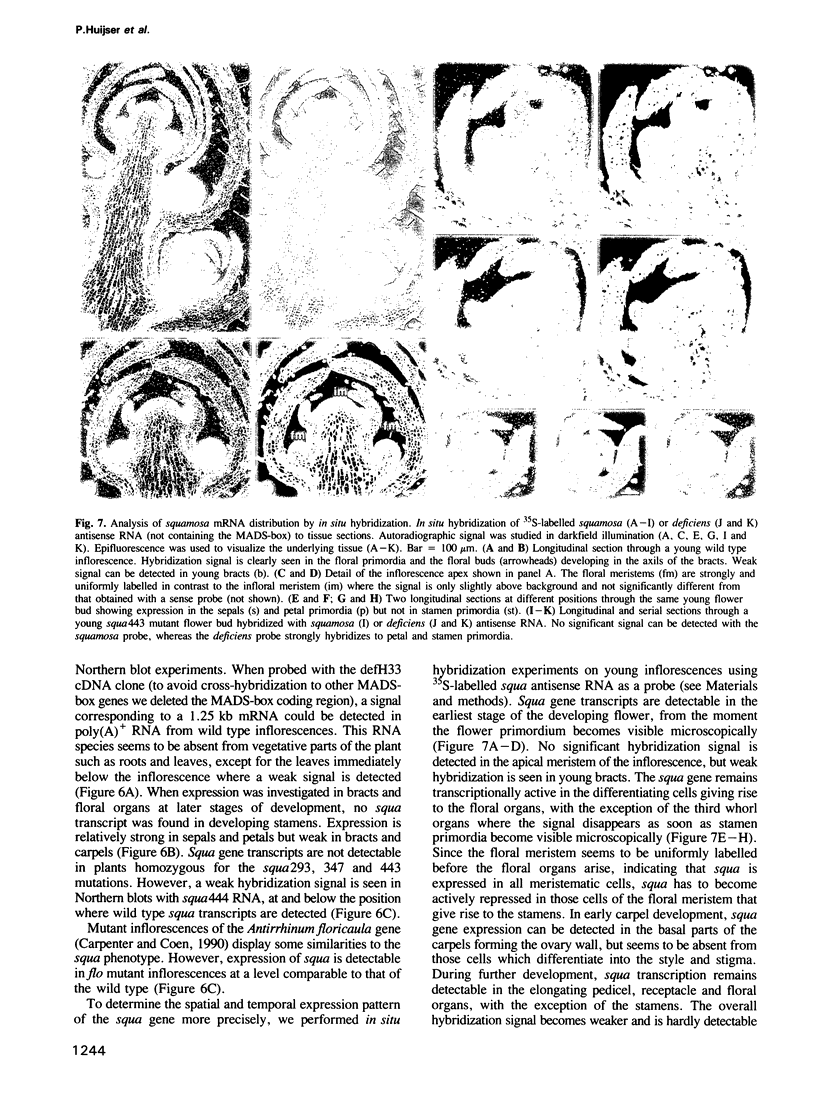
Abstract
Anomalous flowering of the Antirrhinum majus mutant squamosa (squa) is characterized by excessive formation of bracts and the production of relatively few and often malformed or incomplete flowers. To study the function of squamosa in the commitment of an inflorescence lateral meristem to floral development, the gene was cloned and its genomic structure, a well as that of four mutant alleles, was determined. SQUA is a member of a family of transcription factors which contain the MADS-box, a conserved DNA binding domain. In addition, we analysed the temporal and spatial expression pattern of the squa gene. Low transcriptional activity of squa is detectable in bracts and in the leaves immediately below the inflorescence. High squa transcript levels are seen in the inflorescence lateral meristems as soon as they are formed in the axils of bracts. Squa transcriptional activity persists through later stages of floral morphogenesis, with the exception of stamen differentiation. Although necessary for shaping a normal racemose inflorescence, the squa function is not absolutely essential for flower development. We discuss the function of the gene during flowering, its likely functional redundancy and its possible interaction with other genes participating in the genetic control of flower formation in Antirrhinum.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987 Sep;101(1):1–22. [PubMed] [Google Scholar]
- Carpenter R., Coen E. S. Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majus. Genes Dev. 1990 Sep;4(9):1483–1493. doi: 10.1101/gad.4.9.1483. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Meyerowitz E. M. The war of the whorls: genetic interactions controlling flower development. Nature. 1991 Sep 5;353(6339):31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Romero J. M., Doyle S., Elliott R., Murphy G., Carpenter R. floricaula: a homeotic gene required for flower development in antirrhinum majus. Cell. 1990 Dec 21;63(6):1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Cohen P. Protein phosphorylation and hormone action. Proc R Soc Lond B Biol Sci. 1988 Jul 22;234(1275):115–144. doi: 10.1098/rspb.1988.0040. [DOI] [PubMed] [Google Scholar]
- Fishleigh R. V., Robson B., Garnier J., Finn P. W. Studies on rationales for an expert system approach to the interpretation of protein sequence data. Preliminary analysis of the human epidermal growth factor receptor. FEBS Lett. 1987 Apr 20;214(2):219–225. doi: 10.1016/0014-5793(87)80060-1. [DOI] [PubMed] [Google Scholar]
- Hagberg L., Hull R., Hull S., Falkow S., Freter R., Svanborg Edén C. Contribution of adhesion to bacterial persistence in the mouse urinary tract. Infect Immun. 1983 Apr;40(1):265–272. doi: 10.1128/iai.40.1.265-272.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes T. E., Sengupta P., Cochran B. H. The human c-fos serum response factor and the yeast factors GRM/PRTF have related DNA-binding specificities. Genes Dev. 1988 Dec;2(12B):1713–1722. doi: 10.1101/gad.2.12b.1713. [DOI] [PubMed] [Google Scholar]
- Ingham P. W. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988 Sep 1;335(6185):25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Irish V. F., Sussex I. M. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell. 1990 Aug;2(8):741–753. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi C. P. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987 Aug 25;15(16):6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J., Schell J., Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987 May 15;163(1):16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Luo D., Coen E. S., Doyle S., Carpenter R. Pigmentation mutants produced by transposon mutagenesis in Antirrhinum majus. Plant J. 1991 Jul;1(1):59–69. [PubMed] [Google Scholar]
- Ma H., Yanofsky M. F., Meyerowitz E. M. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991 Mar;5(3):484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Norman C., Runswick M., Pollock R., Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988 Dec 23;55(6):989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- Passmore S., Elble R., Tye B. K. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev. 1989 Jul;3(7):921–935. doi: 10.1101/gad.3.7.921. [DOI] [PubMed] [Google Scholar]
- Pnueli L., Abu-Abeid M., Zamir D., Nacken W., Schwarz-Sommer Z., Lifschitz E. The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1991 Sep;1(2):255–266. [PubMed] [Google Scholar]
- Schultz E. A., Haughn G. W. LEAFY, a Homeotic Gene That Regulates Inflorescence Development in Arabidopsis. Plant Cell. 1991 Aug;3(8):771–781. doi: 10.1105/tpc.3.8.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Hue I., Huijser P., Flor P. J., Hansen R., Tetens F., Lönnig W. E., Saedler H., Sommer H. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 1992 Jan;11(1):251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Huijser P., Nacken W., Saedler H., Sommer H. Genetic Control of Flower Development by Homeotic Genes in Antirrhinum majus. Science. 1990 Nov 16;250(4983):931–936. doi: 10.1126/science.250.4983.931. [DOI] [PubMed] [Google Scholar]
- Shannon S., Meeks-Wagner D. R. A Mutation in the Arabidopsis TFL1 Gene Affects Inflorescence Meristem Development. Plant Cell. 1991 Sep;3(9):877–892. doi: 10.1105/tpc.3.9.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H., Beltrán J. P., Huijser P., Pape H., Lönnig W. E., Saedler H., Schwarz-Sommer Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 1990 Mar;9(3):605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky M. F., Ma H., Bowman J. L., Drews G. N., Feldmann K. A., Meyerowitz E. M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990 Jul 5;346(6279):35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]








