.Kruczek C, Qaisar U, Colmer-Hamood JA, Hamood AN. 2014. Serum influences the expression of Pseudomonas aeruginosa quorum-sensing genes and QS-controlled virulence genes during early and late stages of growth. Microbiologyopen 3:64–79. doi: 10.1002/mbo3.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fanali G, di Masi A, Trezza V, Marino M, Fasano M, Ascenzi P. 2012. Human serum albumin: From bench to bedside. Mol Aspects Med 33:209–290. doi: 10.1016/j.mam.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Francis GL. 2010. Albumin and mammalian cell culture: implications for biotechnology applications. Cytotechnology 62:1–16. doi: 10.1007/s10616-010-9263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baroni S, Mattu M, Vannini A, Cipollone R, Aime S, Ascenzi P, Fasano M. 2001. Effect of ibuprofen and warfarin on the allosteric properties of haem-human serum albumin. A spectroscopic study. Eur J Biochem 268:6214–6220. doi: 10.1046/j.0014-2956.2001.02569.x. [DOI] [PubMed] [Google Scholar]
- 36.Bojesen IN, Bojesen E. 1994. Binding of arachidonate and oleate to bovine serum albumin. J Lipid Res 35:770–778. [PubMed] [Google Scholar]
- 37.Kaufmann GF, Sartorio R, Lee SH, Rogers CJ, Meijler MM, Moss JA, Clapham B, Brogan AP, Dickerson TJ, Janda KD. 2005. Revisiting quorum sensing: discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc Natl Acad Sci U S A 102:309–314. doi: 10.1073/pnas.0408639102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struss AK, Nunes A, Waalen J, Lowery CA, Pullanikat P, Denery JR, Conrad DJ, Kaufmann GF, Janda KD. 2013. Toward implementation of quorum sensing autoinducers as biomarkers for infectious disease states. Anal Chem 85:3355–3362. doi: 10.1021/ac400032a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sengupta A, Sasikala WD, Mukherjee A, Hazra P. 2012. Comparative study of flavins binding with human serum albumin: a fluorometric, thermodynamic, and molecular dynamics approach. Chemphyschem 13:2142–2153. doi: 10.1002/cphc.201200044. [DOI] [PubMed] [Google Scholar]
- 40.Sharma R, Choudhary S, Kishore N. 2012. Insights into the binding of the drugs diclofenac sodium and cefotaxime sodium to serum albumin: calorimetry and spectroscopy. Eur J Pharm Sci 46:435–445. doi: 10.1016/j.ejps.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Lu J, Stewart AJ, Sadler PJ, Pinheiro TJ, Blindauer CA. 2012. Allosteric inhibition of cobalt binding to albumin by fatty acids: implications for the detection of myocardial ischemia. J Med Chem 55:4425–4430. doi: 10.1021/jm3003137. [DOI] [PubMed] [Google Scholar]
- 42.Wu LL, Gao HW, Gao NY, Chen FF, Chen L. 2009. Interaction of perfluorooctanoic acid with human serum albumin. BMC Struct Biol 9:31. doi: 10.1186/1472-6807-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis BM, Richens JL, O'Shea P. 2011. Label-free critical micelle concentration determination of bacterial quorum sensing molecules. Biophys J 101:245–254. doi: 10.1016/j.bpj.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peterson MM, Mack JL, Hall PR, Alsup AA, Alexander SM, Sully EK, Sawires YS, Cheung AL, Otto M, Gresham HD. 2008. Apolipoprotein B is an innate barrier against invasive Staphylococcus aureus infection. Cell Host Microbe 4:555–566. doi: 10.1016/j.chom.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicholson JP, Wolmarans MR, Park GR. 2000. The role of albumin in critical illness. Br J Anaesth 85:599–610. doi: 10.1093/bja/85.4.599. [DOI] [PubMed] [Google Scholar]
- 46.Cartotto R, Callum J. 2012. A review of the use of human albumin in burn patients. J Burn Care Res 33:702–717. doi: 10.1097/BCR.0b013e31825b1cf6. [DOI] [PubMed] [Google Scholar]
- 47.Kumar R, Chhibber S, Harjai K. 2009. Quorum sensing is necessary for the virulence of Pseudomonas aeruginosa during urinary tract infection. Kidney Int 76:286–292. doi: 10.1038/ki.2009.183. [DOI] [PubMed] [Google Scholar]
- 48.Hoiby N, Ciofu O, Bjarnsholt T. 2010. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol 5:1663–1674. doi: 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- 49.Delaney AP, Dan A, McCaffrey J, Finfer S. 2011. The role of albumin as a resuscitation fluid for patients with sepsis: a systematic review and meta-analysis. Crit Care Med 39:386–391. doi: 10.1097/CCM.0b013e3181ffe217. [DOI] [PubMed] [Google Scholar]
- 50.Wilkes MM, Navickis RJ. 2001. Patient survival after human albumin administration. A meta-analysis of randomized, controlled trials. Ann Intern Med 135:149–164. [DOI] [PubMed] [Google Scholar]
- 51.Bahemia IA, Muganza A, Moore R, Sahid F, Menezes CN. 2015. Microbiology and antibiotic resistance in severe burns patients: a 5 year review in an adult burns unit. Burns 41:1536–1542. doi: 10.1016/j.burns.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 52.Gupta P, Chhibber S, Harjai K. 2015. Efficacy of purified lactonase and ciprofloxacin in preventing systemic spread of Pseudomonas aeruginosa in murine burn wound model. Burns 41:153–162. doi: 10.1016/j.burns.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 53.Rumbaugh KP, Griswold JA, Iglewski BH, Hamood AN. 1999. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun 67:5854–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller KG, Tran PL, Haley CL, Kruzek C, Colmer-Hamood JA, Myntti M, Hamood AN. 2014. Next science wound gel technology, a novel agent that inhibits biofilm development by gram-positive and gram-negative wound pathogens. Antimicrob Agents Chemother 58:3060–3072. doi: 10.1128/AAC.00108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diggle SP, Winzer K, Lazdunski A, Williams P, Camara M. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J Bacteriol 184:2576–2586. doi: 10.1128/JB.184.10.2576-2586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carlsson M, Shukla S, Petersson AC, Segelmark M, Hellmark T. 2011. Pseudomonas aeruginosa in cystic fibrosis: pyocyanin negative strains are associated with BPI-ANCA and progressive lung disease. J Cyst Fibros 10:265–271. doi: 10.1016/j.jcf.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 57.Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, Greenberg EP. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci U S A 91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holloway BW, Krishnapillai V, Morgan AF. 1979. Chromosomal genetics of Pseudomonas. Microbiol Rev 43:73–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 60.Davinic M, Carty NL, Colmer-Hamood JA, San Francisco M, Hamood AN. 2009. Role of Vfr in regulating exotoxin A production by Pseudomonas aeruginosa. Microbiology 155:2265–2273. doi: 10.1099/mic.0.028373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pearson JP, Pesci EC, Iglewski BH. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol 179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li LL, Malone JE, Iglewski BH. 2007. Regulation of the Pseudomonas aeruginosa quorum-sensing regulator VqsR. J Bacteriol 189:4367–4374. doi: 10.1128/JB.00007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darch SE, West SA, Winzer K, Diggle SP. 2012. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc Natl Acad Sci U S A 109:8259–8263. doi: 10.1073/pnas.1118131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Popat R, Crusz SA, Messina M, Williams P, West SA, Diggle SP. 2012. Quorum-sensing and cheating in bacterial biofilms. Proc Biol Sci 279:4765–4771. doi: 10.1098/rspb.2012.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rumbaugh KP, Diggle SP, Watters CM, Ross-Gillespie A, Griffin AS, West SA. 2009. Quorum sensing and the social evolution of bacterial virulence. Curr Biol 19:341–345. doi: 10.1016/j.cub.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 66.Toder DS, Ferrell SJ, Nezezon JL, Rust L, Iglewski BH. 1994. lasA and lasB genes of Pseudomonas aeruginosa: analysis of transcription and gene product activity. Infect Immun 62:1320–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rampioni G, Pustelny C, Fletcher MP, Wright VJ, Bruce M, Rumbaugh KP, Heeb S, Camara M, Williams P. 2010. Transcriptomic analysis reveals a global alkyl-quinolone-independent regulatory role for PqsE in facilitating the environmental adaptation of Pseudomonas aeruginosa to plant and animal hosts. Environ Microbiol 12:1659–1673. doi: 10.1111/j.1462-2920.2010.02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Kievit TR, Gillis R, Marx S, Brown C, Iglewski BH. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol 67:1865–1873. doi: 10.1128/AEM.67.4.1865-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Connell JL, Wessel AK, Parsek MR, Ellington AD, Whiteley M, Shear JB. 2010. Probing prokaryotic social behaviors with bacterial “lobster traps.” mBio 1:e00202-10. doi: 10.1128/mBio.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, Rice SA, Eberl L, Molin S, Hoiby N, Kjelleberg S, Givskov M. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87–102. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]
- 72.Winson MK, Swift S, Fish L, Throup JP, Jorgensen F, Chhabra SR, Bycroft BW, Williams P, Stewart GS. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett 163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- 73.Swift S, Karlyshev AV, Fish L, Durant EL, Winson MK, Chhabra SR, Williams P, Macintyre S, Stewart GS. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J Bacteriol 179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fletcher MP, Diggle SP, Crusz SA, Chhabra SR, Camara M, Williams P. 2007. A dual biosensor for 2-alkyl-4-quinolone quorum-sensing signal molecules. Environ Microbiol 9:2683–2693. doi: 10.1111/j.1462-2920.2007.01380.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.